SUMMARY
Aminoacyl-tRNA synthetases (AARSs) catalyze aminoacylation of tRNAs in the cytoplasm. Surprisingly, AARSs also have critical extracellular and nuclear functions. Evolutionary pressure for new functions might be manifested by splice variants that skip only an internal catalytic domain (CD) and link non-catalytic N- and C-terminal polypeptides. Using disease-associated histidyl-tRNA synthetase (HisRS) as an example, we discovered an expressed 171 amino acid protein (HisRSΔCD) that deleted the entire CD, and joined an N-terminal WHEP to the C-terminal anticodon-binding domain (ABD). X-ray crystallographic and 3-D NMR methods revealed the first structures of human HisRS and HisRSΔCD. In contrast to homodimeric HisRS, HisRSΔCD is monomeric, where rupture of the ABD’s packing with CD resulted in a dumbbell-like structure of flexibly linked WHEP and ABD domains. In addition, the ABD of HisRSΔCD presents a new local conformation. This natural internally deleted HisRS suggests evolutionary pressure to reshape AARS tertiary and quaternary structures for repurposing.
INTRODUCTION
Although AARSs catalyze the aminoacylation of tRNAs in the first step of protein synthesis in the cytoplasm, numerous reports document their activities in nuclear and extracellular locations, where translation does not occur (Fu et al., 2012; Guo et al., 2010; Kim et al., 2011; Martinis and Joy Pang, 2007; Park et al., 2012; Park et al., 2008; Sajish et al., 2012; Xu et al., 2012). These activities include major roles in regulating angiogenesis (Xu et al., 2012; Yao et al., 2012), inflammatory responses (Arif et al., 2009; Fu et al., 2012; Lee et al., 2012), mTOR signaling (Bonfils et al., 2012; Han et al., 2012), and tumor growth (Dorrell et al., 2007; Park et al., 2012). In at least some instances, a fragment produced by natural proteolysis is the active factor. These fragments typically remove an external N- or a C-terminal peptide and leave intact all or most of the internal catalytic domain. Indeed, a CD pocket is used by a natural tryptophanyl-tRNA synthetase (TrpRS) fragment to bind to the extracellular domain of VE-cadherin on endothelial cells to prevent the assembly of blood vessels (Zhou et al., 2010). These observations suggest evolutionary pressures to expropriate AARSs for functions outside of the cytoplasm, perhaps because of their close association with the origin and creation of the genetic code, and the latter’s capacity to evolve new functions and species in response to changes in the environment (Giegé, 2008).
With that consideration in mind, we felt that a clear manifestation of these selective pressures and their consequences would be the appearance of forms of AARSs that could not arise from proteolysis, but rather from alternative splicing that specifically removed the internal CD and little else. (The one well studied example of an AARS splice variant, mini TrpRS, removes the N-terminal 48 amino acids and leaves the entire CD intact (Wakasugi et al., 2002). It has full catalytic activity.) An exquisitely tailored deletion that only excised the CD would suggest strong selective pressures to create forms that were catalytically inactive and therefore presumably designed for repurposing. The structural consequences of an internal deletion of this sort are unknown. To investigate this question, we chose HisRS, which is associated with idiopathic inflammatory myopathies (IIM) and interstitial lung disease (ILD) (Bernstein et al., 1984; Jura et al., 2007). Our rationale was that the roots of this disease-association could be related to a variant of HisRS that was designed for another function, which itself was connected to inflammatory responses.
We identified the alternatively spliced forms of the gene for human HisRS by the high-throughput deep-sequencing method, and discovered a splice variant HisRSΔCD that skips exons encoding the entire CD. This splice variant encodes an endogenously expressed protein with the N-terminal WHEP-domain joined to the C-terminal ABD. It showed enriched expression in human lung tissue and interacted with Jo-1 antibodies of human myositis patients, implicating the connection to the autoimmune diseases IIM and ILD. Employing crystallographic and NMR techniques, we revealed the first structures of human HisRS and HisRSΔCD. Different from homodimeric HisRS, HisRSΔCD is monomeric. Release of the ABD’s packing with CD resulted in a dumbbell-like structure of flexibly linked WHEP and ABD domains and the ABD presents a new local conformation, readily allowing novel interaction partners and non-conventional biological activities. Our study extends the understanding of structure and function of the AARS family of ancient enzymes and suggests evolutionary pressure to reshape AARS tertiary and quaternary structures for repurposing.
RESULTS AND DISCUSSION
Comprehensive Identification of Alternative Splice Variants of Human HisRS by Deep Sequencing of AARS-Transcriptome Enriched cDNA
Based on its sequence, the 509 amino acid human HisRS is a class II tRNA synthetase composed of a core catalytic domain, a C-terminal anticodon binding domain, and an N-terminal WHEP domain (Figure 1A). The catalytic aminoacylation domain is shared by all class II tRNA synthetases, which have a characteristic 7-stranded β-structure and flanking α-helices, with 3 class-defining conserved sequence motifs (Carter, 1993; Li et al., 2011). The WHEP domain is a 50-amino-acid, helix-turn-helix motif present in one or more copies in tryptophanyl-, histidyl-, glutamyl-prolyl-, glycyl- and methionyl-tRNA synthetases, and takes its name from the first three (W, H and EP). This domain was appended to specific AARSs as the tree of life ascended from lower to higher eukaryotes. It first appeared in HisRS in nematodes and was retained ever since (Guo et al., 2010). Although the functional role of the HisRS WHEP domain remains unclear, it harbors epitope(s) targeted by autoantibodies of IIM/ILD patients (Jura et al., 2007; Martin et al., 1995; Raben et al., 1994). Until now, no crystal structures of human HisRS or expressed splice variant proteins have been reported.
Figure 1. Identification and Validation of a HisRS Splice Variant That Skips the Entire Catalytic Domain.
(A) Schematic illustration of human HisRS protein and the identified exon-skipping splicing events. Human HisRS is composed of an N-terminal WHEP domain, a core catalytic aminoacylation domain and a C-terminal anticodon binding domain. The three conserved sequence motifs in its co re active site are indicated by green, blue and brown bars, respectively. The mRNA transcript of the gene for human HisRS is shown below and aligns with its encoded protein. Canonical exons are drawn in scale to the nucleotide length and are labeled consecutively. The splicing events identified by deep sequencing in the current study are illustrated by dashed lines to indicate non-canonical exon junctions. Novel exon junctions that were not annotated in the AceView database, including ΔE3–10 (HisRSΔCD), are indicated by red lines, and the previously reported ones are shown in grey. Targeting sites of the PCR primers are indicated by blue arrows and those of qPCR primers by green arrows. The primer sequences are shown in Table S2.
(B) Validation by PCR of the splice variant that skips exons 3 to 10. PCR was performed using cDNA of IMR-32 neuronal cells and a pair of primers targeting 5’-UTR/Exon1 (FP) and 3’-UTR (RP) of the gene for HisRS. PCR products were separate by agarose gel electrophoresis. Lane 1: PCR by FP and RP, Lane 2: PCR by primers targeting GAPDH as control.
(C) Sequencing result shows the novel Exon2–11 junction in the HisRSΔCD transcript.
(D) Schematic of protein products of human HisRS FL and HisRSΔCD. The protein product of splice variant HisRSΔCD has the entire aminoacylation domain (aa61–398) removed due to skipping of exons 3 to 10 and therefore directly connects the WHEP domain to the ABD.
(E) Detection of endogenous HisRSΔCD protein by western blot analysis. HisRSΔCD protein was detected in whole extracts of IMR-32 cells using antibodies against, separately, the N- and C-terminus of HisRS (N-mAb, monoclonal antibody against HisRS aa1–97; C-pAb, polyclonal antibody against HisRS C-terminus). Total lysates of 293T cells transiently transfected with a HisRSΔCD construct were run in parallel with IMR-32 cell extracts to serve as a control that shows the size of HisRSΔCD. The expected running position of the HisRSΔCD protein is indicated by an arrow.
See also Figure S1, and Tables S1 and S2.
For a comprehensive identification of alternatively spliced forms of HisRS, we employed the high-throughput transcriptome sequencing technique. Because whole transcriptome sequencing limits the read-depth of the exome of individual genes, we developed an amplification–based transcriptome sequencing method for a more thorough discovery of splice variants. (Previous work annotated a number of alternative mRNA variants of the gene for human HisRS by sequencing of cDNA clones (Thierry-Mieg and Thierry-Mieg, 2006). However, as annotated in the AceView database, these splice variants include truncation of the 5' or 3' ends, overlapping exons with different boundaries, splicing versus retention of introns, and the presence or absence of cassette exons. Also, expression of either recombinant or natural versions of these potential splice variants has never been investigated.) The RNA was reverse transcribed to cDNA by primers specific to the target gene, and then amplified using primers targeting their exon regions at positions close to the exon-exon junctions. This method allowed sensitive detection of low-abundant splice variants and was mainly designed for discovery of splice variants having exon-skipping events.
A complete and more comprehensive description of the AARS-specific transcriptome approach will be published separately. In brief, the transcriptome for HisRS was first enriched with RNA of human adult brain, fetal brain, total leukocytes, Jurkat T, Raji B lymphocytes or THP monocytes, and then followed with deep sequencing. When compared to other human exome sequencing efforts, this method was found to significantly enhance the sequencing depth, yielding a > 1000 fold increase in sequencing reads after enrichment. The exon-skipping events in the gene for HisRS were concentrated on the region of exons 3 to 10 which encode the aminoacylation domain (Figure 1A, Table S1). If these splice variants give rise to protein products, the generated HisRS isoforms are expected to have partially or completely disrupted enzymatic activity. Thus, these splice variants may be endowed with novel biological functions through new domain compositions and structures. Possibly, they could be immunogenic or associated with pathologies when abnormally regulated or secreted.
Validation and Expression Analysis of a Splice Variant HisRSΔCD That Skips the Entire Catalytic Domain
HisRSΔCD, the splice variant with the largest internal deletion, has the skipping of exons 3 to 10 (ΔE3–10) that encode the entire aminoacylation domain (Figure 1A). We found HisRSΔCD with 50 sequencing reads in human adult brain and 7 reads in Jurkat T lymphocyte (Table S1). The putative protein product would carry no aminoacylation activity, but retained the N-terminal 60 amino acids and the C-terminal ABD. To further verify this splice variant and obtain a more complete sequence of its transcript, the polymerase chain reaction (PCR) was performed. Because HisRSΔCD was identified with highest reads in the adult brain, we employed a human neuronal cell line IMR-32 for its validation. The PCR reaction with the cDNA template of mRNA from IMR-32 cells, together with a pair of primers targeting the 5’-UTR/Exon1 and 3’-UTR regions of the gene for HisRS (FP and RP), amplified a product with a size shorter than the expected band of the full-length (FL) transcript (Figures 1A and 1B, Table S2). This shorter PCR product was subjected to sequencing, and confirmed to bear the Exon2–11 junction of HisRSΔCD (Figure 1C). Based on the sequence of the PCR product, the HisRSΔCD transcript has exons 3 to 10 (1014 nt) removed, but still retains the 5’- and 3’-UTR region and the remaining exons of the FL transcript. It is therefore expected to translate into a protein with the in-frame deletion of the entire aminoacylation domain (aa61–398), and thereby join the N-terminal WHEP domain to the C-terminal ABD (Figure 1D).
We employed the SYBR green quantitative real-time PCR (qPCR) method to examine the mRNA expression level of native HisRS and of HisRSΔCD transcripts in various human tissues. We first optimized qPCR reactions to produce specific PCR products with high efficiency. All amplified products were designed to cover exon junctions and to exclude amplicons derived from intronic regions. After optimization, a pair of primers targeting Exon9 and Exon10 (qFP1 and qRP1) was used for amplification of the FL transcripts (Figure 1A, Table S2). For HisRSΔCD a pair of primers targeting Exon2 and Exon11 (qFP2 and qRP2) was employed in qPCR reactions having a short extension time (30 sec), which thereby attenuated amplification of the longer FL transcript. Using the optimized qPCR reactions, the presence of the HisRS transcripts was analyzed across 13 human tissues, including those of the immune system (total leukocytes, bone marrow, spleen), circulatory system (lung, heart, kidney), digestive system (liver, pancreas, small intestine, colon), and others (thyroid, adipose cells, skeletal muscle). The FL transcript for HisRS was found to be evenly distributed among the various tissues that were tested, with no more than 3 times deviation from the median value (Figure S1A). In contrast, HisRSΔCD exhibited a differential mRNA expression profile, with the mRNA level highest in lung (about 11-fold above the median level, Figure S1B), and second highest in spleen (about 5-fold above the median). Although a causal relationship to IIM/ILD cannot be inferred from these results, it is nonetheless of interest to note that the mRNA expression of HisRSΔCD relative to HisRS FL is prominently higher in lung than in other tissues (Figure S1C). It has been suggested that the lung is the site of disease initiation in Jo-1 (HisRS) autoantibody associated ILD (Danoff and Casciola-Rosen, 2011).
Next, we used western blot methods to detect the HisRSΔCD splice variant. For this purpose, we used antibodies raised against the N- and, separately, the C-terminal region. In view of the relatively small amounts of the mRNAs that correspond to these splice variants, and due to the difficulty in obtaining adequate amounts of human tissues, human cell lines cultured in vitro were employed. Because the HisRSΔCD transcript was detected in the total RNA of IMR-32 cells, its protein product was probed using total cell extracts of IMR-32 cells with a monoclonal antibody raised against the N-terminus (aa1–97) of human HisRS, and a polyclonal antibody generated by a peptide from the C-terminus. Both antibodies reacted with the same species having a MW of about 20 kDa (Figure 1E; see also Figure S1D). The size of this protein corresponds to the recombinant HisRSΔCD protein overexpressed in HEK 293T cells. Consistent with the relatively low amounts of mRNA that have been annotated, HisRSΔCD is clearly in much lower abundance (estimated as slightly above 1% of HisRS FL on western blots probed with the same antibodies).
Structure Determination of Human HisRS by X-Ray Crystallography
Crystal structures of E. coli and T. thermophilus and of eukaryotic parasites T. brucei and T. cruzi in apo and histidine- or His-AMP- bound forms have been published (Aberg et al., 1997; Arnez et al., 1995; Merritt et al., 2010). All are α2 dimers and, as expected for a class II synthetase, all have the characteristic and well conserved anti-parallel β-sheet fold flanked by α-helices in the catalytic domain. The HisRSs all have an α/β fold in the anticodon binding domain. The adenine binding pocket and the topology of an extra domain inserted between the characteristic conserved motifs 2 and 3 of the class II AARS catalytic core is substantially different in bacterial and eukaryote parasitic forms of the enzyme. So far no structure has been reported for a higher eukaryote form of HisRS.
Purified recombinant human HisRS protein aggregated in the normal buffer conditions we used. By mapping cysteines in the human HisRS sequence to the known structures of T. brucei and T. cruzi HisRS, we found that two cysteines (C507 and C509) at the very C-terminus may be solvent-exposed and non-specifically form intermolecular disulfide bonds. Therefore, we removed (from the HisRS-encoding recombinant gene) three residues at the C-terminus (Figure 2A). Indeed, this truncation mutant Δ507–509 did not aggregate on PAGE gels, even without added reducing agent such as DTT. Using this C-terminal truncation mutant we obtained large crystals, which diffracted to 3.1 Å resolution (Figure S2A). The space group was determined to be P41212 with the unit cell dimensions a = b = 100.4 Å, c = 257.1 Å (Table 1). The crystal structure was solved by molecular replacement using T. brucei HisRS (PDB: 3HRI) as the template.
Figure 2. Structure Determination of Human HisRS by X-ray Crystallography.
(A) Optimization of the boundary of human HisRS for high quality crystals. The amino acid range included in each mutant and the corresponding crystal resolutions are shown. The three conserved sequence motifs as well as the insertion domain in the aminoacylation domain are indicated by colored bars and labeled below.
(B) Ribbon diagram of the 2.4 Å crystal structure of HisRS Δ1–53_507–509 (blue: CD, green: ABD). The secondary structure α-helices and β-strands are consecutively labeled, and the amino acids composing these structural elements are shown in detail in Figure S2D.
(C) Structure comparison of HisRSs of different species including human, trypanosoma (T. brucei, PDB: 3HRI), archaea (T. acidophilum, 1WU7) and bacterium (S. aureus, 1QE0).
(D) Superposition of the insertion domains of human, trypanosoma and archaea shows differences in the respective orientations of this domain. The α-helices in the insertion domain of human HisRS are labeled, and the corresponding amino acid sequence and the alignment among different species are shown in Figure S2D.
Table 1.
Statistics of Data Collection and Model Refinement of Human HisRS Crystal and NMR Structures
| Crystal structures of HisRS variants | Family of 20 NMR structures of HisRSΔCD*d | |||
|---|---|---|---|---|
| HisRS Δ507–509 | HisRS Δ1–53_ Δ507–509 | NMR distance and dihedral constraints | ||
| Data collection | Total NOE | 2397 | ||
| Space group | P41212 | P41212 | Intra-residue | 978 |
| Unit cell parameters (Å) | a = b = 100.4, c = 257.1 | a = b = 93.5, c = 254.5 | Inter-residue | |
| Resolution range (Å) | 50 − 3.1 (3.15 − 3.1)a | 50 − 2.4 (2.44 − 2.4) | Sequential (|i − j| = 1) | 491 |
| No. of unique reflections | 24210 (1191) | 45571 (2212) | Medium-range (|i − j| < 4) | 404 |
| Redundancy | 5.5 (5.6) | 6.1 (5.8) | Long-range (|i − j| > 5) | 929 |
| I/σ | 22.6 (2.4) | 21.8 (2.3) | Hydrogen bonds | 148 |
| Completeness (%) | 98.8 (99.9) | 99.8 (99.9) | Total dihedral angle restraints | 216 |
| Rmerge (%)b | 6.9 (69.6) | 7.4 (67.9) | Structure statistics | |
| Structure refinement | Violations (mean and s.d.) | |||
| Resolution (Å) | 50 − 3.1 (3.2 − 3.1) | 50 − 2.4 (2.49 − 2.4) | Distance constraints (Å) | 0.001 ± 0.001 |
| Rcryst / Rfree (%)c | 27.1 (34.4) / 32.7 (40.0) | 19.1 (26.8) / 25.0 (35.1) | Dihedral angle constraints (°) | 0.574 ± 0.063 |
| Rmsd bonds (Å) / angles (°) | 0.014 / 1.5 | 0.005 / 0.9 | Deviations from idealized geometry | |
| Average B factor | 55 | 54.4 | Bond lengths (Å) | 0.002 ± 0.000 |
| No. of atoms | Bond angles (°) | 0.429 ± 0.022 | ||
| Protein atoms | 6430 | 6907 | Impropers (°) | 0.347 ± 0.047 |
| Water molecules | 166 | Ramachandran plote (%) | ||
| Other molecules | 21 | Most favorable regions | 79.7 | |
| No. of reflections | Additional allowed regions | 14.2 | ||
| Working set | 22659 | 42096 | Generously allowed regions | 4.3 |
| Test set | 1213 | 2124 | Disallowed regions | 1.8 |
| Ramachandran plot | Coordinate precision | |||
| Most favored regions (%) | 88.2 | 92.7 | Atomic Rmsdf (Å) | |
| Additionally allowed (%) | 11 | 7.3 | Residues 1–45 for the WHEP domain | |
| Generously allowed (%) | 0.8 | 0 | Heavy | 1.081 |
| Backbone | 0.447 | |||
| Residues 69–165 for the ABD | ||||
| Heavy | 1.468 | |||
| Backbone | 0.872 | |||
Numbers in parentheses represent the value for the highest resolution shell.
Rmerge = Σ|Ii − Im|/ΣIi, where Ii is the intensity of the measured reflection and Im is the mean intensity of all symmetry related reflections.
Rcryst = Σ||Fobs| − |Fcalc||/Σ|Fobs|, where Fobs and Fcalc are observed and calculated structure factors. Rfree = ΣT||Fobs| − |Fcalc||/ΣT|Fobs|, where T is a test data set of about 5% of the total reflections randomly chosen and set aside prior to refinement.
None of the structures exhibits distance violations greater than 0.3 Å or dihedral angle violations greater than 4°.
The program Procheck was used to assess the overall quality of the structures.
The precision of the atomic coordinates is defined as the average Rmsd between 20 final structures and the mean coordinates of the protein.
In the crystal structure of human HisRS Δ507–509 (PDB: 4G84), the N-terminal WHEP domain is not visible, indicating that this domain is not tightly packed with the structural core (Figure S2B). The loose packing of the N-terminal domain is also supported by a previous study of the trypanosomal HisRS, which showed that its N-terminus was amenable to enzymatic cleavage during expression (Merritt et al., 2010). To further improve crystal quality, we optimized the boundary of HisRS by removing the flexible N-terminal region in different mutants and achieved a crystal structure at 2.4 Å resolution with the deletion mutant Δ1–53_507–509 (Figures 2A and S2A). The space group was determined to be P41212 with the unit cell dimensions a = b = 93.5 Å, c = 254.5 Å (Table 1). This new crystal form has the same space group and similar unit cell dimensions as the previous one, suggesting that the Δ1–53_507–509 mutant keeps the same conformation and crystal packing as the Δ507–509 mutant. Indeed, its structure is essentially identical to that of the HisRS Δ507–509 (Figures 2B and S2B), and shows a dimeric composition that agrees with the molecular weight determined by size exclusion chromatography (Figure S2C).
According to the structure of human HisRS Δ1–53_507–509 (PDB: 4G85), the overall fold of the CD and ABD of human HisRS is similar to its bacterial, archaeal and T. brucei and T. cruzi orthologs (Figure 2C). The most prominent difference among these structures is in the additional domain inserted between conserved motifs 2 and 3 of the class II catalytic core. This insertion domain increases in size from prokaryotes to eukaryotes, and is not conserved in either sequence or structure between prokaryotic and eukaryotic HisRSs (Figure S2D). With the exception of a missing α9 helix in the insertion domain of eukaryote parasite homologs, the secondary structure elements of human HisRS are similar to those of the parasite homologs. It was previously proposed that the insertion domain may contact the acceptor stem of the tRNA (Freist et al., 1999). Superposition of structural cores of human, T. brucei, T. cruzi and T. thermophilus HisRS also reveals the orientation difference of the insertion domains (Figure 2D).
Structure Determination of the Splice Variant HisRSΔCD by NMR Spectroscopy
Similar to native HisRS, wild-type HisRSΔCD forms oligomers even in the presence of 1 mM DTT (Figure S3A). To avoid the disulfide formation, the C-terminal Cys169 and Cys171 (corresponding to C507 and C509 in HisRS) were mutated to serines (2C2S, Figure 3A). The HisRSΔCD_2C2S proteins were mostly monomeric in solution (Figures S3A and S3B), and the 1H-15N heteronuclear single quantum coherence (HSQC) spectrum showed an increase in peak count and more uniform peak shape compared to that of the wild-type HisRSΔCD (Figures S3C and S3D). But the peak number was still less than expected and, together with the presence of broadened peaks, we concluded that this protein still had non-specific interactions and was not sufficiently homogeneous for structure determination. Based on the solved crystal structure of HisRS, it seemed likely that the absence of the CD in HisRSΔCD exposed the hydrophobic inter-domain interface of the ABD, leading to non-specific hydrophobic interactions and thereby introducing heterogeneity. To decrease the hydrophobicity, Trp94 (corresponding to Trp432 in the numbering of the sequence of HisRS and located at the center of the hydrophobic interface) was substituted by the more hydrophilic Gln (Figure 3A). This substitution greatly improved the protein homogeneity as demonstrated by the 1H-15N HSQC spectrum, which displays well dispersed peaks with a peak yield > 95% (Figure 3B). In addition, comparing HisRSΔCD 2C2S_W94Q mutant (HisRSΔCD*) to the wild-type and 2C_2S mutant, the shared HSQC peaks of the proteins exhibited no obvious chemical shifts, thus indicating that the substitutions did not alter the protein conformation (Figures S3C and S3E). Therefore, the HisRSΔCD* mutant was used for further structural characterizations.
Figure 3. Structure Determination of the Splice Variant HisRSΔCD by NMR Spectroscopy.
(A) Schematic of HisRSΔCD* (2C2S_W94Q) mutant employed for structural characterizations. The mutational sites are labeled in red and the corresponding C507, C509 and W432 residues in the native HisRS sequence are also indicated.
(B) The 1H-15N HSQC spectrum of HisRSΔCD* used for structure determination.
(C) Backbone superimposition of 20 calculated lowest-energy structures of the WHEP domain and the ABD of HisRSΔCD* are shown, and the HisRSΔCD* structure is presented below by ribbon diagram. WHEP domain (red) and ABD (green) are well folded and linked by a flexible loop.
(D) Superposition of NMR structures of HisRSΔCD* and of the WHEP domain alone (PDB: 1X59). Structures are shown in ribbon diagram. Red: WHEP domain of HisRSΔCD*, pink: 1X59.
(E) Superposition of human HisRS Δ1–53_507–509 crystal structure and HisRSΔCD* NMR structure. The W432 in HisRS FL was labeled in red. The circled area including helix α15 and the preceding loop had the most prominent differences (see also Figure S2D). Structures are shown in ribbon diagram. Orange: ABD of HisRSΔCD*, Green: ABD of HisRS FL, grey: CD of HisRS FL.
See also Table 1 and Figures S2 and S3.
We first tried to solve the structure of HisRSΔCD* by X-ray crystallography, but extensive trials failed to obtain well diffracting crystals. Because the 15N-1H HSQC spectrum of HisRSΔCD* is well dispersed and contains only one set of peaks, showing that this protein forms a well-folded structure in solution, we could determine its structure to a high resolution by NMR spectroscopy (Figure 3B, Table 1, PDB: 2LW7). In contrast to homodimeric HisRS, HisRSΔCD* behaved as a monomer in solution, which is probably due to the lack of the CD that facilitates dimerization of HisRS. The ensemble of 20 NMR structures of HisRSΔCD* are well defined, with a RMSD of 0.447 Å for backbone atoms and 1.081 Å for heavy atoms of the WHEP domain, and a RMSD of 0.872 Å for backbone atoms and 1.468 Å for heavy atoms of the ABD (Figure 3C). Its WHEP domain adopts an antiparallel bi-helical structure. (An NMR structure of a 73-aa peptide, comprised of the first 60 amino acids of human HisRS and 13 amino acids derived from a recombinant plasmid construct that was used, is documented in the PDB (1X59). According to our solution structure, the WHEP domain of this peptide is identical to that of the WHEP domain of HisRSΔCD* that is shown in Figure 3D.) The ABD of HisRSΔCD* forms a compact mixed α/β fold. The WHEP and ABD domains are connected by a 27 amino acid, highly flexible linker (Figure 3C). No long-distance NOE couplings between the residues of the two domains were found in the NMR spectra. The lack of couplings indicates that the two domains do not make contacts. As a further support, we purified the ABD (aa398–506) alone and, when comparing its HSQC spectrum with that of the ABD in HisRSΔCD*, these amino acids showed largely the same chemical shifts (Figure S3F). Thus, HisRSΔCD* appears as a dumbbell-like structure with “free-floating” N- and C-terminal domains. This structure appears poised to interact with two different partners and, with the flexible linker, to be able to adapt to a variable separation of these partners.
In HisRSΔCD*, the packing interactions of the ABD with the CD have been released. Comparing the ABDs in the NMR structure of HisRSΔCD* with that of the crystal structure of HisRS Δ1–53_507–509, the overall folds remain the same (Figure 3E). However, a prominent difference was found at helix α15 and the loop preceding it (Figure S2D), which are rigidly packed with the CD in HisRS. In HisRSΔCD* this region becomes flexible and moves inward, due to the lack the interaction with the CD.
Possible Association of HisRSΔCD with IIM and ILD
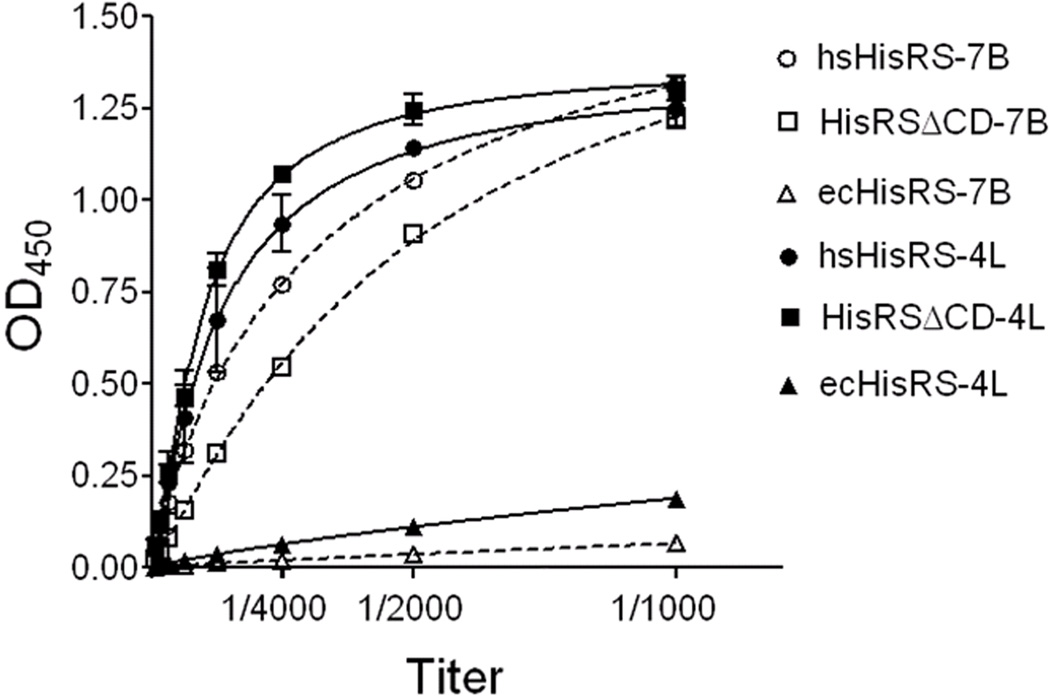
As stated earlier, HisRS is associated with IIM and ILD. HisRS or its constituent peptides have been implicated in the etiology of these diseases. For patients with IIM or ILD, Jo-1 autoantibodies target, in part, the N-terminal region of HisRS (Jura et al., 2007; Martin et al., 1995; Raben et al., 1994). As might be expected from its structure, we observed that Jo-1 antibodies from patients react with HisRSΔCD (Figure 4). In addition, Granzyme B cleavage of autoantigens is suggested to contribute to the initiation and propagation of autoimmunity, and it is noteworthy that HisRS was amenable to Granzyme B cleavage in vitro at the N-terminal sequence LGPD48, a tetrapeptide that is also encompassed by HisRSΔCD (Darrah and Rosen, 2010; Levine et al., 2007). While the purpose of this study was not to investigate the etiology of IIM and ILD that is associated with HisRS, these results suggest that HisRSΔCD should at least be further considered in this regard.
Figure 4. Possible Association of HisRSΔCD with IIM/ILD.
Jo-1 antibodies from two different IIM patients react with recombinant human HisRS FL (hsHisRS) and HisRSΔCD, but not with E. coli HisRS (ecHisRS). The optical density at 450 nm was used to monitor the formation of antibody complexes in the ELISA. The “7B” stands for lot 7B04507 of Jo-1 antibodies and “4L” stands for lot 4L34811. The titration assay by ELISA was performed in duplicate and data are represented as mean ± SEM.
Conclusions
We discovered an internal deletion splice variant of human HisRS that completely and precisely ablates the CD and little else. From investigations of the structures of HisRSΔCD and its parent HisRS, three prominent differences were noted. First, in contrast to the dimeric structure shared by HisRSs throughout evolution, HisRSΔCD is a monomer. Second, the tight domain-domain packing interactions seen in HisRS are absent in HisRSΔCD, which presents two ‘free-floating’ domains loosely tethered together by a 27 amino acid flexible linker. Third, the release of packing interactions for the ABD of HisRSΔCD engenders a specific conformational change in the ABD. The existence of an AARS variant that lacks a catalytic domain, and that has undergone demonstrable changes in the tertiary and quaternary structures, is highly suggestive of strong selective pressures to repurpose HisRS.
EXPERIMENTAL PROCEDURES
The exon-skipping splicing events in the gene for human HisRS were identified by deep sequencing of AARS-transcriptome enriched cDNA of human tissues and cells. The splice variant HisRSΔCD was validated by PCR, and the endogenous protein was detected by western blot analysis. The mRNA expression level of HisRSΔCD in various human tissues was analyzed by the qPCR method. Recombinant human HisRS, HisRSΔCD, and their variants were over-expressed in E. coli, purified, and subjected to structural characterizations using X-ray crystallography and NMR techniques. The binding of the HisRS proteins to patient Jo-1 antibodies were analyzed by an enzyme-linked immunosorbent assay (ELISA). The detailed description of the experimental procedures is provided in Supplemental Information.
Supplementary Material
HIGHLIGHTS.
Identification of a natural internally deleted HisRS splice variant
Crystal structure of human HisRS at a resolution of 2.4 Å
NMR structure of the splice variant shows new conformation
ACKNOWLEDGEMENTS
We thank the BL17U1 beamline of the Shanghai Synchrotron Radiation Facility for the X-ray beamline time. The work was supported in part by the Innovation and Technology Fund of Hong Kong (UIM181, UIM192, and UIM199), aTyr Pharma, grants GM 15539, GM 23562, GM 88278, and CA 92577 from the NIH, and by a fellowship from the National Foundation for Cancer Research. We also thank Drs. Elisabeth Gardiner and Melissa Ashlock for helpful comments and advice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The atomic coordinates and structure factors of HisRS Δ507–509 and Δ1–53_507–509 have been deposited in the Protein Data Bank under the accession codes 4G84 and 4G85 respectively. The atomic coordinates, chemical shifts and restraints of HisRSΔCD* have been deposited in the Protein Data Bank under the accession code 2LW7.
REFERENCES
- Aberg A, Yaremchuk A, Tukalo M, Rasmussen B, Cusack S. Crystal structure analysis of the activation of histidine by Thermus thermophilus histidyl-tRNA synthetase. Biochemistry. 1997;36:3084–3094. doi: 10.1021/bi9618373. [DOI] [PubMed] [Google Scholar]
- Arif A, Jia J, Mukhopadhyay R, Willard B, Kinter M, Fox PL. Two-site phosphorylation of EPRS coordinates multimodal regulation of noncanonical translational control activity. Mol. Cell. 2009;35:164–180. doi: 10.1016/j.molcel.2009.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnez JG, Harris DC, Mitschler A, Rees B, Francklyn CS, Moras D. Crystal structure of histidyl-tRNA synthetase from Escherichia coli complexed with histidyl-adenylate. Embo J. 1995;14:4143–4155. doi: 10.1002/j.1460-2075.1995.tb00088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein RM, Morgan SH, Chapman J, Bunn CC, Mathews MB, Turnerwarwick M, Hughes GRV. Anti-Jo-1 antibody: a marker for myositis with interstitial lung disease. Brit. Med. J. 1984;289:151–152. doi: 10.1136/bmj.289.6438.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfils G, Jaquenoud M, Bontron S, Ostrowicz C, Ungermann C, De Virgilio C. Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol. Cell. 2012;46:105–110. doi: 10.1016/j.molcel.2012.02.009. [DOI] [PubMed] [Google Scholar]
- Carter CW., Jr Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu. Rev. Biochem. 1993;62:715–748. doi: 10.1146/annurev.bi.62.070193.003435. [DOI] [PubMed] [Google Scholar]
- Danoff SK, Casciola-Rosen L. The lung as a possible target for the immune reaction in myositis. Arthritis Res. Ther. 2011;13:230. doi: 10.1186/ar3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrah E, Rosen A. Granzyme B cleavage of autoantigens in autoimmunity. Cell Death Differ. 2010;17:624–632. doi: 10.1038/cdd.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrell MI, Aguilar E, Scheppke L, Barnett FH, Friedlander M. Combination angiostatic therapy completely inhibits ocular and tumor angiogenesis. Proc. Natl. Acad. Sci. USA. 2007;104:967–972. doi: 10.1073/pnas.0607542104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freist W, Verhey JF, Rühlmann A, Gauss DH, Arnez JG. Histidyl-tRNA synthetase. Biol. Chem. 1999;380:623–646. doi: 10.1515/BC.1999.079. [DOI] [PubMed] [Google Scholar]
- Fu G, Xu T, Shi Y, Wei N, Yang XL. tRNA-controlled nuclear import of a human tRNA synthetase. J. Biol. Chem. 2012;287:9330–9334. doi: 10.1074/jbc.C111.325902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé R. Toward a more complete view of tRNA biology. Nat. Struct. Mol. Biol. 2008;15:1007–1014. doi: 10.1038/nsmb.1498. [DOI] [PubMed] [Google Scholar]
- Guo M, Yang XL, Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat. Rev. Mol. Cell Bio. 2010;11:668–674. doi: 10.1038/nrm2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, Ha SH, Ryu SH, Kim S. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149:410–424. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- Jura M, Rychlewski L, Barciszewski J. Comprehensive insight into human aminoacyl-tRNA synthetases as autoantigens in idiopathic inflammatory myopathies. Crit. Rev. Immunol. 2007;27:559–572. doi: 10.1615/critrevimmunol.v27.i6.60. [DOI] [PubMed] [Google Scholar]
- Kim S, You S, Hwang D. Aminoacyl-tRNA synthetases and tumorigenesis: more than housekeeping. Nat. Rev. Cancer. 2011;11:708–718. doi: 10.1038/nrc3124. [DOI] [PubMed] [Google Scholar]
- Lee PS, Zhang HM, Marshall AG, Yang XL, Schimmel P. Uncovering of a short internal peptide activates a tRNA synthetase procytokine. J. Biol. Chem. 2012;287:20504–20508. doi: 10.1074/jbc.C112.369439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine SM, Raben N, Xie D, Askin FB, Tuder R, Mullins M, Rosen A, Casciola-Rosen LA. Novel conformation of histidyl-transfer RNA synthetase in the lung: the target tissue in Jo-1 autoantibody-associated myositis. Arthritis Rheum. 2007;56:2729–2739. doi: 10.1002/art.22790. [DOI] [PubMed] [Google Scholar]
- Li L, Weinreb V, Francklyn C, Carter CW., Jr Histidyl-tRNA synthetase urzymes: Class I and II aminoacyl tRNA synthetase urzymes have comparable catalytic activities for cognate amino acid activation. J. Biol. Chem. 2011;286:10387–10395. doi: 10.1074/jbc.M110.198929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Shulman MJ, Tsui FW. Epitope studies indicate that histidyl-tRNA synthetase is a stimulating antigen in idiopathic myositis. FASEB J. 1995;9:1226–1233. doi: 10.1096/fasebj.9.12.7672516. [DOI] [PubMed] [Google Scholar]
- Martinis SA, Joy Pang YL. Jekyll & Hyde: evolution of a superfamily. Chem. Biol. 2007;14:1307–1308. doi: 10.1016/j.chembiol.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Merritt EA, Arakaki TL, Gillespie JR, Larson ET, Kelley A, Mueller N, Napuli AJ, Kim J, Zhang L, Verlinde CLMJ, et al. Crystal structures of trypanosomal histidyl-tRNA synthetase illuminate differences between eukaryotic and prokaryotic homologs. J. Mol. Biol. 2010;397:481–494. doi: 10.1016/j.jmb.2010.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MC, Kang T, Jin D, Han JM, Kim SB, Park YJ, Cho K, Park YW, Guo M, He W, et al. Secreted human glycyl-tRNA synthetase implicated in defense against ERK-activated tumorigenesis. Proc. Natl. Acad. Sci. USA. 2012;109:E640–E647. doi: 10.1073/pnas.1200194109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SG, Schimmel P, Kim S. Aminoacyl tRNA synthetases and their connections to disease. Proc. Natl. Acad. Sci. USA. 2008;105:11043–11049. doi: 10.1073/pnas.0802862105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raben N, Nichols R, Dohlman J, McPhie P, Sridhar V, Hyde C, Leff R, Plotz P. A motif in human histidyl-tRNA synthetase which is shared among several aminoacyl-tRNA synthetases is a coiled-coil that is essential for enzymatic activity and contains the major autoantigenic epitope. J. Biol. Chem. 1994;269:24277–24283. [PubMed] [Google Scholar]
- Sajish M, Zhou QS, Kishi S, Valdez DM, Kapoor M, Guo M, Lee S, Kim S, Yang XL, Schimmel P. Trp-tRNA synthetase bridges DNA-PKcs to PARP-1 to link IFN-gamma and p53 signaling. Nat. Chem. Biol. 2012;8:547–554. doi: 10.1038/nchembio.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(Suppl 1):S12. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi K, Slike BM, Hood J, Otani A, Ewalt KL, Friedlander M, Cheresh DA, Schimmel P. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc. Natl. Acad. Sci. USA. 2002;99:173–177. doi: 10.1073/pnas.012602099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XL, Shi Y, Zhang HM, Swindell EC, Marshall AG, Guo M, Kishi S, Yang XL. Unique domain appended to vertebrate tRNA synthetase is essential for vascular development. Nat. Commun. 2012;3 doi: 10.1038/ncomms1686. Article number 681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao P, Potdar AA, Arif A, Ray PS, Mukhopadhyay R, Willard B, Xu YC, Yan J, Saidel GM, Fox PL. Coding region polyadenylation generates a truncated tRNA synthetase that counters translation repression. Cell. 2012;149:88–100. doi: 10.1016/j.cell.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou QS, Kapoor M, Guo M, Belani R, Xu XL, Kiosses WB, Hanan M, Park C, Armour E, Do MH, et al. Orthogonal use of a human tRNA synthetase active site to achieve multifunctionality. Nat. Struct. Mol. Biol. 2010;17:57–61. doi: 10.1038/nsmb.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.