Abstract
Brief seizures (epileptic/seizure preconditioning) are capable of activating endogenous protective pathways in the brain which can temporarily generate a damage-refractory state against subsequent and otherwise harmful episodes of prolonged seizures (tolerance). Altered expression of microRNAs, a class of non-coding RNAs that function post-transcriptionally to regulate mRNA translation has recently been implicated in the molecular mechanism of epileptic tolerance. Here we characterized the effect of seizure preconditioning induced by low-dose systemic kainic acid on microRNA expression in the hippocampus of mice. Seizure preconditioning resulted in up-regulation of 25 mature microRNAs in the CA3 subfield of the mouse hippocampus, with the highest levels detected for miR-184. This finding was supported by real-time PCR and in situ hybridization showing increased neuronal miR-184 levels and a reduction in protein levels of a miR-184 target. Inhibiting miR-184 expression in vivo resulted in the emergence of neuronal death after preconditioning seizures and increased seizure-induced neuronal death following status epilepticus in previously preconditioned animals, without altered electrographic seizure durations. The present study suggests miRNA up-regulation after preconditioning may contribute to development of epileptic tolerance and identifies miR-184 as a novel contributor to neuronal survival following both mild and severe seizures.
Keywords: Dicer, Epigenetics, Hippocampus, Status epilepticus, Temporal lobe Epilepsy
Exposure of the brain to a stimulus or stressor that is sub-threshold for injury (preconditioning) activates endogenous programmes of neuroprotection resulting in a damage-refractory phenotype (tolerance) (Chen and Simon, 1997, Dirnagl, et al., 2003, Gidday, 2006). Brain damage caused by prolonged seizures (status epilepticus, SE) can be substantially reduced when preceded by seizure preconditioning, resulting in epileptic tolerance. This has been demonstrated for several preconditioning stimuli including kindling, electroshocks and low doses of chemoconvulsants such as kainic acid (KA) (Jimenez-Mateos and Henshall, 2009).
Although the molecular mechanisms that underlie tolerance are incompletely understood, microarray profiling shows that epileptic tolerance features, like ischemic tolerance (Stenzel-Poore, et al., 2003), large-scale down-regulation of protein-coding genes (Jimenez-Mateos, et al., 2008). Among the pathways most impacted are those associated with neuronal excitation and excitotoxicity, which may contribute to protection against neuronal death and post-SE spontaneous seizures (Jimenez-Mateos, et al., 2008, Jimenez-Mateos, et al., 2010). Work has since focused on the mechanism of suppressed transcription, identifying contributions from DNA methylation (Miller-Delaney, et al., 2012), polycomb group proteins (Stapels, et al., 2010) and microRNAs (miRNAs) (Saugstad, 2010).
MiRNAs are a class of small (~22 nt), non-coding RNAs which mediate post-transcriptional regulation of gene expression. They are predicted to regulate protein levels of about half of all protein-coding genes in mammals (Bartel, 2009, Krol, et al., 2010), with over 1000 miRNAs now identified in humans and over 700 in mice. MiRNAs mainly mediate their effects via imperfect base-pairing with the 3′ untranslated region (UTR) of their target mRNAs, resulting in mRNA degradation or inhibition of translation (Bartel, 2004, Krol, et al., 2010). Their biogenesis begins with transcription from independent genes or from introns of protein-coding genes (Krol, et al., 2010). The primary transcript folds into a hairpin structure that is processed by the Drosha microprocessor complex to generate a pre-miRNA. In the cytoplasm, Dicer generates a ~20-bp miRNA/miRNA* duplex and the mature strand is guided to the RNA-induced silencing complex (RISC) where Argonaute (Ago) proteins mediate translational repression or mRNA decay (Peters and Meister, 2007). miRNAs are critical for normal brain development and function (Fiore, et al., 2008) and loss of miRNA production results in synaptic dysfunction and neuronal apoptosis (Davis, et al., 2008, Schaefer, et al., 2007, Tao, et al., 2011). Dysregulation of miRNAs may contribute to neurologic, psychiatric and neurodegenerative diseases including ischemic and epileptic brain injury (Eacker, et al., 2009, Hebert and De Strooper, 2009, Jimenez-Mateos, et al., 2011, McKiernan, et al., 2012, Saugstad, 2010).
Several recent studies reported expression profiles for miRNAs following SE (Hu, et al., 2011, Liu, et al., 2010, Pichardo-Casas, et al., 2012). MiRNA profiling has also been undertaken for ischemic tolerance (Lusardi, et al., 2010), ischemic preconditioning (Dharap and Vemuganti, 2010, Lee, et al., 2010, Lusardi, et al., 2010) and electroshock (Eacker, et al., 2011), which can model seizure preconditioning (Jimenez-Mateos and Henshall, 2009). We recently contrasted the response of mature miRNAs after SE with the profile in tolerant animals that had been exposed to seizure preconditioning before SE, revealing epileptic tolerance was associated with subdued and occasionally bi-directional changes for miRNAs (Jimenez-Mateos, et al., 2011). The effect of seizure preconditioning alone has not been reported. Since many protein-coding genes are downregulated after SE in preconditioned animals we postulated seizure preconditioning would, broadly, increase miRNA expression. Here we profiled the miRNA response to seizure preconditioning and identify miR-184 as contributing to neuronal survival in this model.
MATERIALS AND METHODS
Animal procedures
Animal experiments were carried out as previously described (Hatazaki, et al., 2007) in accordance with the European Communities Council Directive (86/609/EEC) and were reviewed and approved by the Research Ethics Committee of the Royal College of Surgeons in Ireland, under license from the Department of Health, Dublin, Ireland. Adult male C57BL/6 mice (20–25 g) were obtained from Harlan (Oxon, Bicester, U.K.) and housed in a vivarium on a 12 hour light/dark cycle with access to food and water ad libitum.
Seizure preconditioning was induced by a single intraperitoneal (i.p.) injection of KA (15mg/kg, Ascent Scientific Ltd, Bristol, U.K. in 0.2 ml volume). Control mice received a single i.p. injection of 0.2 ml phosphate buffered saline (PBS) as sham-preconditioning.
For epileptic tolerance, mice were prepared as described (Hatazaki, et al., 2007, Jimenez-Mateos, et al., 2008). Briefly, mice received seizure preconditioning on day 1 followed by intra-amygdala KA (1 μg in 0.2 μl) 24 h later. For the intra-amygdala injections, mice were anesthetized using isoflurane (5 % induction, 1–2 % maintenance) and fixed into a stereotaxic frame and maintained normothermic with a heat pad (Harvard Apparatus, Kent, U.K.). Three partial craniectomies were performed for skull-mounted recordings (Bilaney Consultants, Sevenoaks, Kent, U.K.) and craniectomy for fixation of a guide cannula (coordinates from Bregma (Paxinos and Franklin, 2001) [AP = −0.94 mm, L = −2.85 mm]). The assembly was fixed with dental cement and mice were allowed to recover and move freely in a Perspex chamber. Baseline EEG activity (Grass Comet XL lab-based EEG) was recorded and then an internal injection cannula lowered for injection of KA or vehicle into the basolateral amygdala. Lorazepam (6 mg/kg, i.p.) was administered to all animals 40 min later to reduce morbidity and mortality and animals were removed to a warmed recovery chamber.
All mice were euthanized by pentobarbital overdose and perfused with ice-cold saline to remove intravascular blood components. Brains were then either frozen whole or microdissected to obtain CA1, CA3 and DG enriched portions from the hippocampus, or neocortex, as described (Hatazaki, et al., 2007).
EEG analysis
Using TWin® software, the duration of high amplitude, high frequency discharges (HAHFDs), was recorded for 40 min after KA administration as described (Jimenez-Mateos, et al., 2011). Additional analysis of frequency and amplitude parameters was performed using LabChart Pro v7 software (ADInstruments Ltd, Oxford, U.K.).
Antagomir injections
For intracerebroventricular (i.c.v.) administration of antagomir, mice were affixed with an additional cannula (Coordinates from Bregma: AP = −0.3 mm, L = −1.0 mm, V = −2.0 mm) ipsilateral to the side of KA injection, as described (Jimenez-Mateos, et al., 2011). Scrambled or miRNA-184 antagomirs (Exiqon, LNA- and 3′-cholesterol modified oligonucleotides) were infused in a volume of 2 μl artificial cerebrospinal fluid (aCSF) (Harvard Apparatus) and mice underwent seizure-preconditioning or epileptic tolerance, as above.
Behavioral analysis
Behavioural analysis of mice subject to seizure preconditioning was performed using a modified ethogram (Clifford, et al., 2000). Briefly, mice were observed continuously for 1 h following i.p. injections and the duration of time spent conducting various behaviours scored for exploring, immobility, grooming, socially interacting and twitching.
MiRNA extraction and expression profiling
The two CA3 subfields from each control or seizure preconditioned animal were combined to generate individual samples (n = 4 per group). Total RNA extracted using a miRNA easy kit (Qiagen, West Sussex, UK) as previously described (Jimenez-Mateos, et al., 2011). The quality and quantity of RNA yield was determined using a Nanodrop Spectrophotometer (Thermoscientific, Loughborough, UK) and RNA concentrations were normalised in nuclease-free water. Reverse transcription of 100 ng of miRNA was carried out using stem-loop Multiplex primer pools (Applied Biosystems), allowing reverse transcription of 48 different miRNAs in each of eight RT pools. The miRNA screen was carried out using a 7900HT Fast Realtime System and TaqMan Low-Density Arrays (TLDA) (TaqMan TLDA MicroRNA Assays v1.0 containing 382 human microRNAs assays; Applied Biosystems). A relative fold change in expression of the target gene transcript was determined using the comparative cycle threshold method (2−ΔΔCT). miRNAs were called present when amplified within 35 cycles in 3 out of 4 samples. Data for each miRNA were normalised to the average Ct value for each mouse, as reported (Bray, et al., 2009, Jimenez-Mateos, et al., 2011). A threshold of ≥ 1.5 fold was considered either increased or decreased relative to control.
Stem-loop reverse transcription and real time qPCR of individual miRNAs
Reverse transcription of 100 ng of total RNA from hippocampal CA1 or CA3 was carried out for individual qPCRs using the High-Capacity Reverse Transcription Kit (Applied Biosystems). RT specific primers for miR-184 and miR-204 (Applied Biosystems) were used. Individual qPCRs were carried out on the 7900HT Fast Realtime System (Applied Biosystems) using the complementary Taqman miRNA assay probes (Applied Biosystems). The endogenous control, RNU6B, was used for normalization. Potential miR-184 targets were identified using microrna.org as previously described (Sano, et al., 2012).
Western blotting
Western blotting was performed as previously described (Hatazaki, et al., 2007). Hippocampal subfields from individual mice were homogenized in lysis buffer containing a protease and phosphatase inhibitor cocktail (Sigma-Aldrich) and boiled in gel-loading buffer. Thirty micrograms of protein was separated by SDS-PAGE and transferred onto nitrocellulose membranes. The following primary antibodies were used: Dicer, c-Fos (Santa Cruz Biotechology, Santa Cruz, CA, USA), Drosha, Ago2 (Cell Signaling Technology, Beverly, MA, USA), Cacnb3 (Abcam, Cambridge, MA, USA) and α-Tubulin (Sigma-Aldrich). Membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch, Plymouth, PA, USA) and bands visualized using Supersignal West Pico chemiluminescence (Pierce, Rockford, IL, USA). Images were captured on a Fuji-Film LAS-300 (Fuji, Sheffield, UK), and densitometry was performed using AlphaEaseFC4.0 gel-scanning integrated optical density software (Alpha Innotech, San Leandro, CA, USA).
Histopathology
Brains were sectioned at 12 μm on a cryostat (−20°C) at rostral (AP = −1.58 mm) and medial (AP = −1.82 mm) hippocampal levels (Paxinos and Franklin, 2001). Fluoro-Jade B (FJB) staining was performed as previously described (Jimenez-Mateos, et al., 2011, Sano, et al., 2012). Briefly, tissue sections were allowed to air-dry, post-fixed in formalin (10 %), immersed in 0.006 % potassium permanganate solution, rinsed again and then transferred to FJB solution (0.001% in 0.1% acetic acid) (Chemicon Europe Ltd, Chandlers Ford, U.K.). Sections were rinsed again, dried, cleared and mounted with DPX (Sigma-Aldrich). Analysis of DNA damage was performed on fresh-frozen sections using a fluorescein-based terminal deoxynucleotidyl dUTP nick end labelling (TUNEL) technique, according to manufacturer’s guidelines (Promega, Madison, WI, USA) and as described before (Jimenez-Mateos, et al., 2011). Images were captured using a monochromatic Hamamatsu Orca 285 and image-inverted in Adobe® Photoshop® 6.0 such that FJB-positive cells appear as black on a white-grey background. Counts of FJB and TUNEL-positive cells were performed as previously described (Jimenez-Mateos, et al., 2011, Sano, et al., 2012). Under a 20x lens magnification an observer blinded to experimental treatment counted positive cells along the entire CA3 or CA1 subfields, defined using standard boundaries, for two adjacent tissue sections and an average count was used.
In situ hybridization
Mice were perfused with ice-cold paraformaldehyde (PFA, 4 %) and 12 μm thick sections mounted on SuperFrost-Plus slides (VWR International, Dublin, Ireland). Under RNAse free conditions, slides were washed with PBS and RIPA buffer (150ml NaCl, 1% IGEPAL, 0.5% Na deoxycholate, 0.1%SDS, 1mM EDTA, 50mM Tris pH:8.0) for 5 min and then treated with 4% PFA for 10 min. Sections were washed and treated with 0.25% acetic anhydride/0.1M triethhanolmine, then rinsed with 0.1 % Tween-20/PBS for 5 min, treated with 5μg/ml Proteinase K for 4 min and washed with PBS. Slides were then rinsed in hybridization buffer (1x saline solution, 50% formamide and 1X Denhardt’s) for 1 h at 56°C (Tm-20C, Tm provided by Exiqon). The probes to detect miR-184 and miR-204 were 5′-digoxigenin-labeled, 2′-O,4′-C methylene bicyclonucleoside monomer-containing oligonucleotide (LNA-modified). Sequences possessed reverse complementarity to the mature miRNA. Probes (0.05 μM) were incubated in hybridization buffer overnight at 56°C in a humidified chamber. The following day sections were washed in FAM buffer (2x SSC, 50% formamide and 0.1% Tween 20) for 1 h at 60°C. Then sections were rinsed in B1 buffer (150 mM NaCl, 100 mM Maleic acid and 0.4 % IGEPAL pH, 7.5) for 1 h at room temperature and B2 buffer (2% blocking reagent and 10% goat serum in B1) for 30 min. Anti-DIG-PA antibody (1:1000, Roche) was incubated in B2 overnight at 4°C. The following day sections were washed in B1 buffer and incubated in B3 buffer (100 mM NaCl, 50 mM MgCl2, 0.025% Tween 20 and 100 mM Trizma pH9.5) for 30 min. Then 200 μL of colour substrate solution (CSS: Nitroblue tetrazolium/BCIP stock solution (Roche) diluted 1:50 in B3 buffer) was added to each slide until the signal appeared. Slides were then rinsed, mounted with medium and coverslipped.
Data analysis
All data are presented as mean ± s.e.m. Two group comparisons were made using Student’s t-test, while multi-group comparisons were made using analysis of variance (ANOVA) followed by appropriate post hoc testing. Significance was accepted at p < 0.05.
RESULTS
Seizure preconditioning induces c-Fos in the hippocampus
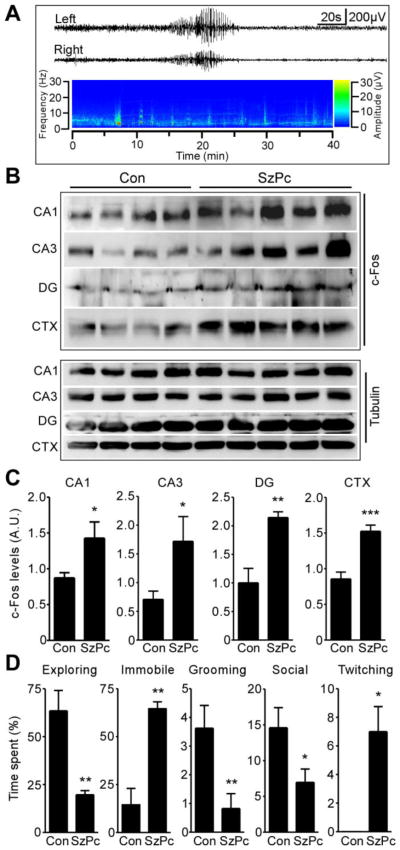
We first characterized the electrographic and behavioural effects of seizure preconditioning and confirmed hippocampal recruitment. Cortical EEG was recorded in mice using skull-mounted electrodes for 40 min following induction of seizure preconditioning (KA injection; 15 mg/kg, i.p.). Seizure preconditioning caused brief episodes of polyspike seizures (HAHFDs) lasting an average of 26 ± 11 s (n = 3–4 per group, Fig. 1A and data not shown). Seizures did not become continuous and severe tonic-clonic seizures were not observed. Analysis of c-Fos protein, a marker of neuronal activity triggered by seizures (Bozzi, et al., 2011), in whole cell lysates from microdissected hippocampus 4 h following KA injection, determined seizure preconditioning caused an increase in c-Fos protein in the CA1, CA3 and dentate gyrus (Fig. 1B, C). Seizure preconditioning also increased c-Fos levels in the neocortex (Fig. 1B, C).
Figure 1. Characterization of seizure preconditioning.
(A) Representative EEG trace and heat-map spectrogram of seizure activity recorded from skull electrodes following injection of KA to induce seizure preconditioning (SzPc). (B) Western blots (n = 1 per lane) showing c-Fos in 4 h samples from hippocampal subfields and the neocortex (CTX) from controls (Con) and after SzPc. α-Tubulin is included as a guide to protein loading. (C) Levels of c-Fos in each sample at 4 h (*p < 0.05; **p < 0.01; ***p < 0.001 vs. Con; n = 4–5 per group) A.U., arbitrary units. (D) Ethogram results showing the percentage time mice spent conducting each behaviour (*p < 0.05; **p < 0.01 vs. Con; n = 4–5 per group).
Behavioral correlates of seizure preconditioning
Next, we assessed the effects of seizure preconditioning on behaviour. Standard Racine-type scales were not sufficiently sensitive to score the behaviour so we employed a scoring system that uses an ethologically-based assessment (ethogram) (Clifford, et al., 2000). During one hour observations, seizure preconditioning resulted in onset of altered behaviour at 6 ± 4 min and resumption of normal behaviour at 31 ± 4 min (n = 4/5 per group). Seizure preconditioning resulted in mice spending significantly less time exploring and engaged in social interaction and significantly more time in an immobile state and twitching (Fig. 1D).
miRNA biogenesis components after seizure preconditioning
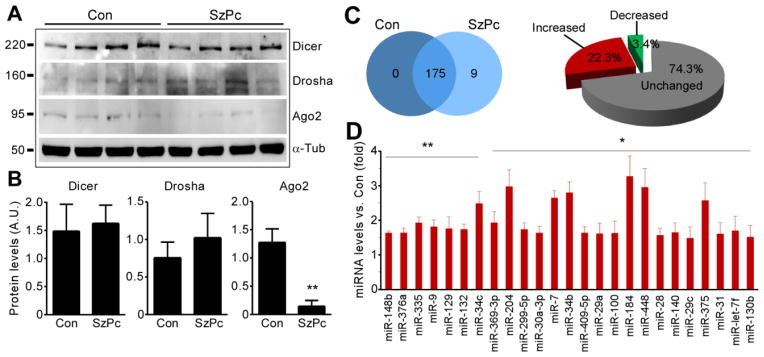
To establish the presence of key miRNA biogenesis components in the model, we examined hippocampal CA3 protein levels of Dicer, Drosha and Ago-2 (Fig. 2A). Levels of Dicer and Drosha were unchanged after seizure preconditioning although Ago-2 levels were reduced (Fig 2A, B).
Figure 2. Modulation of miRNA expression following seizure preconditioning.
(A) Western blots (n = 1 per lane) showing Dicer, Drosha and Ago2 protein levels in CA3 in Con and 8 h following SzPc. (B) Semi-quantification of protein levels (p < 0.01 vs. Con; n = 4 per group). (C) Venn diagram and pie chart showing numbers of miRNAs expressed uniquely in Con and SzPc, those commonly expressed, and the increase or decrease as a percentage. (D) Graph showing expression data for individual miRNAs upregulated after SzPc (*p < 0.05; **p < 0.01; n = 4 per group).
Seizure preconditioning up-regulates hippocampal expression of miRNAs
We next analyzed miRNA expression after seizure preconditioning using Taqman Low Density Arrays (TLDA). An 8 h sampling point was chosen because this is one third of the time interval between preconditioning and when SE is normally given in the model (24 h); this is the equivalent of the 24 h time point between preconditioning and challenge in the ischemic tolerance model in a recent study where microRNAs were profiled (Lusardi, et al., 2010). We note that a shorter interval between seizure preconditioning and SE does not produce as much neuroprotection in the epileptic tolerance model (Supplementary data Figure S1). The CA3 was selected since it is the principal region which undergoes damage after intra-amygdala KA and which is protected in epileptic tolerance (Hatazaki, et al., 2007, Jimenez-Mateos, et al., 2008).
From 380 miRNAs screened, a total of 175 were called present in the CA3 subfield of control mice (Fig. 2C, D). In mice subject to seizure preconditioning a total of 184 miRNAs were called present (Fig. 2C, D). Comparison of the two groups revealed 9 miRNAs (miRs -10a, -155, -185, -193b, -196a, -216, -450, -493, and -494) were uniquely present in seizure preconditioning samples, with 175 miRNAs commonly detected in both groups (Fig. 2C). Of the commonly expressed miRNAs, 74.3% showed no change in expression (RQ < 1.5 ± fold) following seizure preconditioning. Seizure preconditioning increased levels of 39 miRNAs (22.3 % of the commonly expressed miRNAs; RQ > 1.5), with 25 miRNAs significantly increased compared to control (Fig. 2D and Supplementary data Figure S2). The most up-regulated miRNA was miR-184 (3.26 fold), followed by miR-204 (2.96 fold) (Fig. 2D). Only 6 miRNAs showed reduced levels, representing 3.4 % of the commonly expressed miRNAs, and none to a statistically significant level (Supplementary data Figure S2).
Validation of miR-184 up-regulation after seizure preconditioning
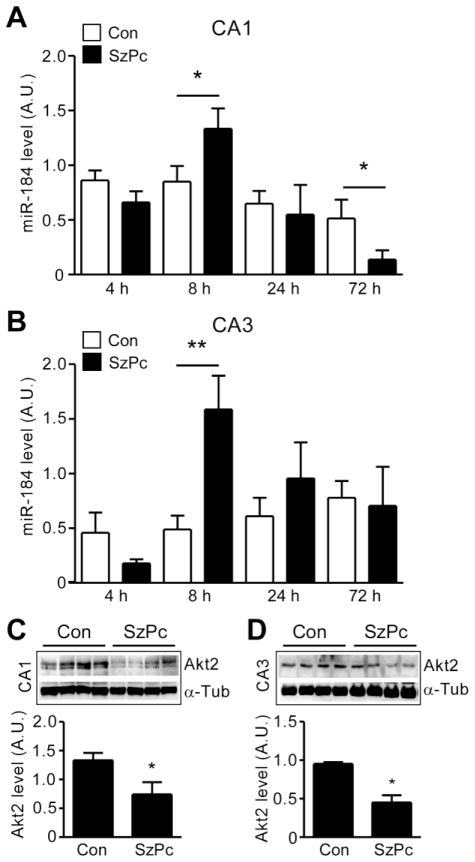
To validate the TLDA results, we undertook individual RT-qPCR analysis of the two most up-regulated miRNAs, miR-184 and miR-204, in separate CA3 and CA1 samples from mice after seizure preconditioning. Of note, basal expression of miR-184 in control animals was significantly higher (P < 0.01) in the CA3 subfield compared to the CA1 subfield (data not shown). Expression of miR-184 was significantly increased in both CA3 and CA1 samples 8 h following seizure preconditioning (Fig. 3A, B). Expression of miR-184 was not different between seizure preconditioning and control at 4 or 24 h in either region, although a small reduction in CA1 levels was noted at 72 h (Fig. 3A, B).
Figure 3. Validation of miR-184 up-regulation after seizure preconditioning.
(A, B) miR-184 level measured by RT-qPCR in CA1 and CA3 samples 4–72 h after SzPc. Expression was corrected to U6B (*p < 0.05 vs. Con; n = 4–7 per group). (C, D) Western blots (n = 1 per lane) and semi-quantification showing reduction in protein levels of Akt2 after SzPc in CA1 and CA3. Levels corrected to α-Tubulin, data presented as arbitrary units (A.U). *p < 0.05 vs. Con (n = 4 per group).
We also performed in situ hybridisation to identify the cell type(s) that expressed miR-184 (Supplementary Data Figure S3). This showed that miR-184 was present within pyramidal neurons of the CA1 and CA3 subfields. We also examined the protein level of a known miR-184 target, Akt2 (Foley, et al., 2010). Akt2 protein was detected in control mouse CA1 and CA3 subfields at ~55 kD (Fig. 3C, D). Protein levels of Akt2 were significantly lower in CA1 and CA3 in samples 8 h after seizure preconditioning (Fig. 3C, D). A bioinformatics analysis of other potential mRNA targets of miR-184 (see Materials and Methods for details) is included in Supplementary Table S1.
While in situ hybridization showed miR-204 was expressed in the pyramidal neurons of the CA1 and CA3 subfields, up-regulation of miR-204 after seizure preconditioning was not validated by RT-qPCR (Supplementary Data Figure S4). Nevertheless, protein levels of a putative target of miR-204 showed reduced levels in samples 8 h after seizure preconditioning (Supplementary data Figure S4).
Antagomir-mediated inhibition of miR-184 in the hippocampus
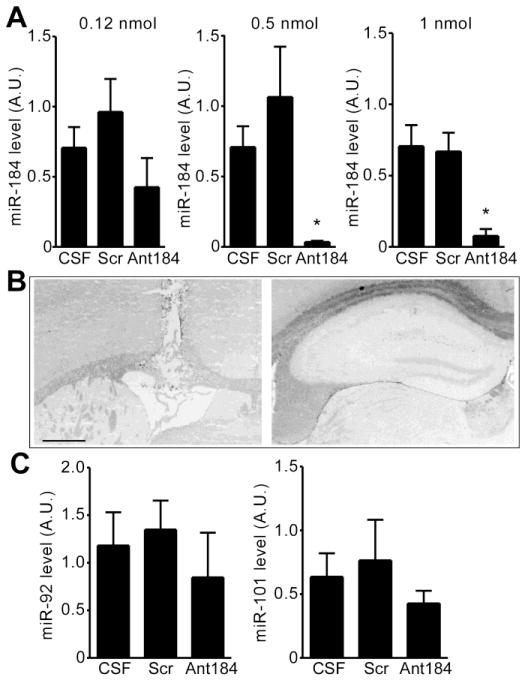
To determine whether miR-184 influenced neuronal survival after seizure preconditioning, we used locked nucleic acid (LNA)-modified and 3′ cholesterol-conjugated anti-miR-184 oligonucleotides (“antagomirs”) (Jimenez-Mateos, et al., 2011, Sano, et al., 2012) to reduce brain levels of miR-184. To establish a suitable dosing regimen, mice were injected i.c.v. with different amounts of miR-184-targeting antagomirs (Ant184) or a scrambled control and assessed 24 h later, in line with the optimal time taken for antagomirs to deplete their target (Jimenez-Mateos, et al., 2012) (Fig. 4A). At the lowest tested dose (0.12 nmol), the antagomir did not significantly alter hippocampal miR-184 levels (Fig. 4A). In contrast, injection of either 0.5 or 1.0 nmol Ant184 significantly reduced miR-184 levels at 24 h (Fig. 4A). The same dose of the scrambled antagomir had no effect on miR-184 levels (Fig. 4A) and neither Ant184 nor the scrambled antagomir altered levels of other measured miRNAs (Fig. 4C).
Figure 4. Effects of antagomirs on miRNA expression.
(A) RT-qPCR analysis showing effect of i.c.v. injection of Ant184 or scrambled (Scr) compared to CSF alone, on hippocampal CA3 levels of miR-184 measured 24 h later. Each group is corrected to U6B and compared to mice who received CSF (*p < 0.05 vs. CSF; n = 3 per group). (B) Photomicrographs showing representative FJB staining (left) at the level of i.c.v. injection, and (right) dorsal hippocampus, 24 h later. No cell death is present in the hippocampus of mice given antagomirs alone. Scale bar, 650 μm. (C) RT-qPCR analysis of hippocampal levels of two unrelated miRNAs 24 hours after a single injection of 0.5 nmol Ant184 (n = 3 per group).
To determine whether Ant184 or scrambled injection had any cytotoxic effect we stained hippocampal sections at 24 h for FJB, a specific marker of degenerating neurons (Schmued and Hopkins, 2000). No FJB-positive cells were observed in sections of the hippocampus from mice treated 24 h earlier with up to 1.0 nmol Ant184 (Fig. 4B).
Seizure preconditioning is injurious in mice given antagomirs targeting miR-184
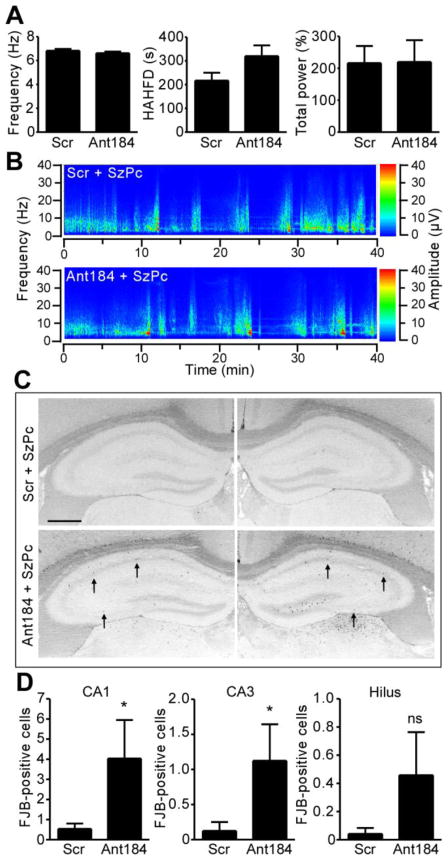
Semi-quantitative analysis of EEG after seizure preconditioning revealed no differences between scrambled and Ant184-injected animals (Fig. 5A, B). However, we noted that the electrographic seizures in mice which had undergone cannula placement and i.c.v. injection the day before were more severe than the ordinary response to seizure preconditioning in non-instrumented mice regardless of whether scrambled or Ant184 were injected (compare Fig. 5A, B to Fig. 1A). Analysis of the hippocampus 24 h after seizure preconditioning in scrambled-injected mice revealed no significant seizure-damage to CA1, CA3 or the hilus (Fig. 5C, D). In contrast, significant numbers of FJB-positive cells were detected in both the CA1 and CA3 subfields, as well as more variably in the hilus, after seizure preconditioning in Ant184-injected mice (Fig. 5C, D). Similar results were obtained in rostral sections stained for FJB and in medial sections stained for DNA fragmentation using TUNEL (data not shown).
Figure 5. Inhibition of miR-184 results in emergence of neuronal death after seizure preconditioning.
(A) Electrographic seizure parameters recorded during 40 min after SzPc in mice injected i.c.v. 24 h earlier with either Scr or Ant184 (0.5 nmol each; n = 6 per group). (B) Representative spectrogram of EEG frequency and amplitude data during the 40 min after SzPc in a mouse that received Scr or Ant184 24 h earlier. (C) Representative bilateral photomicrographs showing FJB staining in the hippocampus 24 h after SzPc in animals that received Scr or Ant184. Dead neurons (black dots) are visible only in mice that received Ant184 before SzPc (arrows). Scale bar, 650 μm. (D) Graphs showing averaged bilateral FJB counts 24 h after SzPc in mice given either Scr or Ant184 (*p < 0.05 vs. Scr; n = 6 per group; ns, non-significant).
Antagomir-mediated inhibition of miR-184 results in increased neuronal death in epileptic tolerance
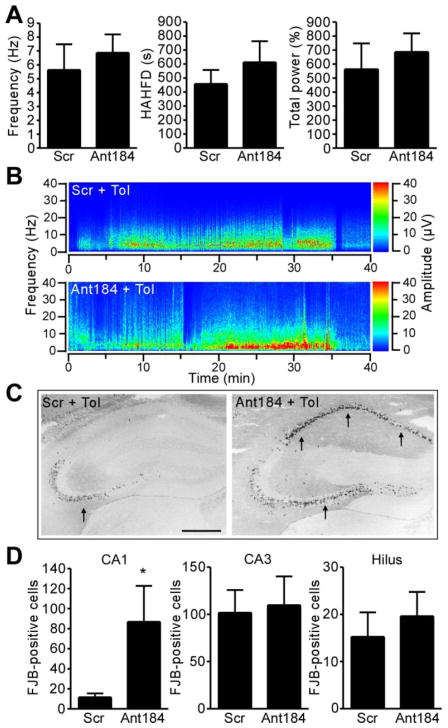
Last, we wanted to investigate whether inhibition of miR-184 before seizure preconditioning also altered damage in epileptic tolerance. Two groups of mice were prepared which received either scrambled or Ant184, followed 24 h later by seizure preconditioning. A day later, mice were subjected to SE via intra-amygdala KA and damage assessed 24 h afterwards. Seizure duration and severity during SE was not different between scrambled and Ant184 animals (Fig. 6A, B). But, Ant184-injected seizure-preconditioned mice displayed significantly more seizure-damage within the CA1 subfield compared to scrambled animals following SE (Fig. 6C, D). No significant difference was found between groups for the CA3 or hilus regions. Similar results were obtained in rostral sections stained for FJB and in medial sections stained for DNA fragmentation using TUNEL (data not shown).
Figure 6. Inhibition of miR-184 increases seizure-induced neuronal death in epileptic tolerance.
Mice received Scr or Ant184 (0.5 nmol i.c.v.) then 24 h later epileptic tolerance (Tol) was modelled by first subjecting mice to seizure preconditioning (15 mg/kg KA, i.p.) followed 24 h later by SE induced by intra-amygdala KA. (A) Electrographic seizure parameters recorded for 40 min after triggering SE in seizure preconditioned mice (n = 6–8 per group). (B) Representative spectrogram of EEG frequency and amplitude data during the 40 min after triggering SE. (C) Photomicrographs showing FJB staining in the ipsilateral hippocampus 24 h after SE in seizure preconditioned mice that received Scr or Ant184. Dead neurons (black dots) are more abundant in area CA1, while both CA3 and hilus are similarly injured (arrows). Scale bar, 650 μm. (D) Graphs showing ipsilateral FJB counts 24 h after SE in seizure preconditioned mice given either Scr or Ant184 (*p < 0.05 vs. Scr; n = 6–8 per group).
Discussion
We report the identification of miR-184 as a seizure-regulated miRNA and demonstrate that inhibition of miR-184 increases neuronal death after seizures. Our study profiled miRNA expression changes brought about by seizure preconditioning and included characterization of the electrographic and behavioral features of seizure preconditioning. These findings identify a novel regulator of seizure-induced neuronal death and further implicate miRNAs in the molecular mechanisms underlying epileptic tolerance.
Epileptic tolerance develops when the brain is pre-exposed to non-harmful seizure activity prior to SE. The underlying molecular mechanisms are incompletely known but microarray profiling has revealed down-regulation of protein-coding genes as a conserved mechanism in tolerance (Jimenez-Mateos, et al., 2008, Stenzel-Poore, et al., 2003). Up-regulation of miRNAs is a plausible mechanistic effector, although changes to transcriptional and epigenetic processes may also be important (Jimenez-Mateos and Henshall, 2009). Here we investigated miRNA expression after preconditioning seizures induced by a single systemic injection of KA (Hatazaki, et al., 2007). Surface EEG revealed that seizure preconditioning caused brief electrographic seizures, indicating the occurrence of low-grade generalized epileptiform activity. In addition, hippocampal recruitment was confirmed by increased c-Fos protein levels, an established index of seizure activity (Bozzi, et al., 2011, Harvey and Sloviter, 2005). The present study also showed that seizure preconditioning does not generate clinical behavior amenable to scoring using mouse-adapted Racine scales (Borges, et al., 2003). However, altered behavior during seizure preconditioning could be scored using an ethogram-based technique, developed to identify alterations in spontaneous behavior (Clifford, et al., 2000). Thus, ethological monitoring could be a means to monitor the induction of seizure preconditioning.
MiRNAs and their biogenesis pathway have emerged as important factors in the pathogenesis of several neurologic and neurodegenerative diseases (Eacker, et al., 2009, Saugstad, 2010). Ischemic and epileptic tolerance both feature changes in miRNA expression (Jimenez-Mateos, et al., 2011, Lusardi, et al., 2010), but the present study is the first transcriptional profiling of miRNA responses to seizure preconditioning. We detected changes to 25 miRNAs and all, remarkably, were upregulated. This included some previously known neuronal activity-regulated miRNAs such as miR-132 (Hu, et al., 2011, Nudelman, et al., 2010, Wayman, et al., 2008) and let-7f (Eacker, et al., 2011). The most up-regulated miRNA in our study was miR-184, another brain-expressed activity-regulated miRNA (Nomura, et al., 2008). In situ hybridization confirmed other work indicating neuronal expression within the hippocampus (Liu, et al., 2010). Thus miR-184 is a novel seizure-regulated miRNA in vivo and identifying its in vivo mRNA targets may provide new directions for research on tolerance and neuroprotection.
We recently reported that epileptic tolerance is associated with differential patterns of DNA methylation, an epigenetic mechanism regulating gene expression (Miller-Delaney, et al., 2012). While protein-coding genes, rather than miRNAs were the focus of this study, it is interesting to note that expression of miR-184 is normally repressed by methyl-CpG-binding proteins in the brain (Liu, et al., 2010, Nomura, et al., 2008). Neuronal depolarization releases this epigenetic brake (Nomura, et al., 2008) and we may speculate that seizure preconditioning increases miR-184 expression via a similar mechanism. More broadly, this emphasizes that analysis of DNA methylation at miRNA regulatory sites can provide further insights into the control of miRNAs after seizure preconditioning or in epileptic tolerance, and the available pharmacologic and genetic tools for modulating DNA methylation (Kelly, et al., 2010, Urdinguio, et al., 2009) may provide new approaches to alter miRNA expression in tolerance.
Our findings contrast the bi-directional response of certain miRNAs following seizures (Hu, et al., 2011, Liu, et al., 2010, Pichardo-Casas, et al., 2012). Also, we did not detect up-regulation of miR-146a or let-7e, both of which have been implicated in post-SE responses (Aronica, et al., 2010, Song, et al., 2011). This is not unexpected because seizure preconditioning has no damaging effects on the hippocampus whereas SE produces profound damage. However, lower-grade seizures triggered by electroshock, also a model of seizure preconditioning (Jimenez-Mateos and Henshall, 2009), produces down- as well as up-regulation of miRNAs (Eacker, et al., 2011). It is therefore likely that bi-directional changes to miRNA occur, perhaps at other time-points. Indeed, we observed a small reduction in miR-184 levels 72 h after seizure preconditioning in the CA1 subfield and other work has shown miRNAs undergo very abrupt induction followed by restitution to baseline or lower levels after seizures (Eacker, et al., 2011, Sano, et al., 2012). Although we detected increased miR-34b/c levels after seizure preconditioning, the levels of miR-34a were not changed. This is consistent with recent work showing miR-34a is upregulated by SE but not following brief seizures (Sano, et al., 2012). Finally, there was no overlap between our profile and the transcriptional response of miRNAs at the same time point after in vivo activation of group 1 metabotropic glutamate receptors (Lusardi, et al., 2012), suggesting seizure preconditioning does not activate this receptor.
As expected, the miRNA expression profile of seizure preconditioning shows major differences from the profile we reported after SE in previously preconditioned animals (Jimenez-Mateos, et al., 2011). Rather, it shares similarities to the response after ischemic preconditioning, where early, mainly increased levels of miRNAs predominate (Lee, et al., 2010). The specific miRNAs altered by ischemic preconditioning are, however, different from those changed by seizure preconditioning. Thus, the preconditioning step drives increased levels of miRNAs in brain which may be specific to the nature of the stimulus. This result is consistent with the idea that preconditioning reprograms the molecular repertoire in a manner tailored to protection against a subsequent and otherwise injurious stressor of a similar nature (Stenzel-Poore, et al., 2007).
Our study also confirmed the presence of key components of the miRNA biogenesis pathway in adult mouse hippocampus. Protein levels for Drosha and Dicer, which regulate primary transcript processing and production of mature miRNA, respectively, were not altered by seizure preconditioning. This agrees with previous work showing these components are not altered after SE or epileptic tolerance (Jimenez-Mateos, et al., 2011). In contrast, a decrease in Ago2 levels occurred. While recent work has demonstrated Ago2 protein is present within mouse hippocampus (Jimenez-Mateos, et al., 2011, Sano, et al., 2012), this is the first study to report a change to Ago2 levels following seizures. Since Ago2 is critical for targeting of mRNAs by miRNAs within the RISC (Peters and Meister, 2007) this change may reduce the capacity of miRNAs to target mRNAs in our model. Ago2 is also capable of directly processing miR-451 (Cheloufi, et al., 2010, Cifuentes, et al., 2010) although this miRNA was not found to be down-regulated in our study. Identification of the mRNA/miRNA contents of the RISC after seizure preconditioning, for example using direct sequencing (Chi, et al., 2009), may provide answers as to whether the drop in Ago2 reduces the capacity of miRNAs to impact their mRNA targets.
Our study supports miR-184 as potentially neuroprotective on the basis that silencing miR-184 made a normally benign episode of seizure preconditioning harmful and increased seizure damage after SE in previously preconditioned animals. The effect was most prominent for CA1, an area that normally undergoes limited cell death after SE induced by intra-amygdala KA (Jimenez-Mateos, et al., 2011), but which displayed extensive degeneration when miR-184 was inhibited. Notably, this was not a consequence of changes to the severity of seizures. Although the CA3 subfield did not appear to be more vulnerable to SE after silencing miR-184, the almost complete damage to this area in the model means a ceiling effect may have prevented an effect of miR-184 antagomirs being detected. Nevertheless, it is notable that basal levels of miR-184 were lower in CA1. Thus, inhibition of miR-184 by antagomirs may have had a stronger effect in this subfield due to reduced miRNA reserve. Taken together, these studies indicate that miR-184, and likely other miRNAs, contribute to the neuroprotection in epileptic tolerance and provide new tools and directions for understanding tolerance in other models.
The finding that miR-184 may be neuroprotective in mouse brain contrasts with work in other cells which identified pro-apoptotic functions of this miRNA via targeting Akt2 (Foley, et al., 2010). Indeed, Akt2 functions as a survival and anti-apoptotic factor (Hers, et al., 2011). Although we did not directly show Akt2 is a target of miR-184 in brain, protein levels of Akt2 followed a reciprocal pattern from miR-184. Down-regulation of Akt2 may, therefore, not be the mechanism by which miR-184 protects neurons. Indeed, the effects of miR-184 may be strongly dependent on the mRNA targets between proliferating versus non-dividing cells and possibly the scale or temporal pattern of induction. Relatively little is known about targets of miR-184. Overexpression of miR-184 in cortical neurons was reported to have no effect on neuronal morphology (Nomura, et al., 2008). However, a recent study reported miR-184 promotes adult neural stem cell proliferation and inhibits differentiation by blocking expression of Numb-like (Liu, et al., 2010). Whether miR-184 contributes to protection via such a mechanism is unknown. Notably, increased neurogenesis is a required component of ischemic tolerance (Maysami, et al., 2008).
In summary, our study provides additional characterization of the electrographic and behavioral features of seizure preconditioning along with the first profile of miRNA expression. We identify miR-184 as a novel regulator of seizure-induced neuronal death. These results contribute to our understanding of the molecular mechanisms underlying epileptic tolerance and may help identify novel neuroprotective strategies to protect against seizure-induced neuronal death.
Supplementary Material
Highlights.
Study is the first to expression profile the microRNA response to epileptic preconditioning in
Identification of miR-184 as a seizure-regulated microRNA in mouse hippocampus
Silencing miR-184 results in increased seizure-induced neuronal death in vivo
Acknowledgments
This work was supported by Science Foundation Ireland grant 08/IN1/B1875, NIH/NINDS award R56 073714, Health Research Board PHD/2007/11, a postdoctoral fellowship from the Irish Research Council for Science Engineering and Technology (to E. J-M.) and Children’s Medical and Research Foundation. The authors would like to thank Daniel Fraughen for technical support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aronica E, Fluiter K, Iyer A, Zurolo E, Vreijling J, van Vliet EA, Baayen JC, Gorter JA. Expression pattern of miR-146a, an inflammation-associated microRNA, in experimental and human temporal lobe epilepsy. Eur J Neurosci. 2010;31:1100–1107. doi: 10.1111/j.1460-9568.2010.07122.x. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges K, Gearing M, McDermott DL, Smith AB, Almonte AG, Wainer BH, Dingledine R. Neuronal and glial pathological changes during epileptogenesis in the mouse pilocarpine model. Exp Neurol. 2003;182:21–34. doi: 10.1016/s0014-4886(03)00086-4. [DOI] [PubMed] [Google Scholar]
- Bozzi Y, Dunleavy M, Henshall DC. Cell signaling underlying epileptic behavior. Front Behav Neurosci. 2011;5:45. doi: 10.3389/fnbeh.2011.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray I, Bryan K, Prenter S, Buckley PG, Foley NH, Murphy DM, Alcock L, Mestdagh P, Vandesompele J, Speleman F, London WB, McGrady PW, Higgins DG, O’Meara A, O’Sullivan M, Stallings RL. Widespread dysregulation of MiRNAs by MYCN amplification and chromosomal imbalances in neuroblastoma: association of miRNA expression with survival. PLoS One. 2009;4:e7850. doi: 10.1371/journal.pone.0007850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Simon R. Ischemic tolerance in the brain. Neurology. 1997;48:306–311. doi: 10.1212/wnl.48.2.306. [DOI] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, Wolfe SA, Giraldez AJ. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford JJ, Usiello A, Vallone D, Kinsella A, Borrelli E, Waddington JL. Topographical evaluation of behavioural phenotype in a line of mice with targeted gene deletion of the D2 dopamine receptor. Neuropharmacology. 2000;39:382–390. doi: 10.1016/s0028-3908(99)00150-1. [DOI] [PubMed] [Google Scholar]
- Davis TH, Cuellar TL, Koch SM, Barker AJ, Harfe BD, McManus MT, Ullian EM. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharap A, Vemuganti R. Ischemic pre-conditioning alters cerebral microRNAs that are upstream to neuroprotective signaling pathways. J Neurochem. 2010;113:1685–1691. doi: 10.1111/j.1471-4159.2010.06735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirnagl U, Simon RP, Hallenbeck JM. Ischemic tolerance and endogenous neuroprotection. Trends Neurosci. 2003;26:248–254. doi: 10.1016/S0166-2236(03)00071-7. [DOI] [PubMed] [Google Scholar]
- Eacker SM, Dawson TM, Dawson VL. Understanding microRNAs in neurodegeneration. Nat Rev Neurosci. 2009;10:837–841. doi: 10.1038/nrn2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eacker SM, Keuss MJ, Berezikov E, Dawson VL, Dawson TM. Neuronal activity regulates hippocampal miRNA expression. PLoS One. 2011;6:e25068. doi: 10.1371/journal.pone.0025068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore R, Siegel G, Schratt G. MicroRNA function in neuronal development, plasticity and disease. Biochim Biophys Acta. 2008;1779:471–478. doi: 10.1016/j.bbagrm.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Foley NH, Bray IM, Tivnan A, Bryan K, Murphy DM, Buckley PG, Ryan J, O’Meara A, O’Sullivan M, Stallings RL. MicroRNA-184 inhibits neuroblastoma cell survival through targeting the serine/threonine kinase AKT2. Mol Cancer. 2010;9:83. doi: 10.1186/1476-4598-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidday JM. Cerebral preconditioning and ischaemic tolerance. Nat Rev Neurosci. 2006;7:437–448. doi: 10.1038/nrn1927. [DOI] [PubMed] [Google Scholar]
- Harvey BD, Sloviter RS. Hippocampal granule cell activity and c-Fos expression during spontaneous seizures in awake, chronically epileptic, pilocarpine-treated rats: implications for hippocampal epileptogenesis. J Comp Neurol. 2005;488:442–463. doi: 10.1002/cne.20594. [DOI] [PubMed] [Google Scholar]
- Hatazaki S, Bellver-Estelles C, Jimenez-Mateos EM, Meller R, Bonner C, Murphy N, Matsushima S, Taki W, Prehn JH, Simon RP, Henshall DC. Microarray profile of seizure damage-refractory hippocampal CA3 in a mouse model of epileptic preconditioning. Neuroscience. 2007;150:467–477. doi: 10.1016/j.neuroscience.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Hers I, Vincent EE, Tavare JM. Akt signalling in health and disease. Cell Signal. 2011;23:1515–1527. doi: 10.1016/j.cellsig.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Hu K, Zhang C, Long L, Long X, Feng L, Li Y, Xiao B. Expression profile of microRNAs in rat hippocampus following lithium-pilocarpine-induced status epilepticus. Neurosci Lett. 2011;488:252–257. doi: 10.1016/j.neulet.2010.11.040. [DOI] [PubMed] [Google Scholar]
- Jimenez-Mateos EM, Bray I, Sanz-Rodriguez A, Engel T, McKiernan R, Mouri G, Tanaka K, Sano T, Saugstad JA, Simon RP, Stallings RL, Henshall DC. MicroRNA expression profile after status epilepticus and hippocampal neuroprotection by targeting miR-132. Am J Pathol. 2011;179:2519–2532. doi: 10.1016/j.ajpath.2011.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Mateos EM, Engel T, Merino-Serrais P, McKiernan RC, Tanaka K, Mouri G, Sano T, O’Tuathaigh C, Waddington JL, Prenter S, Delanty N, Farrell MA, O’Brien DF, Conroy RM, Stallings RL, Defelipe J, Henshall DC. Silencing microRNA-134 produces neuroprotective and prolonged seizure-suppressive effects. Nat Med. 2012 doi: 10.1038/nm.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Mateos EM, Hatazaki S, Johnson MB, Bellver-Estelles C, Mouri G, Bonner C, Prehn JH, Meller R, Simon RP, Henshall DC. Hippocampal transcriptome after status epilepticus in mice rendered seizure damage-tolerant by epileptic preconditioning features suppressed calcium and neuronal excitability pathways. Neurobiol Dis. 2008;32:442–453. doi: 10.1016/j.nbd.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Jimenez-Mateos EM, Henshall DC. Seizure preconditioning and epileptic tolerance: models and mechanisms. Int J Physiol Pathophysiol Pharmacol. 2009;1:180–191. [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Mateos EM, Mouri G, Conroy RM, Henshall DC. Epileptic tolerance is associated with enduring neuroprotection and uncoupling of the relationship between CA3 damage, neuropeptide Y rearrangement and spontaneous seizures following intra-amygdala kainic acid-induced status epilepticus in mice. Neuroscience. 2010;171:556–565. doi: 10.1016/j.neuroscience.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2010;28:1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Lee ST, Chu K, Jung KH, Yoon HJ, Jeon D, Kang KM, Park KH, Bae EK, Kim M, Lee SK, Roh JK. MicroRNAs induced during ischemic preconditioning. Stroke. 2010;41:1646–1651. doi: 10.1161/STROKEAHA.110.579649. [DOI] [PubMed] [Google Scholar]
- Liu C, Teng ZQ, Santistevan NJ, Szulwach KE, Guo W, Jin P, Zhao X. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell. 2010;6:433–444. doi: 10.1016/j.stem.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DZ, Tian Y, Ander BP, Xu H, Stamova BS, Zhan X, Turner RJ, Jickling G, Sharp FR. Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J Cereb Blood Flow Metab. 2010;30:92–101. doi: 10.1038/jcbfm.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusardi TA, Farr CD, Faulkner CL, Pignataro G, Yang T, Lan J, Simon RP, Saugstad JA. Ischemic preconditioning regulates expression of microRNAs and a predicted target, MeCP2, in mouse cortex. J Cereb Blood Flow Metab. 2010;30:744–756. doi: 10.1038/jcbfm.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusardi TA, Thompson SJ, Macdonald IC, Lan JQ, Theofilas P, Saugstad JA. Effect of (S)-3, 5-DHPG on microRNA expression in mouse brain. Exp Neurol. 2012 doi: 10.1016/j.expneurol.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maysami S, Lan JQ, Minami M, Simon RP. Proliferating progenitor cells: a required cellular element for induction of ischemic tolerance in the brain. J Cereb Blood Flow Metab. 2008;28:1104–1113. doi: 10.1038/jcbfm.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKiernan RC, Jimenez-Mateos EM, Bray I, Engel T, Sano T, Delanty N, Farrell MA, O’Brien DF, Meller R, Simon RP, Stallings RL, Henshall DC. Reduced mature microRNA levels in association with Dicer loss in human temporal lobe epilepsy with hippocampal sclerosis. PLoS One. 2012 doi: 10.1371/journal.pone.0035921. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Delaney SF, Das S, Sano T, Jimenez-Mateos EM, Bryan K, Buckley PG, Stallings RL, Henshall DC. Differential DNA methylation patterns define status epilepticus and epileptic tolerance. J Neurosci. 2012;32:1577–1588. doi: 10.1523/JNEUROSCI.5180-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Kimura M, Horii T, Morita S, Soejima H, Kudo S, Hatada I. MeCP2-dependent repression of an imprinted miR-184 released by depolarization. Hum Mol Genet. 2008;17:1192–1199. doi: 10.1093/hmg/ddn011. [DOI] [PubMed] [Google Scholar]
- Nudelman AS, DiRocco DP, Lambert TJ, Garelick MG, Le J, Nathanson NM, Storm DR. Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus. 2010;20:492–498. doi: 10.1002/hipo.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. The mouse brain in stereotaxic coordinates. 2. Elsevier; San Diego, CA: 2001. [Google Scholar]
- Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Pichardo-Casas I, Goff LA, Swerdel MR, Athie A, Davila J, Ramos-Brossier M, Lapid-Volosin M, Friedman WJ, Hart RP, Vaca L. Expression profiling of synaptic microRNAs from the adult rat brain identifies regional differences and seizure-induced dynamic modulation. Brain Res. 2012;1436:20–33. doi: 10.1016/j.brainres.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T, Reynolds JP, Jimenez-Mateos EM, Matsushima S, Taki W, Henshall DC. MicroRNA-34a upregulation during seizure-induced neuronal death. Cell Death Dis. 2012;3:e287. doi: 10.1038/cddis.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saugstad JA. MicroRNAs as effectors of brain function with roles in ischemia and injury, neuroprotection, and neurodegeneration. J Cereb Blood Flow Metab. 2010;30:1564–1576. doi: 10.1038/jcbfm.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, O’Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R, Greengard P. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204:1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmued LC, Hopkins KJ. Fluoro-Jade B: a high affinity fluorescent marker for the localization of neuronal degeneration. Brain Res. 2000;874:123–130. doi: 10.1016/s0006-8993(00)02513-0. [DOI] [PubMed] [Google Scholar]
- Song YJ, Tian XB, Zhang S, Zhang YX, Li X, Li D, Cheng Y, Zhang JN, Kang CS, Zhao W. Temporal lobe epilepsy induces differential expression of hippocampal miRNAs including let-7e and miR-23a/b. Brain Res. 2011;1387:134–140. doi: 10.1016/j.brainres.2011.02.073. [DOI] [PubMed] [Google Scholar]
- Stapels M, Piper C, Yang T, Li M, Stowell C, Xiong ZG, Saugstad J, Simon RP, Geromanos S, Langridge J, Lan JQ, Zhou A. Polycomb group proteins as epigenetic mediators of neuroprotection in ischemic tolerance. Sci Signal. 2010;3:ra15. doi: 10.1126/scisignal.2000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel-Poore MP, Stevens SL, King JS, Simon RP. Preconditioning reprograms the response to ischemic injury and primes the emergence of unique endogenous neuroprotective phenotypes: a speculative synthesis. Stroke. 2007;38:680–685. doi: 10.1161/01.STR.0000251444.56487.4c. [DOI] [PubMed] [Google Scholar]
- Stenzel-Poore MP, Stevens SL, Xiong Z, Lessov NS, Harrington CA, Mori M, Meller R, Rosenzweig HL, Tobar E, Shaw TE, Chu X, Simon RP. Effect of ischaemic preconditioning on genomic response to cerebral ischaemia: similarity to neuroprotective strategies in hibernation and hypoxia-tolerant states. Lancet. 2003;362:1028–1037. doi: 10.1016/S0140-6736(03)14412-1. [DOI] [PubMed] [Google Scholar]
- Tao J, Wu H, Lin Q, Wei W, Lu XH, Cantle JP, Ao Y, Olsen RW, Yang XW, Mody I, Sofroniew MV, Sun YE. Deletion of astroglial dicer causes non-cell-autonomous neuronal dysfunction and degeneration. J Neurosci. 2011;31:8306–8319. doi: 10.1523/JNEUROSCI.0567-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdinguio RG, Sanchez-Mut JV, Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol. 2009;8:1056–1072. doi: 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci U S A. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.