Abstract
Proteorhodopsin (PR) sequences were PCR amplified from three Andean acidic hot spring samples. These sequences were similar to freshwater and marine PRs and they contained residues indicative of proton-pumping activity and of proteins that absorb green light; these findings suggest that PRs might contribute to cellular metabolism in these habitats.
TEXT
Proteorhodopsins (PRs) are retinal-binding bacterial transmembrane light-driven proton pumps that generate energy from light and are considered to play an important role in carbon cycling and energy flux in various aquatic ecosystems (3, 4, 7, 8, 10, 16). PR proteins are widespread and abundant in marine and nonmarine habitats (2, 6, 9, 17, 21, 23) and are tuned to absorb different spectral wavelengths (4, 15, 18). PRs were recently shown to enhance survival and competition of marine Vibrio spp. cells during starvation, consistent with the suggestion that they must have unique functional capabilities (10).
The central Andean mountain range in Colombia is characterized by geothermal activity and the presence of many fumaroles and hot springs located at high altitude (>3,400 m). A recent analysis of the microbial community in one of these springs, El Coquito, showed the presence of chemolithoautotrophic acidophiles and a few phototrophic bacteria, both photoautotrophs and photoheterotrophs, suggesting that primary production can be driven by chemical energy in the water and by solar energy at the surface (5). Given the high exposure to solar light at the surface (approximately 9 to 11 μW/(cm2 nm) UV-B) (12) and the relatively low abundance of phototrophic bacteria identified, we hypothesized that productivity in these ecosystems could also be driven by energy-harvesting bacterial PRs. To explore this possibility, we analyzed the diversity of PRs in several high mountain acidic hot springs by PCR amplification with PR-specific primers.
Surface water samples were collected at four hot springs (A1, A2, A4, and A5) located in the Nevados National Natural Park and processed as described previously (5). These springs are all located at high altitudes and are acidic, but they differ in terms of temperature and water characteristics (Table 1). PCR amplification was carried out using actinorhodopsin primers and six freshwater PR-specific primer combinations (Table 2), as reported elsewhere (2, 20), using both control plasmid DNA (pCD1, pST84 for actinorhodopsins, and ebac31A08 in pBAD.TA for freshwater primer combinations) (3) and hot spring metagenomic DNAs in 50-μl reaction volumes with 1 U of TucanTaq DNA polymerase (CorpoGen, Bogota, Colombia). Amplification products were gel purified (QIAquick gel extraction kit; Qiagen, Germany), cloned into pCR2.1 using the TOPO TA kit (Invitrogen, San Diego, CA), and transformed into Escherichia coli DH5α cells; individual clones were sequenced (Macrogen, South Korea). Amplification products were obtained for three hot spring DNAs (sites A5, A2, and A1) using four different primer combinations. There was no amplification for actinorhodopsins in any of the samples, even though Actinobacteria were detected at least at site A4 by pyrosequencing (5), suggesting that PR sequences were either absent or not amplifiable with the primers used. All DNAs were amplified with 16S rRNA gene primers 27F and 1492R (5), indicating the absence of PCR inhibitors. From 433 cloned amplification products obtained with four primer combinations (mixes 1, 3, 4, and 6), 91 clones (21%) corresponded to PR-like partial genes after sequencing and analysis using blastn and tblastx against information in the GenBank database (based on best hit): 84 from site A5, 5 from site A2, and 2 from site A1 (Table 2). These sequences showed no similarities with other sequences in the databases, and even though we checked for chimeras manually by BLAST analysis of sequence fragments, it is possible that some of the observed variability could still have been due to chimeras and/or amplification errors. The remaining discarded sequences had stop codons or produced hits with very low scores and poor coverage based on comparisons using blastn and tblastx analyses. Previous studies have also shown low recovery of PR genes (5.2%) (14), which may result from the use of degenerate primers.
Table 1.
Characteristics of the sites sampled
| Sample | Site A1 | Site A2 | Site A5 | Site A4 |
|---|---|---|---|---|
| Date of sampling (day/mo/yr) | 07/04/2008 | 07/04/2008 | 08/04/2008 | 09/04/2008 |
| Ecosystema | HAF | HAF-SP | SP | SP |
| Location | 4°58′13.2″ N, 75°22′42″ W | 4°58′10.3″ N, 75°22′38.6″ W | 4°54′32.8″ N, 75°18′19′ W | 04°52′27″N, 75°15′51.4′W |
| Altitude (m) | 3,464 | 3,876 | 4,363 | 3,973 |
| DAPI count (cells/ml) | 2.09 × 105 | 9.16 × 104 | 2.42 × 104 | 2.35 ×105 |
| Temp (°C) | 56.9 | 56.8 | 35 | 28.9 |
| pH | 2.03 | 2.04 | 3.12 | 2.7 |
| Acidity (CaCO3) (mg/liter) | 3,633 | 4,525 | 500 | 500 |
| Chloride (Cl−) (mg/liter) | 653 | 841 | 139 | 56.6 |
| Sulfates (SO42−) (mg/liter) | 2,681 | 3,239 | 1,052 | 1,003 |
| Calcium (Ca2+) (mg/liter) | 195 | 247 | 256 | 320 |
| Sodium (Na+) (mg/liter) | 413 | 531 | 122 | 45.2 |
| Magnesium (Mg2+) (mg/liter) | 282 | 247 | 134 | 55.3 |
| Potassium (K+) (mg/liter) | 60.8 | 74.3 | 13.5 | 9.25 |
| Nitrates (NO3−) (mg/liter) | 2.44 | 0.51 | ≤0.19 | 0.89 |
| Total hardness (CaCO3, mg/liter) | 1,192 | 1,461 | 1,285 | 1,224 |
| Total phosphate (PO4, mg/liter) | 1.89 | 2.76 | 0.25 | 0.1 |
| Total iron (mg/liter) | 56 | 69.4 | 25.8 | 8.27 |
| Total suspended solids (mg/liter) | 13.6 | 138 | 28.4 | ≤8.01 |
| Total solids (mg/liter) | 7,039 | 8,854 | 3,124 | 2,620 |
| Total dissolved solids (mg/liter) | 6,032 | 7,049 | 2,561 | 2,280 |
Ecosystem abbreviations: HAF, high Andean forest; SP, Superpáramo.
Table 2.
Primers used in this study
| Primer type and designation | Forward primer |
Reverse primer |
No. recovered with PCRc | No. of positive PRsc | ||
|---|---|---|---|---|---|---|
| Primer | Sequence (5′–3′) | Primer | Sequence (5′–3′) | |||
| Degenerate primer mixesa | ||||||
| 1 | RYIDW | MGNTAYATHGAYTGG | GWAIYP | GGRTADATNGCCCANCC | A1 (95), A2 (54), A5 (104) | A2 (5), A5 (84) |
| 2 | RYIDW | GWSIYP | GGRTADATNSWCCANCC | None | ||
| 3 | RYIDW | GWAVYP | GGRTANACNGCCCANCC | A1 (120) | A1 (2) | |
| 4 | RYVDW | MGNTAYGTNGAYTGG | GWSIYP | A1 (21) | None | |
| 5 | RYVDW | GWVIYP | GGRTADATNACCCANCC | None | ||
| 6 | RYVDW | GWAVYP | A1 (39) | None | ||
| 7 | YRYVDW | TAYMGNTAYGTNGAYTGG | WGVYPI | ATNGGRTANACNCCCCA | None | |
| 8 | YRYADW | TAYMGNTAYGCNGAYTGG | WGVYPI | None | ||
| Specific primersb | ||||||
| JN648742 | JN648742-F | TTCTACTTAATTCTTGCTGCT | JN648742-R | GCCCAGCCAAAGATGATAATA | A1, A2, A5 | |
| JN648793 | JN648793-F | CTATCTGATCCTTTCCGCCAT | JN648793-R | ATATAGCCCAGCCGAAGGTGA | A1, A2 | |
| JN648744 | JN648744-F | CTGATTACCGTTCCGCTCCTG | JN648744-R | ATCCGATTGTGACGATCCAGC | A5 | |
Degenerate primer mixes used for initial amplifications. The primers were obtained from Atamna-Ismaeel et al. (2) and Sharma et al. (20).
Specific primers designed to amplify PR sequences from metagenomic DNA (the names shown are the PR sequences).
Results shown are the sites with positive findings, with the number of positive samples at each site indicated in parentheses. For the three specific primers, the full-length genes could not be amplified (see text).
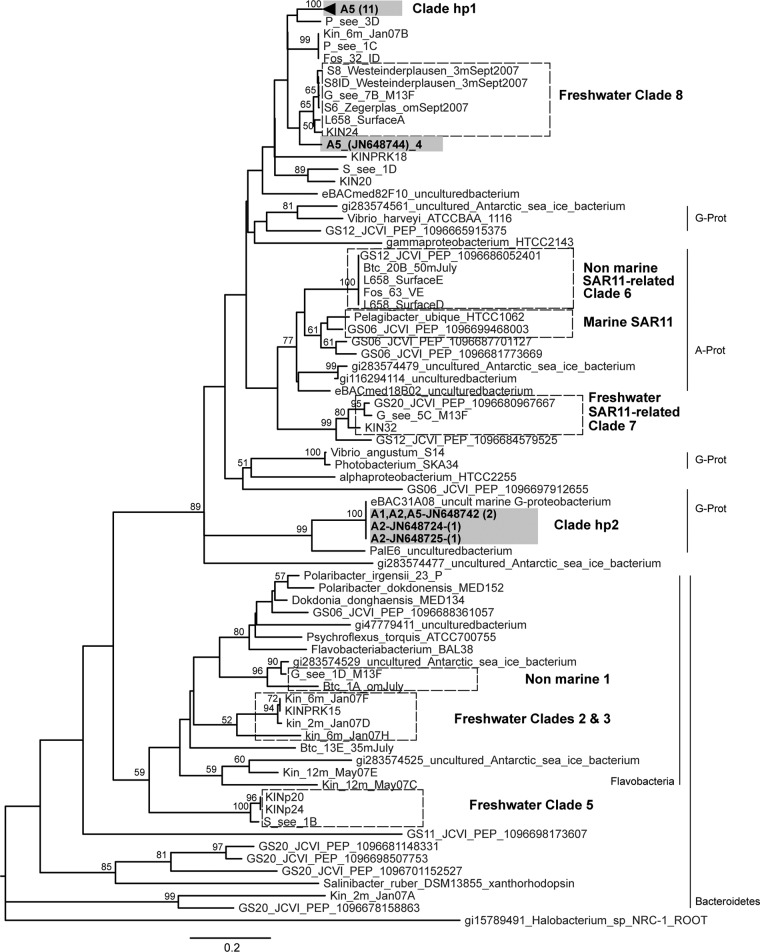
The 91 sequences (accession numbers JN648719 to JN648809) had an average length of 351 nucleotides and similarities across the entire sequence with other putative PRs from environmental uncultured clones. These sequences corresponded to 22 nucleotide sequences and 15 amino acid sequences with 43% identical sites across all sequences, indicating a substantial PR diversity. A phylogenetic reconstruction done with the 15 protein inferred sequences and additional reference sequences showed good bootstrap support, shared features with previously published phylogenetic reconstructions, and clustered our sequences into three groups (Fig. 1). Clade hp1 was formed by 11 different PR sequences from hot spring A5, and clade hp2 clustered 3 PR sequences derived from clones obtained from samples A1, A2, and A5 that were closely related to a coastal water clone, EBAC31A08 (AF279106) (3). The PRs within clades hp1 and hp2 had 91% and 94% identical amino acid sites, respectively. A single PR sequence, represented by 4 different nucleotide sequences from the A5 hot spring, grouped closely with the previously identified “freshwater clade 8” (2). Thus, the PRs identified in these hot springs clustered in different groups and were similar to PRs from both freshwater and marine environments.
Fig 1.
Neighbor-joining phylogenetic tree constructed using 15 inferred PR protein hot spring sequences and 70 reported sequences. Sequences were aligned using ClustalW, and the phylogeny was constructed using the neighbor-joining algorithm with 1,000 bootstrap replicates and the program MEGA (24). Sequences from hot spring samples are shown in gray boxes, indicating the site (A1, A2, or A5) followed by the accession number and number of represented different nucleotide sequences (in parentheses). The number in parentheses for clade hp1 indicates the unique PR amino acid sequences. Hot spring clades (hp) and previously reported clades (hatched boxes) (2) are indicated. Bootstrap values greater than 50% are shown. Known phylogenetic affiliations are indicated. G-Prot, Gammaproteobacteria; A-Prot, Alphaproteobacteria.
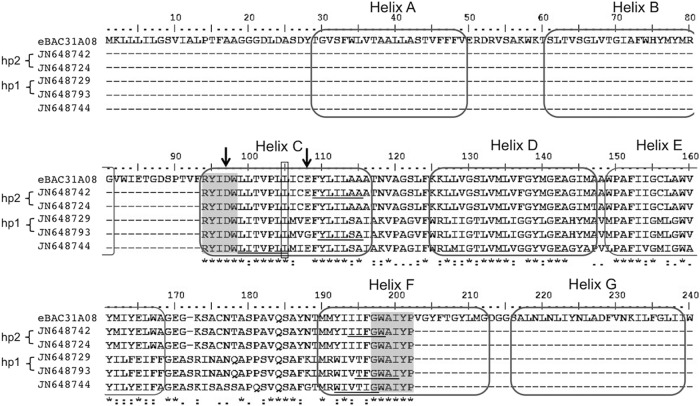
To confirm the PRs detected using degenerate primers, in particular because of the possible errors associated with amplification, cloning, and sequencing, we designed primers to specifically amplify three of the newly identified sequences: JN648793 (clade hp1), JN648742 (clade hp2), and JN648744 (Fig. 2; Table 2). PCR amplification of the same metagenomic DNAs was carried out under more stringent conditions: 35 cycles at 95°C for 1 min, 58°C for 30 s, and 68°C for 45 s. These PCR products were gel purified and sequenced (Macrogen, South Korea), thus avoiding the bias associated with sequencing single cloned sequences that might harbor amplification errors. Amplification was successful with primers for JN648793, using A1 and A2 DNAs, and JN648744, using A5 metagenomic DNA, and the sequences obtained were identical to the original detected sequences, lending support to the idea that these PR sequences were in fact present in these samples and were not simply an artifact of amplification. Primers for JN648742 yielded only very faint amplification bands in all samples (A1, A2, and A5) and insufficient amounts that could not be further sequenced. Efforts to obtain full-length genes from metagenomic DNAs using reported primers (13) or the genome walking technique to amplify flanking regions (1) using a specific primer designed within the sequences reported here were unsuccessful. The lack of amplification of full-length genes could be due to differences in the sequences themselves, preventing correct primer annealing, or to problems stemming from the low quantity of metagenomic DNA and its integrity after prolonged storage, since reamplification with the 16S rRNA gene primers initially used was no longer efficient.
Fig 2.
Multiple alignment of PR amino acid sequences of eBAC31A08 and representative hot spring sequences from each hp clade. Positions are based on the eBAC31A08 protein numbering system (3). Residues 97 and 108 are marked with an arrow, and position 105 is boxed. Transmembrane helices (3) are indicated, as well as the position of the degenerate primers (gray boxes) and the specific primers designed in this study (underlined sequences).
It was interesting that most of the sequences were obtained from site A5. This could be due to inefficient recovery of PR genes from the other samples due to problems inherent to the DNA preparation and/or amplification with the primers used. However, given the fact that with the specific primers designed here we were able to amplify sequences from both sites A1 and A2 that were identical to those originally detected in sample A5 using degenerate primers (JN648793 and JN648744), it seems likely that by changing PCR conditions and primers, more PR genes could be recovered from sites A1 and A2 than originally detected. It is also possible, however, that these springs harbor different microbial communities, as a result of environmental variables such as pH and temperature, and therefore vary in terms of microorganisms harboring PR genes.
The hot spring PR sequences were examined for conservation of two amino acid residues in helix C, Asp at position 97 and Glu at 108, that are key for proton-pumping activity (22). The majority (90 sequences) had a conserved Glu108, and only one sequence had a Gly at this position (Fig. 2), consistent with the vast majority of PRs observed to date (3, 11, 19) and suggesting that PRs that function as light-activated ion pumps involved in phototrophy and not as sensory receptors. However, analysis of full-length genes is required to determine this, since the conservation of Asp97 could not be assessed given that the forward primer used for PCR amplification included this residue close to the 3′ end (20). All of the PR sequences also contained a nonpolar Leu residue at position 105 (Fig. 2), indicative of green-absorbing proteins with absorption maxima (λmax) of 525 nm (15).
In this work we identified diverse and low-abundance PR-like genes in microorganisms from high-mountain, shallow, oligotrophic hot spring waters that have high exposure to UV light (12); these microorganisms are predicted to encode green-absorbing proteins that may harvest the available energy. Energy derived from rhodopsin photosystems, which is very low compared with that from photosynthesis, could be advantageous in these acidic hot springs by contributing to survival in ecosystems that receive abundant sunlight and where alternative energy sources may vary or be scarce (10, 11). Future work aimed at amplification and cloning of a full-length gene will be required, however, to fully assess the functionality of these proteins. This work extends previous inventories of PR genes and shows that they are present in isolated, acidic hot spring communities where energy derived from rhodopsin photosystems may complement a chemotrophic lifestyle and provide an advantage to bacterial cells, helping to compensate for changing environmental conditions.
ACKNOWLEDGMENTS
We thank Jose Salvador Montaña and Gina López for sampling and processing DNA, Luisa Delgado for help with bioinformatics, Nof Atamna-Ismaeel and Howard Junca for help and opinions, Sandra Baena for statistical analysis, Julie Anne Maresca and Asunción Martínez from E. DeLong's lab for providing us with the positive controls for PR genes, and A. Martínez for critical comments on the manuscript.
This work was financed by Colciencias—SENA (project number 6570-392-19990) and was performed under MAVDT contract number 15,2008 for access to genetic resources and UAESPNN research permit number DTNO-N-20/2007.
Footnotes
Published ahead of print 31 August 2012
REFERENCES
- 1. Acevedo JP, et al. 2008. Cloning of complete genes for novel hydrolytic enzymes from Antarctic sea water bacteria by use of an improved genome walking technique. J. Biotechnol. 133:277–286 [DOI] [PubMed] [Google Scholar]
- 2. Atamna-Ismaeel N, et al. 2008. Widespread distribution of proteorhodopsins in freshwater and brackish ecosystems. ISME J. 2:656–662 [DOI] [PubMed] [Google Scholar]
- 3. Beja O, et al. 2000. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289:1902–1906 [DOI] [PubMed] [Google Scholar]
- 4. Beja O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. 2001. Proteorhodopsin phototrophy in the ocean. Nature 411:786–789 [DOI] [PubMed] [Google Scholar]
- 5. Bohorquez LC, et al. 2012. In-depth characterization via complementing culture-independent approaches of the microbial community in an acidic hot spring of the Colombian Andes. Microb. Ecol. 63:103–115 [DOI] [PubMed] [Google Scholar]
- 6. Campbell BJ, Waidner LA, Cottrell MT, Kirchman DL. 2008. Abundant proteorhodopsin genes in the North Atlantic Ocean. Environ. Microbiol. 10:99–109 [DOI] [PubMed] [Google Scholar]
- 7. de la Torre JR, et al. 2003. Proteorhodopsin genes are distributed among divergent marine bacterial taxa. Proc. Natl. Acad. Sci. U. S. A. 100:12830–12835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fuhrman JA, Schwalbach MS, Stingl U. 2008. Proteorhodopsins: an array of physiological roles? Nat. Rev. Microbiol. 6:488–494 [DOI] [PubMed] [Google Scholar]
- 9. Giovannoni SJ, et al. 2005. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature 438:82–85 [DOI] [PubMed] [Google Scholar]
- 10. Gomez-Consarnau L, et al. 2010. Proteorhodopsin phototrophy promotes survival of marine bacteria during starvation. PLoS Biol. 8:e1000358 doi:10.1371/journal.pbio.1000358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gomez-Consarnau L, et al. 2007. Light stimulates growth of proteorhodopsin-containing marine Flavobacteria. Nature 445:210–213 [DOI] [PubMed] [Google Scholar]
- 12. IDEAM, UPME 2005. Atlás de radiación solar de Colombia, p 97–111 Ministerio de Ambiente, Vivienda y Desarrollo Territorial, Bogotá, DC, Colombia [Google Scholar]
- 13. Jung JY, Choi AR, Lee YK, Lee HK, Jung KH. 2008. Spectroscopic and photochemical analysis of proteorhodopsin variants from the surface of the Arctic Ocean. FEBS Lett. 582:1679–1684 [DOI] [PubMed] [Google Scholar]
- 14. Koh EY, et al. 2010. Proteorhodopsin-bearing bacteria in Antarctic sea ice. Appl. Environ. Microbiol. 76:5918–5925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Man D, et al. 2003. Diversification and spectral tuning in marine proteorhodopsins. EMBO J. 22:1725–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Martinez A, Bradley AS, Waldbauer JR, Summons RE, DeLong EF. 2007. Proteorhodopsin photosystem gene expression enables photophosphorylation in a heterologous host. Proc. Natl. Acad. Sci. U. S. A. 104:5590–5595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rusch DB, et al. 2007. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5:e77 doi:10.1371/journal.pbio.0050077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sabehi G, et al. 2007. Adaptation and spectral tuning in divergent marine proteorhodopsins from the eastern Mediterranean and the Sargasso Seas. ISME J. 1:48–55 [DOI] [PubMed] [Google Scholar]
- 19. Sabehi G, et al. 2005. New insights into metabolic properties of marine bacteria encoding proteorhodopsins. PLoS Biol. 3:e273 doi:10.1371/journal.pbio.0030273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sharma AK, et al. 2009. Actinorhodopsin genes discovered in diverse freshwater habitats and among cultivated freshwater Actinobacteria. ISME J. 3:726–737 [DOI] [PubMed] [Google Scholar]
- 21. Sharma AK, Zhaxybayeva O, Papke RT, Doolittle WF. 2008. Actinorhodopsins: proteorhodopsin-like gene sequences found predominantly in non-marine environments. Environ. Microbiol. 10:1039–1056 [DOI] [PubMed] [Google Scholar]
- 22. Spudich JL, Jung K-H. 2005. Microbial rhodopsins: phylogenetic and functional diversity, p 1–24 In Briggs WR, Spudich JL. (ed), Handbook of photosensory receptors. Wiley-VCH Verlag GmbH, Weinheim, Germany [Google Scholar]
- 23. Stingl U, Desiderio RA, Cho JC, Vergin KL, Giovannoni SJ. 2007. The SAR92 clade: an abundant coastal clade of culturable marine bacteria possessing proteorhodopsin. Appl. Environ. Microbiol. 73:2290–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]