Abstract
Many bacteria successfully colonize animals by forming protective biofilms. Molecular processes that underlie the formation and function of biofilms in pathogenic bacteria are well characterized. In contrast, the relationship between biofilms and host colonization by symbiotic bacteria is less well understood. Tsetse flies (Glossina spp.) house 3 maternally transmitted symbionts, one of which is a commensal (Sodalis glossinidius) found in several host tissues, including the gut. We determined that Sodalis forms biofilms in the tsetse gut and that this process is influenced by the Sodalis outer membrane protein A (OmpA). Mutant Sodalis strains that do not produce OmpA (Sodalis ΔOmpA mutants) fail to form biofilms in vitro and are unable to colonize the tsetse gut unless endogenous symbiotic bacteria are present. Our data indicate that in the absence of biofilms, Sodalis ΔOmpA mutant cells are exposed to and eliminated by tsetse's innate immune system, suggesting that biofilms help Sodalis evade the host immune system. Tsetse is the sole vector of pathogenic African trypanosomes, which also reside in the fly gut. Acquiring a better understanding of the dynamics that promote Sodalis colonization of the tsetse gut may enhance the development of novel disease control strategies.
INTRODUCTION
The digestive tract of animals represents an environment that houses a dense population of microbes (18, 28, 36, 52, 53). With the advancement of molecular technologies, our awareness of the diversity and abundance of microbes in this niche has increased dramatically. It is now known that the vast majority of bacteria present in the gut are commensals and thus positively impact the physiology of their hosts (18, 28, 36, 53). However, studies to further decipher the biological mechanisms that underlie digestive tract symbioses are impeded by the taxonomic complexity of the animal microbiome, the presence of unculturable microbes, and the inability to use standard molecular biological tools to genetically manipulate many of these microorganisms. Thus, identification of efficient model systems with less diverse microbial communities can advance our understanding of host-microbe interactions and their effect on pathogenic microbes.
One model system that can be used to study commensal symbioses is the tsetse fly (Diptera: Glossinidae). This fly harbors three distinct bacterial symbionts, including the parasitic Wolbachia, obligate Wigglesworthia, and commensal Sodalis. Tsetse flies are unique in that they exhibit a viviparous mode of reproduction, during which juvenile larvae develop in an intrauterine environment and receive nourishment from maternal milk gland secretions. Wigglesworthia and Sodalis are maternally acquired by intrauterine larvae via milk secretions, while Wolbachia is transmitted through the germ line (4). In addition to residing extracellularly in the female milk gland, Wigglesworthia is also found intracellularly in the tsetse gut-associated bacteriome (3). As a result of the long coevolutionary history of Wigglesworthia and tsetse, this bacterium's genome is reduced in size and highly streamlined in composition (1, 7). Despite these genome reduction processes, Wigglesworthia has retained a number of vitamin biosynthesis pathways, which may provision the tsetse host with metabolites missing from its vertebrate blood-specific diet (2). Tsetse's association with commensal Sodalis is more recent. Sodalis is closely related to several free-living enteric bacteria, including Yersinia and Salmonella (44). Furthermore, phylogenetic analysis of Sodalis from distant tsetse species suggests that these microbes recently descended from a common ancestor and spread via horizontal transfer events (49). Sodalis resides intra- and extracellularly throughout its host and can be cultivated in vitro and genetically manipulated using conventional techniques (8). Tsetse's simple microbial community may enable us to study host-symbiont interactions, including gut colonization processes, host mechanisms that enable tolerance to beneficial microbes, and symbiont mechanisms that enable their persistence against hostile host immune responses.
Functional studies have demonstrated a direct association between bacterial outer membrane protein A (OmpA) and microbial phenotypes in the tsetse. Specifically, young adult flies (3 days posteclosion) tolerate hemocoelic superinfection with Sodalis but quickly perish following the same treatment with normally nonpathogenic Escherichia coli K-12 (50). Analysis of the Sodalis ompA gene sequence indicated that environmentally exposed loop domains of the OmpA protein contain amino acid insertions and/or substitutions compared to their counterparts from E. coli and other closely related free-living and pathogenic bacteria (44, 50). From a functional perspective, OmpA polymorphisms were determined to modulate the infection outcome in tsetse when flies were infected with either E. coli OmpA mutants (E. coli ΔOmpA) or Sodalis that had been genetically modified to express E. coli OmpA (Sodalis pIOE. coli). These infection experiments showed that tsetse flies were able to tolerate E. coliΔOmpA, while Sodalis pIOE. coli exhibited a pathogenic phenotype that caused all infected tsetse flies to perish. These findings suggest that modifications associated with Sodalis OmpA may promote tsetse's tolerance of Sodalis and reflect larger evolutionary adaptations that facilitate bacterial/insect symbioses.
OmpA is also involved in the formation of microbial biofilms (5, 14, 27, 42), which are populations of microbes that adhere to surfaces by producing a polysaccharide extracellular matrix. Biofilms play a role in bacterial colonization dynamics of the mammalian and invertebrate gastrointestinal tracts (29). In E. coli, OmpA influences bacterial colonization of host tissues by promoting biofilm formation on hydrophobic surfaces (27, 41). In fleas (Xenopsylla cheopsis), the causative agent of plague, Yersinia pestis, forms a biofilm that blocks the passage of blood through its host's proboscis. This process starves the host and causes it to take multiple blood meals, which in turn increases the chances of pathogen transmission to the mammalian host (11). Additionally, pathogenic Vibrio cholerae can form biofilms in the rectum of Drosophila melanogaster and eventually kill its insect host (35). It remains to be seen if OmpA also modulates the ability of Yersinia and Vibrio to form biofilms in flea and Drosophila host tissues, respectively.
To date, the role of biofilms with respect to symbiont colonization of the insect gut is poorly understood. In this study, we investigated whether biofilm formation is required for commensal Sodalis to colonize the tsetse gut and whether Sodalis OmpA plays a role in the process of biofilm formation. We also discuss the association between biofilms and symbiont evasion of hostile host immune responses.
MATERIALS AND METHODS
Tsetse and bacterial cultures.
Tsetse flies (Glossina morsitans morsitans) were maintained in Yale University's insectary at 24°C with 55% relative humidity. Unless specifically indicated otherwise, flies received defibrinated bovine blood through an artificial membrane feeding system every 48 h (32, 50). Aposymbiotic tsetse flies (GmmApo) were generated as described previously (48). Sodalis-free tsetse flies (GmmSgm−) were generated by feeding 200 μg of ampicillin to flies two times before bacterial inoculation. Sample sizes for all experiments are indicated in the appropriate figure legends.
All bacterial strains used in this study as described in Table S1 in the supplemental material. Wild-type (WT) Sodalis strains were isolated from tsetse pupae and subsequently maintained in liquid Mitsuhashi-Maramorosch (MM) medium as described previously (50). Brain heart infusion agar plus 10% defibrinated bovine blood (BHIB) was used when Sodalis strains were plated on solid medium. E. coli OmpA mutants (E. coli ΔOmpA) and E. coli OmpA mutants that express Sodalis ompA (E. coli ΔOmpA pIOSodalis) were generated as described previously (50). E. coli strains were grown at 37°C on LB agar plates or in liquid LB medium under the appropriate antibiotic selection. The growth medium was supplemented with designated antibiotics: ampicillin (Amp) (25 μg/ml for Sodalis and 100 μg/ml for E. coli), tetracycline (Tet) (5 μg/ml for Sodalis), and kanamycin (Kan) (50 μg/ml for Sodalis and 100 μg/ml for E. coli).
Construction of 16S clone library.
Genomic DNA (gDNA) was extracted from six female fly guts and one male fly gut (all 8 days old) using the MasterPure DNA purification kit (Epicentre). The 16S clone library was constructed as previously described with the following modification (51): 16S rRNA genes were amplified using the primer set 27F and 1492R. The PCR amplification product was cloned into pGEM-T vector (Promega) and transformed into E. coli DH5α cells, and colony PCR was performed on 320 recombinant clones in 96-well plates, using T7 and SP6 primers. The PCR product was digested with TaqαI and HaeIII (New England BioLabs) restriction enzymes, and the generated profile was visualized on a 2.0% Metaphor agarose gel stained with ethidium bromide. Multiple clones with represented restriction profiles were then sequenced using T7 primer at the DNA analysis facility (Yale University) with BigDye Terminator. Sequences were compared to the NCBI BLAST database and the Ribosomal Database Project II (RDP) (http://rdp.cme.msu.edu/) (10). All oligonucleotide primers and PCR amplification conditions used in this study are summarized in Table S2 in the supplemental material.
Fluorescent in situ hybridization (FISH) and lectin staining.
Guts were microscopically dissected from 4-day-old GmmWT flies and subsequently subjected to FISH analysis and lectin staining as described previously (23, 34). In brief, a 5′-end rhodamine-labeled Sodalis-specific 16S rRNA probe (5′-ACGAGACTCTAGCCTGCCAG-3′) was used for FISH analysis (34). Dewaxed tissue sections, which were produced as described previously (23), were stained in a blocking solution (containing 4 nmol/ml of DAPI [4′,6-diamidino-2-phenylindole]) containing 200 ng of Sodalis-specific probe and 10 μg/ml of one of the following fluorescein-labeled lectins: concanavalin A, Dolichos biflorus agglutinin, peanut agglutinin, Ricinus communis agglutinin I, soybean agglutinin, Ulex europeaus agglutinin, and wheat germ agglutinin (WGA) (lectin kit I, Vector Laboratories, CA). Fluorescent signals were observed with a Zeiss epifluorescence microscope.
Generation of Sodalis ompA mutants.
Sodalis mutants that do not produce OmpA (hereafter referred to as Sodalis ΔOmpA mutants) were generated using the TargeTron gene knockout system (Sigma-Aldrich) according to the manufacturer's protocol. In brief, the group II intron encoded on expression plasmid pACD4K-C was retargeted to insert 540 bp from the ompA start codon. The resulting retargeting plasmid, designated pSOM, was electroporated into Sodalis PT, which harbors a helper plasmid (pAR1219) that encodes the T7 RNA polymerase gene under transcriptional control of the lac UV5 promoter (see Table S2 in the supplemental material). Following electroporation, cells were pelleted and resuspended in MM medium supplemented with chloramphenicol and 1% glucose. Following an overnight incubation, T7 RNA polymerase gene expression was induced for 1 h with the addition of 200 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and cells were pelleted and resuspended in MM medium containing 1% glucose. After 1 h incubation, the cells were similarly pelleted and resuspended and mutant cells were selected on BHIB+Kan plates. Insertion of the intron into the correct chromosomal locus was confirmed by PCR using ompA gene primer pair OmpAF1 and OmpAR1. Amplification primers are listed in Table S3 in the supplemental material. Recombinant OmpA protein production was confirmed by Western blot analysis using an anti-E. coli OmpA antibody at a dilution of 1:20,000 (50).
Sodalis in vitro blood growth assay.
The mammalian complement system was inactivated by heating blood for 1 h at 56°C. Sodalis PT or the Sodalis ΔOmpA mutant at 500 CFU/ml was added to either regular or heat-inactivated bovine blood. Inoculates were maintained at 24°C. Bacterial density was monitored every day for 7 days by serially diluting and subsequently plating samples on BHIB plates. CFU were counted when they became visible (∼7 days after plating).
Biofilm formation assay.
Sodalis PT or Sodalis ΔOmpA cells were inoculated into 5 ml of MM medium and vigorously shaken at an angle in a 15-ml Falcon tube for 7 days. Seven days later, liquid medium was removed from the tubes and the “splash zone” was stained for 15 min with crystal violet. Finally, tubes were gently rinsed with water (2×) and inspected for the presence of a biofilm.
Biofilm formation was also quantified using a previously described microtiter plate biofilm assay (31). A 24-well microtiter plate (Corning Life Science) with 500 μl of MM medium was inoculated with 500 μl of log-phase culture of strains to be tested (at an optical density at 600 nm [OD600] of ≈0.5). The microtiter plate was incubated for 11 days at 25°C, and the OD was measured at 600 nm. The relative biofilm formation was calculated with the following formula: OD of crystal violet/OD of growth yield.
Sodalis OmpA restores the E. coli biofilm formation defect.
E. coli strain MG1655 ΔompA::Tn5Kan-2, which has an ompA transposon-generated mutation (21), was complemented by expressing Sodalis OmpA in this mutant strain. Plasmid pIOSodalis was electroporated into MG1655 ΔompA::Tn5Kan-2, thus generating E. coli strain ΔOmpA pIOSodalis (21). E. coli strain ΔOmpA pIOSodalis was subsequently monitored, as described above, to determine its biofilm formation phenotype.
Per os colonization assay.
To assess the ability of different bacteria to colonize the tsetse gut, 500 CFU of each strain was added to 1 ml of heat-inactivated bovine blood and provided to flies through an artificial membrane system. Following per os inoculation with bacteria, flies were maintained on heat-inactivated blood every 48 h. Gut tissue was microscopically dissected at 1, 3, and 5 days postinoculation with bacteria. Three guts from each group were dissected per time point, homogenized in 0.85% NaCl, serially diluted, and plated on BHIB supplemented with antibiotics. CFU per plate were manually counted. All bacterial strains, tsetse lines, and antibiotics (including concentrations) used for these experiments are indicated in the appropriate figure legends. All per os colonization experiments were performed in duplicate.
Experimental induction of the tsetse immune response.
Tsetse flies were immunologically stimulated as described previously (17). In brief, GmmWT flies were fed either lipopolysaccharide (LPS) and peptidoglycan (PGN) (50 μg of each/ml of blood) or live E. coli (500 CFU/ml blood) prior to inoculation with experimental bacteria. Flies were exposed to E. coli once and LPS and PGN continuously throughout the course of the colonization assay. Colonization assays were performed in duplicate as described above.
RESULTS
Tsetse gut microbiome and bacterial biofilm formation in this niche.
In an effort to analyze the microbial composition of the tsetse gut, we constructed 16S rRNA libraries from this tissue and found that it houses only vertically transmitted Wigglesworthia and Sodalis (Table 1). Gut-associated Wigglesworthia strains are localized exclusively within a specialized organ called the bacteriome. However, Sodalis is found both intra- and extracellularly in several tsetse tissues, including the gut (8). As such, we set out to determine if protective biofilms form the mechanistic basis that underlies the ability of extracellular Sodalis to colonize the gut of wild-type tsetse flies (here designated GmmWT). We used DAPI (Fig. 1A) and fluorescent in situ hybridization (FISH) analysis with Sodalis-specific 16S rRNA probes (Fig. 1B) to detect bacterial clusters in this environment (Fig. 1C is a merged image of Fig. 1A and B). Additionally, these images were enlarged to identify individual bacteria (Fig. 1D to F). These data suggest that Sodalis resides in microcolonies in the luminal fluid of the tsetse gut. Microcolonies, which are considered to be the basic structural unit of a biofilm, were then investigated for the presence of a polysaccharide matrix using lectin staining. Lectins are carbohydrate-binding proteins or glycoproteins that contain at least two carbohydrate-binding sites (40). Out of the 7 different lectins we tested, only wheat germ agglutinin (WGA) reacted positively to 100% of the microcolonies found in the tsetse gut (Fig. 2). Furthermore, DAPI staining revealed that these biofilms may contain DNA in their extracellular matrix. A signal using WGA was detected along the host gut epithelial tissue, indicating that the polysaccharides could be host derived in nature (data not shown). WGA binds specifically to N-acetylglucosamine, a major component of peptidoglycan (39). We also tested whether WGA binds to Sodalis maintained in culture and detected staining in close proximity to the individual Sodalis cells (see Fig. S2 in the supplemental material). These findings suggest that WGA binds polysaccharides, such as the peptidoglycan and/or lipopolysaccharides that are on the bacterial cell membrane.
Table 1.
16S RNA clone libraries of tsetse fly gut
| Source | No. of clones | Species | No. of isolates |
|---|---|---|---|
| Female guts w/bacteriocyte | |||
| Gut 1 | 30 | Wigglesworthia glossinidia | 28 |
| Sodalis glossinidius | 2 | ||
| Gut 2 | 30 | Wigglesworthia glossinidia | 27 |
| Sodalis glossinidius | 3 | ||
| Gut 3 | 20 | Wigglesworthia glossinidia | 18 |
| Sodalis glossinidius | 2 | ||
| Gut 4 | 18 | Wigglesworthia glossinidia | 16 |
| Sodalis glossinidius | 2 | ||
| Female guts w/no bacteriocyte | |||
| Gut 1 | 120 | Wigglesworthia glossinidia | 0 |
| Sodalis glossinidius | 120 | ||
| Gut 2 | 39 | Wigglesworthia glossinidia | 4 |
| Sodalis glossinidius | 35 |
Fig 1.
In situ visualization of Sodalis clusters present in the tsetse gut. (A) Cross sections of tsetse gut tissue stained with DAPI. DNA from host epithelial tissue and Sodalis cells all stains blue. Bar, 50 μm. (B) Visualization of Sodalis clusters in the lumen of the tsetse gut using fluorescent in situ hybridization. Tissues were probed with a rhodamine-labeled oligonucleotide specific for Sodalis 16S rRNA (red). Bar, 50 μm. Autofluorescent blood meal-derived erythrocytes surround Sodalis cells. Bars, 50 μm. (C) Panels A and B merged to better orient the position of Sodalis cell clusters in relation to tsetse gut epithelial tissues. In each of panels A, B, and C, the most prominent cluster of Sodalis cells is indicated by a white arrow. (D to F) Enlarged views of the Sodalis clusters shown in panels A to C, respectively. Bars, 10 μm.
Fig 2.
Lectin staining of a biofilm-associated polysaccharide matrix surrounding a cluster of Sodalis cells. (A) A cross section of the tsetse gut tissue stained with a rhodamine-labeled oligonucleotide specific for Sodalis 16S rRNA. (B) A WGA lectin probe (green) bound to an extracellular polysaccharide matrix. (C) Panels A and B merged. The presence of a polysaccharide matrix surrounding a Sodalis cluster in the lumen of the tsetse gut indicates that these bacterial cells have formed a biofilm. Bars, 50 μm.
Generation of Sodalis OmpA mutants and characterization of their growth phenotype.
The process of biofilm formation in bacteria, including E. coli, Acinetobacter baumannii, and Salmonella enterica serovar Typhimurium, is influenced by OmpA (14, 26, 27). In an effort to determine if OmpA exhibits a homologous function in Sodalis, we generated a mutant strain that does not produce this protein. To do so, an intron was inserted in frame within the Sodalis ompA gene, which is encoded on a single chromosomally located operon. Intron insertion was verified by PCR (Fig. 3A), and OmpA was subsequently undetectable in the mutant strain (the Sodalis ΔOmpA mutant; Fig. 3B). We next characterized the growth phenotype of the Sodalis ΔOmpA mutant in comparison to its parent strain (Sodalis PT). When grown in MM medium, the Sodalis ΔOmpA mutant exhibited no growth defect (Fig. 3C). Tsetse flies feed exclusively on vertebrate blood, and Sodalis resides in the tsetse gut where the blood meal accumulates and undergoes digestion. We first examined the ability of Sodalis PT to grow in vitro in complement-active and -inactive blood. Our data show that vertebrate blood contains sufficient nutrients to maintain Sodalis in vitro but that these bacterial cells are sensitive to the complement system when inoculated into this medium (see Fig. S3 in the supplemental material). We also found that Sodalis ΔOmpA exhibits a similar growth curve when cultured in heat-inactivated blood (Fig. 3D). Interestingly, Sodalis strains found naturally in the tsetse gut are able to survive in the presence of vertebrate complement. This phenomenon suggests that vertebrate complement may be inactivated prior to coming in contact with endogenous Sodalis. Alternatively, Sodalis may utilize alternative mechanisms, such as the formation of protective biofilms, to circumvent this antimicrobial system.
Fig 3.
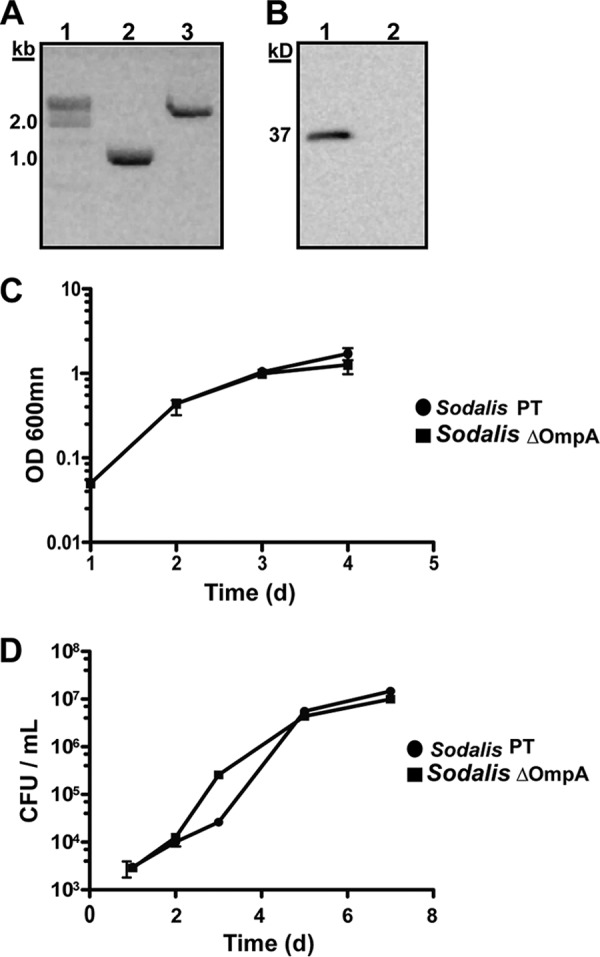
Generation of a Sodalis ompA mutant strain. (A) PCR amplification of the ompA gene in the Sodalis parent strain (Sodalis PT; lane 2) and Sodalis ompA mutants (Sodalis ΔOmpA; lane 3). The ∼1-kb band shift is the result of intron insertion into the ompA locus of the Sodalis chromosome. (B) A Western blot showing OmpA expression in Sodalis PT cells (lane 1) but not in their Sodalis ΔOmpA derivatives (lane 2). (C and D) Sodalis PT and Sodalis ΔOmpA growth curves in MM medium and blood, respectively. Sodalis ΔOmpA does not exhibit a growth defect when cultured in either medium.
OmpA modulates Sodalis biofilm formation in vitro.
The Sodalis ΔOmpA mutant does not exhibit a growth defect in heat-inactivated vertebrate blood and will thus serve as a robust strain to characterize the relationship between OmpA and the ability of this symbiont to colonize its tsetse host. We next set out to characterize the biofilm-associated phenotype of the Sodalis ΔOmpA mutant. To do so, an in vitro assay was performed where Sodalis PT and Sodalis ΔOmpA cultures were placed in a 15-ml Falcon tube at an angle and shaken for 7 days. Subsequently, the tube's splash zone was observed for the presence of a Sodalis biofilm (14, 26, 31). As shown in Fig. 4A, Sodalis PT cells form a distinct biofilm while their Sodalis ΔOmpA counterparts do not. Additionally, a crystal violet microtiter plate biofilm assay was used to quantitate the biofilm formation defect presented by Sodalis ΔOmpA cells. This assay indicated that the Sodalis ΔOmpA mutant has a 91% decrease in biofilm formation compared to Sodalis PT. These results indicate a role for OmpA in Sodalis biofilm formation.
Fig 4.

OmpA regulates bacterial biofilm formation in vitro. (A) Sodalis PT and Sodalis ΔOmpA cells were shaken vigorously at an angle for 7 days in 15-ml Falcon tubes. The bacterial splash zone was subsequently stained with crystal violet to visualize the presence or absence of a biofilm. These structures were present in the tubes containing Sodalis PT cells (shown enlarged in the corresponding box). Sodalis ΔOmpA cells failed to form a biofilm. This experiment was performed in triplicate with 3 distinct clonal populations of each bacterial strain. (B) Spectrophotometric quantification of relative biofilm formation exhibited by Sodalis PT and Sodalis ΔOmpA cells. (C) E. coli ΔOmpA cells exhibit a biofilm formation defect that is restored in their counterparts that were genetically modified to express Sodalis ompA (E. coli ΔOmpA pIOSodalis cells). In panels B, C and D, statistical significance was determined by the Mann-Whitney test.
Sodalis OmpA reverses the biofilm defect of E. coli OmpA mutants.
Data from our experiments using symbiotic Sodalis, as well as data from previous studies using free-living E. coli, suggest a role for OmpA in the ability of these bacteria to form biofilms (14, 27). To further validate the functional role of OmpA in biofilm formation, we expressed the Sodalis OmpA protein in a mutant strain of E. coli (E. coli ΔOmpA) that does not produce this protein and thus is unable to form biofilms (27, 50). Strain E. coli ΔOmpA pIOSodalis was previously generated by transforming E. coli ΔOmpA cells with a construct (pIOSodalis) that encodes the Sodalis ompA gene under transcriptional regulation of the constitutively expressed insulinase promoter (50). We used a crystal violet microtiter plate biofilm assay to monitor the biofilm formation phenotype of E. coli ΔOmpA pIOSodalis cells. We observed that E. coli ΔOmpA pIOSodalis and WT E. coli are equally competent in their ability to form biofilms in vitro, whereas E. coli ΔOmpA mutants are not (Fig. 4D). The results of our complementation assay further accentuate the role of Sodalis OmpA in biofilm formation processes, as biofilm-deficient E. coli strains are able to form these structures in vitro after they are genetically modified to express this protein.
OmpA-mediated bacterial colonization of the tsetse gut.
We next developed a per os colonization assay and used it to examine the role of OmpA in the ability of Sodalis to colonize the tsetse gut. GmmWT and symbiont-cured (here referred to as aposymbiotic, or GmmApo) flies were given a heat-inactivated blood meal supplemented with Sodalis PT. Subsequently, all flies received sterile heat-inactivated blood every other day for the entirety of the 5-day experiment. At 1, 3, and 5 days after feeding with bacterial strains, gut tissues were microscopically harvested from individual flies of each group and the number of Sodalis isolates present was quantified. We determined that Sodalis PT was able to colonize the gut of both GmmWT and GmmApo flies and replicate to a density of ∼106 CFU in this niche (Fig. 5A), a density which is comparable to natural Sodalis levels present in GmmWT guts (see Fig. S1 in the supplemental material). These data suggest that Sodalis PT can survive in the presence of natural Sodalis present in the gut of GmmWT flies and that these exogenous cells grow to densities similar to those of their native counterparts. Sodalis PT can also replace natural infections when provided to aposymbiotic flies. We next set out to determine if recombinant Sodalis strains can produce a biofilm in GmmWT and/or GmmApo flies. We recolonized these tsetse lines per os with a green fluorescent protein (GFP)-expressing strain of this bacterium (SodalispGFP) and then microscopically examined gut fluid for the presence of Sodalis clusters, similar to the procedure for Fig. 1. We discovered that 1 day postfeeding, Sodalis begins to form small microcolonies in the gut of GmmWT flies and that biofilms are present 3 days later. Furthermore, biofilms were also present in the gut of GmmApo flies by 3 days postinoculation (Fig. 5B). Results from a control experiment, in which gut fluid from GmmWT flies (which contained no recombinant Sodalis) was also observed, indicated that natural populations of this symbiont do not autofluoresce (see Fig. S4 in the supplemental material). Taken together, these data demonstrate that our per os colonization assay can be exploited to study the molecular mechanisms that underlie bacterial colonization of the tsetse gut.
Fig 5.
OmpA modulates Sodalis biofilm formation in the tsetse gut. (A) A newly developed per os colonization assay was used to determine if Sodalis PT cells could establish an infection in GmmWT and GmmApo fly guts. Five days postinoculation, Sodalis PT was found at a density of ∼106 cells in both host backgrounds. (B) Biofilm formation in the tsetse gut was visualized by orally inoculating GmmWT and GmmApo flies with a strain of Sodalis (Sodalis pGFP) that expresses green fluorescent protein. Fluorescing microcolonies, indicated by a white arrow, were present in GmmWT flies 1 day postinoculation with Sodalis pGFP. Three days postinoculation, these microcolonies are more pronounced in GmmWT flies. Additionally, Sodalis pGFP colonies are also beginning to form at this time in GmmApo flies. Bars, 50 μm. (C) Colonization of GmmWT fly guts by Sodalis PT and Sodalis ΔOmpA strains. Sodalis ΔOmpA cells exhibit an initial colonization defect but, by 5 days postinoculation, reach a density similar to that of Sodalis PT cells. (D) Colonization of GmmSgm− fly guts by Sodalis PT and Sodalis ΔOmpA strains. Sodalis ΔOmpA cells cannot colonize tsetse when their native counterparts are absent. In panel A, colonization assays were performed in duplicate and data are presented as the means ± standard deviations. In panels C and D, each symbol represents one fly gut, and the dotted line indicates the assay's limit of detection. Statistical significance in C and D was determined by the Mann-Whitney test.
We next investigated whether OmpA regulates the ability of Sodalis to colonize the tsetse gut by feeding GmmWT individuals with either Sodalis ΔOmpA or Sodalis PT cells. In contrast to Sodalis PT, Sodalis ΔOmpA cells exhibited a significant colonization defect 1 and 3 days after being fed to wild-type flies, which still retain their native microbiota. However, by 5 days postinoculation, both Sodalis strains reached similar homeostatic densities in the gut of GmmWT flies (Fig. 5C). These data suggest that OmpA is most important during initial stages of Sodalis gut colonization, as cells that lack this protein can reach normal densities over time. Interestingly, previous studies using other model systems have shown that mutant bacteria can “cheat” their colonization defect by cross-feeding off relevant protein products produced by endogenous bacteria also present in the system (30, 38, 45, 46). We thus investigated whether Sodalis ΔOmpA cells are able to colonize wild-type tsetse by cheating off the Sodalis naturally found in these flies. This could explain the delay in the colonization process we observed with the mutant strain. To do so, we generated Sodalis-free tsetse flies (GmmSgm−) by feeding GmmWT flies two blood meals supplemented with 200 μg of ampicillin per ml of blood. Prior to beginning the experiment, the absence of native Sodalis in GmmSgm− flies was verified by dissecting 4 fly guts per group and measuring the CFU by plating (data not shown). Subsequently, GmmSgm− individuals were orally inoculated with either Sodalis PT or Sodalis ΔOmpA cells. Sodalis PT effectively colonized GmmSgm− flies (Fig. 5D). However, in contrast to what we observed previously in GmmWT flies (which have their natural microbiome), we found that Sodalis ΔOmpA cells exhibited a significant colonization defect in GmmSgm− individuals over the course of the entire 5-day experiment (Fig. 5D). These findings provide further confirmation that Sodalis requires OmpA to colonize the tsetse gut.
The Sodalis ΔOmpA mutant receives protection from host immune responses in biofilms.
We demonstrate that Sodalis ΔOmpA cells cannot form a biofilm in vitro. Furthermore, while these bacterial cells can readily colonize the gut of GmmWT flies, they lack the ability to do so in tsetse that are missing their native microbiota (GmmSgm−). Many bacteria evade host immunity by forming biofilms (13, 22, 43). Thus, we speculated that the biofilm formation defect presented by Sodalis ΔOmpA cells could leave these cells susceptible to the tsetse's innate immune system and result in their elimination from the gut environment. To test this theory, we assessed the ability of Sodalis ΔOmpA cells to colonize tsetse that exhibit a state of heightened immunity. As done previously, we induced a local immune response in the gut of GmmWT flies by feeding distinct groups of flies either lipopolysaccharide (LPS) and peptidoglycan (PGN) or live E. coli. These treatments induce an increase in the expression of a variety of immunity-related effector molecules, including peptidoglycan recognition protein (PGRP) LB and the antimicrobial peptides (AMPs) attacin, defensin, and diptericin (17, 47). These flies were subsequently administered a heat-inactivated blood meal containing either Sodalis PT or Sodalis ΔOmpA cells. We found that host immune stimulation by either LPS and PGN or E. coli had no effect on Sodalis PT, as these cells reached a density of 106 CFU within 5 days (Fig. 6A and B, respectively). These findings confirmed previous results, which showed that Sodalis strains are resistant to the actions of tsetse's innate immune molecules in vitro (16, 20) and in vivo (47). We next tested the ability of Sodalis ΔOmpA cells to colonize GmmWT flies following host immune activation with either LPS and PGN or E. coli. When introduced per os into flies that were immune stimulated by LPS and PGN, Sodalis ΔOmpA strains exhibited an initial colonization defect that was significant compared to that of Sodalis PT. However, by 3 days postinoculation, the Sodalis ΔOmpA colonization defect was less severe than on day 1 (Fig. 6A). These findings indicate that Sodalis ΔOmpA proteins, acquired in the blood meal, are initially susceptible to tsetse's immune responses but can subsequently survive as they incorporate into native biofilms. When host immune challenge was achieved with live E. coli feeding, Sodalis ΔOmpA proteins were not able to colonize wild-type tsetse (Fig. 6B). Administration of live E. coli likely induces a more robust host immune response than LPS and PGN, and this response may clear Sodalis ΔOmpA cells before they are afforded protection by endogenous biofilms.
Fig 6.

Biofilms protect Sodalis from tsetse's innate immune response. Sodalis PT and Sodalis ΔOmpA cell colonization of immune-activated GmmWT fly guts. LPS and PGN (A) or live E. coli (B) was fed to flies prior to bacterial inoculation. Guts were dissected at 1 and 3 days postinoculation to determine the number of CFU present. Each symbol represents one fly gut, and the dotted line indicates the assay's limit of detection. Statistical significance was determined by the Mann-Whitney test.
DISCUSSION
Congregation within biofilms is essential for bacterial colonization of eukaryotic hosts. To date, most studies pertaining to this subject have focused on biofilms as they relate to host colonization by bacterial pathogens (33, 35). However, biofilms are gaining increased recognition as modulators of beneficial symbiotic associations. In this study, we investigated factors that mediate colonization of the tsetse fly gut by its commensal symbiont, Sodalis. We determined that Sodalis forms biofilms in the tsetse gut and that this process is contingent upon the presence of surface-exposed bacterial outer membrane protein A (OmpA). Furthermore, our results show that both the biofilms present in the tsetse gut and the major surface coat OmpA may provide protection to Sodalis against host innate immune responses. We propose that the tsetse-Sodalis symbiosis may serve as a useful model for better understanding the dynamics of symbiont colonization processes in the more complex mammalian systems.
Most insects, including colony-reared individuals and those captured in the field, harbor a taxonomically diverse assemblage of bacteria in their digestive tracts (12). Tsetse flies reared in our laboratory appear unique in that their guts are dominated by symbiotic Wigglesworthia and Sodalis strains. This finding implies that biofilms present in the tsetse gut lumen do not harbor mixed bacterial species like those found in other insects (24, 25). Instead, the tsetse gut lumen harbors Sodalis exclusively, as Wigglesworthia is found only intracellularly in the adjacent bacteriome. Tsetse is the sole vector of pathogenic African trypanosomes, which cause sleeping sickness in humans and nagana in domesticated animals. Laboratory-reared tsetse flies that lack their symbiotic microbes (GmmApo) are unusually susceptible to infection with trypanosomes (47). Our recent investigations indicated that GmmApo flies present a severely compromised immune system that is characterized by a significantly reduced population of phagocytic hemocytes (48, 50). The increased parasite transmission ability of GmmApo flies could be due to the lack of important host immune gene expression or could result from the absence of microbial biofilms in the gut. Interestingly, unlike their colony-reared counterparts, guts from field-captured tsetse flies house a more diverse bacterial population, although information on the densities of the different microbes relative to the symbiotic fauna is lacking (15). Similarly, field-based populations of other insect disease vectors are associated with taxonomically complex gut commensals that are intimately involved in their host's ability to transmit mammalian pathogens. For example, both Anopheles vectors of human malaria and Aedes vectors of dengue fever harbor gut microbiomes that influence the dynamics of parasite transmission processes (9, 37). Future studies using field-captured and laboratory-reared tsetse flies are required to determine if interactions between trypanosomes and bacterial biofilms influence this fly's vector competence.
In this study, we demonstrate that Sodalis colonizes the tsetse gut by forming biofilms and biofilm production is modulated by OmpA. These findings suggest that OmpA facilitates the tsetse-Sodalis symbiosis by promoting this bacterium's ability to reside in its host's gut. Interestingly, we demonstrated previously that OmpA also mediates the ability of Sodalis to reside symbiotically in tsetse hemocoel (50). This tolerance phenomenon was attributed to environmentally exposed OmpA loop domains. Specifically, the Sodalis OmpA strain was found to include amino acid insertions and substitutions in these regions that were absent from their homologs in E. coli. More so, exposed loop domains of OmpA from other insect symbionts also exhibit polymorphisms similar to those found in Sodalis. In tsetse, the distinct Sodalis and E. coli OmpA phenotypes corresponded with infection outcomes, as flies were found to be permissive of thoracic superinfection with Sodalis and E. coli ΔOmpA pIOSodalis but highly susceptible to the same treatment with WT E. coli. Exposed loop domain polymorphisms appeared to differentially regulate tsetse immunity, as avirulent Sodalis activated a robust host immune response while virulent E. coli did not. We speculate that E. coli replication proceeded without impediment and that tsetse perished due to their inability to tolerate high densities of this bacterium in their hemocoel. In contrast, we suspect Sodalis was able to circumvent tsetse's hemocoelic immune response by residing within host hemocytes (50). Taken together, our studies of Sodalis OmpA indicate that this protein exhibits multiple functional roles that are critical to the success of this bacterium's symbiosis with its tsetse host.
Many pathogenic bacteria, including E. coli, Salmonella Typhimurium, and Acinetobacter baumannii, evade host immunity by cloaking themselves within protective biofilms (14, 26, 27). One way these structures function in this manner is by limiting bacterial contact with host leukocytes (13, 22, 43). This process significantly reduces host antibody production and decreases the number of bacterial cells cleared via phagocytosis. An additional way biofilms protect pathogens from their host's immune system is by shielding them from the bactericidal activity of antimicrobial peptides (13, 22, 43). In fact, this phenomenon was recently observed in Drosophila melanogaster following infection with Pseudomonas aeruginosa. In this system, biofilm-encased bacteria within the fly's crop were more resistant to host AMPs than were planktonically grown cells (33). In the tsetse gut, the expression of several potent AMPs increases significantly when flies are fed live E. coli (17). These effector molecules function by punching pores in the outer membrane of bacteria, which causes the cells to lyse and die (6, 17). While previous experiments demonstrated that wild-type Sodalis cells are resistant to host AMPs, we observed in this study that Sodalis ΔOmpA mutant cells are cleared from tsetse flies that exhibited heightened gut immunity (16, 20, 47). This finding suggests that a correlation exists between the susceptibility of Sodalis ΔOmpA cells to the tsetse immune system and this bacterial strain's inability to form biofilms. We suspect that biofilm formation-deficient Sodalis cells perish in the tsetse gut because they are exposed to the bacteriolytic activity of host AMPs. Our findings suggest that OmpA may provide protection to Sodalis cells in the tsetse's pernicious gut environment, thus allowing this bacterium to reside symbiotically in this niche.
Blood-feeding insects such as tsetse flies and mosquitoes vector highly virulent mammalian pathogens. Current methods used to reduce vector populations, such as setting traps and applying pesticides, are limited in their effectiveness (8). Thus, in addition to traditional population suppression techniques, novel control strategies that reduce the vector competence of these insects are desirable. One such approach, called paratransgenesis, involves genetically modifying midgut commensal symbionts so that they produce antiparasitic compounds. These genetically modified symbionts are subsequently passed on to future offspring (8). This potential control strategy is promising in part because these recombinant symbionts colonize their host's gut and subsequently reside in close proximity to pathogenic microbes. This intimate association suggests that effector molecules secreted by genetically altered commensals will encounter and eliminate pathogens. Our characterization of the molecular mechanisms that underlie Sodalis colonization of the tsetse gut will be applicable to other insect-symbiont model systems. Additionally, the newly developed per os colonization assay described herein accurately mimics the route that many commensals utilize to naturally colonize their host. These findings will be of fundamental importance for optimizing the efficacy of paratransgenesis in blood-feeding arthropods.
Supplementary Material
ACKNOWLEDGMENTS
This work was generously funded by grants to S.A. from the National Institute of Allergy and Infectious Diseases (NIAID) (AI051584), the National Institute of General Medical Science (NIGMS) (069449), and the Ambrose Monell Foundation (http://www.monellvetlesen.org/). M.A.M. is the recipient of a NIH Parasitology and Vector Biology training grant (5T32AI007404).
Footnotes
Published ahead of print 31 August 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Akman L, Rio RV, Beard CB, Aksoy S. 2001. Genome size determination and coding capacity of Sodalis glossinidius, an enteric symbiont of tsetse flies, as revealed by hybridization to Escherichia coli gene arrays. J. Bacteriol. 183:4517–4525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Akman L, et al. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32:402–407 [DOI] [PubMed] [Google Scholar]
- 3. Aksoy S, Pourhosseini AA, Chow A. 1995. Mycetome endosymbionts of tsetse flies constitute a distinct lineage related to Enterobacteriaceae. Insect Mol. Biol. 4:15–22 [DOI] [PubMed] [Google Scholar]
- 4. Attardo GM, et al. 2008. Analysis of milk gland structure and function in Glossina morsitans: milk protein production, symbiont populations and fecundity. J. Insect Physiol. 54:1236–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barrios AF, Zuo R, Ren D, Wood TK. 2006. Hha, YbaJ, and OmpA regulate Escherichia coli K12 biofilm formation and conjugation plasmids abolish motility. Biotechnol. Bioeng. 93:188–200 [DOI] [PubMed] [Google Scholar]
- 6. Carlsson A, Nystrom T, de Cock H, Bennich H. 1998. Attacin—an insect immune protein—binds LPS and triggers the specific inhibition of bacterial outer-membrane protein synthesis. Microbiology 144:2179–2188 [DOI] [PubMed] [Google Scholar]
- 7. Chen X, Li S, Aksoy S. 1999. Concordant evolution of a symbiont with its host insect species: molecular phylogeny of genus Glossina and its bacteriome-associated endosymbiont, Wigglesworthia glossinidia. J. Mol. Evol. 48:49–58 [DOI] [PubMed] [Google Scholar]
- 8. Cheng Q, Aksoy S. 1999. Tissue tropism, transmission and expression of foreign genes in vivo in midgut symbionts of tsetse flies. Insect Mol. Biol. 8:125–132 [DOI] [PubMed] [Google Scholar]
- 9. Cirimotich CM, Ramirez JL, Dimopoulos G. 2011. Native microbiota shape insect vector competence for human pathogens. Cell Host Microbe 10:307–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cole JR, et al. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294–D296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Darby C. 2008. Uniquely insidious: Yersinia pestis biofilms. Trends Microbiol. 16:158–164 [DOI] [PubMed] [Google Scholar]
- 12. Dillon RJ, Dillon VM. 2004. The gut bacteria of insects: nonpathogenic interactions. Annu. Rev. Entomol. 49:71–92 [DOI] [PubMed] [Google Scholar]
- 13. Foster TJ. 2005. Immune evasion by staphylococci. Nat. Rev. Microbiol. 3:948–958 [DOI] [PubMed] [Google Scholar]
- 14. Gaddy JA, Tomaras AP, Actis LA. 2009. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect. Immun. 77:3150–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Geiger A, et al. 2011. Bacterial diversity associated with populations of Glossina spp. from Cameroon and distribution within the Campo sleeping sickness focus. Microb. Ecol. 62:632–643 [DOI] [PubMed] [Google Scholar]
- 16. Haines LR, Hancock RE, Pearson TW. 2003. Cationic antimicrobial peptide killing of African trypanosomes and Sodalis glossinidius, a bacterial symbiont of the insect vector of sleeping sickness. Vector Borne Zoonotic Dis. 3:175–186 [DOI] [PubMed] [Google Scholar]
- 17. Hao Z, et al. 2001. Tsetse immune responses and trypanosome transmission: implications for the development of tsetse-based strategies to reduce trypanosomiasis. Proc. Natl. Acad. Sci. U. S. A. 98:12648–12653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hooper LV, Gordon JI. 2001. Commensal host-bacterial relationships in the gut. Science 292:1115–1118 [DOI] [PubMed] [Google Scholar]
- 19. Hu C, Aksoy S. 2006. Innate immune responses regulate trypanosome parasite infection of the tsetse fly Glossina morsitans morsitans. Mol. Microbiol. 60:1194–1204 [DOI] [PubMed] [Google Scholar]
- 20. Hu Y, Aksoy S. 2005. An antimicrobial peptide with trypanocidal activity characterized from Glossina morsitans morsitans. Insect Biochem. Mol. Biol. 35:105–115 [DOI] [PubMed] [Google Scholar]
- 21. Kang Y, et al. 2004. Systematic mutagenesis of the Escherichia coli genome. J. Bacteriol. 186:4921–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kharazmi A. 1991. Mechanisms involved in the evasion of the host defence by Pseudomonas aeruginosa. Immunol. Lett. 30:201–205 [DOI] [PubMed] [Google Scholar]
- 23. Kikuchi Y, Graf J. 2007. Spatial and temporal population dynamics of a naturally occurring two-species microbial community inside the digestive tract of the medicinal leech. Appl. Environ. Microbiol. 73:1984–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kikuchi Y, Hosokawa T, Fukatsu T. 2011. Specific developmental window for establishment of an insect-microbe gut symbiosis. Appl. Environ. Microbiol. 77:4075–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kikuchi Y, Meng XY, Fukatsu T. 2005. Gut symbiotic bacteria of the genus Burkholderia in the broad-headed bugs Riptortus clavatus and Leptocorisa chinensis (Heteroptera: Alydidae). Appl. Environ. Microbiol. 71:4035–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kint G, De Coster D, Marchal K, Vanderleyden J, De Keersmaecker SC. 2010. The small regulatory RNA molecule MicA is involved in Salmonella enterica serovar Typhimurium biofilm formation. BMC Microbiol. 10:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ma Q, Wood TK. 2009. OmpA influences Escherichia coli biofilm formation by repressing cellulose production through the CpxRA two-component system. Environ. Microbiol. 11:2735–2746 [DOI] [PubMed] [Google Scholar]
- 28. Maccaferri S, Biagi E, Brigidi P. 2011. Metagenomics: key to human gut microbiota. Dig. Dis. 29:525–530 [DOI] [PubMed] [Google Scholar]
- 29. Macfarlane S, Dillon JF. 2007. Microbial biofilms in the human gastrointestinal tract. J. Appl. Microbiol. 102:1187–1196 [DOI] [PubMed] [Google Scholar]
- 30. Maltz M, Graf J. 2011. The type II secretion system is essential for erythrocyte lysis and gut colonization by the leech digestive tract symbiont Aeromonas veronii. Appl. Environ. Microbiol. 77:597–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Merritt JH, Kadouri DE, O'Toole GA. 2005. Growing and analyzing static biofilms. Curr. Protoc. Microbiol. Chapter 1:Unit 1B 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moloo SK. 1971. An artificial feeding technique for Glossina. Parasitology 63:507–512 [DOI] [PubMed] [Google Scholar]
- 33. Mulcahy H, Sibley CD, Surette MG, Lewenza S. 2011. Drosophila melanogaster as an animal model for the study of Pseudomonas aeruginosa biofilm infections in vivo. PLoS Pathog. 7:e1002299 doi:10.1371/journal.ppat.1002299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pais R, Lohs C, Wu Y, Wang J, Aksoy S. 2008. The obligate mutualist Wigglesworthia glossinidia influences reproduction, digestion, and immunity processes of its host, the tsetse fly. Appl. Environ. Microbiol. 74:5965–5974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Purdy AE, Watnick PI. 2011. Spatially selective colonization of the arthropod intestine through activation of Vibrio cholerae biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 108:19737–19742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Qin J, et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ramirez JL, et al. 2012. Reciprocal tripartite interactions between the Aedes aegypti midgut microbiota, innate immune system and dengue virus influences vector competence. PLoS Negl. Trop. Dis. 6:e1561 doi:10.1371/journal.pntd.0001561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sandoz KM, Mitzimberg SM, Schuster M. 2007. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc. Natl. Acad. Sci. U. S. A. 104:15876–15881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sizemore RK, Caldwell JJ, Kendrick AS. 1990. Alternate gram staining technique using a fluorescent lectin. Appl. Environ. Microbiol. 56:2245–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Slifkin M, Doyle RJ. 1990. Lectins and their application to clinical microbiology. Clin. Microbiol. Rev. 3:197–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smith SG, Mahon V, Lambert MA, Fagan RP. 2007. A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol. Lett. 273:1–11 [DOI] [PubMed] [Google Scholar]
- 42. Snoussi S, et al. 2012. Adaptation of Salmonella enterica Hadar under static magnetic field: effects on outer membrane protein pattern. Proteome Sci. 10:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thurlow LR, et al. 2011. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J. Immunol. 186:6585–6596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Toh H, et al. 2006. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 16:149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Venturi V, Bertani I, Kerenyi A, Netotea S, Pongor S. 2010. Co-swarming and local collapse: quorum sensing conveys resilience to bacterial communities by localizing cheater mutants in Pseudomonas aeruginosa. PLoS One 5:e9998 doi:10.1371/journal.pone.0009998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vulic M, Kolter R. 2001. Evolutionary cheating in Escherichia coli stationary phase cultures. Genetics 158:519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang J, Wu Y, Yang G, Aksoy S. 2009. Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proc. Natl. Acad. Sci. U. S. A. 106:12133–12138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Weiss BL, Maltz M, Aksoy S. 2012. Obligate symbionts activate immune system development in the tsetse fly. J. Immunol. 188:3395–3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weiss BL, et al. 2006. Interspecific transfer of bacterial endosymbionts between tsetse fly species: infection establishment and effect on host fitness. Appl. Environ. Microbiol. 72:7013–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Weiss BL, Wu Y, Schwank JJ, Tolwinski NS, Aksoy S. 2008. An insect symbiosis is influenced by bacterium-specific polymorphisms in outer-membrane protein A. Proc. Natl. Acad. Sci. U. S. A. 105:15088–15093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Worthen PL, Gode CJ, Graf J. 2006. Culture-independent characterization of the digestive-tract microbiota of the medicinal leech reveals a tripartite symbiosis. Appl. Environ. Microbiol. 72:4775–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xu J, et al. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074–2076 [DOI] [PubMed] [Google Scholar]
- 53. Xu J, Chiang HC, Bjursell MK, Gordon JI. 2004. Message from a human gut symbiont: sensitivity is a prerequisite for sharing. Trends Microbiol. 12:21–28 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





