Abstract
Environmental surfaces contaminated with pathogens can be sources of indirect transmission, and cleaning and disinfection are common interventions focused on reducing contamination levels. We determined the efficacy of cleaning and disinfection procedures for reducing contamination by noroviruses, rotavirus, poliovirus, parechovirus, adenovirus, influenza virus, Staphylococcus aureus, and Salmonella enterica from artificially contaminated stainless steel surfaces. After a single wipe with water, liquid soap, or 250-ppm free chlorine solution, the numbers of infective viruses and bacteria were reduced by 1 log10 for poliovirus and close to 4 log10 for influenza virus. There was no significant difference in residual contamination levels after wiping with water, liquid soap, or 250-ppm chlorine solution. When a single wipe with liquid soap was followed by a second wipe using 250- or 1,000-ppm chlorine, an extra 1- to 3-log10 reduction was achieved, and except for rotavirus and norovirus genogroup I, no significant additional effect of 1,000 ppm compared to 250 ppm was found. A reduced correlation between reduction in PCR units (PCRU) and reduction in infectious particles suggests that at least part of the reduction achieved in the second step is due to inactivation instead of removal alone. We used data on infectious doses and transfer efficiencies to estimate a target level to which the residual contamination should be reduced and found that a single wipe with liquid soap followed by a wipe with 250-ppm free chlorine solution was sufficient to reduce the residual contamination to below the target level for most of the pathogens tested.
INTRODUCTION
Viruses are the most common cause of infectious disease acquired in the indoor environment in hospitals, schools, and households (4), causing considerable impact on human health. Transmission of enteric and respiratory viruses is assumed to occur predominantly directly from person to person, followed by indirect transmission through contaminated surfaces (7, 40, 47, 53). The risk of infection resulting from transmission through contaminated surfaces depends on a number of factors, including the level of shedding of infective particles, their stability on surfaces and resistance to decontamination procedures, and the low dose required for infection. Among the enteric viruses, human noroviruses (NoVs) and rotaviruses are most notorious for causing outbreaks of gastroenteritis within hospitals, nursing homes, and cruise ships and are a significant cause of hospitalization (12, 36, 44). Human NoV outbreaks are often prolonged and reoccurring (5) due to the high levels of shedding of over 107 NoV particles/g in stool (45) or vomitus (43) and the low number of particles required for infection (52). Noroviruses are found on different types of surfaces (floors, tables, doorknobs, handles, bed rails, carpets, and curtains) in health care facilities, food production facilities, schools, and the community (7, 25, 59). Moreover, NoVs, and many other enteric viruses, stay infectious for up to several weeks (19, 38, 56), which is considered another important factor in environmental transmission.
Besides human NoV, other enteric viruses like poliovirus and rotavirus and respiratory viruses like influenza virus and adenovirus may also be transmitted through contaminated surfaces (7). Influenza A virus has been frequently associated with epidemics and occasional pandemics. Adenovirus type 5 is a recommended test organism for testing disinfectants (22) as well as an interesting virus since it can be detected in respiratory excretions and in feces (48). Parechovirus infections have commonly been associated with mild gastrointestinal symptoms in young children, and parechoviruses are excreted in feces as well (6). The transmission of parechovirus through contaminated surfaces has not been reported yet, but an indirect transmission route is likely to play a role in its spreading, given its similarities to enteroviruses.
Cleaning and disinfection of contaminated surfaces are one of the frequently implemented measures to control transmission of pathogens in indoor environments (15, 24, 31). The effectiveness of cleaning and disinfection practices is often monitored by determining reductions for bacteria such as Gram-positive Staphylococcus aureus in hospital settings and Staphylococcus aureus and Gram-negative Salmonella enterica serovar Enteritidis in food preparation facilities (14, 16). Additionally, the importance of environmental cleaning to control NoV outbreaks in health care settings is widely accepted (5, 28, 33), and decontamination of food production facilities may reduce the number and size of food-borne outbreaks (8, 17). However, the reduction levels achieved for bacterial contaminations do not necessarily correlate with reduction levels for viral contaminations, and as recently reported by Greig and Lee (30), the scientific proof supporting the effectiveness of implemented intervention measures is limited. Therefore, effective science-based control measures to reduce environmental contamination are urgently needed to reduce the burden of disease of these viruses.
To be able to implement the most effective viral decontamination method, it is necessary to have quantitative data on residual contamination levels after commonly applied cleaning and disinfection practices for some of the most relevant viruses, and preferably, these data should be comparable to data for some bacteria. Thus, in the present study, we assessed the effects of different cleaning and disinfection procedures on stainless steel carriers that were artificially contaminated with poliovirus Sabin 1, parechovirus 1, NoV GI.4, NoV GII.4 and its cultivable surrogate murine norovirus 1 (MNV1) (10), simian rotavirus SA 11, influenza A (H1N1) virus, adenovirus type 5, and the bacteria S. aureus and S. Enteritidis. The experiments were designed to reflect the order of magnitude of the levels of contamination that may result from common events such as toilet flushing (3), poor hygiene, or environmental dispersal of viral particles through droplets generated during a vomiting accident (11) or remain after removal of visible contamination.
The residual contamination was quantified by (cell) culture and PCR assays. As human NoV cannot be cultured (21), the residual contamination by these viruses was determined by quantitative PCR only.
MATERIALS AND METHODS
Test organisms and stocks.
Viruses used for the test were poliovirus Sabin 1 (vaccine strain), simian rotavirus SA 11 (ATCC no. VR-1565), adenovirus type 5 (Hu/adenovirus/type 5/6270/1988/Ethiopia), influenza A (H1N1) virus (Hu/influenza A/266/2008/Netherlands [H1N1] virus), parechovirus 1 (Hu/parechovirus/type 1/147/2008/Netherlands), MNV1 (Mu/NoV/GV/MNV1/2002/USA), human NoV GI.4 (Hu/NoV/GI.4/946/2009/Netherlands), and human NoV GII.4 (Hu/NoV/GII.4/1803/2008/Netherlands). The bacterial test organisms were Staphylococcus aureus (196E, toxin producer, human isolate) and Salmonella enterica serovar Enteritidis (phage type 4).
Virus stocks were prepared as described before (57) and stored at −80°C. The stocks used contained the following: poliovirus Sabin 1, 7.2 × 108 50% tissue culture infective doses (TCID50)/ml and 5.3 × 1011 PCR units (PCRU)/ml; adenovirus type 5, 2.8 × 107 TCID50/ml and 6.7 × 109 PCRU/ml; parechovirus 1, 3.9 × 108 TCID50/ml and 6.7 × 1010 PCRU/ml; rotavirus 5, 1.4 × 108 TCID50/ml and 6.7 × 109 PCRU/ml; influenza A (H1N1) virus, 2.3 × 107 TCID50/ml and 2.0 × 109 PCRU/ml; and MNV1, 4.9 × 106 TCID50/ml and 1.2 × 109 PCRU/ml. The human NoV GI.4 and GII.4 stocks were 6.6 × 108 and 1.1 × 108 PCRU/ml, respectively.
S. aureus and S. Enteritidis were cultured in brain heart infusion broth (Difco) and enumerated on tryptone soy agar (Oxoid, England) as described before (37). Bacterial stocks contained 8.8 × 109 CFU/ml of S. aureus and 4.2 × 108 CFU/ml of S. Enteritidis, and the detection limit of both bacteria was 10 CFU per contaminated spot.
Preparation of sterile stool suspension.
The sterile stool suspension from a healthy volunteer was prepared (57), and the suspension was free of rotaviruses, enteric adenoviruses, astroviruses, and sapoviruses as determined by PCR (50).
Cleaning and disinfection experiments.
The cleaning and disinfection experiments were performed on 2.2-cm by 2.2-cm stainless steel carriers (AISI type 304 standard; Netherlands). The carriers were degreased by being dipped into acetone for 10 min, followed by being rinsed five times under running tap water. Thereafter, the carriers were soaked in 70% alcohol and dried. The carriers were then sterilized by autoclaving (121°C for 15 min). The viscose wiping cloth was cut into pieces (approximately 4 cm by 3.5 cm) and sterilized by autoclaving.
One chlorine tablet (Suma Tab D4; Germany) was dissolved in 1,000 ml sterile water. From this solution, 250- and 1,000-ppm chlorine solutions were freshly prepared and free chlorine concentrations were measured using a Hatch colorimeter kit (Hanna HI 96771; Romania).
Bovine serum albumin (BSA) (3% [wt/vol] in water) or sterile stool suspension (20% [wt/vol]) was added to the virus stock to perform the experiments under clean and dirty conditions. Final concentrations were 0.03% BSA and 1% stool, respectively. Since human stool is not the natural matrix for influenza A virus, this experiment was performed under clean conditions only. The human NoVs were used as 10% (wt/vol) stool suspensions, and no extra feces was added. Stainless steel carriers were contaminated by spreading 30 μl of each virus suspension in 0.03% (wt/vol) BSA or 1% (wt/vol) stool separately (contaminated spot) and thereafter dried inside a biosafety cabinet for 1 h at room temperature (22 to 25°C, 40 to 45% relative humidity [RH]). Then the following cleaning and disinfection procedures were applied.
(i) Single wiping.
One thousand milliliters of each cleaning and disinfection solution was prepared. The cloth pieces were soaked in water, water with liquid soap, or 250-ppm or 1,000-ppm free chlorine solutions separately, and excess liquid was squeezed out by hand. With this wet cloth, the contaminated carriers were wiped once by hand and sampled 20 min after wiping.
(ii) Double wiping.
The carriers contaminated with viruses and bacteria were wiped once with the cloth soaked in water with liquid soap as described for the single-wiping procedure and wiped once again with cloths that were soaked in 250- or 1,000-ppm free chlorine solution and wrung. The carriers were sampled after 20 min. Gloves were worn throughout the cleaning process and changed after each wiping.
For sampling, the carrier was kept in a sterile flat-bottomed tube (Sarstedt 60.597.001; Germany) with the wiped surface facing upwards, and 2 ml cold Dulbecco's modified Eagle medium (4 to 8°C) with 10% fetal bovine serum (DMEM-FBS) was added for neutralization. For the carriers that were wiped with chlorine solutions, 500 μl of 7% (wt/vol) sodium thiosulfate solution in water was added for neutralization first and then 1,500 μl DMEM-FBS was added. Thereafter, the virus was extracted by vortexing at maximum speed for 30 s and flushing the carrier with the medium several times. The suspensions were then collected, and infective viruses were enumerated by cell culture assays. Additionally, quantitative PCR assays were performed on samples obtained from wiping with water with liquid soap or 1,000-ppm free chlorine solution (single wiping) and on samples obtained from wiping with water with liquid soap followed by wiping with 1,000-ppm free chlorine solution (double wiping) to quantify the genomic copies left.
Spot disinfection.
If infective virus could still be detected after wiping with water with liquid soap followed by wiping with 1,000-ppm free chlorine solution, virus inactivation was further tested by spot disinfection under dirty conditions to determine if extra contact time with the chlorine solution would result in lower residual contamination levels. After the contaminated carrier was wiped with water with liquid soap, 800 μl of 1,000-ppm free chlorine solution was added onto the carrier so that the carrier was completely covered with the chlorine solution for 5, 10, and 20 min. After the exposure time, the chlorine solution was neutralized with an equal volume of 7% (wt/vol) sodium thiosulfate solution in water and 400 μl DMEM-FBS was added to make a total volume of 2 ml. Untreated carriers were kept as control. For a neutralization control, compounds (liquid soap or chlorine solutions) were diluted with DMEM-FBS or neutralized with 7% sodium thiosulfate solution before addition to the virus. The experiments were also performed with S. aureus and S. Enteritidis. Neutralized bacteriological peptone water (Oxoid, England) was used instead of DMEM-FBS. As stool is not the natural matrix for S. aureus, the experiment was done only under clean conditions.
TCID50 determination.
The viruses were enumerated by titration in 96-well plates on sensitive cells as described before (57).
Real-time PCR.
To allow comparison of virus reductions between the cultivable viruses and the noncultivable human NoVs (21), quantitative PCR assays were performed. Viral nucleic acid extraction was performed using the Magna Pure total nucleic acid extraction kit as described before (50). Real-time PCR assays were performed as described before for poliovirus Sabin 1 (18), adenovirus type 5 (34), rotavirus SA 11 (50), parechovirus 1 (55), MNV1 (2), and human NoV GI.4 (50) and NoV GII.4 (57). Amplifiable PCRU were determined by slopes of standard curves made for each virus. The standard curve was made by plotting cyclic threshold (CT) values versus log PCRU of 10-fold dilutions of the virus stocks. The highest dilution giving a positive result was assigned a value of 1 PCRU.
Residual contamination.
In order to provide data that will allow for risk assessments, we present data on the basis of residual contamination instead of pathogen reduction. The number of pathogens present on the carrier after cleaning or after cleaning and disinfection was considered the residual contamination. The reduction of the pathogens was calculated as (log10 pathogens on the control carrier) − (log10 pathogens on wiped carrier). The control carriers were contaminated and dried but not subjected to the treatments. All the experiments were performed in triplicate and repeated for confirmation (n = 6).
Data analysis.
Statistical analysis was performed by using Student's t test. The log10 values of infectivity (x) and PCRU (y) reduction for cleaning with liquid soap or 1,000-ppm chlorine solution and wiping with liquid soap followed by wiping with 1,000-ppm chlorine solution were plotted to compare with the line of equality y = x.
RESULTS
Calculation of the residual contamination target level.
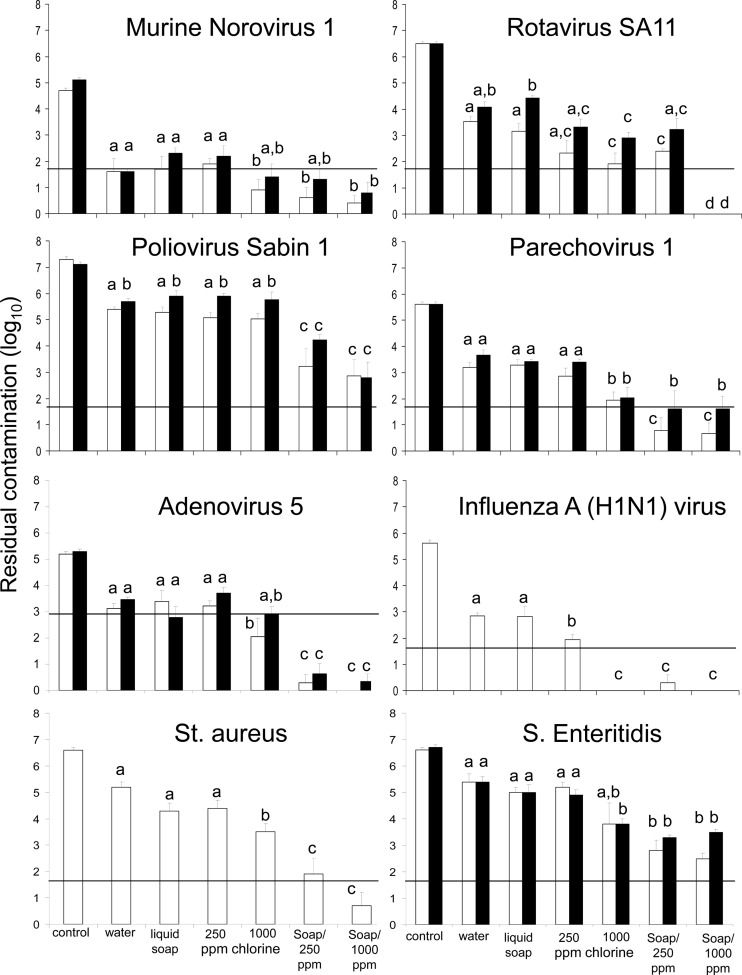
The residual contamination on the carrier after cleaning and disinfection poses a risk when enough infectious microorganisms can be transferred to individuals either to cause infection or to continue transmission indirectly through handling. The data for transfer of microorganisms from contaminated surfaces to the human hand (finger pads) have been determined for rotavirus and hepatitis A virus (1, 42) and shown to be approximately 20% after 20 min of drying (1). The number of viruses required for peroral infection is estimated as 10 to 100 infectious particles for rotavirus, norovirus, poliovirus, parechovirus, and influenza A virus (23, 32, 51, 60), and approximately 150 infectious particles for adenovirus (29). An estimated 10 to 100 cells are required for peroral S. Enteritidis infection (49) and S. aureus infection. If we assume 20% transfer from fomite to fingers for all microorganisms tested, then the risk of infection will be small if the residual contamination is less than 5 times the number of particles required for infection; this level may result in an infection only in the unlikely event that a contaminated finger is directly put in the mouth. We therefore assumed that at residual contamination levels of infective particles of less than 50 (1.7 log10) for rotavirus, MNV1, poliovirus, parechovirus, influenza A (H1N1) virus, S. Enteritidis, and S. aureus and less than 750 (2.9 log10) for adenovirus type 5, per contact spot, the probability of continued transmission or becoming infected is low (but not zero). On the basis of this assumption, lines indicating the residual contamination target levels were drawn in Fig. 1.
Fig 1.
Residual contamination of different pathogens on stainless steel carriers under clean (white bars) and dirty (black bars) conditions after different cleaning and disinfection methods. Control is the recovery after 1 h of drying. Water, liquid soap, 250-ppm chlorine, and 1,000-ppm chlorine indicate the suspensions used to wet a wipe for the one-wipe (cleaning) procedure. Soap/250 ppm and Soap/1,000 ppm indicate the consecutive suspensions used to wet wipes for the two-step (cleaning and disinfection) procedure. Error bars indicate standard deviations of the means, and means with different letters differ significantly (P < 0.05) (n = 6). The horizontal lines in the figures indicate the residual contamination target levels.
Residual contamination after cleaning—single wiping.
The recovery of the viruses and bacteria from the stainless steel carriers after drying for 1 h ranged from 24 to 76%. After wiping, the surfaces were visibly dry within 3 min. The residual contamination levels of infective viruses and bacteria under clean and dirty conditions after single and double wiping are shown in Fig. 1. There was no significant difference in residual contamination after wiping with water and after wiping with water with liquid soap. For poliovirus and rotavirus only, there was a minor but significantly higher residual contamination when feces were present than under clean conditions. We found little or no effect of the use of 250-ppm chlorine solutions instead of liquid soap in the cleaning step; for rotavirus under dirty conditions and influenza A virus only (i.e., in only 2 out of 14 pathogen-matrix combinations tested), a lower residual contamination was seen when 250-ppm chlorine was used.
The residual contamination after wiping with 1,000-ppm chlorine solution was significantly lower (P < 0.05) than that after wiping with water or liquid soap in 10 out of 14 pathogen-matrix combinations. Additionally, in 7 out of 14 pathogen-matrix combinations the wipe with 1,000-ppm chlorine solutions resulted in a significantly lower residual contamination than did wiping with 250-ppm chlorine solution.
Residual contamination after cleaning and disinfection—double wiping.
The residual contamination after wiping with liquid soap followed by wiping with 250-ppm chlorine solution (double wiping) was significantly lower (P < 0.05) than that after wiping with liquid soap alone (single wiping) for most of the viruses (except MNV1 and rotavirus) and bacteria tested (Fig. 1). After the double wiping procedure, there was no significant difference (P > 0.05) in residual contamination between 250- and 1,000-ppm chlorine solutions in 12 out of 14 pathogen-matrix combinations. For rotavirus only, the reduction achieved with 1,000 ppm was better than the reduction achieved with 250-ppm chorine solution, resulting in a residual contamination of less than 2 infectious particles per spot (detection limit; >6-log10 reduction).
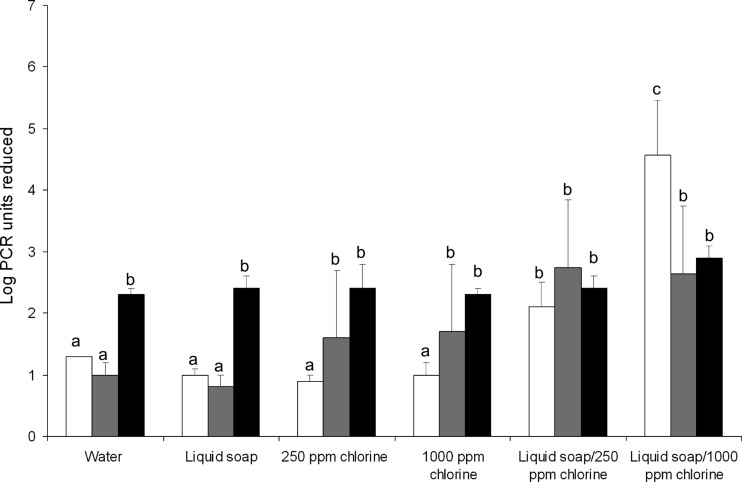
Reduction of genomic copies of norovirus after cleaning and disinfection.
As human NoVs could not be cultured, the reductions in genomic copies were quantified by PCR assays. The reductions in genomic copies of NoV GI.4, GII.4, and MNV1 are shown in Fig. 2. For MNV1, all the treatments resulted in a comparable reduction, while for NoV GI.4 and GII.4, we observed a significantly higher reduction in PCRU with the double wiping protocol. In 5 out of 6 treatments, the reductions in PCRU differed between NoV GI.4 and MNV1; in 3 out of 6, they differed between NoV GI.4 and NoV GII.4; and in 2 out of 6, they differed between NoV GII.4 and MNV1.
Fig 2.
Reduction of genomic copies of human NoVs GI.4 (white bars) and GII.4 (gray bars) and MNV1 (black bars) under dirty conditions after different cleaning methods. Water, liquid soap, 250-ppm chlorine, and 1,000-ppm chlorine indicate the suspensions used to wet a wipe for the one-wipe (cleaning) procedure. Liquid soap/250-ppm chlorine and liquid soap/1,000-ppm chlorine indicate the consecutive suspensions used to wet wipes for the two-step (cleaning and disinfection) procedure. Error bars indicate standard deviations of the means, and means with different letters differ significantly (P < 0.05) (n = 6).
Reduction of infective load and genomic copies.
The PCRU reductions of poliovirus Sabin 1, adenovirus type 5, parechovirus 1, MNV1, rotavirus SA 11, and influenza A (H1N1) virus by wiping with water with liquid soap or with 1,000-ppm chlorine solution and wiping with water with liquid soap followed by wiping with 1,000-ppm chlorine solution were also determined. The equality between reduction of genomic copies and reduction of infectivity of the tested viruses under clean conditions is shown in Fig. 3. After wiping with water with liquid soap, there was a correlation between the infectivity and PCRU reduction except for rotavirus SA 11 and influenza A (H1N1) virus. The infectivity reduction was higher than the PCRU reduction (i.e., deviating from the equality line) on wiping with 1,000-ppm chlorine solution and with liquid soap followed by wiping with 1,000-ppm chlorine solution in the case of parechovirus 1, rotavirus SA 11, MNV1, adenovirus type 5, and influenza A (H1N1) virus.
Fig 3.
Correlation between PCR units and infectivity reduction by different cleaning methods under clean and dirty conditions of poliovirus Sabin 1 (diamonds), adenovirus type 5 (squares), parechovirus 1 (circles), rotavirus SA 11 (triangles), MNV1 (rectangles), and influenza A (H1N1) virus (asterisks) (n = 6). White, gray, and black symbols represent the reductions by wiping with liquid soap or 1,000-ppm chlorine solution and wiping with liquid soap followed by wiping with 1,000-ppm chlorine solution, respectively. In some cases, only one data point is visible due to overlap of data points.
Residual contamination after spot disinfection.
Since there was residual contamination by MNV1, poliovirus Sabin 1, adenovirus type 5, parechovirus 1, and S. Enteritidis after the double wiping procedure using 1,000-ppm chlorine, spot disinfection of the bacteria and viruses under dirty conditions by 1,000-ppm chlorine solution after cleaning with water with liquid soap was tested to determine if this treatment would result in a residual contamination that is below the detection limit. The residual contamination was reduced to below the detection limit of 10 particles of MNV1 in 5 min (a reduction of 5 log10) and of poliovirus Sabin 1 (6.9 log10) and adenovirus type 5 (5.3 log10) in 10 min. The infective loads of parechovirus 1 and S. Enteritidis were reduced by 3.2 ± 0.1 and 4.9 ± 0.4 log10, respectively, within 20 min of disinfection. Genomic copies of NoVs GI.4 (6.7 log10 PCRU) and GII.4 (5.2 log10 PCRU) were reduced to below the detection limit of 60 PCRU/spot after 10 and 5 min, respectively. MNV1 was reduced by 6.9 ± 0.7 log10 PCRU within 20 min of disinfection with 1,000-ppm free chlorine solution.
DISCUSSION
Our data indicate that in the case of an outbreak of gastroenteritis, by either NoV, rotavirus, or Salmonella, a cleaning step with liquid soap followed by a wipe using a 1,000-ppm chlorine solution most consistently results in the lowest residual contamination level of all treatments tested. However, if we assume that an equivalent of 1 in 2 NoV PCRU is infectious (data for NoV GI.1 [52]), the residual infectivity of NoV GI.4 and GII.4 will be approximately 5 × 102 or 5 × 103 infectious particles (approximately 1 × 103 or 1 × 104 PCRU), respectively, per contaminated spot, which is well above the level that we defined as target level. Increasing the contact time between the pathogen and the 1,000-ppm chlorine solution to at least 5 min (as studied by spot disinfection) did result in residual contamination below the target levels of NoV and rotavirus and may be considered an effective intervention strategy in controlling gastroenteric pathogen transmission via hard surfaces, although it may be impractical. Our data suggest that S. Enteritidis may still be present at loads above our target levels; however, the low prevalence of S. Enteritidis in nonfood and health care-related outbreaks (58) suggests that transmission via hard surfaces is not a main route of transmission for this pathogen. We did not find clear differences in the reductions in infective enteric viruses or viable bacteria in our experiments, indicating that the apparently greater outbreak potential of NoV and rotavirus is not due to a higher resistance to cleaning and disinfection but more likely due to the extremely high infectivity of NoV and the high levels of shedding for rotavirus.
Due to the inability to cultivate the human NoVs in vitro, several cultivable viruses such as feline calicivirus (FCV), canine calicivirus (CaCV), MS2 bacteriophage, and MNV1 have been used as surrogates to study NoV inactivation (10, 20, 46). However, NoV GI and GII viruses differ in binding properties for shellfish tissues and lettuce surfaces, for example (41, 54), and also in resistance to freeze-drying and heat treatment (9, 35), making it unlikely that one model virus will be a valuable surrogate for NoV GI and NoV GII. This was confirmed in our studies that showed inconsistencies in the level of correlation of MNV results with those for NoV GII.4 and GI.4 in complex situations such as this study where removal and disinfection were combined. In the absence of a cultivation method for the human NoV, we postulate that especially for quantitative risk assessment purposes, the use of any model virus should be accompanied by a PCR-based method to allow comparison.
The two picornaviruses tested (poliovirus and parechovirus) showed remarkable differences in residual contamination and thus risk of infection remaining after cleaning; however, this was mainly caused by a 2-log10 difference in starting contamination level. Since differences in levels of shedding do occur (13, 39), these data may reflect real variation in levels of contamination after cleaning and disinfection. Spot disinfection showed a remarkably resistant parechovirus fraction, as some parechoviruses could still be cultured after 20 min of exposure to 1,000-ppm chlorine solution. Such a very resistant virus fraction, representing 0.01% of the stock suspensions used, was also shown to exist during thermal inactivation at 73°C (55). Due to the low infectious dose, these resistant fractions may represent a risk when present in foods or on surfaces when very high levels are shed.
In this study, we confirmed the higher sensitivity of the enveloped respiratory influenza A virus than of the nonenveloped enteric viruses to disinfection (56), and the complete removal of infectious influenza virus after a single wipe with a 1,000-ppm chlorine solution confirms a recent study that showed complete inactivation of human influenza A viruses by wipes containing 1% bleach (sodium hypochlorite and sodium hydroxide) (27). The two-step procedure consisting of a single wipe with liquid soap followed by a disinfection step using 250-ppm chlorine solution is likely to be a good intervention strategy in cases of viral respiratory disease outbreaks since it reduced the infectivity of both respiratory viruses tested to well below the target level.
The efficacy of cleaning and disinfection is determined not only by the intrinsic effectiveness of the method applied but also by the appropriateness of the surfaces treated. Cleaning and disinfection should be focused on the critical spots, i.e., the surfaces mainly involved in transmission. Reducing the infective load on critical spots such as doorknobs, handles, light switches, and other frequently touched surfaces is more likely to have a profound impact on transmission than is treating rarely touched surfaces. Interestingly, a recent study on the removal of viruses from hard surfaces found a reduction of infective MNV1 after wiping the surfaces 6 times (26) comparable to what we found after a single wipe, indicating that surface cleaning and disinfection can be performed quite efficiently. Nonetheless, manual cleaning and disinfection procedures will always be more labor-intensive than, for example, room disinfection using hydrogen peroxide vapor (57), and for the control of outbreaks, a combination of the two methods is most likely needed.
In this study, we performed cleaning and disinfection by wiping as it may be carried out in health care settings. Since these procedures will be carried out by different individuals, variability in residual contamination levels is likely. Additional variation will occur due to differences in levels of shedding, differences in temperature and humidity, and types of contaminated surfaces. However, tests like these, even if describing just one scenario, provide the scientific background for evidence-based cleaning and disinfection guidelines or protocols.
In health care facilities, cleaning may be performed according to different protocols: general cleaning performed on a day-to-day basis and more stringent cleaning, often in combination with disinfection procedures, during outbreaks. Our findings show that in all cases a single wipe with a wet cloth with either water or liquid soap resulted in a significant reduction (>1 log10) of the infective load of all pathogens tested, but the residual contaminations indicate that further transmission may still occur. Adding a wiping step with 250- or 1,000-ppm chlorine solution resulted in an additional reduction of the infective load, most likely through inactivation of the pathogens rather than by particle removal, as indicated by the discrepancy between infectivity and PCRU reduction. Precleaning before disinfection of the contaminated surfaces is recommended, and the removal and disinfection together will often result in residual contamination levels below the target levels of residual contamination.
ACKNOWLEDGMENT
We thank Rina Puspitasari for an excellent contribution to the laboratory work.
There are no conflicts of interest to declare.
Footnotes
Published ahead of print 31 August 2012
REFERENCES
- 1. Ansari SA, Sattar SA, Springthorpe VS, Wells GA, Tostowaryk W. 1988. Rotavirus survival on human hands and transfer of infectious virus to animate and nonporous inanimate surfaces. J. Clin. Microbiol. 26:1513–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bae J, Schwab KJ. 2008. Evaluation of murine norovirus, feline calicivirus, poliovirus, and MS2 as surrogates for human norovirus in a model of viral persistence in surface water and groundwater. Appl. Environ. Microbiol. 74:477–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barker J, Jones MV. 2005. The potential spread of infection caused by aerosol contamination of surfaces after flushing a domestic toilet. J. Appl. Microbiol. 99:339–347 [DOI] [PubMed] [Google Scholar]
- 4. Barker J, Stevens D, Bloomfield SF. 2001. Spread and prevention of some common viral infections in community facilities and domestic homes. J. Appl. Microbiol. 91:7–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barker J, Vipond IB, Bloomfield SF. 2004. Effects of cleaning and disinfection in reducing the spread of norovirus contamination via environmental surfaces. J. Hosp. Infect. 58:42–49 [DOI] [PubMed] [Google Scholar]
- 6. Benschop K, Molenkamp R, van der Ham A, Wolthers K, Beld M. 2008. Rapid detection of human parechoviruses in clinical samples by real-time PCR. J. Clin. Virol. 41:69–74 [DOI] [PubMed] [Google Scholar]
- 7. Boone SA, Gerba CP. 2007. Significance of fomites in the spread of respiratory and enteric viral disease. Appl. Environ. Microbiol. 73:1687–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boxman IL, et al. 2009. Environmental swabs as a tool in norovirus outbreak investigation, including outbreaks on cruise ships. J. Food Prot. 72:111–119 [DOI] [PubMed] [Google Scholar]
- 9. Butot S, Putallaz T, Amoroso R, Sanchez G. 2009. Inactivation of enteric viruses in minimally processed berries and herbs. Appl. Environ. Microbiol. 75:4155–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cannon JL, et al. 2006. Surrogates for the study of norovirus stability and inactivation in the environment: a comparison of murine norovirus and feline calicivirus. J. Food Prot. 69:2761–2765 [DOI] [PubMed] [Google Scholar]
- 11. Caul EO. 1994. Small round structured viruses: airborne transmission and hospital control. Lancet 343:1240–1242 [DOI] [PubMed] [Google Scholar]
- 12. Cheesbrough JS, Green J, Gallimore CI, Wright PA, Brown DW. 2000. Widespread environmental contamination with Norwalk-like viruses (NLV) detected in a prolonged hotel outbreak of gastroenteritis. Epidemiol. Infect. 125:93–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chung PW, Huang YC, Chang LY, Lin TY, Ning HC. 2001. Duration of enterovirus shedding in stool. J. Microbiol. Immunol. Infect. 34:167–170 [PubMed] [Google Scholar]
- 14. Dancer SJ. 2004. How do we assess hospital cleaning? A proposal for microbiological standards for surface hygiene in hospitals. J. Hosp. Infect. 56:10–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dancer SJ. 1999. Mopping up hospital infection. J. Hosp. Infect. 43:85–100 [DOI] [PubMed] [Google Scholar]
- 16. Dancer SJ, White LF, Lamb J, Girvan EK, Robertson C. 2009. Measuring the effect of enhanced cleaning in a UK hospital: a prospective cross-over study. BMC Med. 7:28–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Wit MA, et al. 2007. Large outbreak of norovirus: the baker who should have known better. J. Infect. 55:188–193 [DOI] [PubMed] [Google Scholar]
- 18. Donaldson KA, Griffin DW, Paul JH. 2002. Detection, quantitation and identification of enteroviruses from surface waters and sponge tissue from the Florida Keys using real-time RT-PCR. Water Res. 36:2505–2514 [DOI] [PubMed] [Google Scholar]
- 19. D'Souza DH, et al. 2006. Persistence of caliciviruses on environmental surfaces and their transfer to food. Int. J. Food Microbiol. 108:84–91 [DOI] [PubMed] [Google Scholar]
- 20. Duizer E, et al. 2004. Inactivation of caliciviruses. Appl. Environ. Microbiol. 70:4538–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Duizer E, et al. 2004. Laboratory efforts to cultivate noroviruses. J. Gen. Virol. 85:79–87 [DOI] [PubMed] [Google Scholar]
- 22. European Committee for Standardisation 2005. Chemical disinfectants and antiseptics. Virucidal quantitative suspension test for chemical disinfectants and antiseptics used in human medicine. Test methods and requirements (phase 2/step 1). European Standard NEN-EN 14476. European Committee for Standardisation, Brussels, Belgium [Google Scholar]
- 23. Food and Agriculture Organization/World Health Organization 2008. Viruses in food. Microbiological risk assessment series no. 13. Scientific advice to support risk management activities: meeting report. Food and Agriculture Organization/World Health Organization, Rome, Italy [Google Scholar]
- 24. Fraise AP. 2007. Decontamination of the environment. J. Hosp. Infect. 65(Suppl 2):58–59 [DOI] [PubMed] [Google Scholar]
- 25. Gallimore CI, et al. 2006. Environmental monitoring for gastroenteric viruses in a pediatric primary immunodeficiency unit. J. Clin. Microbiol. 44:395–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gibson KE, Crandall PG, Ricke SC. 2012. Removal and transfer of viruses on food contact surfaces by cleaning cloths. Appl. Environ. Microbiol. 78:3037–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Greatorex JS, et al. 2010. Effectiveness of common household cleaning agents in reducing the viability of human influenza A/H1N1. PLoS One 5:e8987 doi:10.1371/journal.pone.0008987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Green J, et al. 1998. The role of environmental contamination with small round structured viruses in a hospital outbreak investigated by reverse-transcriptase polymerase chain reaction assay. J. Hosp. Infect. 39:39–45 [DOI] [PubMed] [Google Scholar]
- 29. Greening G. 2006. Viruses in foods, p 5–42 In Goyal SM. (ed), Human and animal viruses in food. Springer, New York, NY [Google Scholar]
- 30. Greig JD, Lee MB. 2012. A review of nosocomial norovirus outbreaks: infection control interventions found effective. Epidemiol. Infect. 140:1151–1160 [DOI] [PubMed] [Google Scholar]
- 31. Griffith CJ, Cooper RA, Gilmore J, Davies C, Lewis M. 2000. An evaluation of hospital cleaning regimes and standards. J. Hosp. Infect. 45:19–28 [DOI] [PubMed] [Google Scholar]
- 32. Hall AJ. 2012. Noroviruses: the perfect human pathogens? J. Infect. Dis. 205:1622–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Harris JP, Lopman BA, O'Brien SJ. 2010. Infection control measures for norovirus: a systematic review of outbreaks in semi-enclosed settings. J. Hosp. Infect. 74:1–9 [DOI] [PubMed] [Google Scholar]
- 34. Heim A, Ebnet C, Harste G, Pring-Akerblom P. 2003. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J. Med. Virol. 70:228–239 [DOI] [PubMed] [Google Scholar]
- 35. Hewitt J, Rivera-Aban M, Greening GE. 2009. Evaluation of murine norovirus as a surrogate for human norovirus and hepatitis A virus in heat inactivation studies. J. Appl. Microbiol. 107:65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ho MS, et al. 1989. Viral gastroenteritis aboard a cruise ship. Lancet ii:961–965 [DOI] [PubMed] [Google Scholar]
- 37. Kusumaningrum HD, et al. 2003. Tolerance of Salmonella Enteritidis and Staphylococcus aureus to surface cleaning and household bleach. J. Food Prot. 66:2289–2295 [DOI] [PubMed] [Google Scholar]
- 38. Lee J, Zoh K, Ko G. 2008. Inactivation and UV disinfection of murine norovirus with TiO2 under various environmental conditions. Appl. Environ. Microbiol. 74:2111–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lodder WJ, et al. 2012. Feasibility of quantitative environmental surveillance in poliovirus eradication strategies. Appl. Environ. Microbiol. 78:3800–3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lopman B, et al. 2012. Environmental transmission of norovirus gastroenteritis. Curr. Opin. Virol. 2:96–102 [DOI] [PubMed] [Google Scholar]
- 41. Maalouf H, et al. 2011. Strain-dependent norovirus bioaccumulation in oysters. Appl. Environ. Microbiol. 77:3189–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mbithi JN, Springthorpe VS, Boulet JR, Sattar SA. 1992. Survival of hepatitis A virus on human hands and its transfer on contact with animate and inanimate surfaces. J. Clin. Microbiol. 30:757–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miller M, et al. 2002. Norwalk-like virus outbreak in Canberra: implications for infection control in aged care facilities. Commun. Dis. Intell. 26:555–561 [PubMed] [Google Scholar]
- 44. Morter S, et al. 2011. Norovirus in the hospital setting: virus introduction and spread within the hospital environment. J. Hosp. Infect. 77:106–112 [DOI] [PubMed] [Google Scholar]
- 45. Ozawa K, Oka T, Takeda N, Hansman GS. 2007. Norovirus infections in symptomatic and asymptomatic food handlers in Japan. J. Clin. Microbiol. 45:3996–4005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Park GW, Linden KG, Sobsey MD. 2011. Inactivation of murine norovirus, feline calicivirus and echovirus 12 as surrogates for human norovirus (NoV) and coliphage (F+) MS2 by ultraviolet light (254 nm) and the effect of cell association on UV inactivation. Lett. Appl. Microbiol. 52:162–167 [DOI] [PubMed] [Google Scholar]
- 47. Repp KK, Keene WE. 2012. A point-source norovirus outbreak caused by exposure to fomites. J. Infect. Dis. 205:1639–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rosario RF, Kimbrough RC, Van Buren DH, Laski ME. 2006. Fatal adenovirus serotype-5 in a deceased-donor renal transplant recipient. Transpl. Infect. Dis. 8:54–57 [DOI] [PubMed] [Google Scholar]
- 49. Scheil W, Cameron S, Dalton C, Murray C, Wilson D. 1998. A South Australian Salmonella Mbandaka outbreak investigation using a database to select controls. Aust. N. Z. J. Public Health 22:536–539 [DOI] [PubMed] [Google Scholar]
- 50. Svraka S, et al. 2009. Novel approach for detection of enteric viruses to enable syndrome surveillance of acute viral gastroenteritis. J. Clin. Microbiol. 47:1674–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tellier R. 2009. Aerosol transmission of influenza A virus: a review of new studies. J. R. Soc. Interface 6(Suppl 6):S783–S790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Teunis PF, et al. 2008. Norwalk virus: how infectious is it? J. Med. Virol. 80:1468–1476 [DOI] [PubMed] [Google Scholar]
- 53. Thornley CN, Emslie NA, Sprott TW, Greening GE, Rapana JP. 2011. Recurring norovirus transmission on an airplane. Clin. Infect. Dis. 53:515–520 [DOI] [PubMed] [Google Scholar]
- 54. Tian P, Yang D, Mandrell R. 2011. Differences in the binding of human norovirus to and from romaine lettuce and raspberries by water and electrolyzed waters. J. Food Prot. 74:1364–1369 [DOI] [PubMed] [Google Scholar]
- 55. Tuladhar E, Bouwknegt M, Zwietering MH, Koopmans M, Duizer E. 2012. Thermal stability of structurally different viruses with proven or potential relevance to food safety. J. Appl. Microbiol. 112:1050–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tuladhar E, de Koning MC, Fundeanu I, Beumer R, Duizer E. 2012. Different virucidal activities of hyperbranched quaternary ammonium coatings on poliovirus and influenza virus. Appl. Environ. Microbiol. 78:2456–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tuladhar E, Terpstra P, Koopmans M, Duizer E. 2012. Virucidal efficacy of hydrogen peroxide vapour disinfection. J. Hosp. Infect. 80:110–115 [DOI] [PubMed] [Google Scholar]
- 58. van Duynhoven YT, et al. 2005. A one-year intensified study of outbreaks of gastroenteritis in The Netherlands. Epidemiol. Infect. 133:9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wu HM, et al. 2005. A norovirus outbreak at a long-term-care facility: the role of environmental surface contamination. Infect. Control Hosp. Epidemiol. 26:802–810 [DOI] [PubMed] [Google Scholar]
- 60. Yezli S, Otter JA. 2011. Minimum infective dose of the major human respiratory and enteric viruses transmitted through food and the environment. Food Environ. Virol. 3:1–30 [DOI] [PMC free article] [PubMed] [Google Scholar]