Abstract
The prevalence of Vibrio vulnificus on the external surfaces of fish from the northern Gulf of Mexico was determined in this study. A collection of 242 fish comprising 28 species was analyzed during the course of 12 sampling trips over a 16-month period. The prevalence of V. vulnificus was 37% but increased up to 69% in summer. A positive correlation was found between the percentages of V. vulnificus-positive fish and water temperatures, while salinity and V. vulnificus-positive fish prevalence were inversely correlated. A general lineal model (percent V. vulnificus-positive fish = 0.5930 − 0.02818 × salinity + 0.01406 × water temperature) was applied to best fit the data. Analysis of the population structure was carried out using 244 isolates recovered from fish. Ascription to 16S rRNA gene types indicated that 157 isolates were type A (62%), 72 (29%) were type B, and 22 (9%) were type AB. The percentage of type B isolates, considered to have greater virulence potential, was higher than that previously reported in oyster samples from the northern Gulf of Mexico. Amplified fragment length polymorphism (AFLP) was used to resolve the genetic diversity within the species. One hundred twenty-one unique AFLP profiles were found among all analyzed isolates, resulting in a calculated Simpson's index of diversity of 0.991. AFLP profiles were not grouped on the basis of collection date, fish species, temperature, or salinity, but isolates were clustered into two main groups that correlated precisely with 16S rRNA gene type. The population of V. vulnificus associated with fishes from the northern Gulf of Mexico is heterogeneous and includes strains of great virulence potential.
INTRODUCTION
Vibrio vulnificus is a Gram-negative bacterium commonly found in estuarine and coastal habitats throughout the northern Gulf of Mexico (18, 28). This species is an opportunistic human pathogen that can cause primary septicemia, wound infection, and gastroenteritis in susceptible individuals (46). Gastroenteritis is the more benign but less common clinical syndrome associated with V. vulnificus infections that typically courses as a self-limited illness. Conversely, primary septicemia is the most common and severe manifestation of V. vulnificus-associated illnesses, having a mortality rate of more than 50% (8, 21, 46). Both gastroenteritis and primary septicemia are associated with the consumption of raw shellfish harboring the pathogen, particularly the eastern oyster (Crassostrea virginica) (46). In addition, V. vulnificus can produce severe skin and soft tissue infections in patients with preexisting wounds who come in contact with the bacterium via seawater or by handling seafood or who sustain an injury while exposed to those sources (22).
Ecological studies have shown a seasonal pattern wherein the number of V. vulnificus bacteria in oysters, seawater, and sediments increases with warmer temperatures (7, 28, 34, 37). Predictably, the incidence of wound infections has been found to be positively correlated with warm temperatures (16, 33). Because nearly all septicemia cases are associated with the consumption of raw eastern oysters (46), it has been possible to establish a risk assessment model for this pathogen in the eastern oyster (15, 49). However, on the basis of epidemiological data, a direct, robustly documented linkage between the specific source of the pathogen and subsequent manifestation of V. vulnificus-associated wound infection has not been well documented in all cases (10, 21, 35, 38).
According to the Cholera and Other Vibrio Illnesses Surveillance (COVIS), a Vibrio sp. wound infection is recorded as such when the pathogen is cultured from a wound and the patient is reported to have sustained a wound or had a preexisting one while exposed to marine or estuarine water or during physical contact with marine wildlife in the 7 days prior to illness onset (11). In most clinical cases, patients reported that they had handled seafood prior to the onset of the disease, but the data do not specify the kind of handled seafood (e.g., shellfish, crustaceans, or fish) (16, 21, 35). However, the only two documented outbreaks of V. vulnificus involving wound infections were attributed to handling of farm-raised fish in Israel (5) or from injuries sustained prior to or during a fishing contest in Texas (33), resulting in 62 and 5 cases, respectively.
Recreational fishing is a main service industry for the United States, generating large revenues for local coastal communities (1). The northern Gulf of Mexico is a top destination for recreational anglers, where an estimated >2.8 million anglers participate in more than 7 million fishing trips annually (29). Although many people recreate in marine and estuarine waters and handle fish there, little information is available regarding the prevalence and distribution of V. vulnificus in Gulf of Mexico fishes. DePaola et al. (17) enumerated the density of V. vulnificus in the intestine of estuarine fishes of Mississippi and Alabama, reporting higher levels (105 of 108 CFU/g) in fish intestine than in the surrounding seawater and sediments, suggesting that fish may be reservoirs for V. vulnificus. However, it is noteworthy that those authors did not analyze the external surfaces of those fish. Anglers may sustain puncture wounds, lacerations, or bites from live fish, or they may be incidentally punctured or cut by dead fish during routine recreational angling activities (e.g., dehooking and filleting of fish) (45). During these handling events, anglers may be exposed to bacteria present in the skin and mucus of their catch.
The purpose of this study was (i) to document the prevalence of V. vulnificus on the body surface of a group of estuarine fishes commonly caught by recreational fishermen in the Gulf of Mexico and (ii) to characterize the population structure of V. vulnificus in those fishes.
MATERIALS AND METHODS
Sample collection.
Sampling began during November 2009 and continued through March 2011 at regular intervals except during December 2009 and January and February 2010. Sampling sites were selected on the basis of accessibility and considered to be representative of public fishing piers in Alabama and Mississippi. Locations included Dauphin Island and Gulf Shores in Alabama and Ocean Springs in Mississippi (Fig. 1). Table 1 summarizes the collection dates and locations and the numbers of fish analyzed per collection event. Seawater surface temperature (at a 1-m depth) was measured in situ using a mercury-in-glass thermometer (SargentWelch). Salinities were measured with a handheld refractometer (model SR-6; Vital Sine). Fishing efforts lasted between 4 and 8 h. Fish were captured using standard baited hooks and standard 20-pound-test monofilament fishing line on standard spinning reels. Hooked fish were deliberately exhausted in ambient water before being raised from the water, secured, and suspended in air by the angler grasping the leader base or hook shaft and then touched only by a second worker donning sterile surgical gloves and equipped with flamed and ethanol-rinsed, heavy-gauge scissors. In coordination with raising the exhausted, immobilized fish from the water, the second worker approached and immediately excised a portion (∼1 cm2) of the dorsal fin and placed the excised tissue in a tube containing 10 ml of alkaline peptone water (APW). Hence, no sampled fish was placed on any surface or touched by a second person before each sample was collected by the worker wearing surgical gloves. Each sample was enriched overnight in APW at room temperature (approximately 25°C). All fish were identified as described by Carpenter (9), ordinal classification of fishes was as described by Nelson (30), and common names for fishes were as described by Eschmeyer (19).
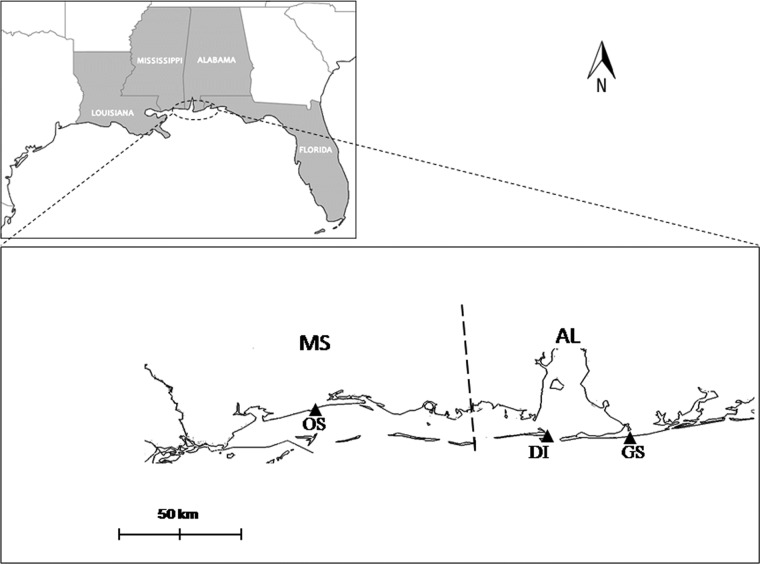
Fig 1.
North-central Gulf of Mexico showing collecting sites: Dauphin Island (DI), Gulf Shores (GS), and Ocean Springs (OS).
Table 1.
Temporal and spatial distributions of fishing efforts summarizing number of fish analyzed and number of fish positive for Vibrio vulnificus
| Date (mo-day-yr) | Sitea | No. of fish | No. of V. vulnificus-positive fish |
|---|---|---|---|
| 11-17-2009 | OS | 19 | 9 |
| 03-26-2010 | OS | 30 | 10 |
| 06-02-2010 | DI | 20 | 12 |
| 06-16-2010 | DI | 25 | 18 |
| 07-18-2010 | OS | 23 | 18 |
| 08-18-2010 | DI | 23 | 6 |
| 09-18-2010 | OS | 18 | 11 |
| 09-18-2010 | DI | 10 | 4 |
| 10-16-2010 | GS | 23 | 0 |
| 11-12-2010 | OS | 18 | 0 |
| 11-28-2010 | OS | 25 | 2 |
| 03-17-2011 | GS | 8 | 0 |
| Total | 242 | 90 |
OS, Ocean Springs, MS; DI, Dauphin Island, AL; GS, Gulf Shores, AL.
Bacteriological analysis.
Upon arrival to the laboratory, 100 μl of each of the APW cultures was plated onto modified cellobiose-polymyxin B-colistin (mCPC) (47) and thiosulfate citrate bile salts sucrose (TCBS; Becton, Dickinson and Co. [BD], Franklin Lakes, NJ) agar plates and incubated overnight at 30°C. Three colonies displaying the typical V. vulnificus morphology (27) were randomly selected from each selective medium and reisolated on marine agar (MA; BD). Putative isolates recovered from TCBS and mCPC agar were subjected to colony dot blot hybridization according to the protocol described by Wright et al. (50). Briefly, putative isolates were cultured in marine broth (MB; BD) overnight in a 96-well microtiter plate, and approximately 5 μl of each culture was transferred to mCPC using a multiple-channel replicator and allowed to grow overnight at 35°C. Colonies were lifted onto Whatman 541 filter papers, followed by hybridization using an alkaline phosphatase-conjugated oligonucleotide (5′-GAGCTGTCACGGCAGTTGGAACCA-3′; DNATechnology A/S, Risskov, Denmark) that recognizes a specific sequence in the V. vulnificus hemolysin gene. Positive isolates were stored at −80°C as glycerol stocks (MB supplemented with 20% glycerol) for further testing.
Ascription of V. vulnificus isolates to biotypes.
A total of 251 V. vulnificus isolates recovered from fish and 8 reference strains (Table 2) were included in the genetic analysis. DNA was extracted from all isolates using standard protocols (36). All V. vulnificus isolates were subjected to a multiplex PCR assay for biotype ascription as described by Sanjuán et al. (43). In short, PCR was performed in a 25-μl reaction volume containing 1× PCR buffer, 1.5 mM MgCl2, 200 μM each deoxynucleoside, 0.1 μM primers vvhA-F (5′-CGCCACCCACTTTCGGGCC-3′) and vvhA-R (5′-CCGCGGTACAGGTTGGCGC-3′), 0.2 μM primers Bt2-F (5′-AGAGATGGAAGAAACAGGCG-3′) and Bt2-R (5′-GGACAGATATAAGGGCAAATGG-3′), 1.5 U GoTaq DNA polymerase, 1 μl DNA template (20 ng), and distilled H2O (dH2O) up to 25 μl. Unless stated otherwise, all molecular reagents were purchased from Promega (Madison, WI). PCR was carried out on a Bio-Rad PTC-0200 DNA engine cycler (Bio-Rad, Valencia, CA) with the following cycling profile: an initial denaturation step of 94°C for 10 min; 35 cycles of 94°C for 30 s, 60°C for 45 s, and 72°C for 1 min; and a final extension step of 72°C for 10 min. The PCR products were analyzed by electrophoresis on a 2% agarose gel and visualized with UV light by staining with ethidium bromide. Ascription to biotype 1 or 3 or biotype 2 was based on amplicon(s) size (the method does not discriminate between biotypes 1 and 3).
Table 2.
Reference strains
| Strain in this study | Straina | Source | Origin | Date | 16S rRNA typed | vcge | Virulencef |
|---|---|---|---|---|---|---|---|
| R-1 | CDC 9060-96 | Clinicalb | TX | 1996 | B | vcgC | Virulent |
| R-2 | CDC 9070-96 | Clinicalc | TX | 1996 | B | vcgC | Virulent |
| R-3 | ATL-9824 | Clinicalc | TX | 1994 | B | vcgC | Virulent |
| R-4 | 98-640 DP-E9 | Oyster | LA | 1998 | A | vcgE | Virulent |
| R-5 | 99-625 DP-D8 | Oyster | TX | 1999 | AB | vcgE | Virulent |
| R-6 | 246-0058 | Clinical | FL | A | vcgE | Virulent | |
| R-7 | 99-609 DP-A4 | Oyster | OR | 1999 | A | vcgE | Virulent |
| R-8 | 99-537 DP-G7 | Oyster | MD | 1999 | A | vcgE | Virulent |
16S rRNA RFLP typing.
The 16S rrn genotype was determined by restriction fragment length polymorphism (RFLP) as described by Nilsson et al. (31). A 492-bp region of the 16S rRNA gene of V. vulnificus was amplified using primers UFUL (5′-GCCTAACACATGCAAGTCGA-3′) and URUL (5′-CGTATTACCGCGGCTGCTGG-3′). PCR was performed as described above, with the only difference being the use of an annealing temperature of 57°C. PCR products were verified by 2% agarose gel electrophoresis. Restriction endonuclease digestion of amplified product was performed in a 20-μl reaction volume including 10 μl of amplicon, 2 μl of 10× restriction buffer B (Promega), 0.2 μl of acetylated bovine serum albumin (10 μg/ml), 0.5 μl of AluI (10 U/μl), and sterile dH2O up to 20 μl. Digestion was carried at 37°C for 2 h, after which DNA fragments were separated by electrophoresis on a 4% agarose gel. 16S rrn types were ascribed on the basis of the profiles described by Nilsson et al. (31).
AFLP.
Amplified fragment length polymorphism (AFLP) reactions were carried out as described by Arias et al. (3). Briefly, 100 ng of RNase-treated genomic DNA was double digested with TaqI and HindIII. Following digestion, specific TaqI and HindIII adaptors were ligated to the restriction fragments and subsequently amplified by PCR using primers T000 (5′CGATGAGTCCTGACCGAA-3′) and H00A (5′-GAACTGCGTACCAGCTTA-3′), where the selective bases at the 3′ ends are underlined. The HindIII primer (H00A) was labeled with an IR700 fluorochrome from LI-COR (Lincoln, NE). PCR amplifications were performed with the following cycle profile: cycle 1, 60 s at 94°C, 30 s at 65°C, and 60 s at 72°C; cycles 2 to 12, 30 s at 94°C, 30 s at an annealing temperature 0.7°C lower than that used for each previous cycle, starting at 64.3°C, and 60 s at 72°C; cycles 13 to 24, 30 s at 94°C, 30 s at 56°C, and 60 s at 72°C. After completion of the cycling program, 5 μl of AFLP Blue stop solution (LI-COR) was added to the reaction mixtures. Prior to gel loading, the samples were heated for 5 min at 94°C and then rapidly cooled on ice to prevent reannealing. The PCR products were electrophoresed on an NEN global edition IR2 DNA analyzer (LI-COR) following the manufacturer's instructions.
Data analysis.
The BioNumerics (version 6.6) software suite (Applied Maths, Saint Martens-Latem, Belgium) was used for AFLP data analysis. Pairwise similarities were calculated using the Pearson correlation coefficient with a 0.5% optimization. Using the similarity matrix as an input, a dendrogram was constructed with the arithmetic averages (unweighted-pair group method using average linkages) algorithm. The jackknife group separation method (based on maximal similarities between isolates) was used to assess the fidelity of the clustering analysis. Only bands within the range of 100 to 530 bp and with at least 8% of the minimum profiling were considered in the analysis. Transversal clustering was performed on the basis of the swapped data matrix of profile similarities (fingerprint patterns, horizontal cluster) and characters (band classes, vertical cluster). In transversal clustering, the isolates were grouped by fingerprint patterns (similarities based on band position and intensity, Pearson correlation coefficient), while the characters were sorted by means of value of band classes (0 for absent and 1 for present, Jaccard coefficient). The diversity of the V. vulnificus isolates from each fish species was compared by generating rarefaction curves using the software Diversity (version 1.4; Hunt Mountain Software, Athens, GA).
The SAS software 9.2 version (SAS Institute, Cary, NC) was applied to analyze the relationship between environmental factors and the percentages of fish harboring Vibrio vulnificus. The percentage, namely, is the frequency of fish fin clips that yielded confirmed V. vulnificus isolates and was calculated for each sampling event. The Pearson correlation coefficient was used to estimate the relationship between percentages of V. vulnificus-positive fish and environmental factors (water temperature and salinity). A general linear regression analysis was used to quantify the trends observed between V. vulnificus-positive fish and variation in salinity and water temperature.
RESULTS
Prevalence of V. vulnificus in fish.
A collection of 242 fish was randomly sampled during 12 sampling trips. Overall, 90 (37%) individual fish (each represented by an excised dorsal fin sample) yielded V. vulnificus isolates and were therefore considered positive for harboring the bacterium (Table 1). The prevalence of V. vulnificus in fish varied between sampling events from 0 to 78%. Vibrio vulnificus was recovered from a large diversity of fish species (Table 3), with prevalence ranging from 0% to 58%. However, the numbers of analyzed fish were not equal across all species due to variable capture success. Because of this limitation, comparing the prevalence of V. vulnificus among fish species was not statistically feasible. Vibrio vulnificus was documented from any fish species wherein more than 3 individuals of that species were sampled. The order Perciformes was best represented in our sampling, with 63 of 171 (37%) individual fish harboring V. vulnificus.
Table 3.
Occurrence of Vibrio vulnificus in fish collected during the study
| Fish species (order: family), common name | No. of V. vulnificus-positive fish/total no. of fish tested | No. of recovered isolates | AFLP type(s)a |
|---|---|---|---|
| Dasyatis sabina (Myliobatiformes: Dasyatidae), Atlantic stingray | 1/3 | 3 | 55 |
| Elops saurus (Elopiformes: Elopidae), ladyfish | 3/9 | 11 | 6, 36, 62, 64, 79, 95, 100 |
| Brevoortia patronus (Clupeiformes: Clupeidae), Gulf menhaden | 0/1 | 0 | |
| Dorosoma petenense (Clupeiformes: Clupeidae), threadfin shad | 0/1 | 0 | |
| Bagre marinus (Siluriformes: Ariidae), gafftopsail sea catfish | 2/2 | 2 | 2 |
| Ariopsis felis (Siluriformes: Ariidae), hardhead sea catfish | 6/9 | 16 | 7, 11, 12, 14, 26, 37, 43, 59, 94, 98 |
| Opsanus beta (Batrachoidiformes: Batrachoididae), Gulf toadfish | 0/1 | 0 | |
| Mugil cephalus (Mugiliformes: Mugilidae), flathead gray mullet | 6/24 | 16 | 5, 82, 84, 86, 111, 117, 118 |
| Strongylura marina (Beloniformes: Belonidae), Atlantic needlefish | 0/2 | 0 | |
| Prionotus tribulus (Scorpaeniformes: Triglidae), bighead searobin | 0/2 | 0 | |
| Echeneis naucrates (Perciformes: Echeneidae), live sharksucker | 0/2 | 0 | |
| Caranx sp. (Perciformes: Carangidae) | 0/1 | 0 | |
| Selene vomer (Perciformes: Carangidae), lookdown | 0/1 | 0 | |
| Orthopristis chrysoptera (Perciformes: Haemulidae), pigfish | 2/7 | 9 | 21, 80 |
| Archosargus probatocephalus (Perciformes: Sparidae), sheepshead | 1/6 | 2 | 116 |
| Lagodon rhomboides (Perciformes: Sparidae), pinfish | 7/23 | 18 | 1, 4, 25, 28, 39, 47, 69, 73, 82, 97, 102, 103 |
| Bairdiella chrysoura (Perciformes: Sciaenidae), silver perch | 5/24 | 13 | 60, 81, 85, 94, 99, 100, 106 |
| Cynoscion arenarius (Perciformes: Sciaenidae), sand weakfish | 11/28 | 32 | 13, 17, 22, 26, 29, 43, 44, 50, 52, 57, 63, 75, 79, 89, 94, 96, 117, 120 |
| Cynoscion nebulosus (Perciformes: Sciaenidae), spotted weakfish | 7/16 | 23 | 10, 15, 17, 33, 34, 35, 45, 66, 90, 91, 121 |
| Leiostomus xanthurus (Perciformes: Sciaenidae), spot croaker | 0/3 | 0 | |
| Menticirrhus sp. (Perciformes: Sciaenidae) | 2/9 | 8 | 36, 51, 109 |
| Micropogonias undulatus (Perciformes: Sciaenidae), Atlantic croaker | 22/38 | 67 | 3, 8, 9, 10, 11, 15, 16, 19, 20, 23, 24, 38, 40, 41, 46, 48, 50, 54, 58, 61, 65, 67, 68, 70, 72, 77, 78, 82, 87, 92, 101, 104, 105, 108, 113 |
| Pogonias cromis (Perciformes: Sciaenidae), black drum | 0/1 | 0 | |
| Sciaenops ocellatus (Perciformes: Sciaenidae), red drum | 3/5 | 7 | 15, 18, 64, 85, 112 |
| Chaetodipterus faber (Perciformes: Ephippidae), Atlantic spadefish | 3/3 | 8 | 27, 28, 49, 71, 114 |
| Scomberomorus cavalla (Perciformes: Scombridae), king mackerel | 0/1 | 0 | |
| Scomberomorus maculatus (Perciformes: Scombridae), Atlantic Spanish mackerel | 0/3 | 0 | |
| Paralichthys lethostigma (Pleuronectiformes: Paralichthyidae), southern flounder | 9/17 | 16 | 12, 31, 74, 76, 81, 83, 88, 93 |
| Total | 90/242 | 251 |
Only 239 out of 251 isolates were typeable by AFLP.
Among fishes wherein >10 individuals were analyzed, Atlantic croaker and southern flounder had the highest prevalence at 58% and 53%, respectively, while striped mullet had the lowest at 25%.
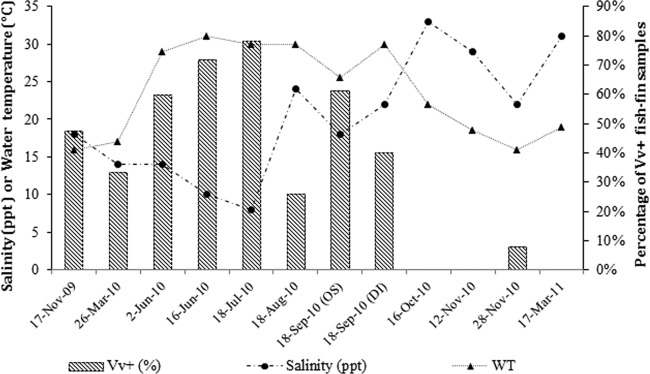
Environmental parameters fluctuated as expected during the study, with water temperatures ranging from 16°C to 31°C and salinity ranging from 8 ppt to 33 ppt (Fig. 2). The percentage of V. vulnificus-positive fish and salinity were inversely correlated (Pearson's correlation coefficient r = −0.91077, n = 12, P < 0.0001). In contrast, a positive correlation was found between V. vulnificus-positive fish and water temperature (Pearson's correlation coefficient r = 0.62481, n = 12, P = 0.0298). General linear models were applied to describe the changes in percentages of V. vulnificus-positive fish and the environmental factors (salinity and water temperature) analyzed in the study. The linear regression model identified these two environmental factors as having a significant effect on the percentage of V. vulnificus-positive fish. The model with two significant factors that best fits the data is as follows: percent V. vulnificus-positive fish = 0.5930 − 0.02818 × salinity + 0.01406 × water temperature. Parameter estimate statistics are included in Table 4, and the surface plot for the model equation is shown in Fig. S1 in the supplemental material. The coefficient of determination (r2) for the model was 0.9041, which denotes that 90% of the observed variation is explained by the independent variables. When an interaction term between salinity and water temperature was introduced into the model, the P value indicated that the interaction term was not significant (P = 0.9571). Essentially, this indicates that the number of V. vulnificus-positive fish was independently affected by salinity and temperature but that both parameters were not linked to each other. In addition, and to test if the collections sites influenced the percentages of V. vulnificus-positive fish, two dummy variables (S1 and S2) accounting for two of the collection areas (Dauphin Island and Ocean Springs) were brought into the model. The result of t test on parameters S1 (P = 0.6413) and S2 (P = 0.9199) indicated that site was not significant.
Fig 2.
Distribution of Vibrio vulnificus-positive (Vv+) fish throughout the study (bars) in relationship to salinity and water temperature (WT).
Table 4.
Statistical values for the multiple linear regression model
| Parameter | Estimate | SE | t value | Pr > ¦t¦a |
|---|---|---|---|---|
| Intercept | 0.5930 | 0.1786 | 3.3200 | 0.0089 |
| Salinity | −0.0282 | 0.0041 | −6.9500 | <.0001 |
| Water temp | 0.0141 | 0.0053 | 2.6500 | 0.0266 |
Pr, probability.
Population structure of V. vulnificus on fish.
Out of more than 500 putative V. vulnificus colonies recovered from selective media, 270 isolates were positive by colony dot blot hybridization, and out of those, 251 were positive by vvhA-specific PCR. None of 251 isolates were biotype 2, based on multiplex PCR (43). 16S rRNA gene RFLP typing classified the isolates into 3 types (16S rRNA types A, B, and AB), as previously reported (31). According to this classification method, 157 isolates (62%) were 16S rRNA type A, 72 (29%) were type B, and 22 (9%) were type AB. Representatives of 16S rRNA types A and B were recovered in every sampling, except for that in March 2010, where only 16S rRNA type A isolates were obtained. Figure 3 shows the temporal distribution of 16S rRNA types. The percentage of 16S rRNA type B isolates varied from 0% to 50%, but no clear correlation between salinity, water temperature, or fish species and 16S rRNA type could be inferred from our data.
Fig 3.
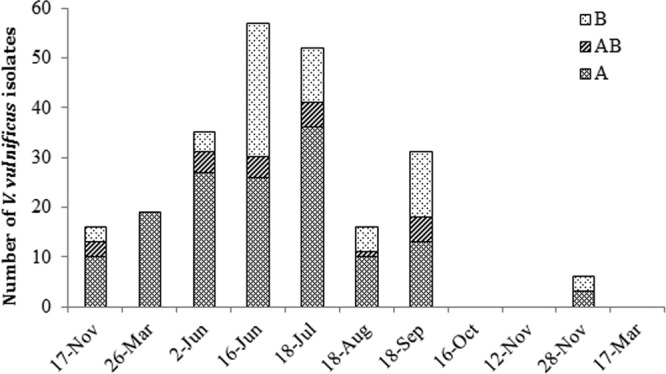
Distribution of Vibrio vulnificus 16S rRNA types A, AB, and B across the study.
Eight additional strains, proved to be virulent in a mouse model (48), were included as references in the AFLP analysis. All but 12 fish isolates were typeable by AFLP (untypeable isolates consistently produced profiles of weak intensity). AFLP profiles were highly informative, with an average of 95 bands ranging from 50 to 700 bp per profile. In order to test the reproducibility of the AFLP method, we performed 3 independent AFLP experiments using 10 fish isolates (data not shown). Based on the variability observed, we selected 90% as our threshold for considering an AFLP type unique, as previously described (4). Profiles displaying more than 90% similarity were ascribed to the same AFLP type, while similarities lower than 90% indicated different AFLP profiles. A total of 121 unique AFLP types were defined within the 239 isolates of V. vulnificus (Table 3). Rarefaction curves were generated for each fish species for which 10 or more isolates from the same fish species had been typed (data not shown). In all cases, the slope of the curves was similar and the number of AFLP types observed linearly increased with the total number of V. vulnificus isolates analyzed. These results suggest that the genetic diversity of the isolates was similar in all fishes.
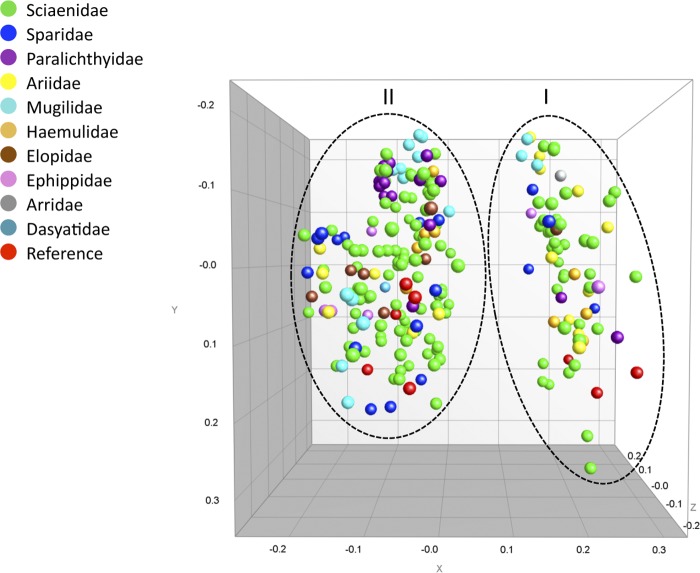
Figure 4 displays a multiscaling dimensional (MSD) analysis of the similarity among all isolates derived from the AFLP cluster analysis. Two nonoverlapping clusters that correlated with 16S rRNA type ascription could be clearly delineated. Cluster I grouped all 16S rRNA type B isolates at 70% similarity (see Fig. S2 in the supplemental material), while cluster II consisted of 148 isolates, including all 16S rRNA type A and type AB isolates at 71% similarity. All V. vulnificus isolates shared a similarity of 65% or higher. Cluster I and cluster II were comprised of 36 and 85 unique AFLP types, respectively. Group separation statistics based on jackknife analysis confirmed the statistically significant correlation between AFLP clusters I and II and 16S rRNA types B and A-AB. Resampling of the data showed a 100% agreement between assigned 16S rRNA type B and randomly selected subgroups (for types A and AB, the levels of agreement were 96% and 83%, respectively).
Fig 4.
MDS representation of the similarity matrix generated by AFLP cluster analysis. Each of 251 V. vulnificus isolates is represented by a dot, and the distance between dots represents relatedness obtained from the similarity matrix. Isolates are colored on the basis of origin (fish family or reference strains). Dotted lines highlight the two main clusters observed in the analysis. Cluster II groups all V. vulnificus 16S rRNA of types A and AB, and cluster I groups 16S rRNA of type B.
Overall, isolates did not cluster on the basis of the collection date, geographic location, or fish species (data not shown). Moreover, V. vulnificus isolates recovered from the same individual fish were not more related to each other than to those isolates from other fish. As an example of how V. vulnificus types did not show any specificity for fish species, isolates from Atlantic croaker (the fish species that yielded the most V. vulnificus isolates) were distributed across the entire AFLP-based dendrogram (see Fig. S2 in the supplemental material).
The band-matching analysis revealed a total of 119 polymorphic bands responsible for AFLP cluster ascription out of 123 total observed bands. Transversal clustering identified the AFLP bands, or markers, characteristic of 16S type B isolates. In Fig. S2 in the supplemental material, the isolates were clustered on the basis of band profiles, while the character was grouped according to values in the band table which reflected band classes of individual band profiles (1 for present, 0 for absent). In the two-dimensional clustering (data not shown), the isolates and band classes were arranged according to their relatedness. For example, band classes (labeled by band length) 116 bp, 156 bp, 200 bp, 253 bp, 343 bp, 386 bp, and 491 bp were more abundant in cluster I (16S rRNA type B isolates). These band classes are AFLP markers for 16S rRNA type B isolates (highlighted area in Fig. S2 in the supplemental material).
DISCUSSION
Few studies have documented the prevalence and distribution of V. vulnificus in wild fish (17), despite the fact that wound infections caused by this bacterium have been reported in fishermen worldwide (5, 14, 16, 23). Approximately 30 confirmed cases occurred annually in the Gulf Coast region (11), and most of them were purportedly linked to exposure to seawater or seafood (16). However, it is not possible to account for how many cases of wound infections were acquired by direct contact with fish on the basis of epidemiological studies. Our data showed that V. vulnificus commonly occurred on the fins of fishes from the northern Gulf of Mexico, particularly during the summer months. In fact, we were able to isolate V. vulnificus at all sampling events from June through September. In addition, this pathogen was isolated from a wide taxonomic and phylogenetic spectrum of fishes, including 16 species representing 15 genera, 8 families, and 6 orders (Table 3). Interestingly, the highest prevalence of V. vulnificus was found in two bottom-feeder fish (Atlantic croaker and southern flounder) which have been shown to have high densities of this bacterium in their gut (17). Despite being commonly associated with fish, our findings suggest that V. vulnificus is a transient member of the fish external microbiota (39), as it was not found on each fish even when several individuals from the same species were collected at the same time.
The occurrence of V. vulnificus on fish fins may be higher than that reported here since we used a culture-based method to estimate prevalence. The success rate of recovering environmental bacteria with culture-based methods varies depending on several factors, but it tends to underestimate original densities. For instance, both APW and mCPC have been reported to favor the growth of V. vulnificus biotype 1 over biotype 2 (42). Similarly, Chase and Harwood (12) showed than biotype 1 grows better than biotypes 2 and 3 under identical conditions. These factors may have negatively influenced the recovery of biotype 2 in our study. In addition, culture methods disallow recovery of the viable but not culturable (VBNC) form of V. vulnificus, a well-described life stage of this bacterium (32), although the abundance of VBNC forms in the environment is not well-known (37). The protocol used herein (enrichment in APW, followed by plating onto mCPC) could have preferentially enriched for V. vulnificus biotype 1, but overall, it provided a simple and inexpensive methodology for successfully recovering V. vulnificus from fish.
The frequency of detection of V. vulnificus in fish was a factor of salinity and temperature. These environmental factors are known to influence V. vulnificus abundance and distribution in the marine environment (7, 25, 28). Temperature is positively correlated with V. vulnificus presence, while salinity is negatively correlated. However, it has been suggested that salinity becomes the main factor influencing the abundance of V. vulnificus in oysters when salinities exceed 30 ppt, irrespective of water temperature (49). Our field data support this hypothesis, since we failed to recover V. vulnificus from fish when salinities were higher than 29 ppt, even when water temperatures exceeded 18°C. On the basis of our data, the prevalence of V. vulnificus in fish fins was more affected by salinity than by water temperature, and this was particularly noticeable in summer where water temperatures remained over 25°C. Our model accounts for 83% of the variability observed due to salinity and only 7% due to temperature. Nevertheless, our model may not be accurate when water temperatures and salinities are outside the ranges observed during our study.
Epidemiological data suggest that most V. vulnificus strains must present little risk to the susceptible population because of its abundance in marine samples and the small number of clinical cases observed per year (6, 40, 46). The first marker to be used as an indicator for virulence was a polymorphism present in the 16S rRNA gene that classifies the isolates as 16S rRNA type A and 16S rRNA type B; in addition, some strains can present both 16S rRNA gene alleles and are classified as 16S rRNA type AB. On the basis of epidemiological data, up to 75% of clinical V. vulnificus isolates are 16S rRNA type B (this percentage increased up to 94% when clinical fatalities were considered) (31). Similarly, Rosche et al. (41) identified an additional marker referred to as virulence-correlated gene (vcg) that can be used to infer clinical (vcgC) and environmental (vcgE) isolates. Both markers, 16S rRNA type and vcg, correlate well with each other (44). The majority (62%) of the fish isolates recovered in this study were classified as 16S rRNA type A (the proportion is up to 71%, if we include type AB), but at 29%, the number of type B isolates was higher than expected on the basis of previous studies from the same area (13, 20, 31). Although significant correlations with any environmental parameter were not noted, the highest proportion of type B relative to types A and AB was observed during the warmest months, which is consistent with similar observations of V. vulnificus populations in Galveston Bay, TX (26).
In terms of strain characterization, AFLP provided a high level of resolution and confirmed the high genetic diversity present in the species, with a calculated Simpson's index of diversity of 0.991 (24). This value is in range with that previously reported by Arias et al. (2) when isolates from a broader geographic area were characterized by AFLP. Vibrio vulnificus isolates associated with fish were highly heterogeneous, and we could not determine spatiotemporal- or fish-specific patterns that linked V. vulnificus types with environmental parameters or fish species. The inclusion of reference strains (proved to be virulent in a mouse model [48]) further confirmed the lack of uniqueness among fish isolates, since some of them shared high similarities with reference strains. This indicates that fish from the nearshore waters of the northern Gulf of Mexico harbor V. vulnificus genotypes of great virulence potential for humans.
In summary, this study presents new data on the prevalence of V. vulnificus on fish and presents a novel statistical model for predicting its occurrence. Our data indicate that this bacterium has a broad, seasonally dependent distribution among fishes ranging in coastal areas of the north-central Gulf of Mexico. Potentially pathogenic strains of V. vulnificus were recovered from fish, based on 16S rRNA ascription and AFLP similarity with clinical strains. Although reported cases of wound infections caused by this pathogen are clearly low in proportion to the numbers of anglers, fish captured, and fishing trips taken in the Gulf of Mexico, our distributional data show that fishermen could likely be inoculated by V. vulnificus during fish handling and processing, particularly if the fish was captured in an estuary. Finally, the correlation found between the two AFLP-delineated main clusters and 16S rRNA types supports the hypothesis stated by Rosche et al. (40) that V. vulnificus is in the process of diverging into two separate species.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the National Oceanic and Atmospheric Administration (NOAA-NA08NMF4720545), Marine Environmental Science Consortium, Gulf of Mexico Research Institute, and National Science Foundation (NSF DEB-1048523). Zhen Tao is the recipient of a graduate research fellowship funded by the Chinese Scholarship Council and the Ocean University of China.
Footnotes
Published ahead of print 24 August 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Adams CM, Hernandez E, Lee J. 2009. An economic overview of selected industries dependent upon the Gulf of Mexico, p 28–46 In Cato JC. (ed), Gulf of Mexico: origin, waters, and biota, vol 2 Texas A&M University Press, Corpus Christi, TX [Google Scholar]
- 2. Arias CR, Verdonck L, Swings J, Aznar R, Garay E. 1997. A polyphasic approach to study the intraspecific diversity amongst Vibrio vulnificus isolates. Syst. Appl. Microbiol. 20:622–633 [Google Scholar]
- 3. Arias CR, Verdonck L, Swings J, Garay E, Aznar R. 1997. Intraspecific differentiation of Vibrio vulnificus biotypes by amplified fragment length polymorphism and ribotyping. Appl. Environ. Microbiol. 63:2600–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arias CR, Welker TL, Shoemaker CA, Abernathy JW, Klesius PH. 2004. Genetic fingerprinting of Flavobacterium columnare isolates from cultured fish. J. Appl. Microbiol. 97:421–428 [DOI] [PubMed] [Google Scholar]
- 5. Bisharat N, et al. 1999. Clinical, epidemiological, and microbiological features of Vibrio vulnificus biotype 3 causing outbreaks of wound infection and bacteraemia in Israel. Lancet 354:1421–1424 [DOI] [PubMed] [Google Scholar]
- 6. Bisharat N, Amaro C, Fouz B, Llorens A, Cohen DI. 2007. Serological and molecular characteristics of Vibrio vulnificus biotype 3: evidence for high clonality. Microbiology 153:847–856 [DOI] [PubMed] [Google Scholar]
- 7. Blackwell KD, Oliver JD. 2008. The ecology of Vibrio vulnificus, Vibrio cholerae, and Vibrio parahaemolyticus in North Carolina estuaries. J. Microbiol. 46:146–153 [DOI] [PubMed] [Google Scholar]
- 8. Blake PA. 1979. Disease caused by a marine vibrio—clinical characteristics and epidemiology. N. Engl. J. Med. 300:1–5 [DOI] [PubMed] [Google Scholar]
- 9. Carpenter KE. 2002. The living marine resources of the western central Atlantic, p 1374 Food and Agriculture Organization of the United Nations, Rome, Italy [Google Scholar]
- 10. Castillo LE, Winslow DL, Pankey GA. 1981. Wound infections and septic show due to Vibrio vulnificus. Am. J. Trop. Med. Hyg. 30:844–848 [DOI] [PubMed] [Google Scholar]
- 11. CDC 2011. National enteric disease surveillance: COVIS annual summary. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/nationalsurveillance/PDFs/CSTEVibrio2009.pdf [Google Scholar]
- 12. Chase E, Harwood VJ. 2011. Comparison of the effects of environmental parameters on growth rates of Vibrio vulnificus biotypes I, II, and III by culture and quantitative PCR analysis. Appl. Environ. Microbiol. 77:4200–4207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chatzidaki-Livanis M, Hubbard MA, Gordon K, Harwood VJ, Wright AC. 2006. Genetic distinctions among clinical and environmental strains of Vibrio vulnificus. Appl. Environ. Microbiol. 72:6136–6141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dalsgaard A, Frimodt-Moller N, Bruun B, Hoi L, Larsen JL. 1996. Clinical manifestations and molecular epidemiology of Vibrio vulnificus infections in Denmark. Eur. J. Clin. Microbiol. Infect. Dis. 15:227–232 [DOI] [PubMed] [Google Scholar]
- 15. DaSilva L, et al. 2012. Development and validation of a predictive model for the growth of Vibrio vulnificus in postharvest shellstock oysters. Appl. Environ. Microbiol. 78:1675–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dechet AM, Yu PA, Painter J. 2008. Nonfood Vibrio infections: an important cause of morbidity and mortality in the United States, 1997–2006. Clin. Infect. Dis. 46:970–976 [DOI] [PubMed] [Google Scholar]
- 17. DePaola A, Capers GM, Alexander D. 1994. Densities of Vibrio vulnificus in the intestines of fish from the U.S. Gulf Coast. Appl. Environ. Microbiol. 60:984–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. DePaola A, McLeroy S, McManus G. 1997. Distribution of Vibrio vulnificus phage in oyster tissues and other estuarine habitats. Appl. Environ. Microbiol. 63:2464–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eschmeyer WN. 2010. Catalogue of fishes. California Academy of Sciences, San Francisco, CA: http://research.calacademy.org/ichthyology/catalog [Google Scholar]
- 20. Han FF, Pu Sh Hou AX, Ge BL. 2009. Characterization of clinical and environmental types of Vibrio vulnificus isolates from Louisiana oysters. Foodborne Pathog. Dis. 6:1251–1258 [DOI] [PubMed] [Google Scholar]
- 21. Hlady WG, Klontz KC. 1996. The epidemiology of Vibrio infections in Florida, 1981–1993. J. Infect. Dis. 173:1176–1183 [DOI] [PubMed] [Google Scholar]
- 22. Howard RJ, Lieb S. 1988. Soft-tissue infections caused by halophilic marine vibrios. Arch. Surg. 123:245–249 [DOI] [PubMed] [Google Scholar]
- 23. Hsueh PR, et al. 2004. Vibrio vulnificus in Taiwan. Emerg. Infect. Dis. 10:1363–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hunter PR. 1990. Reproducibility and indices of discriminatory power of microbial typing methods. J. Clin. Microbiol. 28:1903–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaspar CW, Tamplin ML. 1993. Effects of temperature and salinity on the survival of Vibrio vulnificus in seawater and shellfish. Appl. Environ. Microbiol. 59:2425–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin M, Schwarz JR. 2003. Seasonal shifts in population structure of Vibrio vulnificus in an estuarine environment as revealed by partial 16S ribosomal DNA sequencing. FEMS Microbiol. Ecol. 45:23–27 [DOI] [PubMed] [Google Scholar]
- 27. Massad G, Oliver J. 1987. New selective medium for Vibrio cholerae and Vibrio vulnificus. Appl. Environ. Microbiol. 53:2262–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Motes ML, et al. 1998. Influence of water temperature and salinity on Vibrio vulnificus in northern Gulf and Atlantic Coast oysters (Crassostrea virginica). Appl. Environ. Microbiol. 64:1459–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. National Marine Fisheries Service 2011. Fisheries of the United States 2010. National Marine Fisheries Service, NOAA, Silver Spring, MD: http://www.st.nmfs.noaa.gov/st1/fus/fus10/FUS_2010.pdf Accessed 20 September 2011 [Google Scholar]
- 30. Nelson JS. 2006. Fishes of the world, 4th ed John Wiley & Sons, Inc., Hoboken, NJ [Google Scholar]
- 31. Nilsson WB, Pranjype RN, DePaola A, Strom MS. 2003. Sequence polymorphism of the 16S rRNA gene of Vibrio vulnificus is a possible indicator of strain virulence. J. Clin. Microbiol. 41:442–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oliver JD. 2005. The viable but nonculturable state in bacteria. J. Microbiol. 43:93–100 [PubMed] [Google Scholar]
- 33. Oliver JD. 2005. Wound infections caused by Vibrio vulnificus and other marine bacteria. Epidemiol. Infect. 133:383–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oliver JD, Warner RA, Cleland DR. 1983. Distribution of Vibrio vulnificus and other lactose-fermenting vibrios in the marine environment. Appl. Environ. Microbiol. 45:985–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Penman AD, et al. 1995. Vibrio vulnificus wound infections from the Mississippi Gulf coastal waters: June to August 1993. South. Med. J. 88:531–533 [DOI] [PubMed] [Google Scholar]
- 36. Pitcher D, Saunders N, Owen R. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151–156 [Google Scholar]
- 37. Randa MA, Polz MF, Lim E. 2004. Effects of temperature and salinity on Vibrio vulnificus population dynamics as assessed by quantitative PCR. Appl. Environ. Microbiol. 70:5469–5476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rhoads J. 2006. Post-Hurricane Katrina challenge: Vibrio vulnificus. J. Am. Acad. Nurse Pract. 18:318–324 [DOI] [PubMed] [Google Scholar]
- 39. Roeselers G, et al. 2011. Evidence for a core gut microbiota in the zebrafish. ISME J. 5:1595–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rosche TM, Binder EA, Oliver JD. 2010. Vibrio vulnificus genome suggests two distinct ecotypes. Environ. Microbiol. Rep. 2:128–132 [DOI] [PubMed] [Google Scholar]
- 41. Rosche TM, Yano Y, Oliver JD. 2005. A rapid and simple PCR analysis indicates there are two subgroups of Vibrio vulnificus which correlate with clinical or environmental isolation. Microbiol. Immunol. 49:381–389 [DOI] [PubMed] [Google Scholar]
- 42. Sanjuán E, Amaro C. 2004. Protocol for specific isolation of virulent strains of Vibrio vulnificus serovar E (biotype 2) from environmental samples. Appl. Environ. Microbiol. 70:7024–7032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sanjuán E, Amaro C. 2007. Multiplex PCR assay for detection of Vibrio vulnificus biotype 2 and simultaneous discrimination of serovar E: development and field studies. Appl. Environ. Microbiol. 73:2029–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sanjuán E, Fouz B, Oliver JD, Amaro C. 2009. Evaluation of genotypic and phenotypic methods to distinguish clinical from environmental Vibrio vulnificus strains. Appl. Environ. Microbiol. 75:1604–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Scharf MJ. 2002. Cutaneous injuries and envenomations from fish, sharks and rays. Dermatol. Ther. 15:47–57 [Google Scholar]
- 46. Strom MS, Paranjpye RN. 2000. Epidemiology and pathogenesis of Vibrio vulnificus. Microbes Infect. 2:177–188 [DOI] [PubMed] [Google Scholar]
- 47. Tamplin ML, Martin AL, Ruple AD, Cook DW, Kaspar CW. 1991. Enzyme immunoassay for identification of Vibrio vulnificus in seawater, sediment, and oyster. Appl. Environ. Microbiol. 57:1235–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thiaville PC, et al. 2011. Genotype is correlated with but does not predict virulence of Vibrio vulnificus biotype 1 in subcutaneously inoculated, iron dextran-treated mice. Infect. Immun. 79:1194–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. WHO-FAO 2005. Risk assessment of Vibrio vulnificus in raw oysters. FAO, Rome, Italy, and WHO, Geneva, Switzerland [Google Scholar]
- 50. Wright AC, et al. 1993. Rapid identification of Vibrio vulnificus on nonselective media with an alkaline phosphatase-labeled oligonucleotide probe. Appl. Environ. Microbiol. 59:541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.