Abstract
Dairy cows have been identified as common carriers of Campylobacter jejuni, which causes many of the human gastroenteritis cases reported worldwide. To design on-farm management practices that control the human infection sourced from dairy cows, the first step is to acquire an understanding of the excretion patterns of the cow reservoir. We monitored the same 35 cows from two dairy farms for C. jejuni excretion fortnightly for up to 12 months. The objective was to examine the concentration of C. jejuni and assess the genetic relationship of the C. jejuni populations excreted by individual cows. Significant differences (P < 0.01) in C. jejuni fecal concentration were observed among the 35 cows, with median concentrations that varied by up to 3.6 log10 · g−1 feces. A total of 36 different genotypes were identified from the 514 positive samples by using enterobacterial repetitive intergenic consensus (ERIC)-PCR. Although 22 of these genotypes were excreted by more than one cow, the analysis of frequencies and distribution of the genotypes by model-based statistics revealed a high degree of individuality in the C. jejuni population in each cow. The observed variation in the frequency of excretion of a genotype among cows and the analysis by multilocus sequence typing (MLST) of these genotypes suggest that excretion of C. jejuni in high numbers is due to a successful adaptation of a particular genotype to a particular cow's gut environment, but that animal-related factors render some individual cows resistant to colonization by particular genotypes. The reasons for differences in C. jejuni colonization of animals warrant further investigation.
INTRODUCTION
Campylobacter is the most common bacterial cause of gastroenteritis worldwide, and it can cause severe postinfection neuropathies, including Guillain-Barré and Miller Fisher syndromes. In New Zealand, a country which has a high rate of reported campylobacteriosis compared to other developed countries, Campylobacter infections and sequelae have been recently estimated to cost the public health system NZ $74 million annually (23). In the United States, Campylobacter infections cause US $1.7 billion in annual costs of illness (2).
The majority (>90%) of the infections are caused by Campylobacter jejuni subsp. jejuni (here C. jejuni). Many risk factors for human infection have been identified and include ingestion of contaminated food, use of contaminated cooking utensils, consumption of raw milk, consumption of water contaminated by agricultural wastes, or contact with infected animals (4, 18). A wide range of host animals, including cattle, poultry, sheep, pigs, wild-living birds, and mammals, carry C. jejuni asymptomatically and excrete it in their feces. Therefore, they can be a source for food or water contamination and subsequently a risk for human health. The bovine reservoir has been associated repetitively with the presence of Campylobacter in lakes, streams, and rivers running through agricultural areas (1, 16). Contamination of water by cattle is due to direct fecal deposition into waterways, overflows from pastures, and recycling or overflows of farm dairy effluents or livestock manure to agricultural land (for a review, see reference 30). With decreases in human campylobacteriosis notifications coinciding with the recent changes in practices in poultry industry production (36), it is likely that the contribution of ruminants as sources for human infection through water, but also through raw milk, contaminated vegetables, or direct contact with animals, will proportionally increase.
Quantitative information on C. jejuni shedding by cattle is important for assessing the risk of contamination sourced from farms to drinking or recreational water resources. Data on the reservoir size have been provided by several cross-sectional surveys on dairy cows from a single or a large group of farms at particular times of year (19, 27). Those studies provided a snapshot of on-farm prevalence and magnitude of shedding in one area at one particular point in time (i.e., sampling event). They have been complemented by data from longitudinal surveys that captured the variability of C. jejuni excretion within a single herd (38). In contrast, data on the carriage and excretion of C. jejuni by individual cows and on the transmission among cows are scant. In a study of 60 beef cattle in feedlots, Inglis et al. (21) observed that seven animals shed Campylobacter at more than three sampling times during a 5-month period. One animal in particular was observed to shed very large numbers of C. jejuni (>5 log C. jejuni cells per g feces) at all five sampling times (21). Animals that shed particular fecal bacteria at levels higher than those of other animals could be keys for the maintenance and transmission of the bacteria in the herd (25). Determining the prevalence of excretion and whether a cow becomes a high shedder of C. jejuni thus has important consequences for the design and implementation of management strategies that might reduce carriage and transmission and that would result in reduced exposure for humans.
The objective of this study was to establish characteristic excretion patterns in individual cows with regard to frequency of excretion and range of concentration. A strategy of sampling the same 35 cows every 2 weeks for a 10- to 12-month period was used; the studied cows belonged to two dairy farms with different management practices, one a pasture-based system with a seasonal maize or grass silage supply and the other a more intensive farm system where the animals had access to off-pasture housing and where a high proportion of the animal diet was consumed as supplementary feedstuffs. An additional aim was to obtain an indication of the diversity and stability of C. jejuni genotypes in individual cows to determine any relationship between excretion pattern and C. jejuni populations that were present in the cow's feces.
MATERIALS AND METHODS
Study animals and farms.
Two dairy farms located in the Waikato region of New Zealand were chosen for the study. Eighteen cows on one farm and 17 cows on the other farm were selected to represent the most frequent ages of the cows of the herds (4, 5, and 6 years old). Each cow was identified by its unique farm identification number. The cows were kept with the main herd and were subjected to normal husbandry practices. On the first farm (designated the pasture farm), which followed a typical rotational pasture-grazing management system, the cows were grazed on pasture but in the autumn and winter also received various forms of supplementary feed to ca. 20% of their food intake. The second farm (designated the housing farm) used a herd home-like housing facility. As well as grazing pasture, the cows received various forms of supplementary feedstuff (including maize silage, potato, palm kernel, soya, straw, tapioca, and kiwifruit) to 50 to 80% of their food intake in all seasons. Lactating cows spent 3 to 4 h each day in the animal housing facility during summer, 4 to 7 h during autumn, and 6 h each day during winter. Nonlactating cows spent 4 to 18 h each day in the animal housing facility. On both farms, cows drank untreated water sourced from a bore.
Sampling design and feces collection.
The experimental design was a longitudinal study carried out during two consecutive years. Experimental cows from the pasture farm were sampled between September 2008 and September 2009 and those from the housing farm between December 2009 and September 2010. Farms were visited every 2 weeks for a total of 25 samplings for the pasture farm and 20 samplings for the animal housing facility farm. Any cows that either farmer removed from the herd for commercial reasons were not replaced. At each visit, a fecal sample (50 to 200 g) was collected from each cow by direct retrieval in accordance with standard animal welfare practices. Sampling took place either during morning milking or early in the morning during the nonlactating season. Cross contamination during sampling was avoided by using single-use plastic gloves. Samples were transported to the laboratory in insulated containers and analyzed within 2 h of collection.
Enumeration of C. jejuni.
C. jejuni was enumerated in each homogenized fecal sample by a three-tube, three-dilution most-probable-number (MPN) technique using selective Campylobacter Exeter broth and mCCDA agar as described in the NZ reference method (9). The presence of C. jejuni was confirmed from every mCCDA plate showing colonial growth by PCR with a primer pair specific for C. jejuni (designated Map A) (3). The limit of detection was 0.3 C. jejuni organisms (MPN) g−1 feces. The concentration of confirmed C. jejuni in the original sample was determined by reference to a three-tube MPN probability table (42). For each fecal sample positive for C. jejuni, the colonial growth from two different dilutions within the MPN series was stored in charcoal Amies transport medium (Fort Richard Laboratory Ltd., Auckland, New Zealand) at −80°C until purification of single-colony isolates.
Purification and DNA extraction of C. jejuni single-colony isolates.
The colonial growth of samples positive for C. jejuni was resuscitated in Exeter broth media for 48 h. The isolation of single colonies was done by spread plating a loopful of the growth onto a fresh mCCDA agar plate, followed by incubation at 42°C for 24 h. Two isolates per sample were purified. The amount of DNA extracted by boiling from the isolates was determined photometrically using a NanoPhotometer (Implen, GmbH, Munich, Germany). The confirmation that the single-colony isolates were C. jejuni was by PCR as described by Vandamme et al. (39).
Subspecies typing by ERIC-PCR.
The single-colony isolates that were confirmed as being C. jejuni were subjected to PCR using enterobacterial repetitive intergenic consensus (ERIC) 2 primers (40) and the protocol of Weijtens et al. (41). The similarities and differences in the ERIC-PCR banding patterns were analyzed by using Quantity-One software, version 4.5.2 (Bio-Rad Laboratories, Hercules, CA). The banding patterns were compared only for isolates that were processed in the same PCR run (26, 33). Three stages of comparison were monitored. The DNA of the single-colony isolates from each individual animal was first compared to determine the occurrence of ERIC types in individual cows. DNA of representative single-colony isolates from an individual cow was then successively compared to those of the other individuals of the same farm. Finally, representative isolates obtained from both farms were compared. Cluster analysis was used to determine relationships between isolates. Isolates that grouped at a ≥90% similarity level were considered the same type. Different types were assigned a number (i.e., ERIC type 1 to ERIC type 36).
Microbiological data analysis.
Campylobacter jejuni concentrations were expressed as log10(1 + MPN) per gram of (fresh weight) feces. When C. jejuni was not detected (i.e., concentration below 0.3 C. jejuni organisms g−1 feces), a value of 0 was assigned to the MPN count. Box plots were drawn for visual comparison of ranges in C. jejuni concentration in each cow using R (version 2.10.0, 2009; R Foundation for Statistical Computing, Vienna, Austria), and statistical analysis was performed by analysis of variance (ANOVA) (GenStat for Windows, 10th ed., version 10.2.0.175, 2008; VSN International, Oxford, United Kingdom). For ease of reading, the cows were classified in three groups (high-shedder, low-shedder, and intermediate-shedder groups) determined by both the median concentration and the incidence of C. jejuni in their feces. The cutoff values of 3 log10 C. jejuni organisms per g feces for the median concentration and 50% prevalence were chosen arbitrarily.
Molecular data analysis.
Standard diversity indices were used to obtain the probabilities that two randomly chosen C. jejuni isolates would be of different ERIC types when selected from (i) the full data set or (ii) the subsets of data from both farms (29). A nonparametric analysis of molecular variance (AMOVA) test was used to partition the total variance of ERIC frequencies within the full data set into the variance components due to differences in the frequencies of ERIC types isolated from the individual cows over time, between cows within a farm, and between farms (10). The significance of the variance components was tested using nonparametric permutation (using 5,000 permutations) to obtain a null distribution for the given hierarchy. Population pairwise fixation indices (FST) were calculated for every possible pair of cows using pairwise differences as the method for calculating the genetic distance between each pair. In order to simultaneously visualize the genetic distances between C. jejuni isolates from all cows, an unrooted split network was constructed using the diagonal matrix of pairwise FST estimates (20). The split network was constructed using the NeighborNet method. The diversity indices, AMOVA, and fixation indices were calculated using the population genetics software package Arlequin (version 3.5.1.2) (11). The split network was produced using SplitsTree 4 (version 4.12.3) (20).
Subspecies genotyping by MLST.
To allow for linkages with clinical cases and potential sources of the C. jejuni genotypes observed in this study, multilocus sequence typing (MLST) of 20 and 24 C. jejuni isolates representative of all of the ERIC types obtained on the pasture farm and on the housing farm, respectively, were conducted using the protocols developed by Dingle et al. (8). Sequence data were collated and alleles assigned as sequence type (ST) and clonal complex (CC) using the Campylobacter PubMLST database (http://pubmlst.org/campylobacter/).
RESULTS
Fecal concentration of C. jejuni excreted by individual cows.
The C. jejuni concentration obtained from the feces collected every 2 weeks for 10 or 12 months varied between 0 and 6.0 log10 C. jejuni organisms per g on the pasture farm and between 0 and 5.7 log10 C. jejuni organisms per g on the housing farm. There was a significant difference (P < 0.01) in the frequency and range of C. jejuni fecal concentration among the 35 studied cows (Fig. 1).
Fig 1.
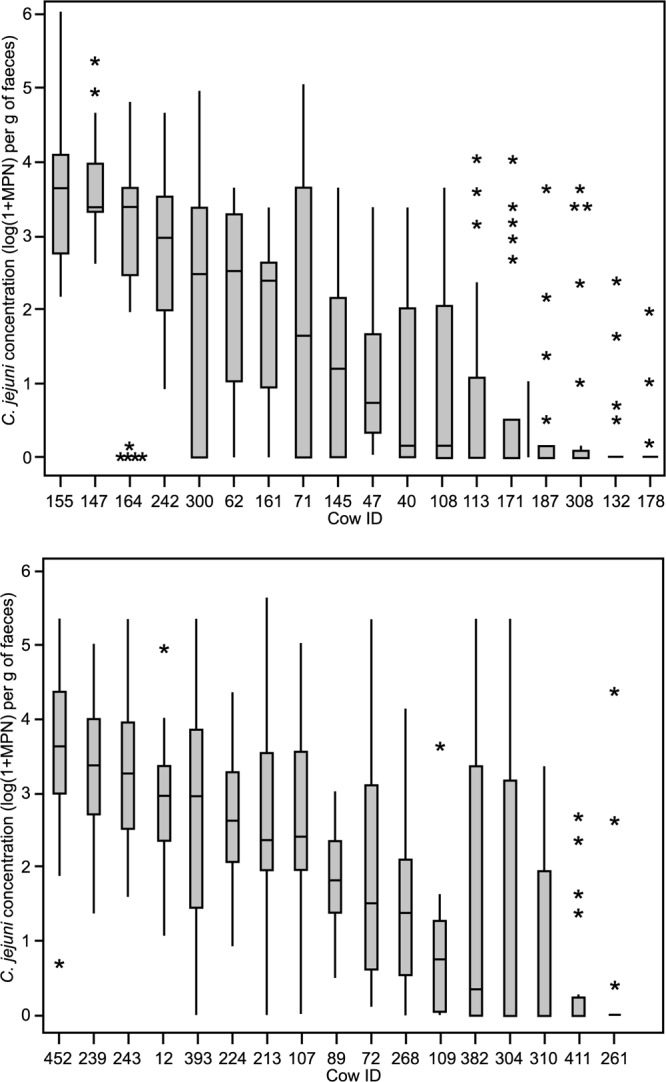
Box plots showing the concentration of C. jejuni in the feces excreted by each cow during the 12 months of sampling on the pasture farm (top) and during the 10 months of sampling on the housing farm (bottom). The median is shown as the horizontal line inside each box; top and bottom edges of the box correspond to the 75th and 25th percentiles, respectively (i.e., each box shows the middle of 50% of the data). Whiskers show the maximum and minimum values (excluding outliers), while asterisks are outlier samples whose values are more than 1.5 times the difference between 3rd and 1st quartiles. n = 25 for each cow on the pasture farm, and n = 20 for each cow on the housing farm, except for cows 108 (n = 23), 304, 12 (n = 19), 62 (n = 16), 40 (n = 13), and 109 (n = 12).
Three cows on the pasture farm (164, 147, and 155) and three cows on the housing farm (243, 239, and 452) that excreted C. jejuni in high numbers (3.3 log10 ≤ median concentration ≤ 3.6 log10 C. jejuni per g fresh feces; interquartile range, 2.4 to 4.4 log10) were grouped together in the high-shedder group. Eighty percent or more of the samples obtained from these cows during the study were positive for C. jejuni.
Six cows on the pasture farm (178, 132, 308, 187, 171, and 113) and four cows on the housing farm (261, 411, 310, and 304) excreted C. jejuni infrequently: between 12 and 47% of the samples collected from these cows were found to contain C. jejuni. When detected, C. jejuni was typically found in low concentration (nil median concentration). There were, however, exceptions to the pattern, with sporadic excretion of C. jejuni in concentrations of above 3 log C. jejuni organisms per g feces in 10 out of 150 (7%) and 11 out of 79 (14%) of the samples collected from these cows on the pasture farm and on the housing farm, respectively. These cows were designated low shedders.
The remaining 9 cows on the pasture farm and 10 cows on the housing farm (designated intermediate shedders) excreted C. jejuni in intermediate concentrations (0.1 log10 ≤ median concentration ≤ 3.0 log10 C. jejuni organisms per g fresh feces) and with frequency that varied between 50 and 100% of the samples.
Genetic structure of C. jejuni population between and within the farms.
A total of 999 C. jejuni isolates (498 from the pasture farm and 501 from the housing farm) were obtained from the 514 samples in which C. jejuni had been detected. Nine percent (22/258) and 19% (49/256) of the samples which contained C. jejuni and which were collected from the cows on the pasture farm and on the housing farm, respectively, contained 2 different ERIC types. Twenty-nine isolates (3%) were either not identified as C. jejuni or failed to revive from frozen. There were 36 ERIC types, of which 22 were found on at least one occasion in more than one cow. Standard diversity calculations found a 0.884 probability of selecting different ERIC types from two randomly chosen C. jejuni isolates in the complete data set. Although there was some conservation of ERIC types within both farms, the isolates from the housing farm showed a within-farm diversity index (0.872) which was higher than that of the isolates from the pasture farm (0.808).
The AMOVA model fitted to analyze the variance of ERIC-type frequencies within the specified hierarchical framework of cows within farms (Table 1) also found significant between-farm differences, but the variance at this level only accounted for 7.3% of the total variance within the data set. There was strong evidence of within-herd diversity, with the variation accounting for a further variance; however, the majority of variation (59.2%) occurred within an individual cow over time.
Table 1.
Variance in ERIC types of C. jejuni isolates originating from cows from pasture and housing farmsa
| Source of variation | Variance factor |
P value | |||
|---|---|---|---|---|---|
| Degree(s) of freedom | Sum of squares | Variance components | % Variation | ||
| Between farms | 1 | 22.6 | 0.03 | 7.3 | 0.002 |
| Between cows | |||||
| Within farms | 33 | 153.3 | 0.15 | 33.5 | <0.0001 |
| Within cows | 963 | 264.7 | 0.28 | 59.2 | <0.0001 |
| Total | 997 | 440.6 | 0.46 | 100 | |
Data are AMOVA model results showing the hierarchical partitioning of the variance in ERIC types of C. jejuni isolates originating from 35 cows across the pasture farm and the housing farm over the sampling period.
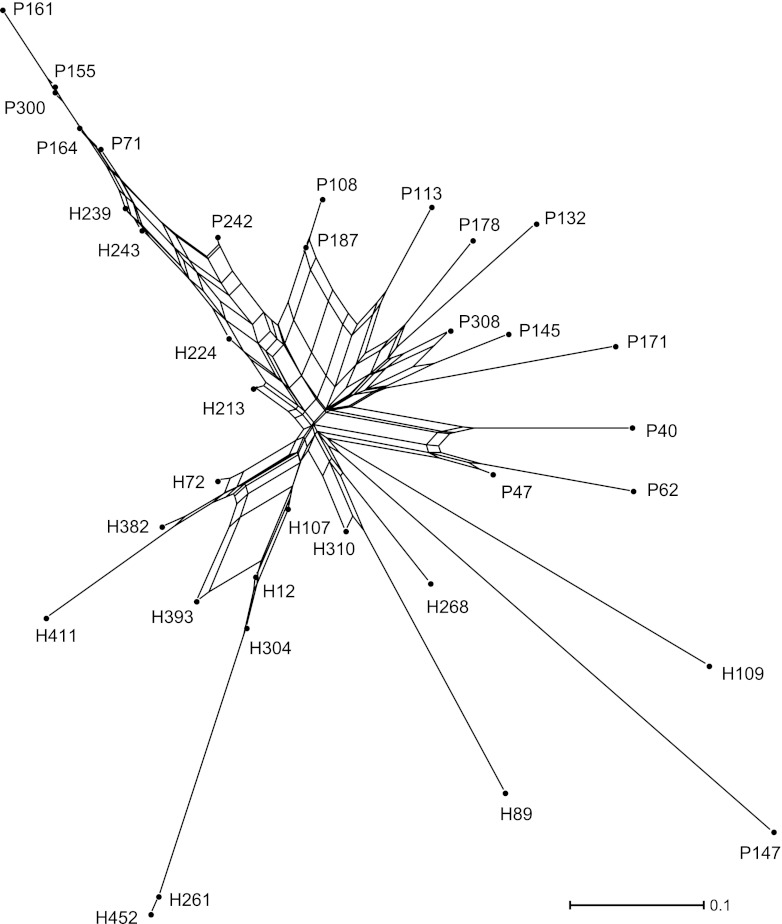
The similarities and dissimilarities between the ERIC type profiles of individual cows are depicted in the split network shown in Fig. 2. The lines of a network are known as edges, the connection points are known as nodes, and the length of an edge is proportional to the genetic distance between nodes. Thus, the sum of the lengths of the edges between two cows shows the relative number of pairwise differences between the ERIC types isolated from those cows. In general, the cows on the pasture farm occur within the top right half of the plot, while those on the housing farm are grouped within the lower left half, which confirms the presence of between-farm differences in ERIC types. However, in the top left corner there is a long edge, which is a closely connected group of animals from both farms (with cow 161 at the terminal node). The cows along this edge are those that were commonly excreting ERIC type 7. In the bottom left section there is an edge that terminates with cow 452; the cows along this edge are all from the housing farm, and they were regularly excreting ERIC type 5. In contrast, the ERIC types isolated from cows positioned in the bottom right corner are much more distinct than those isolated from other cows; for example, cow 147 from the pasture farm predominantly shed ERIC type 35 throughout the study, but this ERIC type was only sporadically isolated from two other cows on the same farm and was never detected on the housing farm.
Fig 2.
Unrooted split network of FST estimates showing the relative diversity of C. jejuni ERIC types isolated from 35 dairy cows on two dairy farms. The alphanumeric labels for each cow denote its farm of origin (P for the pasture farm, H for the housing farm) and farm-allocated cow identity number.
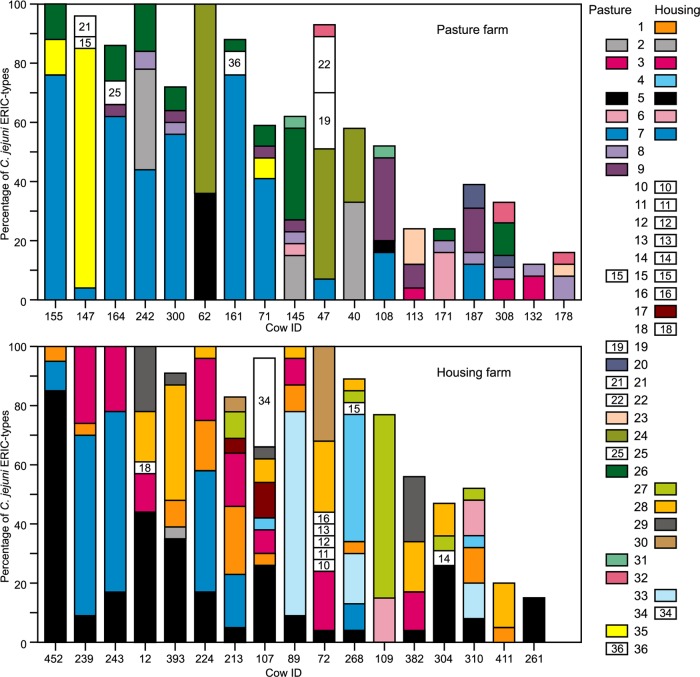
Relating the fecal concentration of C. jejuni shed by individual cows to the ERIC types detected showed that the cows belonging to the high-shedder group each excreted 3 to 4 different ERIC types (Fig. 3). For each of the high-shedder cows, one ERIC type was isolated from at least half of the samples collected, whereas the remaining 2 to 3 types occurred less frequently and were found in 4 to 30% of the samples collected from that cow. ERIC types 5, 7, and 35 were the ones that were most frequently observed in the high-shedder cows. The frequency of occurrence of individual ERIC types was, however, cow dependent. ERIC type 7, which was found in each of the six high-shedder cows, occurred frequently in four cows but less often in the other two cows. ERIC type 35, which was recovered from two high-shedder cows from the pasture farm, occurred frequently in only one of them. ERIC type 5, which was recovered from each of the three high-shedder cows on the housing farm, occurred frequently in only one of them.
Fig 3.
Bar plots showing the frequency (%) of detection of C. jejuni ERIC types in the feces of dairy cows on the pasture farm (top) and on the housing farm (bottom). The 36 different ERIC types detected in the positive samples are depicted using different colors and numbers. The number of samples for each cow is as described for Fig. 1.
Each of the cows belonging to the intermittent-shedder and low-shedder groups excreted 1 to 8 different ERIC types. Each cow excreted its individual combination of C. jejuni types. Five intermittent-shedder cows had one frequently occurring ERIC type, which was either ERIC type 2 (cow 40), 7 (cows 161 and 300), 24 (cow 62), or 33 (cow 89). In the intermittent- and low-shedder cows, types 5, 7, and 35 were identified in 17, 31, and 2%, respectively, of the samples that contain C. jejuni in concentrations greater than 103 g−1.
Characterization of the C. jejuni population by MLST analysis.
To determine the relationship between the C. jejuni populations isolated from dairy cows and those of human origin, MLST analysis was performed on 44 isolates that included at least one isolate from each ERIC type identified. Twelve of the 15 C. jejuni sequence types (ST) and clonal complexes (CC) which were identified (Table 2) have been previously reported in clinical cases recorded in the PubMLST database. The three sequence types which have not been linked to human cases (CC-49, ST-u52, and ST-177) corresponded to ERIC types that were not frequently excreted by the cows (<4% of the samples which contained C. jejuni). The ERIC types for which no ST or CC was identified represented <2% of the samples positive for C. jejuni.
Table 2.
MLST sequence type (ST) and clonal complex (CC) obtained from representative C. jejuni isolates that had been previously characterized by ERIC-PCR
| ERIC type | MLST |
|
|---|---|---|
| ST | CC | |
| 2 | 53 | 21 |
| 3 | 50 | 21 |
| 4 | 177 | 177 |
| 5, 34 | 45 | 45 |
| 6, 23 | 257 | 257 |
| 7 | 2026 | 403 |
| 9, 29, 30, 31, 33 | 61 | 61 |
| 15, 35 | 520 | 21 |
| 17 | 474 | 48 |
| 19 | u52 | 52 |
| 26 | 1517 | 354 |
| 27 | 3610 | 21 |
| 28 | 4337 | 21 |
| 1, 8 | 42 | |
| 20 | 49 | |
| 22, 36 | 21 | |
| 24, 25 | 48 | |
| 32 | 61 | |
| 10, 11, 12, 13, 14, 16, 18, 21 | Unknown | Unknown |
DISCUSSION
In this study, a regular and intensive monitoring of the same cows has been conducted. The feces were collected by direct retrieval from the same 35 cows from two commercially operating dairy herds every 2 weeks for up to 12 months. The range of concentrations of C. jejuni in feces (0 to 6 log10 C. jejuni organisms per g feces) is similar to that reported for fresh feces collected randomly from the pasture on other New Zealand dairy farms (13, 27). It is also comparable to that reported by Häkkinen and Hänninen for dairy cows in Finland (15). Our data further revealed marked variation in the frequency and range of C. jejuni concentrations between individual cows within the same herd.
The majority of the 35 studied cows excreted C. jejuni intermittently, which is consistent with other studies that concluded that cows are intermittent shedders (15, 19). There were six (17%) cows that excreted C. jejuni at much higher numbers and frequency than the other cows. These cows were found on both farms and were referred to as high shedders. High shedding of pathogens such as Escherichia coli O157 has been shown to coincide with peak of infection (34). This implies that there is a short period of time during which an animal excretes a high number of the pathogen and thus qualifies for the definition of a high shedder. Because infection by C. jejuni is asymptomatic in cows, C. jejuni, being a commensal resident in the bovine gastrointestinal tract, we defined the high-shedder cows as those that excreted C. jejuni with a frequency that was >80% and in counts that were ≥3 log10 per g of feces in half of the samples collected. Previous evidence that some cows excrete C. jejuni in high numbers and high frequency were based on observations from consecutive samplings occurring with at least a month-long interval or from medium-term studies (<6 months). For example, Inglis et al., who monitored 60 cows monthly for a 5-month period, observed that only one cow excreted C. jejuni at all of the sampling occasions and often in high concentrations (>5 log10 · g−1) (21). In another study, Hänninen et al. observed 29 dairy cows five times during 1 year and reported that just one cow shed C. jejuni at all five sampling times (16). No concentration was measured. Our finding that cows which excreted C. jejuni in high numbers and frequency were present on both farms suggests that New Zealand dairy herds contain cows that are long-term high shedders for C. jejuni.
Interrogation of the Campylobacter MLST database revealed that most of the isolates excreted by the studied cows have been previously associated with human cases, reinforcing the hypothesis that cows are a reservoir for human infection. Shedding of C. jejuni by cows has the potential to contaminate pastures and subsequently surface waters. The impact of the few high-shedder cows on the total load of C. jejuni excreted can largely exceed the load from the other shedding cows. If the loading of C. jejuni in excreta was calculated by using a published value of 2.1 kg feces per individual excretion (17), the high-shedder cows in our study could release into the environment between 6.6 and 9.4 log10 C. jejuni organisms at any of the 20 or 25 sampling occasions. Because of their frequency of excretion, the high-shedder cow would maintain the environmental loads of C. jejuni. A report prepared for the New Zealand Ministry of Health demonstrated that even a modest reduction (1 log) in the shedding of C. jejuni by cows that excrete a large number of C. jejuni can have a large effect on the environmental loading (24). The authors highlighted the importance of the high shedders as a possible target for control on farms. Nonetheless, the intermittent shedders in our study were found to excrete C. jejuni sporadically in high concentrations, particularly on the housing farm. This indicates that the intermittent shedder cows also have an effect on the environment, although sporadically. Development of interventions for reducing the prevalence and shedding of C. jejuni at the farm level may comprise decisions affecting the animal (e.g., vaccination, culling high-shedder cows, and selection of low-shedder cows), the bacteria (e.g., probiotics), or the environment. Critical evaluation of the most effective strategy to apply will depend on gaining understanding of the drivers for C. jejuni excretion in high numbers.
The genetic diversity of the isolates obtained from each of the 35 cows that were investigated revealed a high degree of individuality in the C. jejuni population present in the feces of each cow. The presence of one specific, persistent (found in more than half of the fecal samples collected) ERIC type in each of the long-term high-shedder cows suggests that the fecal excretion of C. jejuni in high numbers is due to successful adaptation of a particular genotype to a particular cow's gut environment. Genotyping by MLST of a consistently detected ERIC type in this study, ERIC type 7, revealed this type to be ST-2026, which is an ST type that has been previously associated with cattle (14, 22, 28). The presence of a ruminant type in the feces which contain C. jejuni in high concentration is consistent with the hypothesis that successful adaptation of a genotype to the gastrointestinal tract of an individual cow led to high shedding of C. jejuni by that cow. The variation in the frequency of C. jejuni genotypes among cows suggests, however, that animal-related factors render individual cows more resistant than others to colonization by a particular genotype. Ragimbeau et al. (32) observed that samples obtained from different animals on the same cattle farm tended to yield different strains of C. jejuni. Although some degree of overlap was apparent between C. jejuni populations among the different cows, particularly on the pasture farm, our data obtained from an extensive period of time confirmed the specificity of the C. jejuni population present in each cow. It is possible that one or more factors are important to explain the specificity of the C. jejuni populations in cows of the same age group and under the same management practices. A number of other thermophilic Campylobacter species are known to be associated with bovine feces, and in our study C. lari and C. upsaliensis were frequently detected (data not shown). Competition with the other Campylobacter species or other microorganisms present in cow intestine could have affected the fecal concentration and the genotypes of the C. jejuni populations. Another factor could be physiological stress of the cow, hence decreased immunity, due to short calving intervals or high milk production. Further investigations to better define the effect of cow-related factors on C. jejuni excretion are desirable. Interestingly, other ruminant-associated lineages (ST-53, ST-38, ST-61, and ST-42) that have been previously reported to be common in New Zealand dairy herds (28) or overseas (22, 31, 37) were not endemic to the farms of the studied cows. This could be due to the use of an enrichment step prior to the purification process, which might have favored the recovery of those types of C. jejuni that were most adapted to the media or were most concentrated in the feces. An exhaustive assessment of the within-cow diversity would require the inclusion of direct plating, the analysis of more isolates per positive sample, or the use of several media, in particular for studies in which a limited number of fecal samples per cow are analyzed. Also, as observed in our study, the C. jejuni population in a bovine herd has been reported to differ among individual farms (22, 35). Thus, determining whether there are more ruminant types that are shed at high concentration requires the analysis of more animals sourced from a larger number of farms.
The excretion patterns with regard to the frequency of excretion and range of concentration might also be affected by the environment in which the cows are farmed. The C. jejuni populations excreted by the studied cows were more diverse in the housing farm than in the pasture farm. High prevalence and great sequence type diversity of C. jejuni in geographical areas with a high presence of wildlife has shown that the sources of exposure are important to explain the occurrence and maintenance of C. jejuni in the herds (14). C. jejuni has the ability to colonize multiple hosts (5). In our study, C. jejuni genotypes found in the cows include types that are usually not associated with ruminants. For example, ERIC type 4, which was revealed by MLST analysis to be ST-177, was previously reported to be found in birds (6). It was only found in two cows on the pasture farm, suggesting a transfer between birds and cows. Another genotype that was found in the studied cows to be ST-45 (ERIC type 5) has been reported in bovine fecal isolates (7, 14), but it has been associated primarily with poultry and wildlife (12). In our study, this type is widely spread among the cows in the housing farm, suggesting the existence of on-farm sources from which this C. jejuni genotype spread in the dairy cow herd and established in the cows that were most susceptible to colonization. Another explanation for the occurrence of ERIC type 5 is a cow-to-cow transfer, as long-term colonization of a particular bacterium in a cow increases the chance of transmission of this bacterium from one cow to another. More work is required to understand the on-farm cycling of C. jejuni in different farm systems to evaluate the most appropriate farm management strategy that controls the persistence of C. jejuni in dairy herds and environmental loadings.
Conclusion.
The frequency of excretion and range of concentrations was found to be variable among the studied cows on both farms. Although long-term high shedders of C. jejuni were identified on both farms, other cows would excrete C. jejuni in high concentrations more sporadically. This suggests that a mitigation strategy targeting only the consistently high shedders might not completely address the environmental loading of C. jejuni from the herd. Reasons for excretion of C. jejuni in high numbers may include host adaptation of specific C. jejuni genotypes to a specific bovine gut. Our data suggest that animal-related factors render individual cows more resistant than others to colonization by a particular C. jejuni genotype. The challenge of the cows by the genotypes present in the environment might also affect the excretion pattern of the cows. Before the results can be applied to design management strategies that reduce the carriage of C. jejuni in dairy cows, further experimental work is required on the infection process for high-shedder cows in particular and on the influence of on-farm sources on the infection of cows.
ACKNOWLEDGMENTS
We are grateful for the cooperation and advice of the farmers involved in this study; to B. Wise and A. McGowan (AgResearch) and E. Blythe and P. Martin (Agriscience Consulting), who helped with the samplings; to N. P. French and members of mEpiLab (Hopkirk Research Institute, Massey University) for their help in the MLST work; and to A. M. Donnison and J. Mills (AgResearch) for their valuable comments and helpful discussions.
The project was funded by the New Zealand Foundation for Research, Science and Technology under contract number C10x1006.
Footnotes
Published ahead of print 17 August 2012
REFERENCES
- 1. Bates P, Phillips CA. 2005. Agricultural practices as a source of Campylobacter spp. in river water. J. Environ. Health Res. 4:17–23 [Google Scholar]
- 2. Batz MB, Hoffmann S, Glenn Morris J., Jr 2011. Ranking the risks: the 10 pathogen-food combinations with the greatest burden on public health. Emerging Pathogens Institute, University of Florida, Gainesville, FL: http://www.epi.ufl.edu/sites/www.epi.ufl.edu/files/RankingTheRisksREPORT.pdf [Google Scholar]
- 3. Best EL, Powell EJ, Swift C, Grant KA, Frost JA. 2003. Applicability of a rapid duplex real-time PCR assay for speciation of Campylobacter jejuni and Campylobacter coli directly from culture plates. FEMS Microbiol. Lett. 229:237–241 [DOI] [PubMed] [Google Scholar]
- 4. Clark CG, et al. 2003. Characterization of waterborne outbreak-associated Campylobacter jejuni, Walkerton, Ontario. Emerg. Infect. Dis. 9:1232–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Colles FM, Jones K, Harding RM, Maiden MCJ. 2003. Genetic diversity of Campylobacter jejuni isolates from farm animals and the farm environment. Appl. Environ. Microbiol. 69:7409–7413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Colles FM, et al. 2011. Dynamics of Campylobacter colonization of a natural host, Sturnus vulgaris (European Starling). Microbiology. 11:258–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. De Haan CPA, Kivitő RI, Hakkinen M, Corander J, Hänninen MJ. 2010. Multilocus sequence types of Finnish bovine Campylobacter jejuni isolates and their attribution to human infections. BMC Microbiol. 10:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dingle K, et al. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Donnison A. 2003. Isolation of thermotolerant Campylobacter—review and methods for New Zealand laboratories. Ministry of Health, Wellington, New Zealand: http://www.moh.govt.nz/notebook/nbbooks.nsf/0/73166EB251837F95CC257834000271DB/$file/IsolationOfThermotolerantCampylobacter.pdf [Google Scholar]
- 10. Excoffier L, Smouse PE, Quattro JM. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Excoffier L, Laval G, Schneider S. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1:47–50 [PMC free article] [PubMed] [Google Scholar]
- 12. French N, et al. 2009. Molecular epidemiology of Campylobacter jejuni isolated from wild bird fecal material in children's playgrounds. Appl. Environ. Microbiol. 75:779–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gilpin BJ, et al. 2008. Comparison of Campylobacter jejuni genotypes from dairy cattle and human sources from the Matamata-Piako district of New Zealand. J. Appl. Microbiol. 105:1354–1360 [DOI] [PubMed] [Google Scholar]
- 14. Grove-White DH, Leatherbarrow AJ, Cripps PJ, Diggle PJ, French NP. 2011. Molecular epidemiology and genetic diversity of Campylobacter jejuni in ruminants. Epidemiol. Infect. 139:1661–1671 [DOI] [PubMed] [Google Scholar]
- 15. Häkkinen M, Hänninen ML. 2009. Shedding of Campylobacter spp. in Finnish cattle on dairy farms. J. Appl. Microbiol. 107:898–905 [DOI] [PubMed] [Google Scholar]
- 16. Hänninen ML, Niskanen M, Korhonen L. 1998. Water as reservoir for Campylobacter jejuni infection in cows studied by serotyping and pulsed-field gel electrophoresis (PFGE). J. Vet. Med. B 45:37–42 [DOI] [PubMed] [Google Scholar]
- 17. Haynes RJ, Williams PH. 1993. Nutrient cycling and soil fertility in the grazed pasture ecosystem. Adv. Agron. 49:119–199 [Google Scholar]
- 18. Heuvelink AE, et al. 2009. Two outbreaks of campylobacteriosis associated with the consumption of raw cows' milk. Int. J. Food Microbiol. 134:70–74 [DOI] [PubMed] [Google Scholar]
- 19. Humphrey TJ, Beckett P. 1987. Campylobacter jejuni in dairy cows and raw milk. Epidemiol. Infect. 98:263–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267 [DOI] [PubMed] [Google Scholar]
- 21. Inglis GD, Kalischuk LD, Busz HW. 2004. Chronic shedding of Campylobacter species in beef cattle. J. Appl. Microbiol. 97:410–420 [DOI] [PubMed] [Google Scholar]
- 22. Kwan PLS, et al. 2008. Longitudinal study of the molecular epidemiology of Campylobacter jejuni in cattle on dairy farms. Appl. Environ. Microbiol. 74:3626–3633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lake RJ, Cressey PJ, Campbell DM, Oakley E. 2010. Risk ranking for foodborne microbial hazards in New Zealand: burden of disease estimates. Risk Anal. 30:743–752 [DOI] [PubMed] [Google Scholar]
- 24. Marshall J, French N. 2010. Modelling of Campylobacter carriage and transmission between and within animal groups. Ministry of Agriculture and Forestry, Wellington, New Zealand: http://www.maf.govt.nz/news-resources/publications.aspx [Google Scholar]
- 25. Matthews L, et al. 2006. Super-shedding cattle and the transmission dynamics of Escherichia coli O157. Epidemiol. Infect. 134:131–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meacham KJ, Zhang L, Foxman B, Bauer RJ, Marrs CF. 2003. Evaluation of genotyping large numbers of Escherichia coli isolates by enterobacterial repetitive intergenic consensus-PCR. J. Clin. Microbiol. 41:5224–5226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moriarty EM, Sinton LW, Mackenzie ML, Karki N, Wood DR. 2008. A survey of enteric bacteria and protozoans in fresh bovine feces on New Zealand dairy farms. J. Appl. Microbiol. 105:2015–2025 [DOI] [PubMed] [Google Scholar]
- 28. Müllner P, et al. 2009. Assigning the source of human campylobacteriosis in New Zealand: a comparative genetic and epidemiological approach. Infect. Genet. Evol. 9:1311–1319 [DOI] [PubMed] [Google Scholar]
- 29. Nei M. 1987. Molecular evolutionary genetics. Columbia University Press, New York, NY [Google Scholar]
- 30. Oliver DM, Heathwaite AL, Hodgson CJ, Chadwick DR. 2007. Mitigation and current management attempts to limit pathogen survival and movement within farmed grassland. Adv. Agron. 93:95–152 [Google Scholar]
- 31. Oporto B, Juste RA, Lüpez-Portolés JA, Hurtado A. 2011. Genetic diversity among Campylobacter jejuni isolates from healthy livestock and their links to human isolates in Spain. Zoonoses Public Health 58:365–375 [DOI] [PubMed] [Google Scholar]
- 32. Ragimbeau C, Schneider F, Losch S, Even J, Mossong J. 2008. Multilocus sequence typing, pulsed-field gel electrophoresis, and fla short variable region typing of clonal complexes of Campylobacter jejuni strains of human, bovine, and poultry origins in Luxembourg. Appl. Environ. Microbiol. 74:7715–7722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rasschaert G, et al. 2005. Comparison of five repetitive-sequence-based PCR typing methods for molecular discrimination of Salmonella enterica isolates. J. Clin. Microbiol. 43:3615–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Robinson SE, Wright EJ, Hart CA, Bennett M, French NP. 2004. Intermittent and persistent shedding of Escherichia coli O157 in cohorts of naturally infected calves. J. Appl. Microbiol. 97:1045–1053 [DOI] [PubMed] [Google Scholar]
- 35. Rotariu O, et al. 2009. Spatiotemporal homogeneity of Campylobacter subtypes from cattle and sheep across NE and SW Scotland. Appl. Environ. Microbiol. 75:6275–6281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sears A, et al. 2011. Marked campylobacteriosis decline after interventions aimed at poultry, New Zealand. Emerg. Infect. Dis. 17:1007–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sproston EL, et al. 2011. Temporal variation and host association in the Campylobacter population in a longitudinal ruminant farm study. Appl. Environ. Microbiol. 77:6579–6586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stanley KN, Wallace J, Currie JE, Diggle PJ, Jones K. 1998The seasonal variation of thermophilic campylobacters in beef cattle, dairy cattle and calves. J. Appl. Microbiol. 85:472–480 [DOI] [PubMed] [Google Scholar]
- 39. Vandamme P, et al. 1997. Campylobacter hyoilei Alderton et al. 1995 and Campylobacter coli Véron and Chatelain 1973 are subjective synonyms. Int. J. Syst. Bacteriol. 47:1055–1060 [DOI] [PubMed] [Google Scholar]
- 40. Versalovic J, Koeuth T, Lupski R. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weijtens MJBM, Reinders RD, Urlings HAP, Van der Plas J. 1999. Campylobacter infections in fattening pigs; excretion pattern and genetic diversity. J. Appl. Microbiol. 86:63–70 [DOI] [PubMed] [Google Scholar]
- 42. World Health Organization 1984. Guidelines for drinking-water quality, vol. 1: recommendations. World Health Organization, Geneva, Switzerland [Google Scholar]