Abstract
High-throughput, culture-independent surveys of bacterial and archaeal communities in soil have illuminated the importance of both edaphic and biotic influences on microbial diversity, yet few studies compare the relative importance of these factors. Here, we employ multiplexed pyrosequencing of the 16S rRNA gene to examine soil- and cactus-associated rhizosphere microbial communities of the Sonoran Desert and the artificial desert biome of the Biosphere2 research facility. The results of our replicate sampling approach show that microbial communities are shaped primarily by soil characteristics associated with geographic locations, while rhizosphere associations are secondary factors. We found little difference between rhizosphere communities of the ecologically similar saguaro (Carnegiea gigantea) and cardón (Pachycereus pringlei) cacti. Both rhizosphere and soil communities were dominated by the disproportionately abundant Crenarchaeota class Thermoprotei, which comprised 18.7% of 183,320 total pyrosequencing reads from a comparatively small number (1,337 or 3.7%) of the 36,162 total operational taxonomic units (OTUs). OTUs common to both soil and rhizosphere samples comprised the bulk of raw sequence reads, suggesting that the shared community of soil and rhizosphere microbes constitute common and abundant taxa, particularly in the bacterial phyla Proteobacteria, Actinobacteria, Planctomycetes, Firmicutes, Bacteroidetes, Chloroflexi, and Acidobacteria. The vast majority of OTUs, however, were rare and unique to either soil or rhizosphere communities and differed among locations dozens of kilometers apart. Several soil properties, particularly soil pH and carbon content, were significantly correlated with community diversity measurements. Our results highlight the importance of culture-independent approaches in surveying microbial communities of extreme environments.
INTRODUCTION
Bacteria and archaea are integral and diverse components of microbial communities in soil. Although these prokaryotes are ubiquitous in soil, their distribution, diversity, and community composition vary widely depending on both environmental and biotic factors (27, 49, 70). At large spatial scales, on the order of tens to thousands of kilometers, microbial community structure is correlated to edaphic variables, such as soil pH (26, 30) and moisture content (4). More locally, plant communities affect the contingent of soil microbes through interactions within the rhizosphere, the region of soil where microbial communities are directly influenced by plant root systems (9, 12). Unfortunately, the vast majority of soil microbes are recalcitrant to traditional isolation and culturing techniques (60), thereby preventing accurate examination of the diversity and degree of influence between biotic and abiotic factors on soil microbial communities. The advent of next-generation, multiplexed pyrosequencing techniques (40) enables more complete characterizations and comparisons of microbial communities (43, 50, 70).
Soil microbial communities are enormously diverse (27, 60) and show regional and environmental specificity in the prevalence of different bacterial groups (26, 50, 70). Numerous recent studies have highlighted how bacterial communities from soils formed under markedly different ecosystems harbor identifiably distinct communities (4, 7, 26, 50, 70). At smaller scales (tens of meters), different plant species within a single environment, indeed even cultivars of the same species, have been shown to have a strong influence on the diversity of microbial communities (9, 12, 55; reviewed in reference 41), suggesting the possibility that stochastic sampling of “bulk” soils may be influenced by the plants in the immediate sampling vicinity. In order to assess the influence of locality, plant species, rhizosphere association, and stochastic variation exerted on bacterial diversity, we designed a sampling regimen that takes advantage of multiplexed pyrosequencing approaches to increase the number of well-sampled biological replicates from different locales within a single ecosystem.
Arid regions, including deserts, arguably represent the single largest terrestrial ecosystem by surface area (25), yet little is known about the microbial communities inhabiting them. In arid environments, bacterial diversity is thought to be influenced by both abiotic factors, such as extreme fluctuations in temperature, elevated UV radiation, low-nutrient content, and low-soil-moisture content (15, 46), and biotic factors, such as plant abundance and species composition. In nutrient-poor desert soils, plants provide discrete, resource-rich habitats (2, 42, 71, 72). Plants are further known to exert a selective influence on soil microbes, harboring species-specific (1, 55) and cultivar-specific (32, 53, 73, 77) bacterial populations. In water- and nutrient-limited environments, the resource island hypothesis suggests that bacterial diversity should be greater in the rhizosphere than in the surrounding interplant soil (42), due to the accumulation of nutrients at the interface of root and soil (72). This rhizosphere effect is thought to be accentuated in deserts, both qualitatively and quantitatively, for nearly all metabolic types of bacteria (10), but few studies have directly tested this association. To further complicate the matter, plants also produce secondary metabolites that function to hinder the success of certain bacteria and likely reduce bacterial diversity to a selected subset of the microbial populations present in the surrounding soil (41, 54). Desert plants may therefore exert conflicting influences on microbial communities, simultaneously providing valuable nutritive resources and growth-inhibiting compounds, in addition to perpetuating beneficial microbes not necessarily present in the local soil microbial community. The current study aims to determine to what extent dominant desert cacti species influence the diversity of native microbial populations.
In this study, we survey bacterial communities in the Sonoran Desert by characterizing the relationship between bacterial diversity and abundance associated with dominant, long-lived desert plants (61). We determined the rhizosphere bacterial communities of the two largest species of Sonoran Desert columnar cacti of the Cactoideae subfamily: (i) the saguaro cactus, Carnegiea gigantea, with a natural range in southwestern Arizona and Sonora, Mexico (64); and its ecological equivalent, (ii) the cardón cactus, Pachycereus pringlei, with a disjointed distribution in Baja California and Sonora, Mexico. These two closely related cactus species have an overlapping geographic distribution in a small area of the gulf coast of western Sonora, Mexico, and display similar columnar growth, reproductive adaptations (including the same bat pollinators), and biochemical composition (20, 28, 29). Related to the resource island hypothesis, seedlings from both species are commonly found shielded by nurse plants (e.g., mesquite trees of the genus Prosopis), and it has been suggested that a rich bacterial diversity in such niches promotes the growth of the cacti (6). Also, other studies have suggested that root-colonizing endophytic bacteria help cacti thrive in inhospitable substrates, such as barren rocks (23, 68). These observations have suggested a symbiotic relationship between soil microbes and cactus hosts in extreme desert environments connecting the contribution of microorganisms in soil formation and ecological succession dynamics, including the establishment of these dominant cacti in natural and human altered desert environments.
In the present study, a multiplexed pyrosequencing approach was employed to examine the bacterial communities of interplant bulk soil and rhizosphere soil associated with the roots of saguaro cacti at two natural Sonoran Desert sites and from cardón cacti in the artificial desert environment in the coastal desert biome at Biosphere2 (59).
MATERIALS AND METHODS
Study site description and sample collection.
Samples were collected at three locations in the Sonoran Desert around Tucson, Arizona, an arid region with annual average air temperatures of 20.4°C (range, 6 to 38°C) and 30.9 cm of annual rainfall (http://www.wrh.noaa.gov/twc/climate/tus.php) (see Fig. S1A in the supplemental material). The first site, Tumamoc Hill (TH) (elevation, 737 m; 32°22′35.5″ N, 111°01′13.7″ W), is a 370-ha reserve located on a national historic landmark that encompasses the Desert Laboratory, an ecological research station and preserve with a 100-year legacy of research in desert ecology (34, 37). The second site, Finger Rock (FR) (elevation, 988 m; 32°34′21.7″ N, 110°90′90.7″ W), is located on a privately owned land parcel adjacent to Coronado National Forest in the Santa Catalina Mountains. The third site was located within the coastal fog desert biome of the Biosphere2 research facility (B2) (elevation, 1,165 m; 32°34′43.6″ N, 110°51′2.14″ W), an artificial environment that aims to reproduce natural conditions in a partially enclosed system (http://www.b2science.org). The coastal fog desert of B2 is a 1,400-m2 desert area containing 4,000 m3 of desert grassland soil from surrounding locales (59, 78).
Soil and rhizosphere samples were collected for microbial DNA extractions from three saguaro cacti (Carnegiea gigantea) in a randomly chosen 100-m2 plot from the FR and TH sites and from three cardón cacti (Pachycereus pringlei) in the coastal fog desert biome of B2. For each cactus, three replicate soil samples of approximately 100 g each were collected in sterile 50-ml Falcon tubes at a soil depth of 5 to 10 cm. We collected each sample from one of three equally spaced points located on the circle with a radius of 2.5 m circumscribing the base of the cactus, where no roots were visible (see Fig. S1B in the supplemental material). Additionally, one large bulk soil sample (∼500 g) was collected for each cactus and stored at 4°C to be analyzed for moisture content, grain size, pH, total organic carbon, and nitrogen. The direct pH and electrical conductivity of soil samples were measured by dissolving the soil sample in water at a 1:1 (wt/vol) ratio. A second measurement of pH using a 1:1 (wt/vol) suspension in a 0.01 M CaCl2 solution was also employed, because it gives a value similar to that for natural soil solution (containing dissolved Ca2+ and other ions that might displace the H+ ions attached to soil particles) (56). Percent carbon was determined by the amount of released CO2, and percent nitrogen was determined by the release of NO2 after combustion at 900°C. The cation exchange capacity (CEC) was determined from the decrease in Mg2+ concentration of a solution of MgSO4 after equilibration with the soil that was exchanged with Ba2+ (69).
Rhizosphere samples were collected by obtaining three small roots (∼3 to 5 mm in diameter and ∼10 cm in length) from each cactus 0.5 m away from the main trunk. Rhizosphere samples comprised 1 to 10 g of soil directly adhering to these roots. All samples were collected in sterile 50-ml Falcon tubes on ice and later stored at −20°C in the laboratory prior to DNA extraction. This sampling regimen resulted in 54 individual samples (three soil samples and three rhizosphere biological replicates from each of three cacti at three different sites) that were processed through multiplex sequencing.
DNA extraction.
For both soil and rhizosphere samples, 0.25 g of soil was placed in sterile 1.5-ml tubes and visually inspected to remove rocks and plant tissue. To maximize the lysis of Gram-positive bacteria, samples were resuspended in 150 μl sterile water, mixed with lysozyme (Sigma Co., St. Louis, MO) (final concentration of 0.1 μg/μl), and incubated at 37°C for 1 h. DNA was extracted using the PowerSoil DNA isolation kit (MOBIO Laboratories, Carlsbad, CA) following the manufacturer's protocol. Briefly, samples were homogenized and lysed by a combination of chemical agents and mechanical shaking via vortexing for 10 min. PCR inhibitors, such as humic acid, were removed by precipitation with the PowerSoil DNA isolation kit (MOBIO), and total genomic DNA was captured on a silica membrane and then washed and eluted in a volume of 100 μl.
PCR amplification and sequencing.
PCR amplification of the 16S rRNA gene and subsequent pyrosequencing were carried out at the Australian Centre for Ecogenomics (http://www.ecogenomic.org/) as previously described (21). Briefly, broad-specificity oligonucleotide primers 926F (F stands for forward) (5′-AAACTYAAAKGAATTGACGG-3′) and 1392R (R stands for reverse) (5′-ACGGGCGGTGTGTRC-3′) containing multiplex identifiers and LibL adaptor sequences (see Table S1 in the supplemental material) were used to generate amplicons spanning the hypervariable regions of the 16S rRNA gene from V6 to V9 (22). PCR was performed on 20 ng of sample DNA using a PCR mix containing 0.2 μl Taq polymerase, 5 μl buffer, and a 10 μM concentration of both forward and reverse primers. Denaturation was initiated at 95°C for 3 min, followed by 30 cycles, with 1 cycle consisting of 90°C for 30 s, annealing at 55°C for 30 s, and elongation at 74°C for 30 s, with a final extension step held at 74°C for 10 min. Amplicon PCR products were purified using QIAquick PCR purification columns (Qiagen Inc., Valencia, CA) and pooled in equimolar concentrations. The amplicon library was purified and sequenced on a Genome Sequencer FLX Titanium pyrosequencer using the manufacturer's protocols (Roche, Brandford, CT).
Sequence analysis.
16S rRNA amplicon sequences were initially processed and analyzed using Pyrotagger (http://pyrotagger.jgi-psf.org/cgi-bin/index.pl) as described by Kunin and Hugenholtz (48). Briefly, the barcodes and amplicon primer sequences were removed, and reads with more than one unknown nucleotide (N), reads with ≥3% of bases with Phred values of <27, and reads with a length greater than 2 standard deviations away from the mean read length were removed. At this point, custom PERL scripts were used to determine the number of reads from each multiplexed sample. We discovered that the number of reads recovered from two of our soil samples (those from one cactus each at the FR and B2 sites) were significantly fewer than expected and as observed in other samples. Accordingly, we excluded all reads from these two cacti, both soil and rhizosphere in subsequent analyses. We were left, therefore, with two sites (FR and B2) that had samples from only two cacti, and a site (TH) with samples from three cacti for a total of 42 individual multiplexed samples from seven cacti. The remaining multiplexed reads were pooled together based on the soil versus rhizosphere status of the sample and the individual cactus from which the sample was collected. This resulted in 14 pooled samples, a soil and rhizosphere sample from each of seven cacti (3 cacti from TH, 2 cacti from B2, and 2 cacti from FR) (Table 1). Subsequent analyses on the resulting pooled reads were carried out using QIIME (version 1.2.1) (13).
Table 1.
Pyrosequencing statistics and alpha diversity measures of pooled samplesa
| Sample type | Sample site | Sample designationb | No. of reads after QCc | No. of nonchimeric reads | No. of observed OTUs | Alpha diversity measuresd |
Estimated coverage (%) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chao1 | Chao1 range (95% CI) | ACE | Phylogenetic diversity | Shannon diversity index (H′) | |||||||
| Soil | Tumamoc Hill | ST1 | 15,378 | 14,940 | 4,786 | 11,606 | 10,935–12,351 | 12,582 | 385.0 | 10.23 | 41.2 |
| ST2 | 10,384 | 10,088 | 3,372 | 8,434 | 7,853–9,092 | 9,619 | 282.3 | 9.74 | 40.0 | ||
| ST3 | 10,187 | 9,871 | 3,499 | 10,323 | 9,532–11,218 | 11,403 | 293.2 | 9.84 | 33.9 | ||
| Finger Rock | SF1 | 11,233 | 10,660 | 3,486 | 8,182 | 7,648–8,784 | 8,891 | 293.7 | 10.09 | 42.6 | |
| SF2 | 14,320 | 13,714 | 4,376 | 9,533 | 9,005–10,120 | 10,477 | 347.8 | 10.36 | 45.9 | ||
| Biosphere 2 | SB1 | 15,506 | 14,801 | 5,569 | 15,049 | 14,186–15,999 | 16,897 | 470.8 | 10.74 | 37.0 | |
| SB2 | 13,446 | 12,895 | 4,603 | 11,712 | 10,999–12,506 | 12,741 | 392.9 | 10.60 | 39.3 | ||
| Rhizosphere | Tumamoc Hill | RT1 | 15,772 | 15,214 | 4,787 | 12,960 | 12,153–13,857 | 14,196 | 410.1 | 9.87 | 36.9 |
| RT2 | 13,310 | 12,778 | 4,340 | 11,848 | 11,084–12,699 | 13,872 | 371.3 | 9.70 | 36.6 | ||
| RT3 | 16,984 | 16,064 | 5,395 | 17,196 | 16,078–18,430 | 19,414 | 464.6 | 9.70 | 31.4 | ||
| Finger Rock | RF1 | 14,496 | 13,791 | 4,844 | 14,444 | 13,475–15,523 | 14,978 | 394.9 | 10.53 | 33.5 | |
| RF2 | 10,857 | 10,548 | 3,515 | 9,533 | 8,837–10,319 | 10,173 | 273.6 | 9.85 | 36.9 | ||
| Biosphere 2 | RB1 | 14,771 | 14,291 | 5,127 | 13,470 | 12,675–14,350 | 14,553 | 433.1 | 10.50 | 38.1 | |
| RB2 | 14,592 | 13,665 | 5,495 | 15,700 | 14,759–16,737 | 17,791 | 476.5 | 10.91 | 35.0 | ||
| Mean | 13,660 | 13,094 | 4,514 | 12,142 | 13,399 | 377.8 | 10.19 | 37.7 | |||
| Total | 191,236 | 183,320 | 36,162 | ||||||||
Three biological replicates were combined for each sample. Coverage is the percentage of estimated diversity, as measured by Chao1, represented in the observed number of OTUs.
The samples are named as follows: the first letter indicates whether the sample is from soil (S) or rhizosphere (R), the second letter indicates the site (B for Biosphere2, T for Tumamoc Hill, or F for Finger Rock), and the number indicates the sample number (first, second, or third sample from that source and site).
QC, quality control.
95% CI, 95% confidence interval; ACE, abundance-based coverage estimators.
Pooled sequences were denoised using Acacia (version 1.5), which algorithmically corrected pyrosequencing errors and removed reads with a length more than 2 standard deviations away from the mean read length (11). Of the original 198,550 pyrosequencing reads, 7,314 (3.68%) were removed due to aberrant read lengths, and a further 35,693 (17.98%) were corrected for pyrosequencing errors based on the Acacia algorithm. Operational taxonomic units (OTUs) for the remaining 191,236 usable, high-quality, denoised reads were clustered at 97% similarity using the uclust OTU picking method with the clustering algorithm set to furthest. The most-abundant reads from each OTU were aligned using the PyNAST algorithm (13), and taxonomic affiliations were assigned with the naïve Bayesian classifier of the Ribosomal Database Project (RDP) classifier (18, 79) with an 80% bootstrap confidence cutoff. Chimeric OTUs were detected with ChimeraSlayer (38) as implemented in QIIME and removed from subsequent analyses. Nonchimeric sequences were then filtered in QIIME using the greengenes core set lanemask to remove poorly aligned sequences, and a phylogenetic tree of the remaining OTUs was built using FastTree (67). We then generated an OTU table to show the relative abundances and RDP taxonomic assignments of sequences.
Diversity estimates.
To test for sampling effectiveness, rarefaction analysis was performed on the resulting OTU table. For alpha diversity (α diversity) measurements, we produced five replicate subsamples of the OTU table using the multiple_rarefaction.py script in QIIME under otherwise default parameters. We calculated ACE, Chao1, Shannon, and Simpson indices, and the phylogenetic distance (PD) measure along with the observed number of OTUs per sample. Community comparisons were performed with weighted and unweighted UniFrac (39) and Bray-Curtis distances. To remove the inherent heterogeneity of sampling depth, we first performed a single rarefaction that sampled 9,500 sequences from each pooled set of reads. This number was chosen as it is slightly less than the pooled sample with the fewest reads (i.e., Tumamoc Hill soil from cactus three, which had 9,871 reads), ensuring equal sampling among all communities. Principal coordinate analysis was performed in QIIME on both weighted and unweighted UniFrac distance matrices. To determine the robustness of clustering by the unweighted-pair group method using arithmetic means (UPGMA), jackknife beta diversity (β diversity) and clustering analyses were performed using 1,000 permutations sampling slightly less than 75% of the number of sequences from the least well surveyed sample (i.e., 7,500 reads). Samples were clustered using UPGMA, and a tree that included the bootstrap support values from the jackknife analysis was constructed.
Correlation and OTU significance tests.
Pairwise weighted and unweighted UniFrac distances between samples were correlated with the corresponding differences between physical and chemical soil characters using Spearman's rank test implemented in the R statistical environment. The OTU significance by category command in QIIME was used to test for correlations between OTU abundance and soil characteristics using either the analysis of variance (ANOVA) test for discrete variables (e.g., location and type) or Pearson correlation for continuous variables (e.g., pH and percent carbon). P values adjusted by Bonferroni's correction were used to show statistical significance in OTU abundance correlation tests performed in QIIME.
Accession number.
The 454 FLX Titanium flowgrams (sff files) have been submitted to the National Center for Biotechnology Information (NCBI) Sequence Read Archive database (accession no. SRA055936).
RESULTS
Sample collection and pyrosequencing.
We collected a total of 54 soil and rhizosphere samples (see Materials and Methods for a description of our hierarchical and replicate sampling strategy) from three sites: two outdoor locations in southern Arizona, Finger Rock (FR) and Tumamoc Hill (TH), and one inside the Biosphere2 (B2) desert biome. An analysis of soil properties from each location uncovered a wide range of values for soil pH, water content, nutrient availability, and particle size (see Table S2 in the supplemental material).
After pyrosequencing, all data for two cacti (one cactus each from FR and B2) were excluded from the analysis due to a disproportionately low number of reads (data not shown). From the remaining 42 multiplexed samples, we obtained 198,550 raw reads. A total of 7,314 of these reads were removed for having aberrant read lengths, and a further 35,693 reads were corrected for pyrosequencing errors via Acacia (11). ChimeraSlayer, as implemented in QIIME, removed a further 7,916 reads found in 3,234 chimeric OTUs, leaving 183,320 high-quality, denoised, nonchimeric reads, with a median coverage of 4,365 reads per sample (range, 3,396 to 5,661). We combined the three replicates from each cactus to obtain pooled samples in the range of 9,871 to 16,064 reads (median, 13,094) (Table 1). With a sequence similarity of 97%, we recovered 36,162 OTUs, of which 34,959 (96.7%) could be classified. Rarefaction analysis showed even sampling efforts between pooled rhizosphere and soil samples (see Fig. S2A in the supplemental material), with a small difference in the number of observed OTUs between samples pooled by the sample collection location (Fig. S2B). Here, B2 had a larger number of observed OTUs at comparable sampling efforts. Between samples pooled by individual cactus plants within a single location, we observed even sampling (Fig. S3). There was no identifiable bias in recovery of OTUs from different sites (range, 3,372 to 6,569), or between rhizosphere and soil samples (Fig. 1 and Table 1). By far, the dominant phylum across all pooled samples was Crenarchaeota, specifically from the class Thermoprotei, which accounted for 18.7% of all combined pyrosequencing reads (34,367 reads). Other highly represented phyla included Actinobacteria, Proteobacteria, Acidobacteria, Bacteroidetes, Planctomycetacia, and Firmicutes, which constituted an additional 47.4% of the remaining reads. Members of the phyla Euryarchaeota, Chloroflexi, Gemmatimonadetes, Nitrospira, Cyanobacteria, Thermomicrobia, and Verrucomicrobiae were also present in most samples, but at low abundance. The remaining reads comprised 10 additional phyla present at a very low abundance.
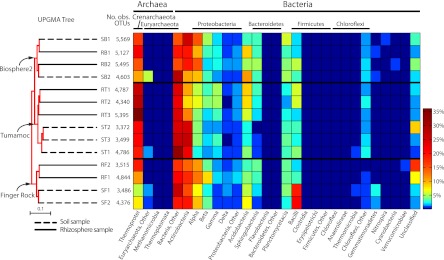
Fig 1.
Distribution heatmap of microbial orders arranged by hierarchical clustering of pooled soil and rhizosphere samples. The bootstrap tree on the left was created by the unweighted-pair group method using arithmetic means (UPGMA) and shows that the primary clustering of samples follows the collection site, with the samples from each site clustering together. The samples are named as follows: the first letter indicates whether the sample is from soil (S) or rhizosphere (R), the second letter indicates the site (B for Biosphere2, T for Tumamoc Hill, or F for Finger Rock), and the number indicates the sample number (first, second, or third sample from that source and site). The number of observed operational taxonomic units (No. obs. OTUs) is shown to the right of the UPGMA tree and to the left of the heatmap. Within the Finger Rock and Tumamoc Hill sites, rhizosphere and soil samples cluster together with 100% bootstrap support. Each pooled sample shows a high percentage of reads belonging to Thermoprotei and Actinobacteria. Similar distributions of bacterial and archaeal orders can be seen in the heatmaps of closely clustered samples, for example, the preponderance of bacilli in the soil samples at the Finger Rock site.
Diversity measures.
To compensate for stochastic sampling efforts and reduce the effects of variation among replicates (81), we pooled the reads from each of three biological replicates from both the rhizosphere and surrounding soil of individual cacti. This organization of data resulted in 14 pooled read sets—a rhizosphere set and soil set from each of seven cacti at the three locations (Table 1). Alpha diversity measurements of these pooled samples demonstrated a range of estimated diversities (see Fig. S4 in the supplemental material), but no obvious correlation between the type (i.e., rhizosphere versus soil) or site (Tumamoc Hill, Finger Rock, or Biosphere2) of samples and estimated levels of α diversity (Table 1). We recovered an average of 37.7% (coverage range, 31.4 to 45.5%) (Table 1) of the total estimated OTUs from each sample, suggesting that the true abundance of soil microbes in desert soils is far greater than what we recovered.
To compare communities between pooled samples, we used both weighted and unweighted UniFrac (51, 52). An initial indication of driving force for diversity measures can be seen in the hierarchical clustering tree produced from the unweighted UniFrac distance matrix (Fig. 1). The tree produced by the unweighted-pair group method using arithmetic means (UPGMA) was produced from 1,000 jackknife iterations, and the UPGMA tree shows that the principal driving force, i.e., the first level of branching organization on the tree, is the location from where the samples were taken. For example, all of the samples from the Finger Rock site (both soil and rhizosphere) group together with strong bootstrap support. With the exception of the samples taken from the Biosphere2 site, the next level of organization, as illustrated by the UPGMA tree, is whether a sample was taken from the rhizosphere or soil. This can readily be seen in the Tumamoc Hill and Finger Rock samples, where individual rhizosphere samples cluster together at the exclusion of soil samples and vice versa (Fig. 1). This suggests that a community of rhizosphere microbes is more similar to another sampled rhizosphere community in the near vicinity (on the order of ∼10 m) than the community of microbes present in the immediately surrounding soil (<1 m). We observed this trend with each pairwise metric of β diversity we calculated, including phylogenetic (weighted and unweighted UniFrac) and nonphylogenetic measures (Spearman rank distance, Bray-Curtis distance, and binary Jaccard distance) for the Tumamoc Hill and Finger Rock samples (see Fig. S5 in the supplemental material). Biosphere2 samples did not follow this trend; instead, the soil and rhizosphere samples from individual cacti clustered together.
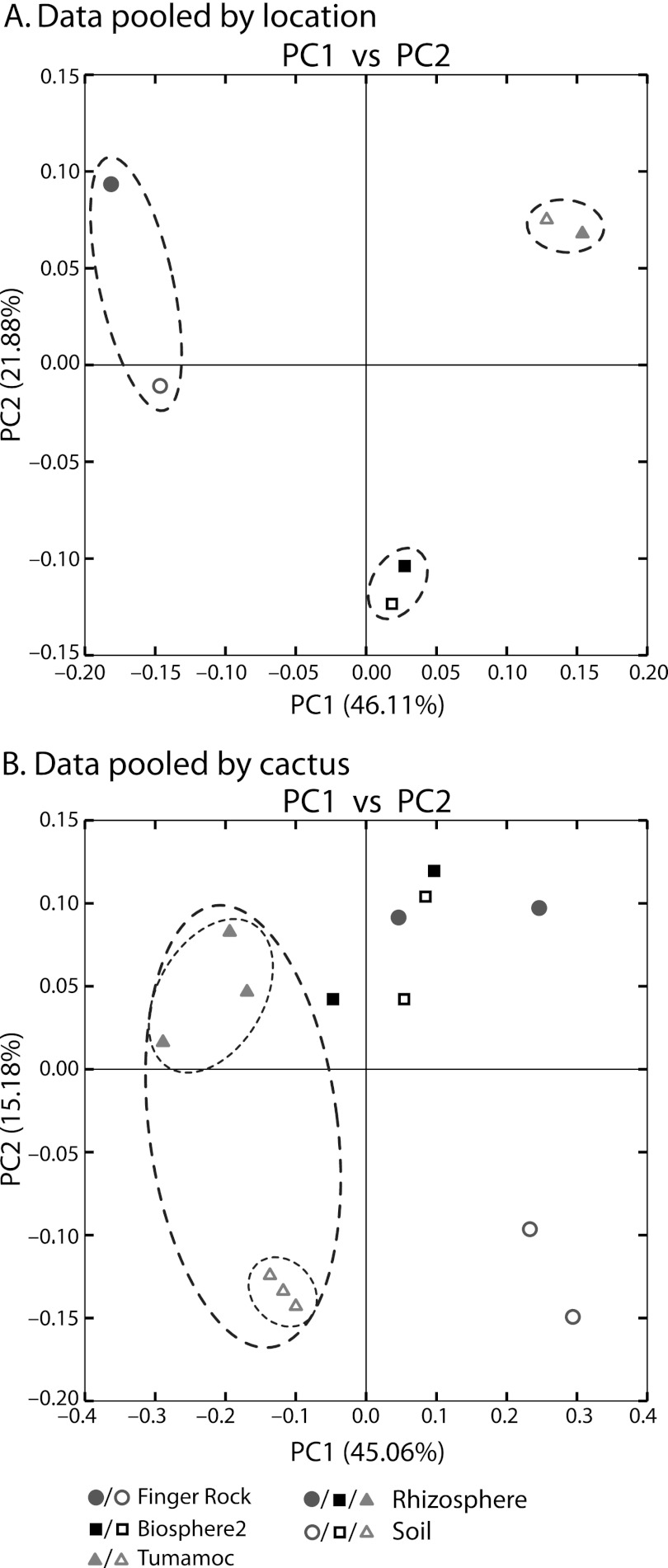
We pooled our samples in silico to determine the relative influences of sample location and type (i.e., rhizosphere or soil). The results of principal coordinate analysis on the weighted UniFrac distance matrix support our results from UPGMA clustering when data are pooled by location (Fig. 2A) and by individual cacti (Fig. 2B). When rhizosphere and soil samples are pooled by location, the first principal coordinate separates samples based on the geographic locale from which they were sampled (Fig. 2A). That is, the rhizosphere and soil samples from one location are more similar to each other than those of the same type collected several kilometers away. As expected, the same trend prevails, although with less stringency, with unpooled data as well—the first principal component separates samples based on location, and the second principal component separates samples based on soil or rhizosphere designation (see Fig. S6 in the supplemental material). When the data are pooled by the cactus from which samples were taken, the same general trend prevails (Fig. 2B), and location is the variable most explained by the principal coordinates. With the data pooled by cacti, the first principal component separates each sample based on location with the exception of one Finger Rock rhizosphere sample that nests within Biosphere2 samples (Fig. 2B). Replicating the pattern illustrated with UPGMA clustering, for samples from the Tumamoc Hill and Finger Rock locations, the individual rhizosphere and soil samples cluster together (Fig. 2B, ovals around Tumamoc Hill samples). This is less obvious with the samples from Biosphere2, which do not show the secondary level of organization that differentiates rhizosphere from soil samples (Fig. 2B).
Fig 2.
Principal coordinate analysis plots of data pooled in silico by location and by cactus. (A) The first principal coordinate (PC1) clearly separates pooled samples by the location from which they were collected, as indicated by the ovals around pooled samples from the same collection site. (B) Soil and rhizosphere samples were pooled by the cactus from which they were collected and show separation based primarily on geographic location and secondarily by soil type (i.e., rhizosphere or soil). PC1 separates the samples first by collection site, with the exception of one sample from Finger Rock that nests within those from Biosphere2. PC2 separates samples by their association with the rhizosphere or bulk soil at both the Tumamoc Hill site (indicated by the small ovals within the large oval) and the Finger Rock site, but not from those samples collected at Biosphere2.
Shared OTUs and the core desert microbiome.
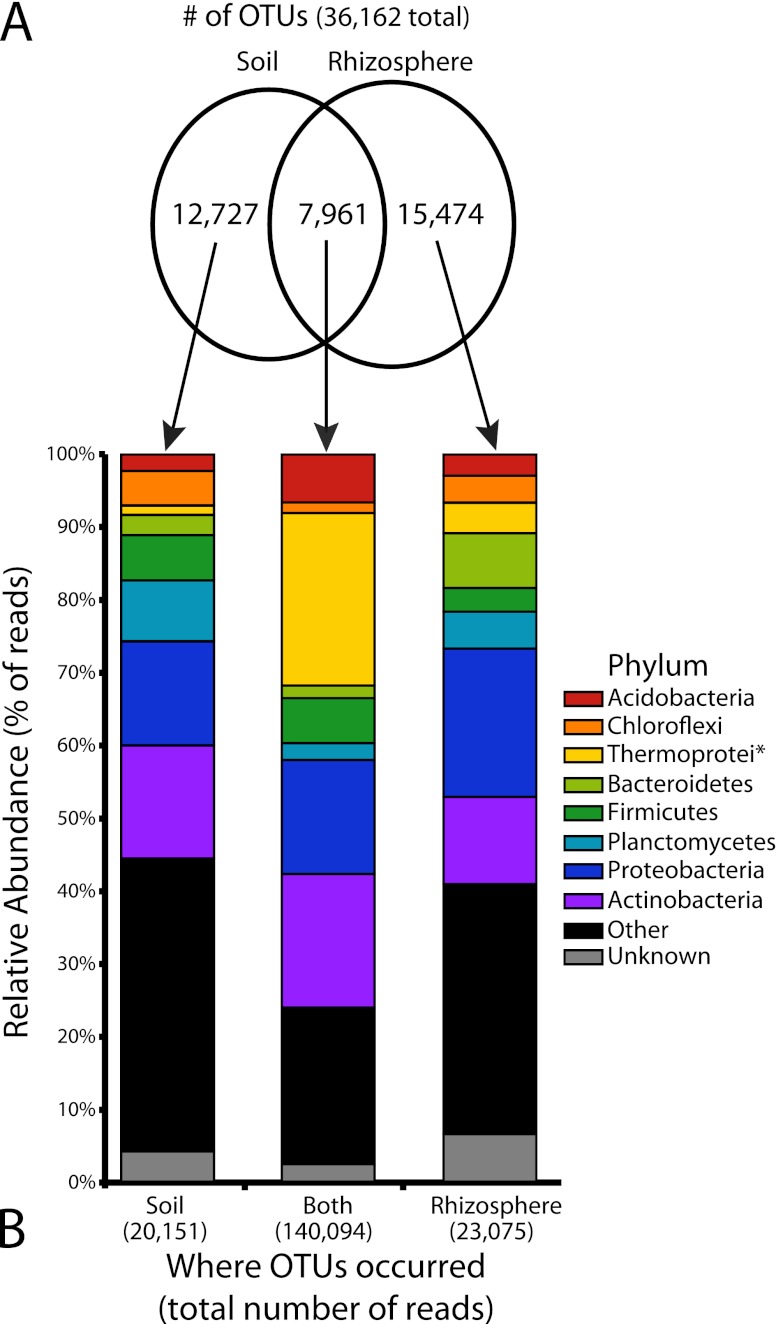
Of the 36,162 total observed OTUs, only 7,961 (22.0%) were found in both soil and rhizosphere samples. The soil and rhizosphere harbored 12,727 (35.2%) and 15,474 (42.8%) unique OTUs, respectively (Fig. 3A). Despite the fact that fewer than a quarter of the OTUs were found in both the soil and rhizosphere, these OTUs comprise 76.4% (140,094 of 183,320) of the total number of reads. This suggests that those OTUs shared between the soil and rhizosphere comprise very common desert microbial species, with a diverse fraction unique to each habitat and represented by rarer, habitat-specific taxa. The distribution of common phyla shows a dramatic increase in the percentage of reads attributed to Crenarchaeota, specifically the class Thermoprotei, compared to the soil and rhizosphere unique fractions (Fig. 3B). There is also an increase, albeit less pronounced, in the percentage of Acidobacteria present in the shared fraction. Other phyla show less remarkable differences between shared and unique microbes.
Fig 3.
Comparison of pooled soil and rhizosphere samples. (A) A proportional Venn diagram showing the number of operational taxonomic units (OTUs) found only in the soil (12,727), only in the rhizosphere (15,474), or in both locations (7,961). These OTUs were used to create the abundance bar graphs in panel B as indicated by the arrows. (B) The relative abundance of raw reads for different phyla belonging to the OTUs depicted in panel A. Note that the relatively smaller number of OTUs containing both soil and rhizosphere reads (7,961 or 22.0% of all OTUs) contain a disproportionately large percentage of the raw reads (140,094 of the 183,320 reads or 76.4%). The class Thermoprotei (phylum Crenarchaeota) is notably abundant and disproportionately overrepresented in this core set of OTUs. The asterisk after Thermoprotei indicates that this designation is at the level of class, not phylum like the other taxa listed.
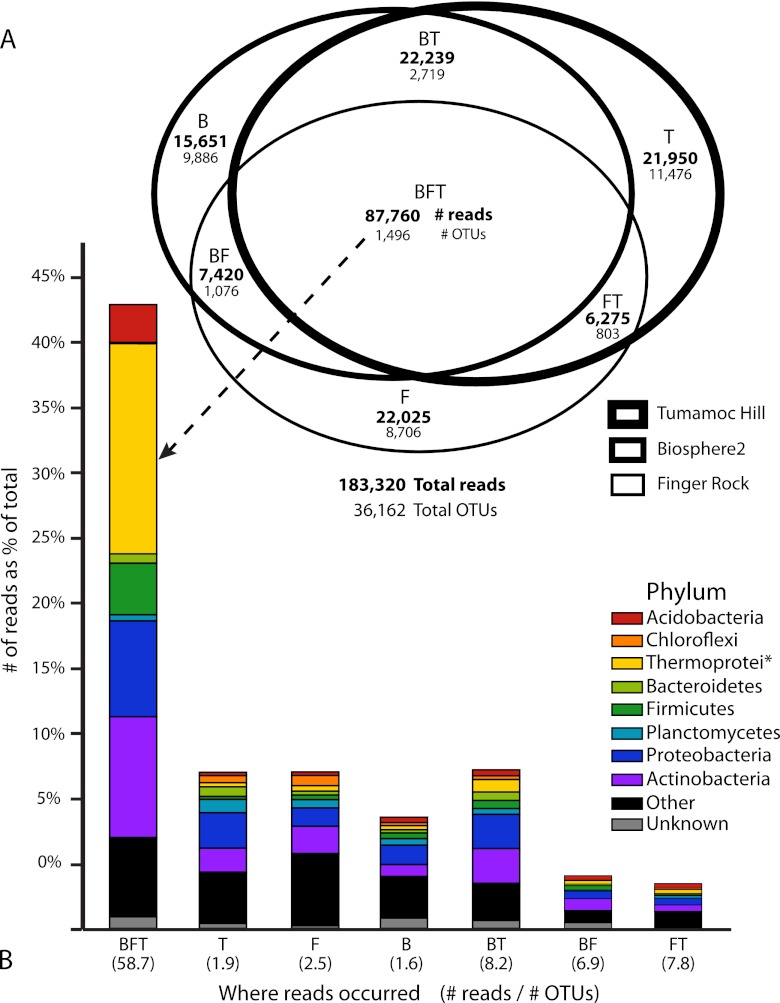
When we combined rhizosphere and soil samples together from each sampling location, we saw a similar trend (Fig. 4). A mere 4.1% (1,496 of 36,162) of the OTUs were shared between all three sampling locations but comprised 47.9% (87,760 of 183,320) of the total number of reads (Fig. 4A). This large fraction of common OTUs comprises a core desert microbiome for this study and is again dominated by the Archaea class Thermoprotei (Fig. 4B). The unique fractions, those found in only one of the three locations, are composed of the very rare OTUs. This can be seen by comparing the ratio of the number of reads to the number of OTUs for each portion of the Euler diagram shown in Fig. 4A. The core microbiome averages 58.7 reads per OTU, compared to the unique fractions that average around 2 reads per OTU (with a range of 1.6 at Biosphere2 to 2.5 at Finger Rock) (Fig. 4B). These rare variants comprise the vast majority (83.1%) of all OTUs (30,068 of 36,162) but only 32.5% of the total reads (59,626 of 183,320). The small numbers of both OTUs and reads associated with only two sites suggest that the core microbiome constitutes most of the shared taxa within the region as a whole. The exceptions are the taxa shared between the Tumamoc Hill and Biosphere2 sites, which are a sizeable portion of the total number of reads (22,239 or 12.1%).
Fig 4.
OTUs and raw reads pooled by the collection location. The collection locations were Biosphere2 (B), Finger Rock (F), and Tumamoc Hill (T). (A) A Venn diagram with set intersections proportional to the number of raw reads shows that the largest portion of reads is found in OTUs present at each collection site. Although this core set (BFT) contains only 1,496 OTUs (4.1% of the total number of OTUs), it contains the largest portion of reads (87,760 or 47.9%). (B) The number of reads for different phyla as a percentage of the total number of reads for each combination of sets as shown in panel A. The height of each bar is proportional to the number of reads. The class Thermoprotei (phylum Crenarchaeota) comprises 18.7% of all raw reads, the vast majority of which are in OTUs present at each site. The number in parentheses below each bar represents the ratio of raw reads to the number of OTUs for a given set. The OTUs unique to a particular site (i.e., B, F, or T) have few reads on average (1.6 to 2.5), suggesting that these unique OTUs represent a rare fraction of soil microbes. The asterisk after Thermoprotei indicates that this designation is at the level of class, not phylum like the other taxa listed.
Soil properties correlate with microbial diversity.
Several of the measured soil properties were significantly correlated (P < 0.05) with UniFrac distances of both soil and rhizosphere samples (Table 2). Two parameters of the surveyed desert soils that showed the most significant correlation with β diversity distances were pH (P < 0.00001 for soil and P < 0.01 for rhizosphere) and percent carbon (%C) (P < 0.05 for soil and P < 0.001 for rhizosphere). Interestingly, water content and the carbon-to-nitrogen ratio (C:N) showed few significant correlations, with the exception of only a few abundant OTU classifications that differed between cacti. Although the differences in pH between samples was significantly correlated with UniFrac distances (Table 2), only a few individual OTUs significantly correlated with pH, including several classes in the phylum Acidobacteria (P < 0.05). The archaeal family Desulfurococcaceae, one of the most highly abundant OTUs in each sample, was well correlated with percent carbon (P < 0.01).
Table 2.
Correlation of diversity measures of soil- and rhizosphere-associated microbes with soil characteristics
| Soil characteristic |
P value for correlationa |
|
|---|---|---|
| Soil | Rhizosphere | |
| % Clay | 0.11 | <0.01 |
| % Silt | <0.01 | <0.05 |
| % Sand | 0.06 | <0.01 |
| % Stones | 0.47 | 0.37 |
| pH | <0.00001 | <0.01 |
| pH (in 0.01 M CaCl2) | ≪0.00001 | <0.01 |
| EC waterb | <0.05 | <0.01 |
| Cation exchange capacity | <0.05 | <0.01 |
| H2O content | 0.41 | 0.38 |
| %C | <0.05 | <0.001 |
| %N | 0.28 | 0.85 |
| C:N | 0.62 | 0.89 |
Correlation of weighted UniFrac distances with differences in soil characteristics between samples. Significant values, as measured by Spearman's rank test, are indicated in boldface type.
EC water, electrical conductivity of the water in the soil.
DISCUSSION
Although amplicon pyrosequencing approaches have revolutionized studies on microbial community composition and diversity, it should be noted that studies employing this approach are subject to an ever-growing list of experimental and interpretive caveats. For example, pyrosequencing error rates (47) and clustering methods (44) have been shown to artificially inflate representatives of the rare biosphere, and PCR primer choice can influence diversity estimates (22). Moreover, a recent study by Zhou et al. (81) showed that experimental reproducibility using amplicon-based methods is low, with very little taxonomic overlap between technical replicates, and difficulties in reliably comparing communities using common β diversity measures. Our study aimed to alleviate some of these concerns and reduce the variability inherent in stochastic sampling by employing a collection scheme that pooled biological replicates in silico from the three soil or rhizosphere samples from each cactus. The use of biological replicates has been shown to reduce sampling artifact and improve quantitative analyses of α diversity and β diversity distances between samples (81).
In contrast to studies that rely on standard cloning methods and that consequently reveal modest levels of taxonomic diversity of microorganisms in desert soils (for example, see references 19, 21, 31, 57, 58, and 66 but see reference 26), our results showed that microbial communities in this extreme environment are both abundant and diverse. This is perhaps unsurprising, as other typically extreme environments likewise harbor diverse microbial communities (26, 45, 50, 70). Both α and β diversity measures are much higher than previous estimates for desert soil (e.g., compare Shannon diversity measures between the current results and the 2005 study of Nagy and colleagues [57]). Further, rarefaction analysis indicated that even by applying culture-independent high-throughput methods, we detected less than half of the species diversity likely present (see Fig. S1 and S2 in the supplemental material; coverage in Table 1). These diverse bacterial communities have potentially important implications for the abundance of bacterial pathogens in desert soils (76) and are likely to play an important role in soil formation, nutrient cycling, plant diversity, and primary productivity of desert environments (24, 45).
Our experimental design aimed to determine the relative influence of location, associated soil properties, and plant associations on microbial communities in the Sonoran Desert of southern Arizona at two different scales, tens and thousands of meters. Our replicate sampling approach enabled us to determine that the primary driver of microbial diversity and community associations is the general location from which samples are chosen. All of our samples were more similar to those collected within the surrounding 100-m2 collection area, regardless of whether they were collected from the rhizosphere or from the bulk soil. An ANOVA test for OTU abundance by location shows that only a few abundant OTUs are significantly associated with the principal location of samples, including the Thermoprotei order Desulfurococcales (P < 0.001) and the class Alphaproteobacteria (P < 0.05). This observation supports the idea that local soil characteristics shape the bulk soil and rhizosphere communities comparably by influencing several common taxa and that cacti have relatively less influence on the community of microbes found in the rhizosphere. This scale has implications for microorganisms affecting human health transported by wind in deserts (35) and for estimating and conserving the desert microbiome (33, 36, 63). Indeed, the microbial communities from the Tumamoc Hill and Biosphere2 sites, despite sampling the rhizosphere from two different cactus species, showed more similarities to each other than either did to those samples from the Finger Rock site, which sampled the same cactus species as at Tumamoc Hill.
Because of our limited sampling depth and high diversity of OTUs in our samples, we were unable to determine whether these spatial differences in microbial communities are due merely to differences in the relative abundance of OTUs between sites or actual differences in the contingent of OTUs. A recent deep sequencing study of marine microbial communities shows that differences in community composition in that environment is driven by changes in the relative abundance of a widely shared and dynamic set of OTUs, and not by temporal differences in the presence or absence of OTUs (14). Deeper sequencing will be necessary to determine whether similar dynamics determine community differences in Sonoran Desert soils.
Several physical and chemical soil properties were correlated with the differences between communities as measured by UniFrac, including pH, percent carbon, electric conductivity, cation exchange capacity, and even soil particle size distribution (e.g., percent silt and sand). Some of these characteristics have previously been implicated in correlations with diversity, particularly pH (17, 26, 50). Although the percent carbon in soil significantly correlated with β diversity measurements, neither the percent nitrogen (%N) nor the ratio of carbon to nitrogen (C:N) available in the soil showed significant correlation between rhizosphere or soil communities (in the rhizosphere, P = 0.85 for %N and P = 0.89 for C:N; in soil, P = 0.28 for %N and P = 0.62 for C:N). These observations suggest that available carbon, not nitrogen, is a limiting factor in driving local microbial diversity in these environments. Interestingly, given the scarcity of water in this desert ecosystem, available water content was not correlated with microbial diversity.
Only within the two natural sites surveyed did the presence of plant roots drive community structure enough to group rhizosphere and interplant samples into distinct communities based on UPGMA clustering and principal coordinate analysis. This suggests that, at the scale of a few meters, bacterial communities found in soil and associated with roots are not random samples from a common pool of species but that these two habitats differ in their microbial composition. As stated above, however, this conclusion may be premature and predicated on our limited sequencing depth. The failure of the artificial environment of Biosphere2 to replicate the pattern of more-natural environs hints that the latter scenario may be the case. It should also be noted that the soil characteristics of the two cacti sampled at Biosphere2 were very different from each other (see Table S2 in the supplemental material), as this artificial environment was created using nonhomogenous soil sources and is daily exposed to high levels of traffic, experimentation, and other human influences.
We did not find support for the resource island hypothesis related to species diversity or abundance. Although we observed relatively more OTUs in rhizosphere samples than in soil samples, the difference was not significant. This contradicts the view that a high nutrient concentration in the vicinity of roots results in higher abundance and diversity of microorganisms compared to interplant soil. Our results, however, show that rhizosphere and soil communities sustain distinct microbial taxa, as the vast majority of OTUs are found exclusively in one or the other communities, but not both. These observations suggest that seedlings of cacti growing under trees might benefit from many types of bacteria unique to the rhizosphere but found in low abundance, rather than by bacterial abundance or diversity per se, or by other physical factors (e.g., shade, temperature, nutrient availability) not related to the microorganisms.
Although archaea are now recognized as important and diverse contingents of many types of soil (4, 62), to our knowledge this is the first report to describe such a large fraction of Thermoprotei in desert soil samples. The direct contradiction to prior studies on soils similar to those from our study area (57), or in cold deserts (65), which failed to observe a significant proportion of Archaea, highlights the ability of pyrosequencing to more completely capture the diversity of nonculturable microbes. Our data suggest that Thermoprotei, particularly the family Desulfurococcaceae, may play an important role in the soils of the Sonoran Desert. Further experiments are necessary to confirm the veracity of our observations (i.e., test the possibility of primer bias for increased Thermoprotei amplification) and to fully elucidate the impact of Thermoprotei on the microbial communities of this region.
One observation consistent to all samples was the dominance of a core set of abundant taxa. This was true whether we were considering the overlap of soil and rhizosphere samples or geographic groupings. The core microbiome of this study has a different composition than those reads found only in individual samples, with the aforementioned preponderance of Thermoprotei. This is perhaps not unexpected, as Crenarchaeota contains radio- and thermotolerant species detected previously in other extreme environments, including desert soils (16, 21), hot springs (75), permafrost (80), and marine environments near the Sonoran Desert (8). The rare OTUs appear to be relatively similar in terms of the proportion of various phyla between sample sites. However, there are a large number of rare phyla that appear to be particular for a specific sample site. For example, the Firmicutes and Gemmatimonadetes taxa were far more abundant in soil samples at the Finger Rock site than other sites, suggesting that these rare variants exhibit a more stochastic distribution than the common and abundant core microbiome. This observation is in agreement with other recent pyrosequencing studies that have found soils to be dominated by a relatively small number of microbial taxa but that harbor a wide array of rare, yet highly diverse microbes (74).
Despite constituting an artificial environment primarily enclosed and isolated from the surrounding desert where the cardón cacti were introduced (3, 78), the contingent of soil microbes found in the coastal fog desert biome of Biosphere2 closely resemble natural surrounding communities dominated by saguaro cacti. Indeed, Biosphere2 shared a larger number of OTUs with the more distantly sampled and natural setting of Tumamoc Hill (22,239 reads from 2,719 OTUs) than the latter did with the other natural sampling site in our study, Finger Rock (6,275 reads from 803 OTUs). Interestingly, there was no more apparent significant differences between the soil and rhizosphere samples from Biosphere2 and the natural collection sites based on weighted UniFrac β diversity significance test, and overall OTU diversity measures were higher than at the natural sites. These two observations are consistent with the altered nature of this human-made environment. For instance, the diversity of the B2 samples could have been enhanced by microorganisms from a remote site where the cardóns were located before they were transplanted in addition to microorganisms from a diverse mix of local soils used to create the desert fog biome of B2. Members of the Thermoprotei were no less prevalent in Biosphere2 than in natural settings, highlighting the importance of this class in distinct locations of the Sonoran Desert ecosystem and on the two dominant columnar cacti species. These observations bode well for previous and future ecological studies in this large synthetic community (78), as soil microbial populations are known to exert a wide range of effects on plant communities and their ecological success (5).
Our results clearly indicate that culture-independent approaches are able to reveal large portions of previously undetected microbial communities and highlight the potential importance of these previously cryptic taxa to extreme environments. By far, the largest percentage of reads from both soil and rhizosphere samples were from unclassified Thermoprotei, suggesting that this abundant class of archaea plays a crucial, if previously unrecognized role in soil and rhizosphere ecology of the Sonoran Desert. The influence of one dominant plant type of the Sonoran Desert, the columnar saguaro and cardón cacti, on microbial communities was slight in comparison to local soil influences on abundant taxa, suggesting that the extreme physical factors in this environment play critical roles in shaping microbial communities. This study reveals a clear gap in the current understanding of desert microbial ecology and stresses that future studies should focus their efforts on isolating and classifying the abundant Thermoprotei, particularly the Desulfurococcales, so dominant in this community.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the support of the National Science Foundation (NSF)-funded Integrative Graduate Education and Research Training (IGERT; http://www.igert.org/) Program in Comparative Genomics at the University of Arizona, which funded this project. The researchers and staff at Biosphere2 were integral in providing funding for this work.
The advice and guidance of Matthew Sullivan were integral to this project's success. The Desert Laboratory of the University of Arizona was kind enough to provide us access to sampling sites on Tumamoc Hill. The researchers and staff at Biosphere2 provided support and access to sampling sites in the desert biome of B2. Gene Tyson of the University of Queensland's Australian Centre for Ecogenomics provided much needed expertise in multiplexed pyrosequencing. Finally, the advice and suggestions of Alejandro Lopez Cortes and three anonymous reviewers greatly improved the manuscript.
Footnotes
Published ahead of print 10 August 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Acosta-Martínez V, Dowds S, Sun Y, Allen V. 2008. Tag-encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biol. Biochem. 40:2762–2770 [Google Scholar]
- 2. Aguilera LE, Gutierrez JR, Meserve PL. 1999. Variation in soil microorganisms and nutrients underneath and outside the canopy of Adesmia bedwellii (Papilionaceae) shrubs in arid coastal Chile following drought and above average rainfall. J. Arid Environ. 42:61–70 [Google Scholar]
- 3. Allen JP, Nelson M, Alling A. 2003. The legacy of Biosphere 2 for the study of biospherics and closed ecological systems. Space Life Sci. 31:1629–1639 [DOI] [PubMed] [Google Scholar]
- 4. Angel R, Soares MIM, Ungar ED, Gillor O. 2010. Biogeography of soil archaea and bacteria along a steep precipitation gradient. ISME J. 4:553–563 [DOI] [PubMed] [Google Scholar]
- 5. Badri DV, Weir TL, van der Lelie D, Vivanco JM. 2009. Rhizosphere chemical dialogues: plant-microbe interactions. Curr. Opin. Biotechnol. 20:642–650 [DOI] [PubMed] [Google Scholar]
- 6. Bashan Y, Salazar B, Puente ME, Bacilio M, Linderman R. 2009. Enhanced establishment and growth of giant cardón cactus in an eroded field in the Sonoran desert using native legume trees as nurse plants aided by plant growth-promoting microorganisms and compost. Biol. Fertil. Soils 45:585–594 [Google Scholar]
- 7. Bates ST, et al. 2011. Examining the global distribution of dominant archaeal populations in soil. ISME J. 5:908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beman JM, Popp BN, Francis CA. 2008. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2:429–441 [DOI] [PubMed] [Google Scholar]
- 9. Berg G, Smalla K. 2011. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68:1–13 [DOI] [PubMed] [Google Scholar]
- 10. Bhatnagar A, Bhatnagar M. 2005. Microbial diversity in desert ecosystems. Curr. Sci. 89:91–100 [Google Scholar]
- 11. Bragg L, Stone G, Imelfort M, Hugenholtz P, Tyson GW. 2012. Fast, accurate error-correction of amplicon pyrosequences using Acacia. Nat. Methods 9:425–426 [DOI] [PubMed] [Google Scholar]
- 12. Buée M, De Boer W, Martin F, van Overbeek L, Jurkevitch E. 2009. The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and some of their structuring factors. Plant Soil 321:189–212 [Google Scholar]
- 13. Caporaso JG, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Caporaso JG, Paszkiewicz K, Field D, Knight R, Gilbert JA. 2012. The Western English Channel contains a persistent microbial seed bank. ISME J. 6:1089–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cary SG, McDonald IR, Barrett JE, Cowan DA. 2010. On the rocks: the microbiology of Antarctic dry valley soils. Nat. Rev. Microbiol. 129:129–138 [DOI] [PubMed] [Google Scholar]
- 16. Chanal A, et al. 2006. The desert of Tataouine: an extreme environment that hosts a wide diversity of microorganisms and radiotolerant bacteria. Environ. Microbiol. 8:514–525 [DOI] [PubMed] [Google Scholar]
- 17. Chu H, Fierer N, Lauber CL, Knight JG, Grogan P. 2010. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ. Microbiol. 12:2998–3006 [DOI] [PubMed] [Google Scholar]
- 18. Cole JR, et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Connon SA, Lester ED, Shafaat HS, Obenhuber DC, Ponce A. 2007. Bacterial diversity in hyperarid Atacama desert soils. J. Geophys Res. 112:G04S17 doi:10.1029/2006JG000311 [Google Scholar]
- 20. Darling MS. 1989. Epidermis and hypodermis of the saguaro cactus (Cereus giganteus): anatomy and spectral properties. Am. J. Bot. 76:1698–1706 [Google Scholar]
- 21. Direito SOL, et al. 2011. A wide variety of putative extremophiles and large beta-diversity at the Mars Desert Research Station (Utah). Int. J. Astrobiol. 10:191–207 [Google Scholar]
- 22. Engelbrektson A, et al. 2010. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J. 4:642–647 [DOI] [PubMed] [Google Scholar]
- 23. Esther Puente M, Li CY, Bashan Y. 2009. Endophytic bacteria in cacti seeds can improve the development of cactus seedlings. Environ. Exp. Bot. 66:402–408 [Google Scholar]
- 24. Esther Puente M, Li CY, Bashan Y. 2009. Rock-degrading endophytic bacteria in cacti. Environ. Exp. Bot. 66:389–401 [Google Scholar]
- 25. Ezcurra E, et al. 2006. Chapter 1: Natural history and evolution of the world's deserts, p 148 In Ezcurra E. (ed), Global deserts outlook. United Nations Environment Programme, Nairobi, Kenya [Google Scholar]
- 26. Fierer N, Jackson RB. 2006. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U. S. A. 103:626–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fierer N, Lennon JT. 2011. The generation and maintenance of diversity in microbial communities. Am. J. Bot. 98:439–448 [DOI] [PubMed] [Google Scholar]
- 28. Fleming TH. 2006. Reproductive consequences of early flowering in organ pipe cactus, Stenocereus thurberi. Int. J. Plant Sci. 167:473–481 [Google Scholar]
- 29. Fogleman J, Danielson P. 2001. Chemical interactions in the cactus-microorganism-Drosophila model system of the Sonoran Desert. Am. Zool. 41:877–889 [Google Scholar]
- 30. Fulthorpe RR, Roesch LFW, Riva A, Triplett EW. 2008. Distantly sampled soils carry few species in common. ISME J. 2:901–910 [DOI] [PubMed] [Google Scholar]
- 31. Garcia-Pichel F, Johnson SL, Youngkin D, Belnap J. 2003. Small-scale vertical distribution of bacterial biomass and diversity in biological soil crusts from arid lands in the Colorado plateau. Microb. Ecol. 46:312–321 [DOI] [PubMed] [Google Scholar]
- 32. Germida JJ, Siciliano SD. 2001. Taxonomic diversity of bacteria associated with the roots of modern, recent and ancient wheat cultivars. Biol. Fert. Soils 33:410–415 [Google Scholar]
- 33. Gilbert JA, et al. 2010. The Earth Microbiome Project: meeting report of the “1st EMP meeting on sample selection and acquisition” at Argonne National Laboratory October 6th 2010. Stand. Genomic Sci. 3:249–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Goldberg DE, Turner RM. 1986. Vegetation change and plant demography in permanent plots in the Sonoran Desert. Ecology 67:695–712 [Google Scholar]
- 35. Griffin DW. 2007. Atmospheric movement of microorganisms in clouds of desert dust and implications for human health. Clin. Microbiol. Rev. 20:459–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Griffith GW. 2012. Do we need a global strategy for microbial conservation? Trends Ecol. Evol. 27:1–2 [DOI] [PubMed] [Google Scholar]
- 37. Guo Q. 2004. Slow recovery in desert perennial vegetation following prolonged human disturbance. J. Veg. Sci. 15:757–762 [Google Scholar]
- 38. Haas BJ, et al. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21:494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hamady M, Lozupone C, Knight R. 2010. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 4:17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. 2008. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods 5:235–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hartmann A, Schmidt M, van Tuinen D, Berg G. 2009. Plant-driven selection of microbes. Plant Soil 321:235–257 [Google Scholar]
- 42. Herman RP, Provencio KR, Herrera-Matos J, Torrez RJ. 1995. Resource islands predict the distribution of heterotrophic bacteria in Chihuahuan Desert soils. Appl. Environ. Microbiol. 61:1816–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hirsch PR, Mauchline TH, Clark IM. 2010. Culture-independent molecular techniques for soil microbial ecology. Soil Biol. Biochem. 42:878–887 [Google Scholar]
- 44. Huse SM, Welch DM, Morrison HG, Sogin ML. 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 12:1889–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Johnson SL, Budinoff CR, Belnap J, Garcia-Pichel F. 2005. Relevance of ammonium oxidation within biological soil crust communities. Environ. Microbiol. 7:1–12 [DOI] [PubMed] [Google Scholar]
- 46. Jones SE, Lennon JT. 2010. Dormancy contributes to the maintenance of microbial diversity. Proc. Natl. Acad. Sci. U. S. A. 107:5881–5886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. 2010. Wrinkles in the rare biosphere: pyrosequencing errors lead to artificial inflation of diversity estimates. Environ. Microbiol. 12:118–123 [DOI] [PubMed] [Google Scholar]
- 48. Kunin V, Hugenholtz P. 21 February 2010. PyroTagger: a fast, accurate pipeline for analysis of rRNA amplicon pyrosequencing data. The Open Journal, article 1. http://www.theopenjournal.org/toj_articles/1
- 49. Kuske CR, et al. 2002. Comparison of soil bacterial communities in rhizosphere of three plant species and the interspaces in an arid grassland. Appl. Environ. Microbiol. 68:1854–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lauber CL, Hamady M, Knight R, Fierer N. 2009. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75:5111–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lozupone CA, Hamady M, Kelley ST, Knight R. 2007. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73:1576–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Manter DK, Delgado JA, Holm DG, Stong RA. 2010. Pyrosequencing reveals a highly diverse and cultivar-specific bacterial endophyte community in potato roots. Microb. Ecol. 60:157–166 [DOI] [PubMed] [Google Scholar]
- 54. Marilley L, Vogt G, Blanc M, Aragno M. 1998. Bacterial diversity in the bulk soil and rhizosphere fractions of Lolium perenne and Trifolium repens as revealed by PCR restriction analysis of 16S rDNA. Plant Soil 198:219–224 [Google Scholar]
- 55. Marschner P, Yang CH, Lieberei R, Crowley DE. 2001. Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol. Biochem. 33:1437–1445 [Google Scholar]
- 56. McLean EO. 1982. Soil pH and lime requirement, p 199–224 In Page AL. (ed), Methods of soil analysis. Part 2. Chemical and microbiological properties. Soil Science Society of America, Madison, WI [Google Scholar]
- 57. Nagy ML, Perez A, Garcia-Pichel F. 2005. The prokaryotic diversity of biological soil crusts in the Sonoran Desert (Organ Pipe Cactus National Monument, AZ). FEMS Microbiol. Ecol. 54:233–245 [DOI] [PubMed] [Google Scholar]
- 58. Navarro-Gonzalez R, et al. 2003. Mars-like soils in the Atacama desert, Chile, and the dry limit of microbial life. Science 302:1018–1021 [DOI] [PubMed] [Google Scholar]
- 59. Nelson M, Dempster W, Alvarez-Romo N, MacCallum T. 1994. Atmospheric dynamics and bioregenerative technologies in a soil-based ecological life support system: initial results from Biosphere2. Adv. Space Res. 14:417–426 [DOI] [PubMed] [Google Scholar]
- 60. Nocker A, Burr M, Camper AK. 2007. Genotypic microbial community profiling: a critical technical review. Microb. Ecol. 54:276–289 [DOI] [PubMed] [Google Scholar]
- 61. Núñez S, Martínez-Yrízara A, Búrquez A, García-Oliva F. 2001. Carbon mineralization in the southern Sonoran Desert. Acta Oecol. 22:269–276 [Google Scholar]
- 62. Oline DK, Schmidt SK, Grant MC. 2006. Biogeography and landscape-scale diversity of the dominant crenarchaeota of soil. Microb. Ecol. 52:480–490 [DOI] [PubMed] [Google Scholar]
- 63. Parker SS. 2010. Buried treasure: soil biodiversity and conservation. Biodiv. Conserv. 19:3743–3756 [Google Scholar]
- 64. Pierson EA, Turner RM. 1998. An 85-year study of saguaro (Carnegiea gigantea) demography. Ecology 79:2676–2693 [Google Scholar]
- 65. Pointing SB, et al. 2009. Highly specialized microbial diversity in hyper-arid polar desert. Proc. Natl. Acad. Sci. U. S. A. 106:19964–19969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Prestel E, Salamitou S, DuBow MS. 2008. An examination of the bacteriophages and bacteria of the Namib desert. J. Microbiol. 46:364–372 [DOI] [PubMed] [Google Scholar]
- 67. Price MN, Dehal PS, Arkin AP. 2010. FastTree 2 – approximately maximum likelihood trees for large alignments. PLoS One 5:e9490 doi:10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Puente ME, Bashan Y. 2004. Microbial populations and activities in the rhizoplane of rock-weathering desert plants. II. Growth promotion of cactus seedlings. Plant Biol. 6:643–650 [DOI] [PubMed] [Google Scholar]
- 69. Rhoades JD. 1982. Cation exchange capacity, p 149–157 In Page AL. (ed), Methods of soil analysis. Part 2. Chemical and microbiological properties. Soil Science Society of America, Madison, WI [Google Scholar]
- 70. Roesch LF, et al. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1:283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schlesinger WH, Pilmans AM. 1998. Plant-soil interactions in deserts. Biogeochemistry 42:169–187 [Google Scholar]
- 72. Schlesinger WH, Raikes JA, Hartley AE, Cross AF. 1996. On the spatial pattern of soil nutrients in desert ecosystems. Ecology 77:364–374 [Google Scholar]
- 73. Siciliano SD, Germida JJ. 1999. Taxonomic diversity of bacteria associated with the roots of field-grown transgenic Brassica napus cv. Quest, compared to the non-transgenic B. napus cv. Excel and B. rapa cv. Parkland. FEMS Microbiol. Ecol. 29:263–272 [Google Scholar]
- 74. Sogin ML, et al. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. U. S. A. 103:12115–12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Song ZQ, et al. 2010. Diversity of Crenarchaeota in terrestrial hot springs in Tengchong, China. Extremophiles 4:287–296 [DOI] [PubMed] [Google Scholar]
- 76. van Elsas JD, et al. 2012. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. U. S. A. 109:1159–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. van Overbeek L, van Elsas JD. 2008. Effects of plant genotype and growth stage on the structure of bacterial communities associated with potato (Solanum tuberosum L.). FEMS Microbiol. Ecol. 64:283–296 [DOI] [PubMed] [Google Scholar]
- 78. Walter A, Lambrecht SC. 2004. Biosphere 2 Center as a unique tool for environmental studies. J. Environ. Monit. 6:267–277 [DOI] [PubMed] [Google Scholar]
- 79. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wilhelm RC, Niederberger TD, Greer C, Whyte LG. 2011. Microbial diversity of active layer and permafrost in an acidic wetland from the Canadian high arctic. Can. J. Microbiol. 57:303–315 [DOI] [PubMed] [Google Scholar]
- 81. Zhou J, et al. 2011. Reproducibility and quantitation of amplicon sequencing-based detection. ISME J. 5:1303–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.