Abstract
Curli are adhesive fimbriae of Enterobactericaeae and are involved in surface attachment, cell aggregation, and biofilm formation. We reported previously that curli-producing (C+) variants of E. coli O157:H7 (EcO157) were much more acid sensitive than their corresponding curli-deficient (C−) variants; however, this difference was not linked to the curli fimbriae per se. Here, we investigated the underlying molecular basis of this phenotypic divergence. We identified large deletions in the rcsB gene of C+ variants isolated from the 1993 U.S. hamburger-associated outbreak strains. rcsB encodes the response regulator of the RcsCDB two-component signal transduction system, which regulates curli biogenesis negatively but acid resistance positively. Further comparison of stress fitness revealed that C+ variants were also significantly more sensitive to heat shock but were resistant to osmotic stress and oxidative damage, similar to C− variants. Transcriptomics analysis uncovered a large number of differentially expressed genes between the curli variants, characterized by enhanced expression in C+ variants of genes related to biofilm formation, virulence, catabolic activity, and nutrient uptake but marked decreases in transcription of genes related to various types of stress resistance. Supplying C+ variants with a functional rcsB restored resistance to heat shock and acid challenge in cells but blocked curli production, confirming that inactivation of RcsB in C+ variants was the basis of fitness segregation within the EcO157 population. This study provides an example of how genome instability of EcO157 promotes intrapopulation diversification, generating subpopulations carrying an array of distinct phenotypes that may confer the pathogen with survival advantages in diverse environments.
INTRODUCTION
Curli are thin aggregative fimbriae produced in many Enterobactericeae, including both pathogenic and nonpathogenic Escherichia coli strains (49, 74). Curli play an important role in surface attachment, biofilm formation, and induction of the host inflammatory response and confer cell protection from toxic compounds (2, 23, 31, 32, 63, 65). Biogenesis of curli requires the function of two operons, csgBA and csgDEFG, which are transcribed divergently (26). CsgA is the curli subunit protein, whereas CsgD is a transcriptional activator of the csgBA operon. The products of other curli genes are required for assembly of mature curli (11, 27, 28, 53, 69). Transcription of both curli operons is regulated hierarchically by several global transcriptional regulators and two-component signal transduction systems. For example, the alternative sigma factor RpoS activates expression of csgBA directly, and this positive regulation is enhanced by the interaction of RpoS with Crl, a small regulatory protein that functions as a thermal sensor for maximal expression of curli fimbriae at low temperatures (3). RpoS also regulates the transcription of csgD positively through the activation of mlrA (4). Other regulators involved in curli gene expression are histone-like protein (HN-S) and the integration host factor (IHF). IHF enhances csgD expression, whereas HN-S can either activate or repress csgBA expression, depending on the strains used in the study (22, 48). The OmpR/EnvZ two-component system responds to low osmolarity and regulates csgD expression positively (68); the CpxA/R two-component system responds to cell envelope stress and regulates expression of both curli operons negatively (14); similarly, the RcsCDB (Rcs) two-component system responds to cell membrane stress and controls the expression of both curli operons negatively, in an RcsA-dependent manner (67). Importantly, these transcriptional regulators interact with each other and modulate their own expression levels, resulting in a complex regulation network that fine-tunes curli biogenesis in response to various environmental and physiological signals (43, 44). Natural variation in curli expression has been reported for E. coli strains isolated from diverse environments (46, 61). Increased curli production has been linked to mutations in the ompR gene (68) or the csgD promoter (54, 64).
Enterohemorrhagic Escherichia coli (EHEC) strains include a diverse group of Shiga toxin-producing E. coli (STEC) that can cause bloody diarrhea, hemorrhagic colitis, and life-threatening hemolytic uremic syndrome (HUS). EHEC naturally resides in ruminants, primarily cows, and is transmitted to humans mainly through food vehicles. To cause infection, EHEC must survive in an extremely acidic environment (pH < 3) within the host's stomach. Similar to Shigella spp., E. coli can survive at pH 2.0 for several hours (24, 57), which is attributed to at least four well-studied acid resistance systems: the oxidative acid resistance system (AR1) and the glutamate-, arginine-, and lysine-dependent acid resistance systems (AR2, AR3, and AR4, respectively) (19, 73). Additionally, E. coli encodes various acid resistance proteins and chaperones, such as HdeA and HdeB, known to confer protection at low pH (pH < 3) (21, 72). Similar to curli biogenesis, expression of acid resistance systems and proteins is regulated hierarchically by numerous global transcriptional regulators and two-component signal transduction systems. The expression of AR1 requires RpoS, cyclic AMP (cAMP), and the cAMP receptor protein CRP (10, 51). Among the amino acid-dependent acid resistance systems, AR2 is thought the most effective system under an extreme acidic environment (pH < 2.5). It requires a functional glutamate decarboxylase (GadA or GadB) and an antiporter (GadC), which exchanges external glutamate for the intracellular decarboxylation product γ-aminobutyric acid (39). The expression of AR2 is regulated by transcriptional regulators GadE, GadX, GadW, RpoS, H-NS, and CRP and the two-component systems EvgAS and RcsCD (34, 41, 42, 45, 55). Within this regulating network, GadE is a central player. It activates AR2 and genes encoding acid resistance chaperones (HdeA and HdeB) and proteins (YhiD, Slp, and YhiF). Transcription of gadE is positively regulated by GadW, GadX, EvgA, and itself, and the activity of GadE is modulated by RcsB (35). Furthermore, RpoS regulates expression of gadW and gadX positively, and GadX activates gadA, gadBC, hdeD, hdeAB, and sip directly. In contrast, H-NS regulates the transcription of gadE, gadX, gadW, and rcsD negatively. Similarly, CRP regulates gadW, gadX, and gadE expression negatively by repressing rpoS and gadA for cells grown in rich medium (9).
We previously reported that natural curli variants of E. coli O157:H7 displayed distinct acid resistance; however, this difference was not linked to the curli fimbriae per se (7). This characteristic is conserved in all three groups of curli variants: group I variants were isolated from the 2006 U.S. spinach-associated outbreak strains; group II variants were isolated from strains linked to a 2006 U.S. iceberg lettuce-associated outbreak; group III variants were isolated from the 1993 U.S. hamburger-associated outbreak strain. Because there are several common transcriptional regulators between curli biogenesis and acid resistance in E. coli, we hypothesize that natural mutations in genes encoding the common regulators are the cause of the marked difference in acid resistance between the natural curli variants of E. coli O157:H7. In this study, we focused on curli variants isolated from the 1993 U.S. hamburger-associated outbreak strains (group III curli variants). We report that inactivation of the response regulator RcsB of the two-component system RcsCDB in curli-producing (C+) variants is the molecular basis of the observed fitness segregation within the population of E. coli O157:H7.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
The E. coli strains and mutants used in this study and their sources are listed in Table 1. Strains were grown routinely in Luria-Bertani (LB) broth (10 g tryptone, 5 g yeast extract, and 5 g NaCl per liter) or modified LB broth containing various concentrations of salt: LB-full salt (LBFS) contained 10 g of NaCl per liter, or LB-no salt (LBNS), which contained no NaCl. Antibiotics were used at the following concentrations: ampicillin (Amp) at 100 μg ml−1; carbenicillin (Cb) at 50 μg ml−1; kanamycin (Km) at 50 μg ml−1; gentamicin (Gm) at 15 μg ml−1. The E. coli O157:H7 curli variants used in this study were described previously (7). Variants were isolated by plating E. coli O157:H7 on Congo red indicator (CRI) plates (LBNS plate supplemented with 40 μg/ml of CR dye and 10 μg/ml of Coomassie brilliant blue), followed by incubation of plates at 28°C for 2 to 3 days. Colonies with various colors on the CRI plates were picked, and curli production of each variant was determined by immunoblot analysis using an antibody specific to the curli subunit protein CsgA (kindly provided by M. Chapman).
Table 1.
Strains used in this study
| Strain or plasmid | Antibiotic resistancea | Characteristics | Source or reference |
|---|---|---|---|
| E. coli O157:H7 strains | |||
| RM6607 | None | Human isolate linked to 1993 U.S. hamburger-associated outbreak | 70 |
| RM6607R | None | Curli-producing (C+) variant of strain RM6607 | 7 |
| RM6607W | None | Curli-deficient (C−) variant of strain RM6607 | 7 |
| RM6608 | None | Meat isolate linked to 1993 U.S. hamburger-associated outbreak | 70 |
| RM6608R | None | Curli-producing (C+) variant of strain RM6608 | 7 |
| RM6608W | None | Curli-deficient (C−) variant of strain RM6608 | 7 |
| MQC118 | Kmr | csgA deletion mutant of RM6607R | 7 |
| MQC120 | Kmr | csgA deletion mutant of RM6607W | 7 |
| MQC143 | Kmr | hdeAB deletion mutant of RM6607W | 8 |
| MQC560 | Gmr | RM6607R transformed with pBBR1MCS-5 | This study |
| MQC562 | Gmr | RM6607R transformed with pXQ30 | This study |
| E. coli non-O157 strain | |||
| DH5α | None | Host strain for pBBR1MCS-5, pKD46, and pKD4 and their derivatives | S. Lory |
| Plasmids | |||
| pKD46 | Ampr | λ Red recombinase expression plasmid | AY048746b (13) |
| pKD4 | Kmr | Template plasmid for gene deletion | AY048743b (13) |
| pBBR1MCS-5 | Gmr | Expression vector for complementation analysis | U25061b (33) |
| pXQ30 | Gmr | The rcsB gene along with its native promoter of RM6607W was cloned into pBBR1MCS-5 | This study |
Kmr, kanamycin resistance; Ampr, ampicillin resistance; Gmr, gentamicin resistance.
GenBank accession number.
DNA sequencing.
Bidirectional DNA sequencing was performed on PCR-amplified DNA fragments or plasmids. Primers used for amplification and sequencing are listed in Table S1 of the supplemental material. PCR products were purified by using ExoSAP-IT (U.S. Biochemical, Cleveland, OH) and then used directly as templates for the cycle sequencing reactions using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA). For sequencing the cloned DNA fragments, plasmids (∼100 ng) were used directly as templates for dye incorporation reactions. Following sequencing reactions, the excess dye was removed using a BigDye XTerminator kit (Applied Biosystems), and the purified samples were sequenced using an automated fluorescence sequencer (ABI Prism 3730 DNA analyzer; Applied Biosystems). DNA sequences were assembled in SeqMan (Lasergene 8; DNASTAR, Madison, WI), and the consensus sequence was exported to MegAlign (Lasergene 8) for comparison.
Stress fitness tests.
Single colonies of curli variants on LB plates were inoculated in LBNS broth and incubated at 28°C overnight with rotation (150 rpm). The acid challenge was performed as described previously (7). Briefly, 5 μl of overnight culture was added to 5 ml acidified LBNS broth (pH 2.5; ∼106 CFU ml−1) and incubated at 37°C for 2 h. For heat shock conditions, 5 μl of overnight culture was added into 5 ml potassium phosphate (KP) buffer (10 mM; pH 7.0; ∼106 CFU ml−1) and incubated at 55°C for 15 min; for osmotic challenge, 5 μl of overnight culture was added into 5 ml of high-salt LB broth (2.5 M NaCl; ∼106 CFU ml−1) and incubated at 28°C for 6 h. The time zero samples were taken immediately before the incubation. For the oxidative stress test, 50 μl of overnight culture was added into 5 ml of KP buffer (∼107 CFU ml−1). After collection of time zero samples, 3% H2O2 was added immediately to a concentration of 12.5 mM, and the cells were then incubated at 28°C for 30 min. At the end of each stress test, cells were diluted first in KP buffer and plated onto LB plates by using a spiral plater (Autoplate 4000; Spiral Biotech). The stress fitness was expressed as the percentage of cells that recovered on LB plates following each stress challenge. At least three biological replicates were performed for each sample.
Swimming motility assay.
LB agar (0.3%) was used to examine the swimming motility. Individual colonies of curli variants grown on CRI plates at 28°C for 2 days were used to inoculate the swimming agar by using a round toothpick. The plates were incubated at 28°C for 16 to 18 h, and the swimming motility was assessed based on the size of the swimming zone (the diameter of the bacterial colony on the swimming agar plate). At least four replicates were carried out for each variant.
Gene deletion and DNA cloning.
In-frame deletion of the individual gene or gene cluster was achieved by replacing the target gene(s) with a Km cassette using the standard Lambda Red-mediated gene replacement method (13). Briefly, the Km cassette was amplified directly from the template plasmid pKD4 (Table 1) and using the gene knockout primers described in Table S1 of the supplemental material. The PCR products (about 1.6 kb) were agarose gel purified and transformed by electroporation into E. coli strains carrying the helper plasmid pKD46 (Table 1). The Kmr transformants were selected on LB agar plates containing 50 μg ml−1 of Km. The deletion mutants were verified by colony PCR using primers flanking the target genes (see Table S1). Mutants were then subcultured, incubated at 37°C, and tested for Amp sensitivity, which indicated loss of the helper plasmid pKD46.
For complementation analysis, a DNA fragment containing the rcsB gene was PCR amplified, digested with the restriction enzymes KpnI and HindIII, and then cloned into the plasmid pBBR1MCS-5, resulting in plasmid pXQ30 (Table 1). The sequence-confirmed clones were then transformed into the target strains by electroporation, and the transformants were selected on Gm-containing plates and verified by colony PCR.
Mass spectrometry analyses of HdeA protein.
The HdeA protein was detected and identified from unfractionated bacterial cell lysates by using matrix-assisted laser desorption ionization–tandem time of flight postsource decay tandem mass spectrometry (MALDI-TOF/TOF-PSD-MS/MS) and top-down proteomic analysis as previously described (16, 17). Briefly, a 1-μl loop of bacterial cells was lysed by bead beating (BioSpec Products, Bartlesville, OK) for 1 min in 300 μl of extraction solution comprised of 67% high-performance liquid chromatography (HPLC)-grade water, 33% HPLC-grade acetonitrile, and 0.2% trifluoroacetic acid. After sample centrifugation at 16,000 × g for 2 min, 0.5 μl of supernatant was spotted onto a stainless steel MALDI target and allowed to dry at room temperature, followed by spotting on top 0.5 μl of saturated solution of MALDI matrix, α-cyano-4-hydroxycinnamic acid (CHCA) or 3,5-dimethoxy-4-hydroxycinnamic acid (sinapinic acid), and again allowed to dry. Data were acquired using a 4800 Plus MALDI-TOF/TOF mass spectrometer (AB Sciex, Foster City, CA) as previously described (16). HdeA was identified by top-down proteomic analysis using software developed in-house (17).
Transcriptomics study.
Single colonies of curli variants were inoculated in LB broth and incubated at 28°C overnight on a shaker (150 rpm). The cells were collected by centrifugation, washed once in LBNS broth, and inoculated in 25 ml of LBNS broth with a concentration equivalent to 0.001 OD600 units/ml (OD600 is the optical density at 600 nm). The cultures were incubated at 28°C for 24 h with aeration (150 rpm). At the end of incubation, an ice-cold phenol-ethanol (5%:95%) solution was immediately added to the culture at a final concentration of 20% (vol/vol), and the mixture was incubated on ice for 30 min. The cells were collected by centrifugation at 4°C, and the pellets were stored at −80°C until RNA extraction.
RNA extraction, cDNA labeling, and microarray hybridization were performed as described previously with slight modifications (37, 50). Briefly, total RNA was extracted using the Promega SV total RNA kit, with additions of 3 mg ml−1 of lysozyme (Fisherbrand) to a bacterial cell suspension. The quality of RNA was assessed using an Agilent Technologies (Santa Clara, CA) 2100 Bioanalyzer. Genomic DNA (gDNA) was used as a common reference for the microarray-based transcriptional profiling study as described previously (15). Two micrograms of gDNA was labeled via incorporation of Cy3-dCTP (GE Healthcare) by using Klenow fragment (New England BioLabs), whereas 40 μg of total RNA from each sample was labeled via incorporation of Cy5-dCTP (GE Healthcare) into a cDNA product with the Fairplay III microarray labeling kit (Stratagene). Hybridization of Cy3-DNA and Cy5-cDNA was carried out overnight at 42°C. Arrays were scanned on a GenePix 4000B scanner (Axon Instruments, Molecular Devices), and the hybridization intensities of individual spots were analyzed with the GenePix Pro 6.0 software (Axon Instruments). Spots with a reference signal lower than background plus 2 standard deviations or spots covered by an obvious blemish were excluded. The average local background value was subtracted from all spots, and the Cy5/Cy3 ratio was calculated. Further normalization to account for differences in dye incorporation included data centering by setting the median natural logarithm to 0 for each group of spots in one sector (printed by one pin).
Three biological replicates were performed for each curli variant. The cDNA-gDNA mixtures for each biological replicate were hybridized on three separate microarrays, providing three technical replicates. The data from all replicates were analyzed by using an unpaired t test with unequal variance within the Genespring 7.3 program (Agilent). The expression level of each detected gene was compared between the curli variants derived from the same strain. Genes were considered to be expressed differentially if the fold change in expression level (the ratio with the normalized hybridization intensity) was >2 and the Benjamini-Hochberg false discovery rate (FDR)-adjusted P value was <0.05.
Statistical analysis.
Statistical analysis was computed using SigmaPlot version 11.0 (Systat Software, Inc.). An unpaired t test was performed for two-group comparisons, and an analysis of variance (ANOVA) followed by a Bonferroni t test was performed for multiple comparisons.
Microarray data accession number.
Microarray data were deposited in the NCBI GEO omnibus database under accession number GSE39439.
Nucleotide sequence accession numbers.
The sequences of the rcsB genes were deposited in GenBank under accession numbers JX258657 (RM6607R), JX258658 (RM6607W), JX258659 (RM6608R), and JX258660 (RM6608W).
RESULTS
C+ variants of the 1993 hamburger-associated outbreak strains are natural rcsB mutants.
Curli variants isolated from two outbreak strains, RM6607 (clinical isolate) and RM6608 (meat isolate), were used in this study. All four curli variants had identical multilocus variable-number tandem repeat analysis (MLVA) genotypes but differed largely in curli production. Colonies of C+ variants (RM6607R and RM6608R) were red on CRI plates, regardless of incubation temperature, indicating that production of curli in both C+ variants was independent of temperature. This conclusion was further supported by a Western blot analysis with an anti-CsgA antibody (Fig. 1A). Colonies of C− variants were either white or light red, depending on the growth temperature; however, this light color appeared to be a result of nonspecific CR binding, because there were no significant differences in the binding of CsgA-specific antibody to cells of RM6607W or its corresponding curli-deficient mutant (MQC120) at either 28°C or 37°C (Fig. 1A). Therefore, in contrast to C+ variants, C− variants did not produce any detectable curli at either of the two growth temperatures examined (Fig. 1A).
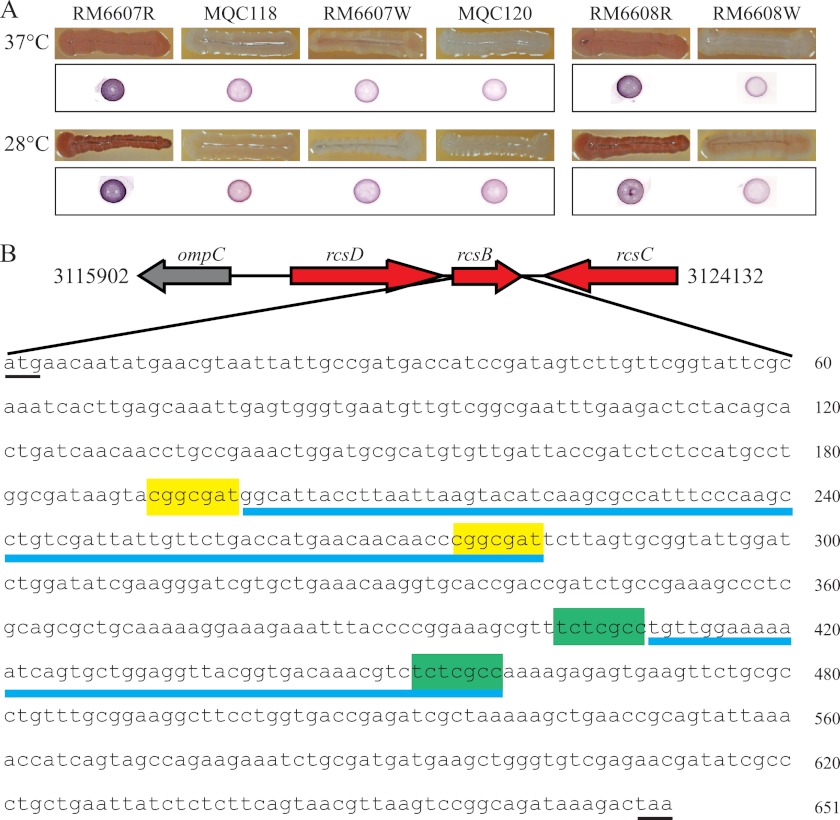
Fig 1.
Sequence analysis of the rcsB gene in E. coli O157:H7 group III curli variants. (A) Group III E. coli O157:H7 curli variants and their corresponding curli-deficient mutants, grown on CRI plates (upper panel), and curli production as measured by Western blotting (low panel). (B) Genomic location of the rcsB gene. The rcsB gene encodes a response regulator, and rcsC encodes a sensor kinase, whereas rcsD encodes a phosphotransfer protein. RcsCDB are essential components of the Rcs signal transduction system. Natural deletions identified in C+ variants of E. coli O157:H7 RM6607R and RM6608R are underlined in blue. The 49-bp DNA fragment bordered by the direct repeat tctcgcc (highlighted in green) was deleted in variant RM6607R, whereas the 83-bp DNA fragment bordered by the direct repeat cggcgat (highlighted in yellow) was deleted in variant RM6608R. Other direct repeats with perfect matches in rcsB are presented in Table S6 of the supplemental material. Both the start (atg) and stop (taa) codons of rcsB genes are underlined.
We previously reported that both C+ variants were significantly more acid sensitive than their corresponding C− variants (7). Sigma factor RpoS, regulatory protein H-NS, and the RcsCDB two-component signal transduction system are known to regulate both curli biogenesis and acid resistance; therefore, we sequenced the genes encoding the above regulatory proteins in curli variants RM6607R and RM6607W. Both variants carried identical DNA sequences of the rpoS, hns, rcsD, and rcsC genes; however, there was a 49-bp deletion (Δ410–458) in the rcsB gene of RM6607R (Fig. 1B). RcsB is the response regulator of the RcsCDB system, which activates GadE, the central regulator of acid resistance, but represses transcription of both curli operons. Therefore, inactivation of RcsB corresponded to increased curli production but deceased acid resistance in RM6607R compared with RM6607W, which had a functional rcsB gene.
To examine if the rcsB mutation was widespread in C+ variants of E. coli O157:H7, we examined the rcsB gene in curli variants RM6608R and RM6608W. Interestingly, RM6608R also carried a large deletion in rcsB (Δ199–281); however, this deletion was different from the deletion detected in the rcsB of variant RM6607R (Fig. 1B). Examination of rcsB genes in C+ variants isolated from 2006 U.S. spinach-associated outbreak strains (group I curli variants) did not reveal any mutations in the rcsB gene of any C+ variant examined (data not shown).
Production of the acid chaperone protein HdeA was blocked in C+ variants.
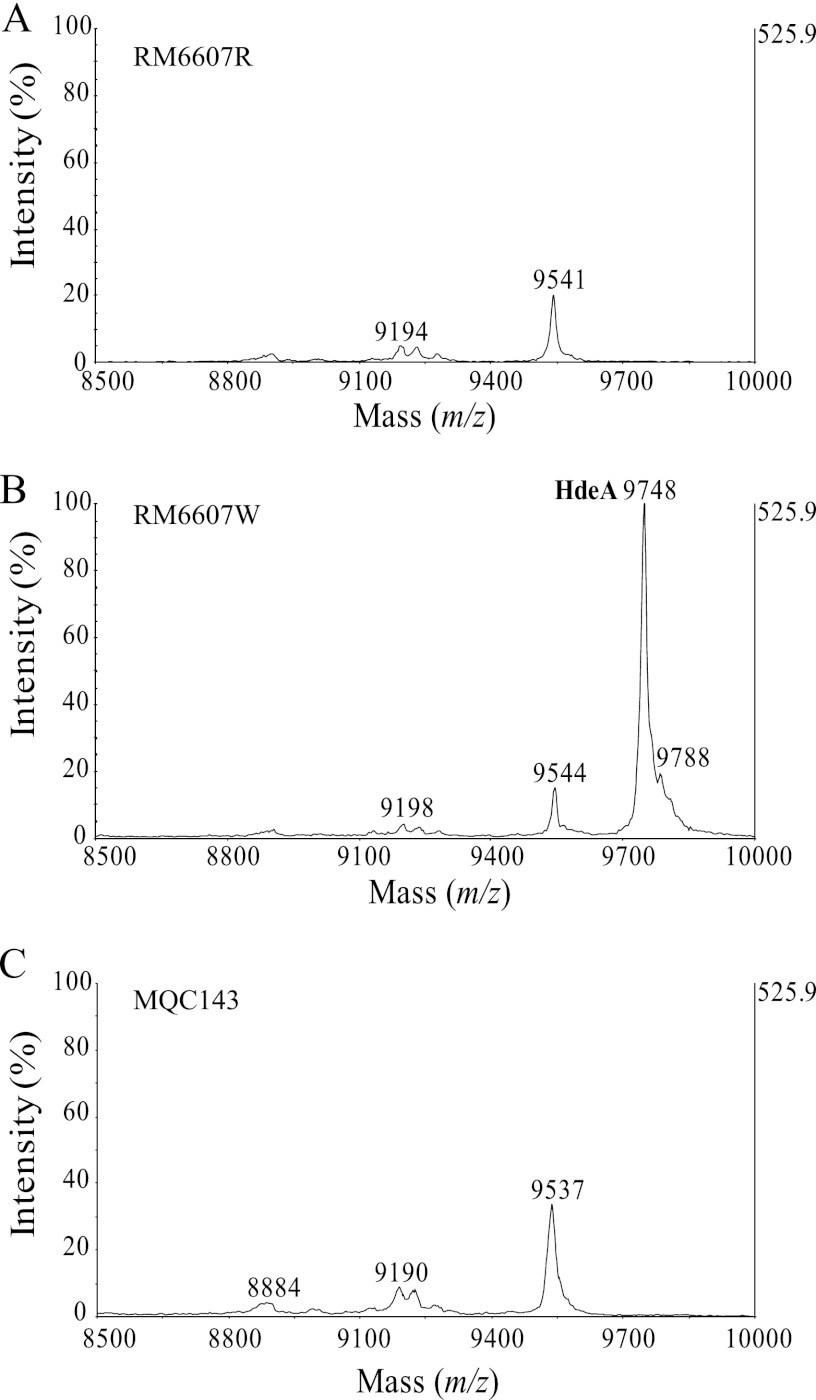
RcsB activates expression of hdeAB via the GadE pathway; therefore, loss of RcsB in a C+ variant would have an impact on HdeA production but not HdeB production, since translation of hdeB is silenced in E. coli O157:H7 due to a point mutation at the start codon of the hdeB gene (8). A distinct HdeA peak was detected in RM6607W, but not in RM6607R (Fig. 2A and B), despite both variants having an identical DNA sequence for the hdeAB genes. Deletion of hdeAB in RM6607W resulted in loss of the HdeA characteristic peak (Fig. 2C), similar to what was observed in RM6607R; thus, RM6607R is naturally deficient in the HdeA chaperone. A similar result was observed for RM6608R (data not shown).
Fig 2.
Detection of HdeA by mass spectrometry. The MS spectra shown are for the extracted cell lysates of E. coli O157:H7 curli variants RM6607R (A), RM6607W (B), and hdeAB deletion mutant strain MQC143 (C), obtained using an α-cyano-4-hydroxycinnamic acid matrix. HdeB biomarker (m/z ∼9,060) was not detected in either variant or mutant strains (8). In variant RM6607W, a distinct HdeA biomarker (m/z 9,748) was detected by MS and identified by MS/MS and top-down analysis, but the corresponding HdeA biomarker was absent in variant RM6607R (A). Deletion of hdeAB in variant RM6607W resulted in loss of the HdeA biomarker (C).
RM6607R was also more sensitive to heat stress than RM6607W.
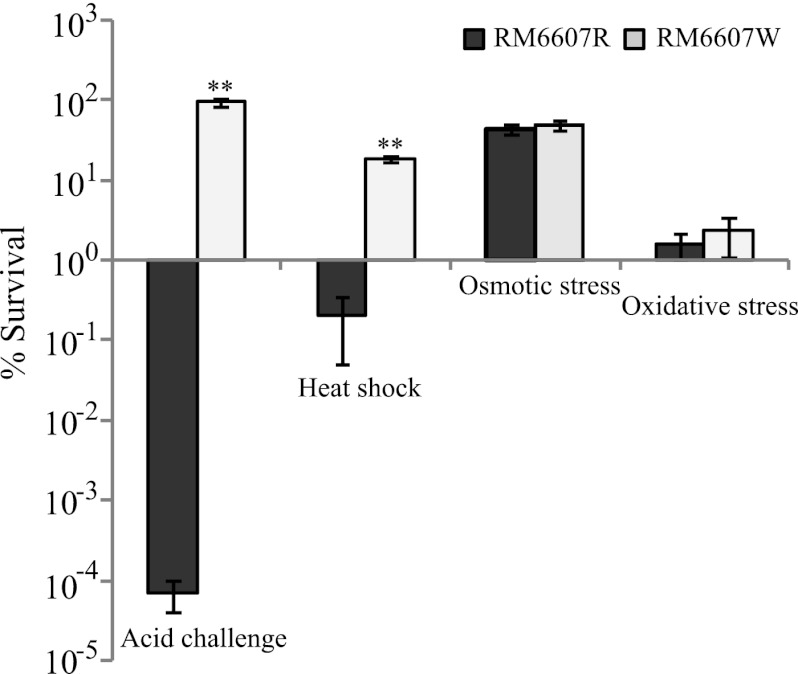
Because RcsB regulates a wide range of cellular process, we investigated if there were any differences between RM6607R and RM6607W in addition to resistance to acid challenge. We compared the survival between RM6607R and RM6607W following exposure of cells to heat shock (55°C for 15 min), osmotic challenge (2.5 M NaCl for 6 h at 28°C), and oxidative stress (12.5 mM H2O2 for 30 min at 28°C). Similar to acid resistance, the heat resistance of RM6607W was significantly greater than that of the RM6607R (Fig. 3, heat shock [t test, P < 0.001]). The average percentages of cells surviving heat shock were 18.2% and 0.2% for RM6607W and RM6607R, respectively. In contrast, there were no significant differences in survival between RM6607R and RM6607W following either osmotic challenge or oxidative stress. The average percentages of cells recovered were 44.5% and 49.4% after the osmotic challenge and 1.6% and 2.3% after the exposure to oxidative stress for RM6607R and RM6607W, respectively (Fig. 3).
Fig 3.
Distinct stress fitness between E. coli O157:H7 group III curli variants. The percentages of curli variant cells that survived following various stress challenge are shown. The number of CFU following each stress challenge was compared with the corresponding CFU at the beginning of the stress challenge to calculate the percent survival. Significant differences in stress fitness (percent survival) between the curli variants RM6607R and RM6607W are indicated (**, P < 0.001 by t test).
Rescue of curli production, HdeA production, and stress fitness in RM6607R by a functional rcsB gene.
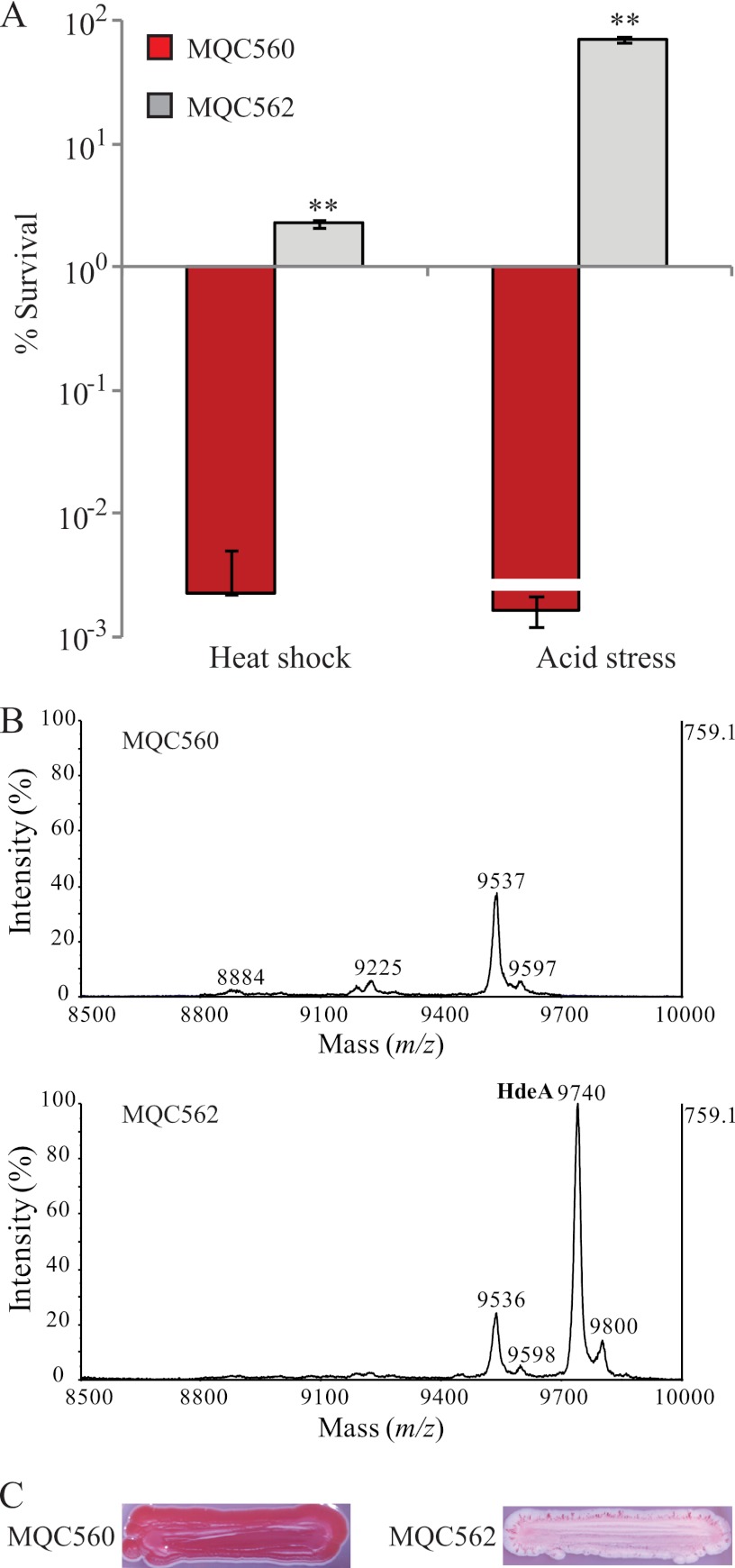
If the natural rcsB mutation were the true cause of distinct stress fitness differences between variants RM6607R and RM6607W, supplying RM6607R with a functional rcsB gene would restore its resistance to heat shock and acid stress. To test this hypothesis, we cloned the rcsB of variant RM6607W and moved it into the variant RM6607R, resulting in strain MQC562 (Table 1). In parallel, we transformed the cloning vector pBBR1MCS-5 into the variant RM6607R, resulting in the control strain MQC560. We then compared the survival of MQC562 with that of strain MQC560 following stress challenges. As expected, the percentage of MQC562 cells surviving (2.3%) after heat shock was over 1,000-fold higher than that of the control strain MQC560 (0.002%). Similarly, the acid survival of complemented strain MQC562 was 69.3%, which was comparable to that of the variant RM6607W; however, the acid survival of the control strain MQC560 was below the detection limit (less than 0.002%, on average) (Fig. 4A). Consistently, expression of rcsB in variant RM6607R restored the production of acid chaperone HdeA, as indicated by the presence of the characteristic HdeA peak in strain MQC562 by MS analysis, but not in the control strain MQC560 (Fig. 4B). Furthermore, expression of rcsB in RM6607R blocked curli production drastically, as evidenced by loss of the red phenotype for strain MQC562 when it was grown on the CRI plates. In contrast, the control strain MQC560 was red on CRI plates, similar to RM6607R (Fig. 4C).
Fig 4.
Complementation of RM6607R with a functional rcsB gene. (A) Rescue of resistance to heat shock and acid challenge in RM6607R. The percentages of curli variant cells that survived heat shock or acid challenge are shown. The number of CFU following each stress challenge was compared with the corresponding CFU at the beginning of the stress challenge to calculate the percent survival. Red columns represent strain MQC560 (RM6607R transformed with the cloning vector pBBR1MCS-5), whereas gray columns represent strain MQC562 (carrying a functional rcsB gene). The line on the red bar indicates that survival of MQC560 was below the value presented here. Differences in stress fitness between the control strain MQC560 and the complemented strain MQC562 are indicated (**, P < 0.001, t test). (B) Restoring HdeA production in RM6607R. The HdeA biomarker (m/z at 9,740) was detected and identified in MQC562 but not in MQC560. (C) Inhibition of curli production in RM6607R expressing the rcsB gene of RM6607W. The colony of control strain MQC560 appeared red on CRI plates, whereas the colony of the complemented strain MQC562 appeared white with red sections at the edges.
Transcriptomic signatures of RM6607 curli variants.
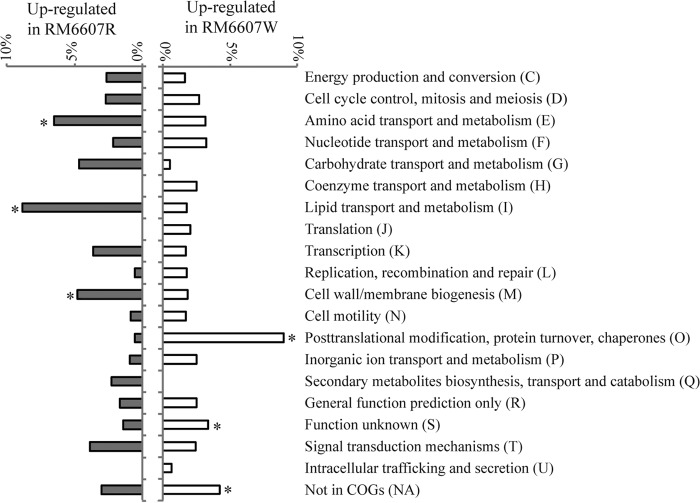
To gain insight into the phenotypic differentiation between natural curli variants of E. coli O157:H7, we first compared the global gene expression profiles between the variants RM6607R and RM6607W. A total of 331 genes showed differential expression between the two curli variants, of which 166 genes were upregulated in RM6607R (ratio of normalized hybridization intensities of RM6607R to RM6607W, >2.0), whereas 165 genes were upregulated in RM6607W (ratio of normalized hybridization intensities of RM6607W to RM6607R, >2.0). We then compared the functional distribution of these differentially expressed genes based on COG (Clusters of Orthologous Groups of proteins) categories after normalizing the upregulated gene over each COG category. The 166 genes with enhanced transcription in RM6607R were distributed across 16 COG categories (Fig. 5; see also Table S2 in the supplemental material). The top three were I (lipid transport and metabolism [8.8%]), E (amino acid transport and metabolism [6.5%]), and M (cell wall and membrane biogenesis [4.8%]), followed by G (carbohydrate transport and metabolism [4.7%]), T (signal transduction mechanisms [3.9%]), and K (transcription [3.6%]) (Fig. 5). Similarly, the 165 upregulated genes in RM6607W were distributed in 18 COG categories (Fig. 5; see also Table S3 in the supplemental material); however, the top functional category was O (posttranslational modification, protein turnover, and chaperones [9.0%]), followed by S (genes with unknown functions [3.3%]). A large number of genes encoding proteins with unknown conserved domains (no assigned COG category[NA]) were also highly expressed in RM6607W compared with RM6607R (NA, 4.2%).
Fig 5.
Transcriptomic analyses of E. coli O157:H7 group III curli variants. Genes that were expressed differentially between the curli variants RM6607R and RM6607W were grouped based on their expression ratios between the two variants. Genes with expression ratios for RM6607R versus RM6607W greater than 2.0 were considered upregulated in RM6607R, whereas genes with expression ratios for RM6607W versus RM6607R greater than 2.0 were considered upregulated in RM6607W. Genes expressed differentially were further categorized based on COG functional assignments. Bars indicate the percentage of genes in a given COG category. NA, no COG category assigned. The top three differentially expressed COGs for each variant are highlighted by an asterisk.
Upregulation of genes involved in curli biogenesis, biofilm formation, and virulence in RM6607R.
The difference in curli production between variants RM6607R and RM6608W was consistent with marked differences in transcriptional levels of curli genes (Table 2). The transcription levels of csgA (encoding the curli subunit protein) and csgB (encoding a nucleator protein) in RM6607R were 93.4- and 97.5-fold above those in RM6607W, respectively. csgC, the third gene in the csgBAC operon, encodes a protein suggested to have a role in redox activity within curli biogensis (62). The expression of csgC in RM6607R was about 8.0-fold above that in RM6607W. Transcription of csgD, csgE, csgF, and csgG in RM6607R were 13.4-, 11.2-, 15.8-, and 7.9-fold above that in RM6607W, respectively. Furthermore, the expression of crl, encoding the small protein Crl, which acts as a transcriptional enhancer and thermal sensor for curli biogenesis, was also higher in RM6607R than RM6607W (Table 2).
Table 2.
Differentially expressed genes related to curli biogenesis and biofilm formation in E. coli O157:H7 curli variants
| Group and gene name | ID | Product | COGa | Fold upregulation in RM6607Rb |
|---|---|---|---|---|
| Curli genes | ||||
| crl | Z0301 | DNA–binding transcriptional regulator Crl | NA | 2.1 |
| csgG | Z1670 | Curli production assembly/transport component | COG1462 M | 7.9 |
| csgF | Z1671 | Curli assembly protein CsgF | NA | 15.8 |
| csgE | Z1672 | Curli assembly protein CsgE | NA | 11.2 |
| csgD | Z1673 | DNA–binding transcriptional regulator CsgD | COG2197TK | 13.4 |
| csgB | Z1675 | Curlin minor subunit, functions as nucleator in assembly of curli on cell surface | NA | 97.5 |
| csgA | Z1676 | Curlin major subunit, coiled surface structures | NA | 93.4 |
| csgC | Z1677 | DsbD–like protein (62) | NA | 8.1 |
| O-antigen genes | ||||
| wbdR | Z3192 | Acetyltransferase, related to O-antigen biosynthesis | COG0110R | 3.1 |
| manB | Z3194 | Phosphomannomutase | COG1109G | 2.0 |
| manC | Z3195 | Mannose-1-P guanosyltransferase | COG0836 M | 2.6 |
| wbdQ | Z3196 | GDP-mannose mannosylhydrolase | COG1051F | 2.1 |
| fcl | Z3197 | Fucose synthetase | COG0451MG | 2.3 |
| gmd | Z3198 | GDP-mannose dehydratase | COG1089 M | 2.2 |
| wbdP | Z3199 | Glycosyl transferase | COG0438 M | 2.3 |
| per | Z3200 | Perosamine synthetase | COG0399 M | 2.7 |
| wzx | Z3201 | O-antigen flippase Wzx | COG2244R | 3.1 |
| wzy | Z3203 | O-antigen polymerase | NA | 2.4 |
| Other genes | ||||
| adrA | Z0481 | Diguanylate cyclase AdrA, catalyzes conversion of 2 GTPs into c-di-GMP | COG2199T | 3.2 |
| flhC | Z2945 | Transcriptional activator FlhC | NA | 2.8 |
| flhD | Z2946 | Transcriptional activator FlhD | NA | 3.8 |
The COG category was assigned based on the annotation for E. coli O157:H7 strain EDL933. NA, no assigned category.
The ratio of normalized hybridization intensities of the gene in RM6607R to that in RM6607W. All genes listed here passed the significance test as detailed in Materials and Methods.
The O-antigen of lipopolysaccharide (LPS) is known as a virulence factor in bacterial pathogens. For example, downregulation of O-antigen in Yersinia enterocolitica strain O:3 has been shown to be required for its efficient internalization (66); Also, downregulation of highly immunogenic EcO157 LPS may minimize the interference of host antibody with colonization or other roles of LPS (38). A slight change in expression of O-antigen genes was observed between RM6607R and RM6607W, including wzy (O-antigen polymerase gene), wzx (O-antigen flippase gene), per (pserosamine synthetase gene), wbdP (glycosyl transferase gene), gmd (GDP-mannose dehydratase gene Z3198), fcl (fucose synthetase gene), wbdQ (GDP-mannose mannosyl hydrolase), manC (mannose-1-P guanosyltransferase gene), manB (phosphomannomutase), and wbdR (acetyltransferase gene) (Table 2). The only two O-antigen genes within the operon that did not pass the cutoff criterion of differentiation (fold change in differentiation of >2.0 and t test P of <0.05) were wbdN (Z3204) and wbdO (Z3202), and both encode a glycosyl transferase.
The adrA gene encodes a diguanylate cyclase that catalyzes the conversion of 2 GTPs into cyclic di-GMP, an important molecular messenger involved in cellulose production, biofilm formation, and bacterial virulence. Transcription of adrA in RM6607R was about 3.2-fold higher than in RM6607W (Table 2). Furthermore, the transcription level of the flhCD operon was slightly higher in RM6607R than in RM6607W (2.8-fold for flhC and 3.8-fold for flhD) (Table 2). flhCD encode the master regulators of flagellar biosynthesis and also regulates genes that contribute to biofilm formation and virulence of E. coli O157:H7 (59). Examination of the swimming motility of both curli variants further revealed that both variants were motile at 28°C; however, the average swimming zone of RM6607R (5.4 ± 0.2 cm) was significantly larger than RM6607W (4.7 ± 0.2 cm; t test, P < 0.003).
Upregulation of genes related to catabolism and transport in RM6607R.
A large number of genes involved in degradation of lipid (I), amino acids (E), and carbohydrates (G) were upregulated in RM6607R compared with RM6607W, suggesting a higher catabolic activity in C+ variant RM6607R. Thus, we examined genes in these functional groups in detail.
Enhanced fatty acid degradation in RM6607R was supported by increased expression of genes related to the fatty acids β-oxidation pathway, including fadL (4.4-fold increase), fadD (5.4-fold), fadE (5.7-fold), and fadAB (4.2- and 5.9-fold) (Table 3). The fadL gene encodes an outer membrane protein that acts as a receptor and transporter for long-chain fatty acids. The fadD gene encodes a fatty acid acyl-CoA synthetase that is activated by the FadL-dependent long-chain fatty acid transport system. FadD converts fatty acids to fatty acyl-CoA, which then enters the β-oxidation cycle: FadE converts acyl-CoA to enoyl-CoA, which is subsequently converted to 3-ketoacyl-CoA by FadB. FadA then cleaves 3-ketoacyl-CoA, generating the acetyl-CoA, which enters the tricarboxylic acid cycle. Additionally, the genes fadJ and fadI, which are involved in the anaerobic degradation of fatty acids via the β-oxidation pathway, were also upregulated in RM6607R (Table 3). FadJ is a multifunctional enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase/3-hydroxybutyryl-CoA epimerase that catalyzes the formation of hydroxyacyl-CoA, whereas FadI is a 3-ketoacyl-CoA thiolase. Similar to the FadA2B2 complex, FadJ and FadI form a heterotetramer in a similar fashion as FadA2B2, yielding acetyl-CoA at the last step of β-oxidation.
Table 3.
Differentially expressed genes that are related to catabolic activity in E. coli O157:H7 curli variants
| Function group and gene name | ID | Product | COGa | Fold upregulation in RM6607Rb |
|---|---|---|---|---|
| Fatty acid uptake or metabolism | ||||
| fadE | Z0278 | Putative acyl-CoA dehydrogenase | COG1960I | 5.7 |
| hdhA | Z2624 | NAD-dependent 7-α-hydroxysteroid dehydrogenase | COG1028IQR | 2.0 |
| fadD | Z2848 | Acyl-CoA synthetase, long-chain fatty acid–CoA ligase | COG0318IQ | 5.4 |
| fadJ | Z3604 | Multifunctional fatty acid oxidation complex subunit α | COG1250I | 3.6 |
| fadI | Z3605 | 3-Ketoacyl–CoA thiolase | COG0183I | 3.1 |
| fadL | Z3608 | Long-chain fatty acid outer membrane transporter | COG2067I | 4.4 |
| ucpA | Z3691 | Short-chain dehydrogenase | COG1028IQR | 4.3 |
| fadA | Z5366 | 3-Ketoacyl–CoA thiolase | COG0183I | 4.2 |
| fadB | Z5367 | Multifunctional fatty acid oxidation complex subunit α | COG1250I | 5.9 |
| acs | Z5668 | Acetyl-CoA synthetase | COG0365I | 5.0 |
| phoH | Z1522 | PhoB dependent, putative ATPase, involved in lipid metabolism and RNA modification | COG1702T | 3.7 |
| Amino acid uptake or metabolism | ||||
| gltL | Z0802 | ATP-binding protein of glutamate/aspartate transport system | COG1126E | 2.4 |
| gltK | Z0803 | Glutamate/aspartate transport system permease | COG0765E | 3.0 |
| gltJ | Z0804 | Glutamate/aspartate transport system permease | COG0765E | 2.5 |
| glnH | Z1033 | Glutamine ABC transporter periplasmic protein | COG0834ET | 2.3 |
| Z2223 | Z2223 | Hemin-binding lipoprotein | COG0747E | 3.8 |
| Z2224 | Z2224 | Transport system permease protein | COG0601EP | 2.4 |
| Z2276 | Z2276 | Transport system permease protein | COG1177E | 6.7 |
| Z2277 | Z2277 | Transport system permease protein | COG1176E | 3.8 |
| Z2278 | Z2278 | ATP-binding component of a transport system | COG3842E | 5.5 |
| Z2279 | Z2279 | Transport protein | COG0687E | 5.7 |
| fliY | Z3010 | Cystine transporter subunit | COG0834ET | 6.4 |
| hisP | Z3568 | Histidine/lysine/arginine/ornithine transporter subunit | COG4598E | 4.2 |
| hisM | Z3569 | Histidine transport, membrane protein M | COG4160E | 2.1 |
| hisQ | Z3570 | Histidine transport system permease protein | COG4215E | 2.7 |
| hisJ | Z3571 | Histidine-binding periplasmic protein of high-affinity histidine transport system | COG0834ET | 2.8 |
| argT | Z3572 | Lysine-, arginine-, and ornithine-binding periplasmic protein | COG0834ET | 3.9 |
| oppD | Z2022 | Oligopeptide ABC transporter ATP-binding protein | COG0444EP | 2.2 |
| dppA | Z4961 | Dipeptide transport protein | COG0747E | 2.4 |
| ylbB | Z0671 | Putative hydantoin utilization protein | COG0624E | 2.9 |
| cstA | Z0740 | Carbon starvation protein, cAMP-CRP dependent, involved in peptide utilization. | COG1966T | 3.0 |
| cstC | Z2780 | Acetylornithine δ-aminotransferase, participates in catabolism of amino acids | COG4992E | 4.9 |
| dadA | Z1952 | d-Amino acid dehydrogenase subunit, catalyzes oxidative deamination of d-amino acids | COG0665E | 2.1 |
| ydjS | Z2776 | Succinylglutamate desuccinylase, hydrolyzes N-succinyl-l-glutamate to succinate and l-glutamate, participates in arginine and proline metabolism | COG2988E | 4.5 |
| Z2777 | Z2777 | Succinylarginine dihydrolase, catalyzes hydrolysis of 2-N-succinylarginine into 2-N-succinylornithine, ammonia, and carbon dioxide in arginine degradation | COG3724E | 6.0 |
| Z2779 | Z2779 | Arginine succinyltransferase, participates in arginine and proline metabolism | COG3138E | 5.8 |
| Z3717 | Z3717 | Putative ethanolamine utilization protein EutP | COG4917E | 2.6 |
| gcvH | Z4241 | Glycine cleavage system protein H | COG0509E | 2.2 |
| gcvT | Z4242 | Glycine cleavage system aminomethyltransferase T | COG0404E | 2.2 |
| aspA | Z5744 | Aspartate ammonia lyase, participates in alanine and aspartate metabolism and nitrogen metabolism | COG1027E | 5.3 |
| Carbohydrate uptake or metabolism | ||||
| nagE | Z0826 | Phosphotransferase system N-acetylglucosamine-specific transporter subunit IIABC | COG1263G | 2.5 |
| Z2192 | Z2192 | ABC transporter ATP-binding protein | COG1129G | 2.5 |
| ycjV | Z2463 | ABC transporter ATP-binding protein | COG3839G | 2.1 |
| ugpE | Z4819 | Glycerol-3-phosphate transporter membrane protein, related to uptake of glycerol-3-phosphate | COG0395G | 2.4 |
| ugpA | Z4820 | Glycerol-3-phosphate transporter permease | COG1175G | 3.1 |
| ugpB | Z4822 | Glycerol-3-phosphate transporter periplasmic binding protein | COG1653G | 3.2 |
| xylF | Z4991 | d-Xylose transporter subunit XylF | COG4213G | 2.0 |
| rbsB | Z5252 | d-Ribose transporter subunit RbsB | COG1879G | 2.3 |
| Z5691 | Z5691 | Ribose ABC transporter ATP-binding protein | COG1129G | 2.1 |
| Z5839 | Z5839 | ABC transporter ATP-binding protein | COG1129G | 2.2 |
| Z2189 | Z2189 | Autoinducer-2 ABC transporter, periplasmic autoinducer-2-binding protein LsrB, sugar ABC transporter | COG1879G | 2.7 |
| ytfQ | Z5838 | Periplasmic binding domain of ABC-type YtfQ-like transport systems | COG1879G | 2.7 |
| mgsA | Z1314 | Methylglyoxal synthase, participates in pyruvate metabolism | COG1803G | 2.4 |
| yneB | Z2188 | Aldolase, catalyzes a reversible aldol reaction | COG1830G | 2.6 |
| manB | Z3194 | Phosphomannomutase, participate in fructose and mannose metabolism | COG1109G | 2.0 |
| yeiQ | Z3431 | Putative oxidoreductase, may be related to metabolic pathways | COG0246G | 3.5 |
| yihM | Z5409 | Hypothetical protein contains conserved domain pfam01261 (AP_endonuc_2) and COG1082 [IolE], may be related to carbohydrate transport and metabolism | COG1082G | 2.6 |
The COG category was assigned based on the annotation for E. coli O157:H7 strain EDL933.
The ratio of normalized hybridization intensities of the gene in RM6607R to that in RM6607W. All genes listed passed the significance test as detailed in Materials and Methods.
Another transcriptomic characteristic of RM6607R was the enhanced expression of transporter genes that are related to the uptake of amino acids, peptides, or carbohydrates and, consequently, genes involved in metabolizing those compounds. For example, increased expression was observed for gltL (2.4-fold), gltK (3.0-fold), and gltJ (2.5-fold), encoding components of the glutamate/aspirate transport system; hisP (4.2-fold), hisM (2.1-fold), hisQ (2.7-fold), and hisJ (2.8-fold), encoding components of high-affinity histidine transport systems; glnH (2.3-fold), encoding a glutamine ABC transporter periplasmic protein; fliY (6.4-fold) and argT (3.9-fold), encoding a cystine transporter subunit protein and a lysine-, arginine-, ornithine-binding periplasmic protein, respectively (Table 3, amino acids related). Genes oppD and dppA are related to peptide transport; two gene clusters, Z2223-Z2224 and Z2276-Z2279, which are involved putatively in amino acids or iron transport, were also upregulated in RM6607R (Table 3). Similarly, increased expression was observed for genes encoding ABC transporter components involved in carbohydrate uptake, such as nagE (2.5-fold), Z2192 (2.5-fold), ycjV (2.1-fold), Z5691 (2.1-fold), Z5839 (2.2-fold), Z2189 (2.7-fold), and ytfQ (2.7-fold), as well as genes related to glycerol-3 phosphate uptake (ugpE, 2.4-fold; ugpA, 3.1-fold; ugpB, 3.2-fold). Correspondingly, a great number of genes related to the metabolism of amino acids or carbohydrates were upregulated in RM6607R. These included cstA (3.0-fold) and cstC (4.9-fold), both of which play a role in amino acid metabolism; aspA (5.3-fold), ydjS (4.5-fold), Z2777 (6.0-fold), and Z2779 (5.8-fold), which encode an aspartate ammonia lyase, a succinylglutamate desuccinylase, a succinylarginine dihydrolase, and an arginine succinyltransferase, respectively, and are involved in metabolism of alanine, aspartate, arginine, and proline, etc. (Table 3). For genes related to carbohydrate metabolism, higher expression was observed for mgsA (2.4-fold), which encodes a methylglyoxal synthase that participates in pyruvate metabolism, yneB (2.6-fold), which encodes an aldolase that generates glyceraldehyde-3-phosphate and dihydroxyacetone phosphate that are involved in glycolysis; manB (2.0-fold), which encodes a phosphomannomutase that is involved in mannose and fructose metabolism; and yeiQ (3.5-fold) and yihM (2.6-fold), which encode an oxidoreductase and a conserved hypothetical protein that are related putatively to carbohydrate metabolism (Table 3).
Upregulation of stress resistance genes in RM6607W.
Consistent with the higher acid resistance of RM6607W compared to RM6607R, transcription of several acid stress resistance genes in this variant were upregulated significantly compared with RM6607R (Table 4). The largest gene expression differences were observed for gadA (100.6-fold) and gadC (99.2-fold), which encode the key components of the glutamate-dependent acid resistance system AR2; the hdeBA operon (100.8- and 83.2-fold), which encodes acid chaperone proteins that are important for cell survival at pH < 3; hdeD (70.6-fold) and yhiD (5.3-fold), which encode proteins that are important for acid resistance of cells under high-density conditions; and yhiM (101.1-fold), which encodes an inner membrane protein that is required for cell survival at pH 2.5.
Table 4.
Differentially expressed stress resistance genes in E. coli O157:H7 curli variants
| Group and gene name | ID | Product | COGa | Fold upregulation in RM6607Wb |
|---|---|---|---|---|
| Acid resistance genes | ||||
| gadC | Z2216 | Acid sensitivity protein | COG0531E | 99.2 |
| yhiM | Z4890 | Hypothetical protein related to acid resistance | NA | 101.1 |
| yhiD | Z4920 | Putative Mg2+ transport ATPase | COG1285S | 5.3 |
| hdeB | Z4921 | Acid resistance protein | NA | 100.8 |
| hdeA | Z4922 | Acid resistance protein | NA | 83.2 |
| hdeD | Z4923 | Acid resistance membrane protein | COG3247S | 70.6 |
| gadA | Z4930 | Glutamate decarboxylase isozyme | COG0076E | 100.6 |
| Heat shock resistance genes | ||||
| dnaK | Z0014 | Chaperone Hsp70, autoregulated heat shock protein | COG0443O | 3.0 |
| dnaJ | Z0015 | Chaperone with DnaK, heat shock protein | COG0484O | 3.0 |
| htpG | Z0590 | Chaperone Hsp90, heat shock protein C 62.5 | COG0326O | 2.7 |
| cbpA | Z1418 | Curved DNA-binding protein, functions closely related to DnaJ | COG0484O | 2.8 |
| grpE | Z3907 | Heat shock protein GrpE | COG0576O | 2.0 |
| groES | Z5747 | Cochaperonin GroES | COG0234O | 3.7 |
| groEL | Z5748 | Chaperonin GroEL | COG0459O | 3.5 |
| ybbN | Z0645 | Trx-like protein, functions as molecular chaperone | COG3118O | 2.4 |
| yrfH | Z4754 | Ribosome-associated heat shock protein Hsp15 | COG1188J | 2.3 |
| lon | Z0545 | DNA-binding, ATP-dependent protease La; heat shock K protein | COG0466O | 2.2 |
| clpB | Z3886 | Protein disaggregation chaperone | COG0542O | 3.5 |
| hslU | Z5478 | ATP-dependent protease ATP-binding subunit | COG1220O | 2.2 |
| hslV | Z5479 | ATP-dependent protease peptidase subunit | COG5405O | 2.6 |
| Other stress resistance genes | ||||
| yecG | Z2948 | Universal stress protein UspC | COG0589T | 2.4 |
| treF | Z4932 | Trehalase | COG1626G | 2.6 |
| katE | Z2761 | Catalase, hydroperoxidase HPII(III) | COG0753P | 2.3 |
| yhjA | Z4931 | Putative cytochrome c peroxidase | COG1858P | 3.2 |
| topA | Z2536 | DNA topoisomerase type I, ω protein | COG0550L | 2.4 |
| mutS | Z4043 | DNA mismatch repair protein | COG0249L | 2.1 |
| exo | Z4115 | Exonuclease IX | COG0258L | 2.6 |
| mutM | Z5059 | Formamidopyrimidine-DNA glycosylase | COG0266L | 3.6 |
| sms | Z5990 | DNA repair protein RadA | COG1066O | 2.5 |
| spy | Z2775 | Periplasmic protein induced by stress response via Cpx and BaeSR system; similar to CpxP | COG3678UNTP | 2.8 |
| yiiO | Z5458 | Periplasmic repressor CpxP related to envelope stress | COG3678UNTP | 2.1 |
| ykfE | Z0277 | Defense, inactivate vertebrate C-type lysozyme | NA | 3.2 |
The COG category was assigned based on the annotation for E. coli O157:H7 strain EDL933. NA, no category assigned.
The ratio of normalized hybridization intensities of the gene in RM6607W to that in RM6607R. All genes listed here passed the significance test as detailed in Materials and Methods.
Consistent with our previous observation that RM6607W survived better than RM6607R following heat shock (Fig. 3), the transcriptional levels of over 10 heat shock genes were higher in RM6607W than in RM6607R (Table 4), including genes encoding the molecular chaperones DnaK (3.0-fold), DnaJ (3.0-fold), HtpG (2.7-fold), CbpA (2.8-fold), GrpE (2.0-fold), GroES (3.7-fold), and GroEL (3.5-fold). Additionally the ybbN gene, which encodes a thioredoxin-like protein that has been shown to enhance the DnaK-DnaJ-GrpE chaperone system but to repress GroESL chaperonin function and ATPase activity in E. coli (40), also was upregulated in RM6607W. yrfH encodes the heat shock protein Hsp15, which plays a role in recycling free 50S subunit to rebuild translation machinery. Transcription levels of ybbN and yrfH in RM6607W were 2.4- and 2.3-fold higher than in RM6607R, respectively (Table 4). In addition to molecular chaperone genes, expression levels of several genes encoding proteases for degradation of denatured proteins, such as lon, clpB, hslU, and hslV, were also significantly higher in RM6607W than in RM6607R (Table 4, heat shock genes).
Other stress-related genes with significantly higher transcription in RM6607W than in RM6607R included yecG, which encodes a general stress protein that is produced under conditions of nutrient deprivation, osmotic shock, and oxidative stress; treF, which encodes a cytoplasmic trehalase that is produced under osmotic stress; oxidative stress genes katE and yhiA, which encode a catalase that converts hydrogen peroxide to water and a cytochrome c peroxidase, a bacterial heme-containing protein that is involved in the peroxide stress response through its ability to convert hydrogen peroxide to water, respectively. Additionally, several genes related to DNA repair and recombination (topA, mutS, mutM, exo, and sms), cell envelope stress (spy and yiiO), or bacterial defense (ykfE) displayed greater expression in RM6607W than RM6607R (Table 4). It is noteworthy that many of these stress-related proteins, especially heat shock proteins, are considered general stress response proteins, because they provide cell protection under various stress conditions.
Core- and strain-specific differentially expressed genes between the curli variants of the 1993 hamburger-associated outbreak strains.
Transcriptomic analyses of differentially expressed genes between the curli variants of strain RM6608, a meat isolate linked to the 1993 U.S. hamburger-associated outbreak, revealed 168 genes that were upregulated in RM6608R and 262 genes that were upregulated in RM6608W. Compared with strain RM6607, there were 76 and 82 common differentially expressed genes between the two C+ variants (RM6607R and RM6608R) and the two C− variants (RM6607W and RM6608W), respectively (Fig. 6A). Similar to RM6607R, genes related to curli production, biofilm formation, and bacterial virulence were also upregulated in RM6608R (see Table S4 in the supplemental material). Although induction of the flhCD operon was not observed in RM6608R, the swimming motility test revealed that, similar to RM6607R, the swimming zone of RM6608R (5.8 ± 0.3 cm) was significantly greater than that of RM6608W (4.9 ± 0.2 cm; t test, P < 0.002). Furthermore, a great number of genes related to lipid transport and metabolism (I), carbohydrate transport and metabolism (G), amino acid transport and metabolism (E), and energy production and conservation (C) displayed higher transcription levels in RM6608R than those in RM6608W, as we observed for RM6607R (Fig. 6B). Similarly to RM6607W, a large number of upregulated genes in RM6608W were related to stress fitness and prophage induction (Fig. 6C; see also Table S5). Among the stress-related genes, the number of upregulated genes conferring resistance to acid stress and heat shock were the two ranked highest, similar to what was observed for RM6607W (Fig. 6C).
Fig 6.
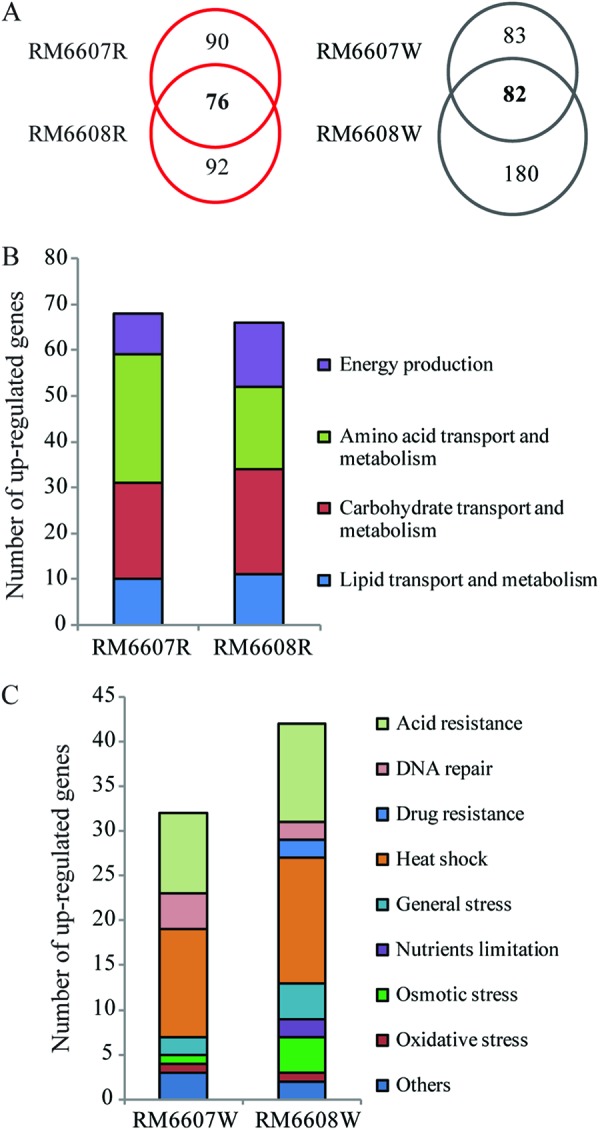
Comparative transcriptomic analyses of E. coli O157:H7 group III curli variants. (A) Number of common and strain-specific differentially expressed genes between the two C+ variants and the two C− variants. (B) Distribution of genes related to energy production and nutrient uptake and metabolism in two C+ variants, RM6607R and RM6608R. (C) Distribution of genes related to stress resistance in two C− variants, RM6607W and RM6608W.
The transcriptomics of group III curli variants also exhibited strain-specific characteristics. For example, unlike in RM6607R, more genes from COG functional categories of cell motility (N), transcription (K), signal transduction (T), and defense (V) were upregulated in C+ variant RM6608R (see Table S4 in the supplemental material). Unlike RM6607W, a large number of genes related to translation (J) and nucleotide transport and metabolism (F) were upregulated in RM6608W compared with its corresponding C+ variant (RM6608R) (see Table S5 in the supplemental material). However, it remains unclear how these strain-specific transcriptional differences impact the physiology of each curli variant (C+ or C−).
DISCUSSION
It has been well documented that intrapopulation diversity, introduced by either genetic modification or phenotypic variations, is an important mechanism of bacterial pathogenesis and niche adaptation (1, 5). In a heterogeneous population, subgroups of bacteria may express certain phenotypes that confer survival fitness in a particular niche, and such phenotypes have been demonstrated for synthesis of cell surface proteins, LPS, capsule, and biogenesis of cell appendages such as pili or flagella (71). We reported previously that intrastrain variations in curli production are widespread in E. coli O157:H7 (7). Additionally, C+ variants displayed better growth under low-nutrient conditions and formed thicker biofilms on glass surfaces under certain growth conditions, but they were significantly more sensitive to acid challenge than their corresponding C− variants (e.g., 100- to 10,000-fold). Unlike the mechanism revealed in Salmonella, or in E. coli O157:H7 strain EDL933, showing that increased curli production was due to mutations in the csgD promoter (54, 64), or in E. coli strain K-12, in which increased curli production resulted from mutations in the ompR gene (68), all 24 E. coli O157:H7 variants examined in our previous study had identical DNA sequences in both curli operons (both genes and the intergenic regions, including the csgD promoter) and the ompR gene. These observations led us to investigate alternative mechanisms for generating inter- and intrastrain variations in curli production.
The group III curli variants (isolated from the 1993 U.S. hamburger-associated outbreak strains) were the focus of this study. The C+ variants of group III were characterized by significantly higher curli production than with group I C+ variants (isolated from the 2006 U.S. spinach-associated outbreak strains) (7). Consistent with our previous phenotypic study, transcriptomic analyses revealed a broad divergence in various cellular processes between the E. coli O157:H7 curli variants, represented by over 300 differentially expressed genes from 20 COG functional categories. The higher growth rate of C+ variants in diluted peptone water (0.01%) was supported by higher expression of a large number of genes related to catabolism and transport systems in these variants; similarly, the greater curli production and biofilm formation by C+ variants were supported by marked increases in transcription of curli biogenesis genes (csgBAC and csgDEFG) and the gene encoding AdrA, a diguanylate cyclase that produces c-di-GMP, an allosteric activator of cellulose synthase. Indeed, enhanced cellulose production in both C+ variants RM6607R and RM6608R was observed on calcofluor plates compared with RM6607W and RM6608W, respectively (data not shown). The group III C− variants are distinguished by their higher resistance to both acid challenge (pH 2.5 for 2 h) and heat shock (55°C for 15 min) compared to their corresponding C+ variants. This stress fitness was supported by the large increase in expression of genes encoding components of AR2 systems and acid resistance proteins and chaperones, as well as several genes encoding molecular chaperones and proteases. Additionally, slight increases in expression of several genes related to DNA damage and repair was observed in C− variants, which suggests additional stress fitness in this group of C− variants. Although increased expression was observed in group III C− variants for several genes related to cell resistance to osmotic challenge, or oxidative stress, there were no marked differences in cell survival after exposure to either NaCl-induced osmotic pressure or H2O2-induced oxidative damage between the C+ and C− variants.
The transcriptomic signature of curli variants derived from strain RM6607 appeared to be conserved in curli variants of strain RM6608 regardless of the fact that more than half of the differentially expressed genes were strain specific. The core transcriptomic features between the two pairs of group III curli variants supported the common phenotypes segregating C+ subpopulation from the C− subpopulation. This set of differentially expressed genes is due most likely to the inactivation of the RcsB regulator in C+ variants of both RM6607 and RM6608. The roles of RcsB in curli production, acid resistance, cell motility, and biosynthesis of colanic acids and type I pili, etc., have been reported previously (20, 30, 35, 56, 67). The results of our study imply that RcsB may regulate heat shock resistance and cell catabolic activity, either directly or indirectly. The RcsCDB regulon has been characterized in E. coli strain K-12 (25). Our data suggest that RcsB also regulates O island genes (E. coli O157:H7-specific genes). For example, there were 9 and 22 common upregulated O island genes in both C+ (see Table S2 in the supplemental material) or both C− variants (see Table S3), respectively. Among the 9 genes that are likely repressed by RcsB, three (Z0463, Z1531, and Z5684) encode a putative transcriptional regulator, one (Z0608) encodes a putative outer membrane export protein, and two (Z3392 and Z3392) encode a protein belonging to the COG category Q (i.e., secondary metabolites for biosynthesis, transport, and catabolism; see Table S2); among the 22 genes that are likely activated by RcsB, 21 are prophage genes (see Table S3). The large number of strain-specific genes expressed differentially also suggests that each pair of curli variants has independently inherited strain-specific characteristics (genetic mutations or epigenetic modifications), which would lead to further physiological divergence among variants of E. coli O157:H7, even though those variants are highly genetically related (identical MLVA types). For example, induction levels for genes related to O-antigen biosynthesis were observed only in RM6607R. It is unclear if this differential expression would lead to a physiologically significant change of O-antigen between RM6607R and RM6607W. Further comparison of genome sequences between the variants will provide insights into the molecular basis of observed strain-specific gene expression profiles.
Genetic modification is known to be an important mechanism that drives population diversification. Recombination between direct repeats often results in deletion of the DNA segment between the repeats, thus generating genetic variants within the population. Phenotypic variants resulting from such deletions have been associated with bacterial virulence, production of LPS or capsule, cell motility, and nutrient uptake in diverse groups of bacteria, including the human pathogens P. aeruginosa, Y. pestis, V. cholerae, H. influenzae, and E. coli (6, 18, 29, 36, 47, 52, 58). Inactivation of RcsB in RM6607R and RM6608R results probably from recombination between the 7-bp direct repeats that border the deleted fragment in each variant. Recombination between short direct repeats has been documented in E. coli via a recA-independent mechanism, where the recombination between the 9-bp or the 13-bp direct repeat led to deletion of Tn5 transposon DNA or of cloned DNA in the pUC18 plasmid, respectively (12, 60). In both C+ variants, the rcsB sequence appeared to be a result of precise excision between the direct repeat TCTCGCC in RM6607R and the direct repeat CGGCGAT in RM6608R (Fig. 1). A search for additional direct repeats within the rcsB gene (651 bp) revealed 12 pairs of direct repeats (7 to 9 bp) that had the perfect match sequence (see Table S6 in the supplemental material); however, further studies are required to determine whether other direct repeats serve as recombination sites for generating rcsB deletion mutations. Examination of the rcsB gene sequence in more C+ variant colonies (n > 20) isolated from strains RM6607 and RM6608 revealed identical deletions in all C+ variants derived from the same strain (data not shown). Supply of a functional rcsB gene in a C+ variant restored its resistance to acid stress and heat shock but blocked curli production, confirming that the observed phenotypic divergence between the group III curli variants of E. coli O157:H7 is indeed due to the loss of RcsB function in the C+ variant.
Our study revealed the molecular basis of fitness segregation between E. coli O157:H7 C+ and C− variants of the 1993 U.S. hamburger-associated outbreak strains. Considering the broad regulatory functions of the response regulator RcsB, inactivation of RcsB would lead to a substantial shift in the global gene expression profile and, consequently, the distinct survival fitness and virulence potential between the variants. This study provides an example of how genomic instability of E. coli O157:H7 promotes intrapopulation diversity, generating population variants carrying distinct phenotypes that might be selected under certain ecological niches, such as animal gastrointestinal tracts, natural environments, food, and infected humans. It remains unclear whether C+ or C− variants are prevalent in an infected host. No significant change was observed in the acid survival of a C+ variant (RM6607R) for cells in the mixed culture with the C− variant (mix ratio of 1:1) compared with RM6607R in pure culture (data not shown), suggesting that C− variants might be dominant during passage through the host stomach. Although an acidic environment (pH 2.5) did not appear to promote the switch between the curli variants, C− variants yielded C+ variants under the anaerobic condition (7). Therefore, it is possible that after passing through the host stomach, the population dynamics of E. coli O157:H7 switch from a low to a high proportion of C+ variants once an anaerobic environment is established. Further studies on the environmental stimulus that promotes inactivation of RcsB would shed light on the role of natural curli variants in survival of E. coli O157:H7 and the relevance of this survival fitness in transmission of the pathogen from farm to food and ultimately to humans.
Supplementary Material
ACKNOWLEDGMENTS
We thank Anne Bates for her technical support and Omar Sultan for his assistance in mass spectrometric data collection. We thank Matthew Chapman for providing anti-CsgA antibody.
This work was supported in part by USDA-ARS CRIS projects 5325-42000-046-00D and 5325-42000-047-00D and by National Research Initiative competitive grants 2006-55212-16927 and 2007-35212-18239 from the USDA National Institute of Food and Agriculture.
Footnotes
Published ahead of print 24 August 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Avery SV. 2006. Microbial cell individuality and the underlying sources of heterogeneity. Nat. Rev. Microbiol. 4:577–587 [DOI] [PubMed] [Google Scholar]
- 2. Bian Z, Brauner A, Li Y, Normark S. 2000. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J. Infect. Dis. 181:602–612 [DOI] [PubMed] [Google Scholar]
- 3. Bougdour A, Lelong C, Geiselmann J. 2004. Crl, a low temperature-induced protein in Escherichia coli that binds directly to the stationary phase sigma subunit of RNA polymerase. J. Biol. Chem. 279:19540–19550 [DOI] [PubMed] [Google Scholar]
- 4. Brown PK, et al. 2001. MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium. Mol. Microbiol. 41:349–363 [DOI] [PubMed] [Google Scholar]
- 5. Brzuszkiewicz E, Gottschalk G, Ron E, Hacker J, Dobrindt U. 2009. Adaptation of pathogenic E. coli to various niches: genome flexibility is the key. Genome Dyn. 6:110–125 [DOI] [PubMed] [Google Scholar]
- 6. Buchrieser C, Prentice M, Carniel E. 1998. The 102-kilobase unstable region of Yersinia pestis comprises a high-pathogenicity island linked to a pigmentation segment which undergoes internal rearrangement. J. Bacteriol. 180:2321–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carter MQ, et al. 2011. Distinct acid resistance and survival fitness displayed by curli variants of enterohemorrhagic Escherichia coli O157:H7. Appl. Environ. Microbiol. 77:3685–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carter MQ, et al. 2012. Evolutionary silence of the acid chaperone protein HdeB in enterohemorrhagic Escherichia coli O157:H7. Appl. Environ. Microbiol. 78:1004–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Castanie-Cornet MP, Foster JW. 2001. Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147:709–715 [DOI] [PubMed] [Google Scholar]
- 10. Castanie-Cornet MP, Penfound TA, Smith D, Elliott JF, Foster JW. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chapman MR, et al. 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295:851–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Collins J, Volckaert G, Nevers P. 1982. Precise and nearly-precise excision of the symmetrical inverted repeats of Tn5; common features of recA-independent deletion events in Escherichia coli. Gene 19:139–146 [DOI] [PubMed] [Google Scholar]
- 13. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dorel C, Vidal O, Prigent-Combaret C, Vallet I, Lejeune P. 1999. Involvement of the Cpx signal transduction pathway of E. coli in biofilm formation. FEMS Microbiol. Lett. 178:169–175 [DOI] [PubMed] [Google Scholar]
- 15. Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103–118 [DOI] [PubMed] [Google Scholar]
- 16. Fagerquist CK, et al. 2010. Rapid identification of protein biomarkers of Escherichia coli O157:H7 by matrix-assisted laser desorption ionization-time-of-flight-time-of-flight mass spectrometry and top-down proteomics. Anal. Chem. 82:2717–2725 [DOI] [PubMed] [Google Scholar]
- 17. Fagerquist CK, et al. 2009. Web-based software for rapid top-down proteomic identification of protein biomarkers, with implications for bacterial identification. Appl. Environ. Microbiol. 75:4341–4353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fetherston JD, Schuetze P, Perry RD. 1992. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol. Microbiol. 6:2693–2704 [DOI] [PubMed] [Google Scholar]
- 19. Foster JW. 2004. Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2:898–907 [DOI] [PubMed] [Google Scholar]
- 20. Francez-Charlot A, et al. 2003. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 49:823–832 [DOI] [PubMed] [Google Scholar]
- 21. Gajiwala KS, Burley SK. 2000. HDEA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria. J. Mol. Biol. 295:605–612 [DOI] [PubMed] [Google Scholar]
- 22. Gerstel U, Park C, Romling U. 2003. Complex regulation of csgD promoter activity by global regulatory proteins. Mol. Microbiol. 49:639–654 [DOI] [PubMed] [Google Scholar]
- 23. Gophna U, et al. 2001. Curli fibers mediate internalization of Escherichia coli by eukaryotic cells. Infect. Immun. 69:2659–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gorden J, Small PL. 1993. Acid resistance in enteric bacteria. Infect. Immun. 61:364–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hagiwara D, et al. 2003. Genome-wide analyses revealing a signaling network of the RcsC-YojN-RcsB phosphorelay system in Escherichia coli. J. Bacteriol. 185:5735–5746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. 1995. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 18:661–670 [DOI] [PubMed] [Google Scholar]
- 27. Hammar M, Bian Z, Normark S. 1996. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 93:6562–6566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hammer ND, Schmidt JC, Chapman MR. 2007. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc. Natl. Acad. Sci. U. S. A. 104:12494–12499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hoiseth SK, Moxon ER, Silver RP. 1986. Genes involved in Haemophilus influenzae type b capsule expression are part of an 18-kilobase tandem duplication. Proc. Natl. Acad. Sci. U. S. A. 83:1106–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johnson MD, Burton NA, Gutierrez B, Painter K, Lund PA. 2011. RcsB is required for inducible acid resistance in Escherichia coli and acts at gadE-dependent and -independent promoters. J. Bacteriol. 193:3653–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kai-Larsen Y, et al. 2010. Uropathogenic Escherichia coli modulates immune responses and its curli fimbriae interact with the antimicrobial peptide LL-37. PLoS Pathog. 6:e1001010 doi:10.1371/journal.ppat.1001010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kikuchi T, Mizunoe Y, Takade A, Naito S, Yoshida S. 2005. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol. Immunol. 49:875–884 [DOI] [PubMed] [Google Scholar]
- 33. Kovach ME, et al. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 34. Krin E, Danchin A, Soutourina O. 2010. Decrypting the H-NS-dependent regulatory cascade of acid stress resistance in Escherichia coli. BMC Microbiol. 10:273 doi:10.1186/1471-2180-10-273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Krin E, Danchin A, Soutourina O. 2010. RcsB plays a central role in H-NS-dependent regulation of motility and acid stress resistance in Escherichia coli. Res. Microbiol. 161:363–371 [DOI] [PubMed] [Google Scholar]
- 36. Kroll JS, Hopkins I, Moxon ER. 1988. Capsule loss in H. influenzae type b occurs by recombination-mediated disruption of a gene essential for polysaccharide export. Cell 53:347–356 [DOI] [PubMed] [Google Scholar]
- 37. Kyle JL, Parker CT, Goudeau D, Brandl MT. 2010. Transcriptome analysis of Escherichia coli O157:H7 exposed to lysates of lettuce leaves. Appl. Environ. Microbiol. 76:1375–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li Y, Frey E, Mackenzie AM, Finlay BB. 2000. Human response to Escherichia coli O157:H7 infection: antibodies to secreted virulence factors. Infect. Immun. 68:5090–5095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lin J, et al. 1996. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 62:3094–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lin J, Wilson MA. 2011. Escherichia coli thioredoxin-like protein YbbN contains an atypical tetratricopeptide repeat motif and is a negative regulator of GroEL. J. Biol. Chem. 286:19459–19469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ma Z, et al. 2003. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol. Microbiol. 49:1309–1320 [DOI] [PubMed] [Google Scholar]
- 42. Ma Z, Richard H, Tucker DL, Conway T, Foster JW. 2002. Collaborative regulation of Escherichia coli glutamate-dependent acid resistance by two AraC-like regulators, GadX and GadW (YhiW). J. Bacteriol. 184:7001–7012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Majdalani N, Chen S, Murrow J, St John K, Gottesman S. 2001. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 39:1382–1394 [DOI] [PubMed] [Google Scholar]
- 44. Majdalani N, Heck M, Stout V, Gottesman S. 2005. Role of RcsF in signaling to the Rcs phosphorelay pathway in Escherichia coli. J. Bacteriol. 187:6770–6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Masuda N, Church GM. 2003. Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 48:699–712 [DOI] [PubMed] [Google Scholar]
- 46. Maurer JJ, Brown TP, Steffens WL, Thayer SG. 1998. The occurrence of ambient temperature-regulated adhesins, curli, and the temperature-sensitive hemagglutinin tsh among avian Escherichia coli. Avian Dis. 42:106–118 [PubMed] [Google Scholar]
- 47. Middendorf B, et al. 2004. Instability of pathogenicity islands in uropathogenic Escherichia coli 536. J. Bacteriol. 186:3086–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Olsen A, Arnqvist A, Hammar M, Sukupolvi S, Normark S. 1993. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol. Microbiol. 7:523–536 [DOI] [PubMed] [Google Scholar]
- 49. Olsen A, Jonsson A, Normark S. 1989. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338:652–655 [DOI] [PubMed] [Google Scholar]
- 50. Parker CT, et al. 2012. Distinct transcriptional profiles and phenotypes exhibited by Escherichia coli O157:H7 isolates related to the 2006 spinach-associated outbreak. Appl. Environ. Microbiol. 78:455–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Price SB, et al. 2000. Role of rpoS in acid resistance and fecal shedding of Escherichia coli O157:H7. Appl. Environ. Microbiol. 66:632–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Qiu X, Gurkar AU, Lory S. 2006. Interstrain transfer of the large pathogenicity island (PAPI-1) of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 103:19830–19835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Robinson LS, Ashman EM, Hultgren SJ, Chapman MR. 2006. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol. Microbiol. 59:870–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Romling U, Sierralta WD, Eriksson K, Normark S. 1998. Multicellular and aggregative behaviour of Salmonella Typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249–264 [DOI] [PubMed] [Google Scholar]
- 55. Sayed AK, Odom C, Foster JW. 2007. The Escherichia coli AraC-family regulators GadX and GadW activate gadE, the central activator of glutamate-dependent acid resistance. Microbiology 153:2584–2592 [DOI] [PubMed] [Google Scholar]
- 56. Schwan WR, Shibata S, Aizawa S, Wolfe AJ. 2007. The two-component response regulator RcsB regulates type 1 piliation in Escherichia coli. J. Bacteriol. 189:7159–7163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski JL. 1994. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J. Bacteriol. 176:1729–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Smirnova NI, Chekhovskaya GV, Davidova NI, Livanova LF, Yeroshenko GA. 1996. Virulence-associated characteristics and phage lysogenicity of two morphologically distinct colonies of Vibrio cholerae O139 serogroup. FEMS Microbiol. Lett. 136:175–180 [DOI] [PubMed] [Google Scholar]
- 59. Sule P, Horne SM, Logue CM, Pruss BM. 2011. Regulation of cell division, biofilm formation, and virulence by FlhC in Escherichia coli O157:H7 grown on meat. Appl. Environ. Microbiol. 77:3653–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sumegi A, Birko Z, Szeszak F, Vitalis S, Biro S. 1997. A short GC-rich sequence involved in deletion formation of cloned DNA in E. coli. Acta Biol. Hung. 48:275–279 [PubMed] [Google Scholar]
- 61. Szabo E, et al. 2005. Curli expression of enterotoxigenic Escherichia coli. Folia Microbiol. (Praha) 50:40–46 [DOI] [PubMed] [Google Scholar]
- 62. Taylor JD, et al. 2011. Atomic resolution insights into curli fiber biogenesis. Structure 19:1307–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tukel C, et al. 2005. CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Mol. Microbiol. 58:289–304 [DOI] [PubMed] [Google Scholar]
- 64. Uhlich GA, Keen JE, Elder RO. 2001. Mutations in the csgD promoter associated with variations in curli expression in certain strains of Escherichia coli O157:H7. Appl. Environ. Microbiol. 67:2367–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Uhlich GA, Keen JE, Elder RO. 2002. Variations in the csgD promoter of Escherichia coli O157:H7 associated with increased virulence in mice and increased invasion of HEp-2 cells. Infect. Immun. 70:395–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Uliczka F, et al. 2011. Unique cell adhesion and invasion properties of Yersinia enterocolitica O:3, the most frequent cause of human yersiniosis. PLoS Pathog. 7:e1002117 doi:10.1371/journal.ppat.1002117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vianney A, et al. 2005. Escherichia coli tol and rcs genes participate in the complex network affecting curli synthesis. Microbiology 151:2487–2497 [DOI] [PubMed] [Google Scholar]
- 68. Vidal O, et al. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 180:2442–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang X, Chapman MR. 2008. Sequence determinants of bacterial amyloid formation. J. Mol. Biol. 380:570–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Westerman RB, He Y, Keen JE, Littledike ET, Kwang J. 1997. Production and characterization of monoclonal antibodies specific for the lipopolysaccharide of Escherichia coli O157. J. Clin. Microbiol. 35:679–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wisniewski-Dye F, Vial L. 2008. Phase and antigenic variation mediated by genome modifications. Antonie van Leeuwenhoek 94:493–515 [DOI] [PubMed] [Google Scholar]
- 72. Wu YE, Hong W, Liu C, Zhang L, Chang Z. 2008. Conserved amphiphilic feature is essential for periplasmic chaperone HdeA to support acid resistance in enteric bacteria. Biochem. J. 412:389–397 [DOI] [PubMed] [Google Scholar]
- 73. Zhao B, Houry WA. 2010. Acid stress response in enteropathogenic gammaproteobacteria: an aptitude for survival. Biochem. Cell Biol. 88:301–314 [DOI] [PubMed] [Google Scholar]
- 74. Zogaj X, Bokranz W, Nimtz M, Romling U. 2003. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 71:4151–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.