Abstract
The Athabasca oil sands deposit is the largest reservoir of crude bitumen in the world. Recently, the soaring demand for oil and the availability of modern bitumen extraction technology have heightened exploitation of this reservoir and the potential unintended consequences of pollution in the Athabasca River. The main objective of the present study was to evaluate the potential impacts of oil sands mining on neighboring aquatic microbial community structure. Microbial communities were sampled from sediments in the Athabasca River and its tributaries as well as in oil sands tailings ponds. Bacterial and archaeal 16S rRNA genes were amplified and sequenced using next-generation sequencing technology (454 and Ion Torrent). Sediments were also analyzed for a variety of chemical and physical characteristics. Microbial communities in the fine tailings of the tailings ponds were strikingly distinct from those in the Athabasca River and tributary sediments. Microbial communities in sediments taken close to tailings ponds were more similar to those in the fine tailings of the tailings ponds than to the ones from sediments further away. Additionally, bacterial diversity was significantly lower in tailings pond sediments. Several taxonomic groups of Bacteria and Archaea showed significant correlations with the concentrations of different contaminants, highlighting their potential as bioindicators. We also extensively validated Ion Torrent sequencing in the context of environmental studies by comparing Ion Torrent and 454 data sets and by analyzing control samples.
INTRODUCTION
Bituminous sands, or oil sands, are unconventional petroleum deposits where bitumen, a dense and extremely viscous form of petroleum, is found in combination with sand, clay, and water. One of the largest bitumen reservoirs, the Athabasca oil sands, is located in northeastern Alberta, Canada, along the Athabasca River. Here, hydrocarbons and associated contaminants occur naturally or anthropogenically. Erosion or groundwater mixing by the Athabasca River and its tributaries (e.g., the Ells River, Steepbank Creek, and Firebag Creek) as they pass through the Athabasca oil sands is responsible for the natural contamination of local aquatic ecosystems (14). With rising gas prices and technological advances, oil sands reserves can now be profitably extracted and upgraded to usable products. Oil sands mining operations in northern Alberta produce over 1.31 million barrels of oil per day, which is expected to rise to 3 million barrels per day by 2018 (http://www.energy.gov.ab.ca/OilSands/oilsands.asp). Obviously, an industrial operation of this scale will have environmental impacts. Environmental impacts, however, are difficult to evaluate, since natural bitumen erosion in the Athabasca River and its tributaries also leads to high levels of hydrocarbon-associated compounds in water and sediments. Although naphthenic acids (NAs) are seen as one of the most important chemical indicators of potential downstream anthropogenic effects (10, 35), they are difficult to measure and can occur naturally. An alternative to chemical indicators would be the use of microorganisms as bioindicators to track this pollution. Benthic microorganisms inhabiting river sediments are ideal for this purpose since they are frequently encountered and constantly exposed to the pollutants. Aquatic microbial communities are also highly sensitive to pollutants, even at very low concentrations (22–25, 46, 47), and as such, they are useful biological indicators of pollution.
Oil sands mining uses large quantities of water during the extraction process (up to 2,000 liters for each barrel of bitumen [6]), and this process water contains polycyclic aromatic hydrocarbons (PAHs) and NAs (18). Mining also produces large amounts of tailings, which consist of sand, clay, organics, and residual bitumen. Oil sands companies in Alberta operate under a zero-discharge policy, such that tailings are accumulated in settling basins. These basins currently contain over 840 million m3 of tailings (http://www.ercb.ca). To reduce water intake, most of the water used for bitumen extraction is recycled from the settling basins, further concentrating contaminants. Oil sands tailings ponds, therefore, are considered a major potential threat. Water in the tailings ponds has been shown to be toxic to a variety of organisms, including birds, fish, amphibians, and plants (5, 9, 27, 31). Of concern is the hydraulic connectivity between the ponds (which are often above grade, setting up a hydraulic head) and the at-grade or subsurface natural water bodies (see reference 17). Other environmental risks include atmospheric release of various trace elements and PAHs and their subsequent deposition in the Athabasca watershed following upgrading of bitumen to crude oil (20, 20a). Although the effects of oil sands mining on various macroorganisms are well documented, evidence on potential effects on aquatic microorganisms is scarce, other than the few studies that reported the toxicity of NAs to Bacteria and algae (14, 16). Microorganisms, which form the base of the food web in aquatic ecosystems, are key players in biogeochemical cycles. Accordingly, their response to pollution could have disproportionate impacts on biodiversity and ecosystem functioning. It is therefore crucial to understand their responses if the environment is to be protected or reclaimed.
For some time it has been known that microorganisms of the Athabasca River watershed have the potential to degrade components of bitumen (43, 44) and NAs (6), suggesting that indigenous microbial communities have the potential to remediate oil sands tailings. Complete remediation and reclamation of tailings ponds, however, are challenging and proceed slowly. This protracted process is mainly due to the low settling rates of the fine tailings and to the recalcitrance of NAs and the lower degradation efficiency of organic contaminants under anaerobic condition typically encountered in tailings ponds. New methods currently being developed might help speed up this process (14, 30). Still, without an in-depth knowledge of the microbial communities inhabiting the tailings ponds and the neighboring natural environments, it would be difficult to develop and evaluate effective bioremediation strategies.
The present study had three main objectives: (i) to evaluate the possible ecological impact of oil sands mining on microbial community structure in the oil sands mining region, (ii) to identify a set of microbial indicator taxa that might be used as indicators of oil sands-related pollution in natural environments, and (iii) to evaluate the accuracy, reliability, and usefulness of Ion Torrent sequencing for environmental microbiology studies. We therefore compared microbial communities in the fine tailings of two tailings ponds with sediments from sites in the Athabasca River and selected tributary (Firebag Creek, Steepbank Creek, and Ells River) sediments. Bacterial and archaeal communities were characterized using next-generation sequencing of 16S rRNA gene amplicons.
MATERIALS AND METHODS
Sampling sites.
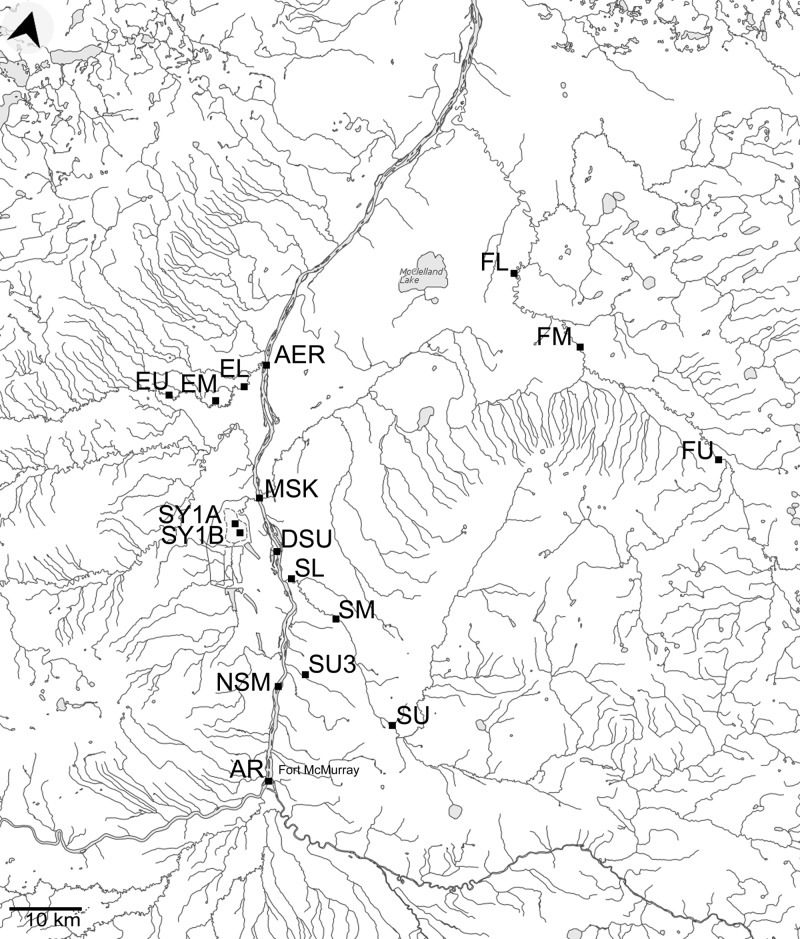
The Athabasca River stretches 1,231 kilometers from the Athabasca Glacier in west-central Alberta to Lake Athabasca in northeastern Alberta. Annual mean discharge downstream of Fort McMurray is 619 cubic meters per second with a maximum of 1,010 cubic meters per second (http://www.ec.gc.ca/eau-water/, station 07DA001; accessed 12 October 2011). The direction of the flow near Fort McMurray is from the south to the north. A single large 10-kg sample was taken from each of the following sites (17 samples): five sampling sites were chosen on the Athabasca River, including a reference site near Fort McMurray, upstream of the oil sands mining activities (AR); a site near the Northland Sawmill, close to mining activities (NSM); a site near Suncor mining activities (DSU); a site near the mouth of the Muskeg River, downstream of the mining activities (MSK); and finally, a site near the mouth of the Ells River, about 25 km downstream of the mining activities (AER). Nine other sampling sites were chosen in three of the Athabasca tributaries: upper (EU), middle (EM), and lower (EL) Ells River (slightly affected by mining activities); upper (FU), middle (FM), and lower (FL) Firebag Creek (not affected by mining activities); and upper (SU), middle (SM), and lower (SL) Steepbank Creek (strongly affected by mining activities). Three samples were taken from oil sands tailings ponds: sample SY1A was taken from the Mildred Lake settling basin at about the midpoint adjacent to the east shore, and sample SY1B was taken parallel to the east shore but further south in the basin. These positions are on a line transect that includes sites sampled by Penner and Foght (29). The third sample (Suncor 8A, sample SU3) was retrieved from a tailings pond adjacent to the Athabasca River which was considered full by 2006 and in the process of being transformed into a fine tailings drying site by 2010. The bottom of the tailings ponds consists of sand, silt, and clay fractions along with some bitumen in contact with overlying waters. Although it is reasonable to consider the geological materials in a tailings pond to be sediment, particularly given that in the region natural stream beds may essentially be bitumen, we note that the tailings being sampled are often referred to as mature fine tailings (MFTs). For simplicity's sake, all samples will further be referred to as “sediment.” See Fig. 1 for a map of the sampling sites and Table 1 for a description and the coordinates of the sampling sites.
Fig 1.
Sampling site locations. AR, Athabasca reference site; NSM, Northland Sawmill; DSU, downstream Suncor; MSK, Muskeg River; AER, mouth of Ells River; EU, EM, and EL, upper, middle, and lower Ells River, respectively; FU, FM, and FL, upper, middle, and lower Firebag Creek, respectively; SU, SM, and SL, upper, middle, and lower Steepbank Creek, respectively; SY1A and SY1B, Syncrude tailings ponds; SU3, Suncor tailings pond. (Template map source: Natural Resources Canada.)
Table 1.
Description, location, and physical characteristics of sediments from the Athabasca River, tributaries, and tailings ponds
| Sample | Description | Latitude | Longitude | Distance to nearest pond (km) | Nearest pond sample | % |
||
|---|---|---|---|---|---|---|---|---|
| Clay | Silt | Sand | ||||||
| AR | Athabasca reference site | 56.72 | 111.40 | 19.41 | SU3 | 19.8 | 12.0 | 68.2 |
| NSM | Athabasca, near mining activities | 56.87 | 111.44 | 4.69 | SU3 | 29.4 | 26.0 | 44.1 |
| DSU | Athabasca, near mining activities | 57.06 | 111.52 | 6.90 | SY1B | 18.6 | 9.2 | 72.2 |
| MSK | Athabasca, downstream of the mining activities | 57.12 | 111.60 | 5.30 | SY1A | 28.4 | 18.9 | 52.8 |
| AER | Athabasca, 25 km downstream of the mining activities | 57.31 | 111.67 | 25.41 | SY1A | 33.3 | 22.1 | 44.6 |
| SU | Tributary, strongly affected by mining | 56.86 | 111.13 | 16.16 | SU3 | 1.8 | 36.7 | 61.4 |
| SM | Tributary, strongly affected by mining | 56.99 | 111.34 | 10.69 | SU3 | 21.0 | 23.1 | 55.9 |
| SL | Tributary, strongly affected by mining | 57.02 | 111.47 | 10.80 | SY1B | 29.2 | 38.5 | 32.3 |
| EU | Tributary, slightly affected by mining | 57.23 | 111.89 | 22.69 | SY1A | 38.1 | 39.5 | 22.3 |
| EM | Tributary, slightly affected by mining | 57.24 | 111.77 | 19.41 | SY1A | 10.9 | 61.0 | 28.1 |
| EL | Tributary, slightly affected by mining | 57.26 | 111.72 | 21.12 | SY1A | 3.8 | 19.3 | 76.9 |
| FU | Tributary, not affected by mining | 57.34 | 110.48 | 73.15 | SU3 | 0.1 | 10.2 | 89.7 |
| FM | Tributary, not affected by mining | 57.44 | 110.89 | 59.67 | SY1A | 2.3 | 56.0 | 41.7 |
| FL | Tributary, not affected by mining | 57.52 | 111.11 | 57.75 | SY1A | 0.1 | 24.7 | 75.1 |
| SU3 | Tailings pond | 56.90 | 111.38 | 0.00 | 30.1 | 15.6 | 54.3 | |
| SY1A | Tailings pond | 57.08 | 111.64 | 0.00 | 27.6 | 15.1 | 57.3 | |
| SY1B | Tailings pond | 57.07 | 111.63 | 0.00 | 57.1 | 34.8 | 8.1 | |
Sediment sampling and physicochemical analyses.
Sampling was carried out between 22 September 2010 and 24 September 2010. At the moment of sampling, the water temperature was 6.5°C. Sediment samples from the Athabasca River and its tributaries were obtained using a stainless steel shovel from shore in less than 1 m of water, while the sediment samples from the tailings ponds were obtained using a long-handled scoop in 1 to 2 m of water. Sediment samples were collected and transferred to plastic bags; excess water was removed, although residual water content was retained. The bags were then closed and placed in plastic pails for transport. Samples were transported in coolers to the laboratory within 48 h, kept at 4°C, and subsampled for analyses within 10 days. Analysis of naphthenic acids in sediment and water matrices was carried out using liquid chromatography-mass spectrometry (LC-MS-MS) (15). Total petroleum hydrocarbons and polyaromatic hydrocarbons were determined for water and sediments using capillary gas chromatography coupled with flame ionization detection (GC-FID) and gas chromatography-mass spectrometry (GC-MS) (13, 38–40). Other sediment and water chemistry data were derived following standard methods for analyses (7).
DNA extraction and PCR amplification.
DNA was extracted from 10 g of sediment using a PowerMax soil DNA isolation kit (MoBio, Carlsbad, CA). Amplification for 454 and for Ion Torrent sequencing was carried out using the Fusion protocol. For 454 sequencing, bacterial 16S rRNA gene amplification was carried out as described by Bell et al. (1) using the forward primer Univ-9F (5′-GAG TTT GAT YMT GGC TC-3′) and the reverse primer BR534/18 (5′-ATT ACC GCG GCT GCT GGC-3′) (42) that were linked to Roche 454 adapters and the multiplex identifiers listed in Table S1 in the supplemental material. For Archaea 16S rRNA gene amplification, we used the following primers: forward primer 5′-CCA TCT CAT CCC TGC GTG TCT CCG ACT CAG XXX XXX XXX XAA TTG GAN TCA ACG CCG G-3′, first reverse primer 5′-CCT CTC TAT GGG CAG TCG GTG ATC GRC GGC CAT GCA CCW C-3′, and second reverse primer 5′-CCT CTC TAT GGG CAG TCG GTG ATC GRC RGC CAT GYA CCW C-3′, where the Xs represent the sample-specific multiplex identifier (listed in Table S1 in the supplemental material), the underlined sequences represent the template-specific sequences (958arcF, 1048arcRmajor, and 1048arcRminor, respectively, from reference 8), and the remaining sequence is the Ion Torrent adapter A (forward) and adapter trP1 (reverse). The two reverse primers were mixed in an equimolar ratio and used as the reverse primer in PCR. Reactions were carried out in 25-μl volumes containing 2 μl of template DNA, a 0.5 μM concentration of each primer, and 12.5 μl of KAPA2G Robust HotStart ReadyMix (KAPA Biosystems, Woburn, MA). Cycling conditions involved an initial 5-min denaturing step at 95°C, followed by 35 cycles of 30 s at 95°C, 30 s at 57°C, and 45 s at 72°C and a final elongation step of 10 min at 72°C. For Ion Torrent bacterial 16S rRNA sequencing, we carried out amplification using the forward primer BSF343/15 (5′-TAC GGR AGG CAG CAG-3′) and the reverse primer BR534/18 linked to the Ion Torrent adapters described above and the multiplex identifier (MID) listed in Table S1 in the supplemental material. We could not use the same primers for bacterial 454 and Ion Torrent sequencing, as the size of the fragments that can be amplified during the emulsion PCR step of the Ion Torrent library preparation is currently limited to ∼330 bp. Therefore, to minimize the amplification biases and maximize comparative power, amplifications were carried out on diluted amplicons that were used for bacterial 454 sequencing. Reactions were performed in 25-μl volumes containing 1 μl of template DNA, a 0.3 μM concentration of each primer, 0.4 mg/ml of bovine serum albumin, 0.2 mM deoxynucleoside triphosphates, and 0.05 U/μl of rTaq DNA polymerase (GE Healthcare, Baie d'Urfé, Canada). Cycling conditions involved an initial 5-min denaturing step at 95°C, followed by 25 cycles of 30 s at 95°C, 30 s at 55°C, and 45 s at 72°C and a final elongation step of 3 min at 72°C. All PCR products were purified on agarose gels using a QIAquick gel extraction kit (Qiagen, Valencia, CA) and quantified using a PicoGreen double-stranded DNA quantitation assay (Invitrogen, Carlsbad, CA). For each sequencing run, all the 17 amplification products from the different samples were pooled in an equimolar ratio, resulting in three pools (two for Bacteria and one for Archaea).
16S rRNA gene amplicon sequencing.
Bacterial 16S rRNA gene amplicons were sequenced on 1/4 of a plate using Roche 454 sequencing technology with FLX chemistry at the DNA Sequencing Facility of the University of Pennsylvania. Bacterial and archaeal 16S rRNA gene amplicons were sequenced in-house. A total of 3.50 × 107 molecules were used in an emulsion PCR using an Ion OneTouch 200 template kit (Life Technologies) and OneTouch and OneTouch ES instruments (Life Technologies, Carlsbad, CA) according to the manufacturer's protocol. The sequencing of the pooled library was performed using an Ion Torrent personal genome machine (PGM) system and a 314 chip (Archaea) or 316 chip (Bacteria) with the Ion Sequencing 200 kit according to the manufacturer's protocol. The bacterial 454 data set was used for most analyses, while the Ion Torrent bacterial data set was used to confirm the reliability of Ion Torrent sequencing of 16S rRNA gene amplicons. To evaluate the classification accuracy for Ion Torrent data, we added a positive control in the PCRs that was carried through the whole process, up to classification in the RDP database (see below). The positive control was genomic DNA extracted from a strain of Methanobrevibacter smithii (for Archaea) and an Alkanindiges sp. (for Bacteria). Amplicons from these positive controls were mixed with amplicons from environmental samples before sequencing, using 10 times fewer molecules than the amount used for environmental samples.
Escherichia coli DH10B resequencing.
Escherichia coli DH10B was sequenced using the Ion Control Materials 100 kit on a 314 chip, following the manufacturer's instructions. The quality metrics used were derived from the automated analysis carried out by the Torrent Suite (version 2.2).
Bioinformatic analyses.
The 16S rRNA gene amplicons were mostly treated using the RDP Pyrosequencing pipeline (http://pyro.cme.msu.edu/) (Bacteria) or a local Perl implementation of it (Archaea). For Bacteria, sequences having an average quality under 20 (45), having unidentified bases (Ns), not exactly matching the MID sequence, or being shorter than 100 bp were discarded. For Archaea, sequences having an average quality under 17, having Ns, not exactly matching the MID sequence, or being shorter than 75 bp were discarded. Following this filtering, the 454 Bacteria data set comprised 460,535 reads of an average of 222 bp for a total of 102 Mbp, the Ion Torrent Bacteria data set comprised 738,123 reads of an average of 173 bp for a total of 125 Mbp, and the Ion Torrent Archaea data set comprised 5,736 reads of an average of 101 bp for a total of 579 kbp. Remaining sequences were then submitted to the RDP classifier using a 0.5 bootstrap cutoff, as recommended for short sequences (4). For Bacteria, sequences matching plant plastids were removed from the data set and not taken into consideration in further analyses. For operational taxonomic unit (OTU) calculations, flowgrams from sff files were denoised and clustered using AmpliconNoise (32). Some parameters were adjusted for Archaea to meet the specifications of the Ion Torrent PGM, mainly the nonrepetitive flow order. Before performing AmpliconNoise calculations, data sets were normalized by randomly selecting 25,000 sequences for Bacteria and 700 sequences for Archaea. A 97% similarity cutoff was used, but similar trends in diversity were observed at lower cutoff values. Weighted-normalized Unifrac distances between each sample pair were calculated using the FastUnifrac website (12), based on the GreenGene core data set.
Statistical analyses.
Canonical correspondence analyses (CCA) were carried out in Canoco for Windows (version 4.5), with phylum/class data entered as supplementary variables (not included in the calculation of the ordination). All other statistical analyses were carried out in R (The R Foundation for Statistical Computing, Vienna, Austria). Pearson (r) and Spearman rank-order (rs) correlations were carried out using the cor.test function. Principal coordinate analyses (PCoA) were carried out using the cmdcsale function. Mantel tests based on Spearman correlations were performed using the mantel function, while the permANOVA program was performed using the adonis function of the vegan package. Geographic distances between sampling sites were derived from Global Positioning System (GPS) coordinates using the earth.dist function of the fossil package.
Nucleotide sequence accession numbers.
Sequence data were deposited in the NCBI Sequence Read Archive under accession number SRA057084.
RESULTS
Sediment physicochemical analyses.
We assessed the following variables: loss on ignition (LOI; percent organic matter), sediment granulometry, total petroleum hydrocarbons (TPHs), total straight-chain hydrocarbons (TSHs), total aromatic hydrocarbons (TAHs), naphthenic acids (NAs), and the sum of the 16 U.S. Environmental Protection Agency priority polycyclic aromatic hydrocarbons (EPA-PAHs; sum of naphthalene, acenaphthylene, acenaphthene, fluorene, phenanthrene, anthracene, fluoranthene, pyrene, benz[a]anthracene, chrysene, benzo[b]fluoranthene, benzo[k]fluoranthene, benzo[a]pyrene, dibenz[a,h]anthracene, benzo[ghi]perylene, and indeno[1,2,3-cd]pyrene). For TPHs, TSHs, TAHs, NAs, and EPA-PAHs, the values were much higher in tailings pond sediments than the other sampled sediments (Table 2). Concomitant higher organic matter in tailings pond sediments, except for the SY1A sample (Table 2), was also noted. The sediment clay percentage, TAHs, and EPA-PAHs were negatively correlated to distance from the closest tailings pond (rs = −0.548, P = 0.023; rs = −0.526, P = 0.030; and rs = −0.523, P = 0.031, respectively), indicating that these variables had higher values in ponds and in sediments found closer to the tailings ponds.
Table 2.
Chemical characteristics and bacterial and archaeal OTUs for sediments from the Athabasca River, tributaries, and tailings pondsa
| Sample | Bacteria OTU | Archaea OTU | LOI (%) | TPH concn (μg/g) | TSH concn (μg/g) | TAH concn (μg/g) | NA concn (mg/liter) | EPA-PAH concn (ng/g) |
|---|---|---|---|---|---|---|---|---|
| AR | 2132 | 611 | 2.1 | 251 | 104 | 147 | ND | 1,176 |
| NSM | 1971 | 628 | 2.2 | 105 | 52 | 53 | 1 | 189 |
| DSU | 2067 | 582 | 1.4 | 69 | 33 | 36 | ND | 128 |
| MSK | 2296 | 627 | 2.4 | 340 | 157 | 183 | 1 | 498 |
| AER | 2381 | 640 | 2.3 | 397 | 170 | 227 | 3 | 762 |
| SU | 1778 | 563 | 5.5 | 121 | 53 | 69 | 4 | 82 |
| SM | 1774 | 505 | 3.7 | 840 | 420 | 420 | 0 | 5,854 |
| SL | 1589 | 590 | 4.3 | 3,880 | 1,948 | 1,932 | 3 | 9,210 |
| EU | 1616 | 592 | 3.2 | 66 | 35 | 31 | 1 | 219 |
| EM | 2177 | 647 | 1.1 | 140 | 70 | 70 | 3 | 434 |
| EL | 1838 | 545 | 2.2 | 2,792 | 1,638 | 1,154 | 4 | 9,497 |
| FU | 2052 | 636 | 2.5 | 34 | 20 | 15 | 1 | 36 |
| FM | 1746 | 624 | 13.0 | 129 | 87 | 42 | 1 | 89 |
| FL | 1954 | 572 | 5.0 | 1,995 | 978 | 1,018 | 3 | 854 |
| SU3 | 1254 | 471 | 56.7 | 254,540 | 111,128 | 143,412 | 6 | 970,169 |
| SY1A | 239 | 549 | 3.5 | 36,200 | 15,900 | 20,300 | 14 | 176,972 |
| SY1B | 442 | 585 | 15.9 | 113,374 | 52,483 | 60,891 | 16 | 600,198 |
OTUs at 97% similarity. LOI, loss on ignition (total organic carbon); TPHs, total petroleum hydrocarbons; TSHs, total straight-chain hydrocarbons; TAHs, total aromatic hydrocarbons; NAs, naphthenic acids; EPA-PAHs, sum of the U.S. Environmental Protection Agency 16 priority polycyclic aromatic hydrocarbons (naphthalene, acenaphthylene, acenaphthene, fluorene, phenanthrene, anthracene, fluoranthene, pyrene, benz[a]anthracene, chrysene, benzo[b]fluoranthene, benzo[k]fluoranthene, benzo[a]pyrene, dibenz[a,h]anthracene, benzo[ghi]perylene, indeno[1,2,3-cd]pyrene); ND, not determined
Microbial community composition and diversity.
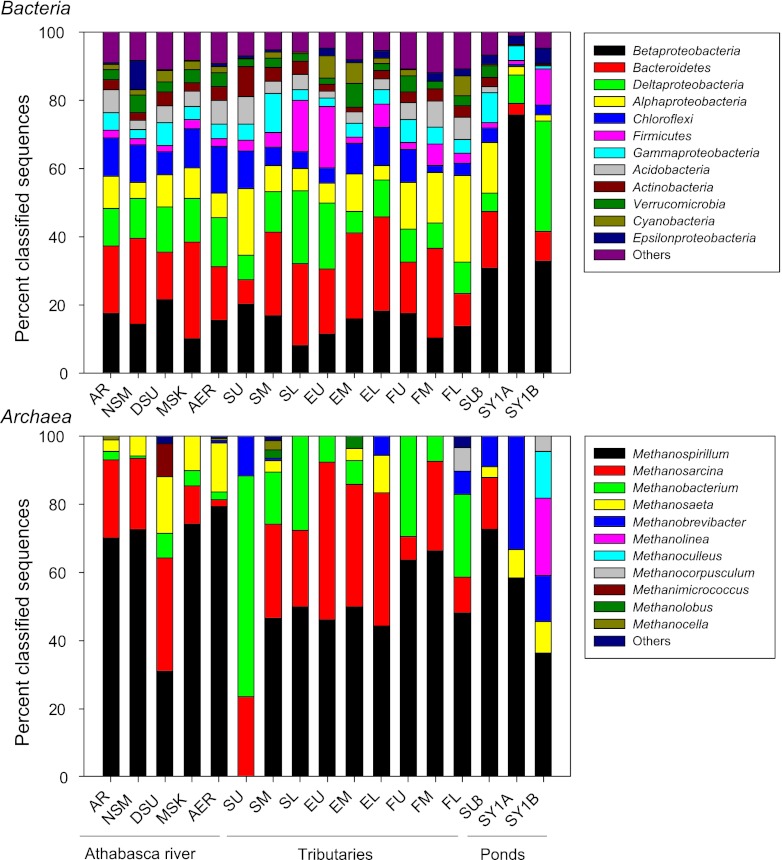
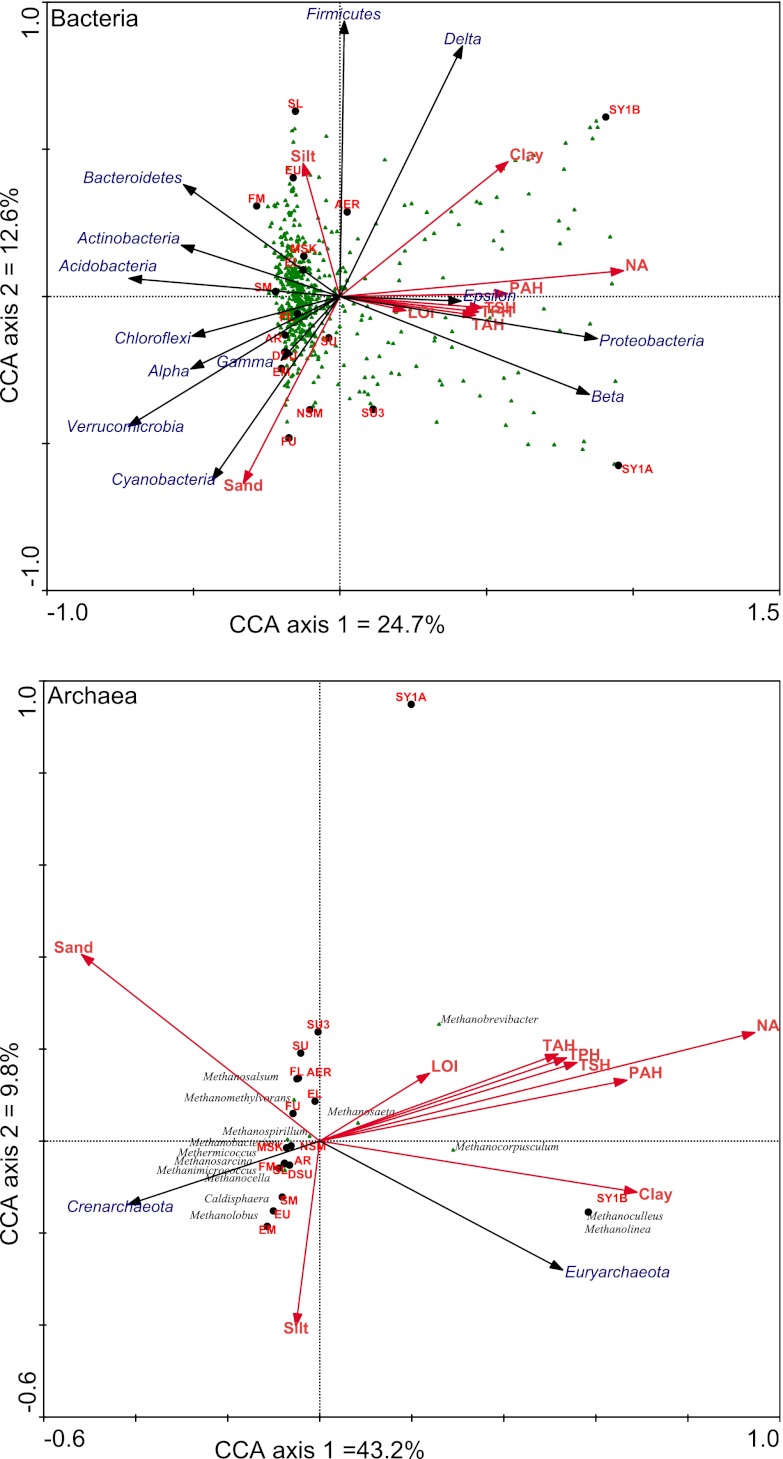
The bacterial 454 data set was used for most of the analyses, while the utility of Ion Torrent sequencing was verified by comparing the 454 and Ion Torrent data sets. The relative abundance at the phylum level varied considerably across the different samples (Fig. 2; see Table S2 in the supplemental material). The largest differences were between the tailings ponds (especially the Syncrude ponds) and the rest of the samples. For all samples, Proteobacteria was the dominant phylum, with Alphaproteobacteria and Betaproteobacteria being the most dominant classes within that phylum, with the exception of samples SL and EU, where Deltaproteobacteria dominated (Fig. 2). Bacteroidetes, Firmicutes, and Chloroflexi were also relatively abundant, but with large sample-to-sample variation (Fig. 2). At this level, no clear grouping of the samples could be observed relative to the sampling location (tributaries versus Athabasca River). Further illustrating the large differences between the tailings ponds and the river sediments, canonical correspondence analysis (CCA) based on genus-level information revealed a clear separation of the Syncrude tailings ponds from all the other samples on the first ordination axis of the plot (Fig. 3). Similar results were obtained by principal coordinate analysis (PCoA) based on Unifrac distances (see Fig. S1 in the supplemental material). This separation was explained by higher clay, NA, PAH, TPH, TSH, and TAH content. The Syncrude samples had relatively more Betaproteobacteria, Epsilonproteobacteria, and Deltaproteobacteria and relatively less of all other groups (Fig. 3). This relative increase in Betaproteobacteria and Deltaproteobacteria was primarily due to three taxa, Rhodoferax, Thiobacillus, and Smithella, which dominated in the Syncrude tailings ponds sediments but were at a lower relative abundance in the other samples (see Table S3 in the supplemental material). When tailings pond sediments were removed and analyses were rerun, sediment samples from the Athabasca River grouped together on the left side of the ordination plot, while all tributary sediment samples grouped on the right side. No other clear grouping, however, was observed (not shown). Bacterial diversity was significantly lower (one-way Kruskal-Wallis rank sum test, P = 0.01759) in the tailings ponds than in the Athabasca River sediments (Table 2). Tributary sediments harbored intermediate bacterial diversity, being lower in the Steepbank Creek sediments and in the upper Ells River sediments (Table 2). In the Athabasca River, sediments taken closest to the mining activities (NSM and DSU) showed the lowest diversity. The number of bacterial OTUs (97% similarity) was significantly correlated with LOI and TSHs (rs = −0.733, P = 0.00081 and rs = −0.525, P = 0.033, respectively).
Fig 2.
Community composition of Bacteria (derived from 454 sequencing) and Archaea (derived from Ion Torrent sequencing) for sediment samples of oil sands tailings ponds and of the Athabasca River and its tributaries. Refer to the legend of Fig. 1 for sample abbreviations. Others for Bacteria are OD1, BRC1, Deferribacteres, Fusobacteria, Caldiserica, TM7, Spirochaetes, Thermotogae, Nitrospira, Gemmatimonadetes, Chlorobi, Deinococcus-Thermus, Synergistetes, WS3, Planctomycetes, Tenericutes, OP11, Fibrobacteres, OP10, Lentisphaerae, Chlamydiae, and SR1. Others for Archaea are Methanosalsum, Caldisphaera, Methermicoccus, and Methanomethylovorans.
Fig 3.
Canonical correspondence analysis of the community structure of Bacteria (derived from 454 sequencing) and Archaea (derived from Ion Torrent sequencing) based on the relative abundance of the genera. Red arrows, environmental variables; black arrows, phylum/class relative abundance (entered as supplementary variables); black dots, samples; green triangles, individual genera. Refer to the legend of Fig. 1 for sample abbreviations.
The archaeal community was mainly composed of Euryarchaeota at the phylum level (58 to 100% of classified hits). The Syncrude tailings ponds were almost or completely devoid of Crenarchaeota (0 to 2.8%), while the remaining samples had relatively more (2.6 to 42%). Only 15 different archaeal genera could be identified, with most belonging to the Methanomicrobia class of the Euryarchaeota (Fig. 2). Two genera, Methanoculleus and Methanolinea, were observed exclusively in the SY1B sample, while Methanobacterium, although absent from the three tailings ponds, was present in almost all other samples (Fig. 2). Methanosarcina was absent from the two Syncrude tailings pond samples but present in all other samples (Fig. 2). CCA based on genus-level data resulted in a clear separation of the samples from the Syncrude tailings ponds from all other sediment samples (Fig. 3). Similar results were obtained by PCoA based on Unifrac distances (see Fig. S2 in the supplemental material). This separation was explained by higher clay, NA, PAH, TPH, TSH, and TAH content, which resulted in these samples having relatively more Methanobrevibacter, Methanocorpusculum, Methanoculleus, and Methanolinea and relatively fewer of the other groups (Fig. 3). Removing tailings ponds samples and rerunning the analyses did not result in clear clustering of samples (not shown). Archaeal diversity was relatively uniform across samples, and no significant differences were observed between tailings ponds, tributaries, and the Athabasca River sites (Table 2). The lowest archaeal diversity was observed in the Suncor tailings pond (SU3), and the highest was observed in the middle Ells River (EM) (Table 2). The number of archaeal OTUs (97% similarity) was significantly correlated with TPHs, TSHs, TAHs, and EPA-PAHs (rs = −0.539, P = 0.028; rs = −0.544, P = 0.026; rs = −0.551, P = 0.024; and rs = −0.544, P = 0.026, respectively).
Relationships with physicochemical and spatial data.
PermANOVA was used to test for significant relationships between physicochemical data and bacterial and archaeal community structure (Unifrac distances). We found similar relationships for Bacteria and Archaea, with clay and naphthenic acids having significant influences on the community structure (Bacteria clay, F = 2.96, P = 0.012; Bacteria NAs, F = 9.29, P = 0.001; Archaea clay, F = 3.06, P = 0.018; Archaea NAs, F = 6.74, P = 0.007). Naphthenic acids had the strongest influence (highest F ratio), followed by clay. None of the other variables significantly influenced the community structure. We also tested for correlations between bacterial and archaeal diversity and physicochemical variables. Bacterial diversity was negatively correlated to LOI and TSH (rs = −0.733, P = 0.00081 and rs = −0.525, P = 0.033, respectively), whereas archaeal diversity was negatively correlated to TPHs, TSHs, TAHs, and EPA-PAHs (rs = −0.539, P = 0.028; rs = −0.544, P = 0.026; rs = −0.551, P = 0.024; and rs = −0.544, P = 0.026, respectively).
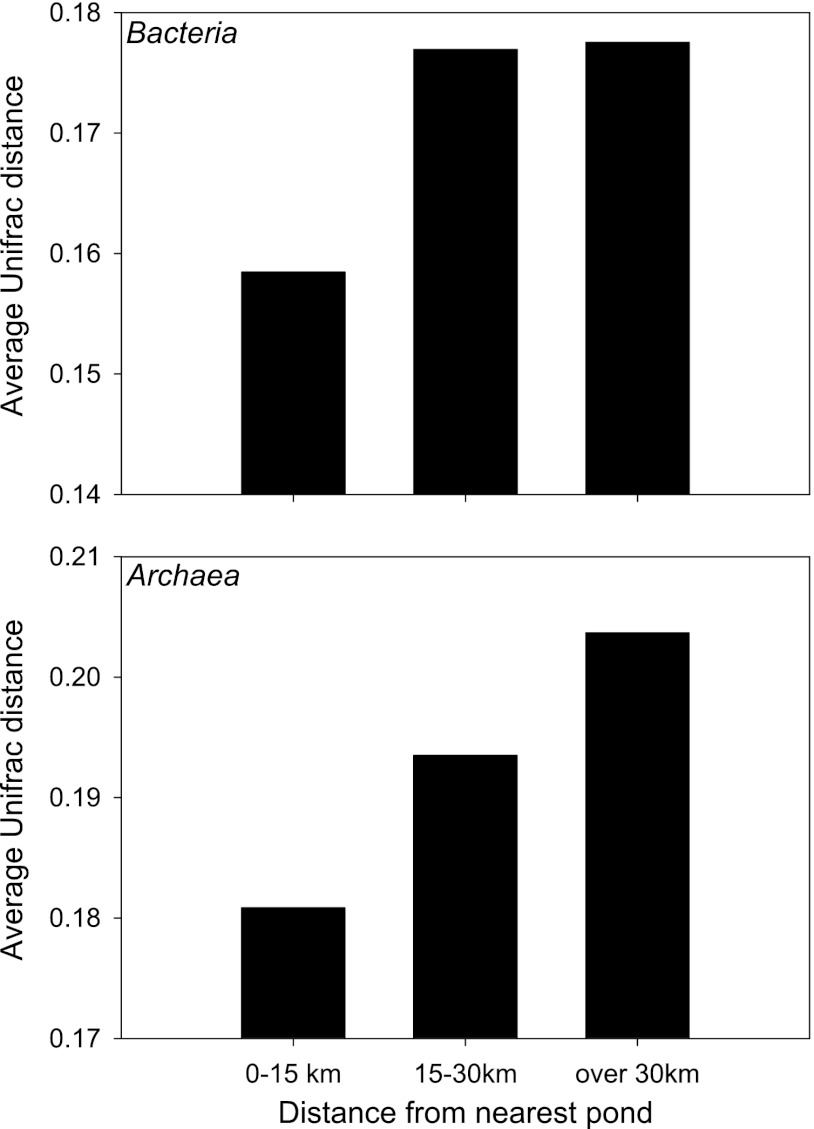
We found a significant relationship between bacterial and archaeal community composition and proximity to the tailings ponds, when proximity was defined as a distance of less than 10 km. This relationship was observed at the phylum and genus levels, as well as for the Unifrac distances calculated from sequence similarity. This observation indicates that the microbial communities from samples taken less than 10 km away from the tailing ponds are more similar to each other and to the tailings ponds communities than to the microbial communities of samples taken more than 10 km away. In contrast, this relationship was no longer significant when defining proximity as less than 25 km from the ponds, suggesting that the tailings ponds effect is highly localized. This effect is illustrated in Fig. 4, where the average Unifrac distance between a river or tributary sample and the closest tailings ponds is lower (indicating higher similarity) for samples taken less than 15 km away from the ponds than for samples taken 15 to 30 km or more than 30 km away from them. Although these differences were not significant because of the small number of samples in each class, this suggests that samples taken less than 15 km away from a tailings pond have microbial communities that are more similar to the ones inhabiting the tailings ponds sediments than samples taken further away.
Fig 4.
Average Unifrac distance between the bacterial communities (derived from 454 sequencing) of the Athabasca River and tributary sediment samples and the bacterial community of the nearest tailings pond sample. Each river-pond sample pair was classified into one of the three different geographic distance classes.
There was no correlation between geographic distance and similarity between microbial communities at any taxonomic level. This analysis is based purely on geographic distance derived from the GPS coordinates of the sampling sites (flying distance), and it indicates that the communities are not gradually getting more different with increasing distance. There were no significant correlations between bacterial or archaeal diversity and the geographic distance to the nearest tailings pond.
Potential indicator phyla/genera.
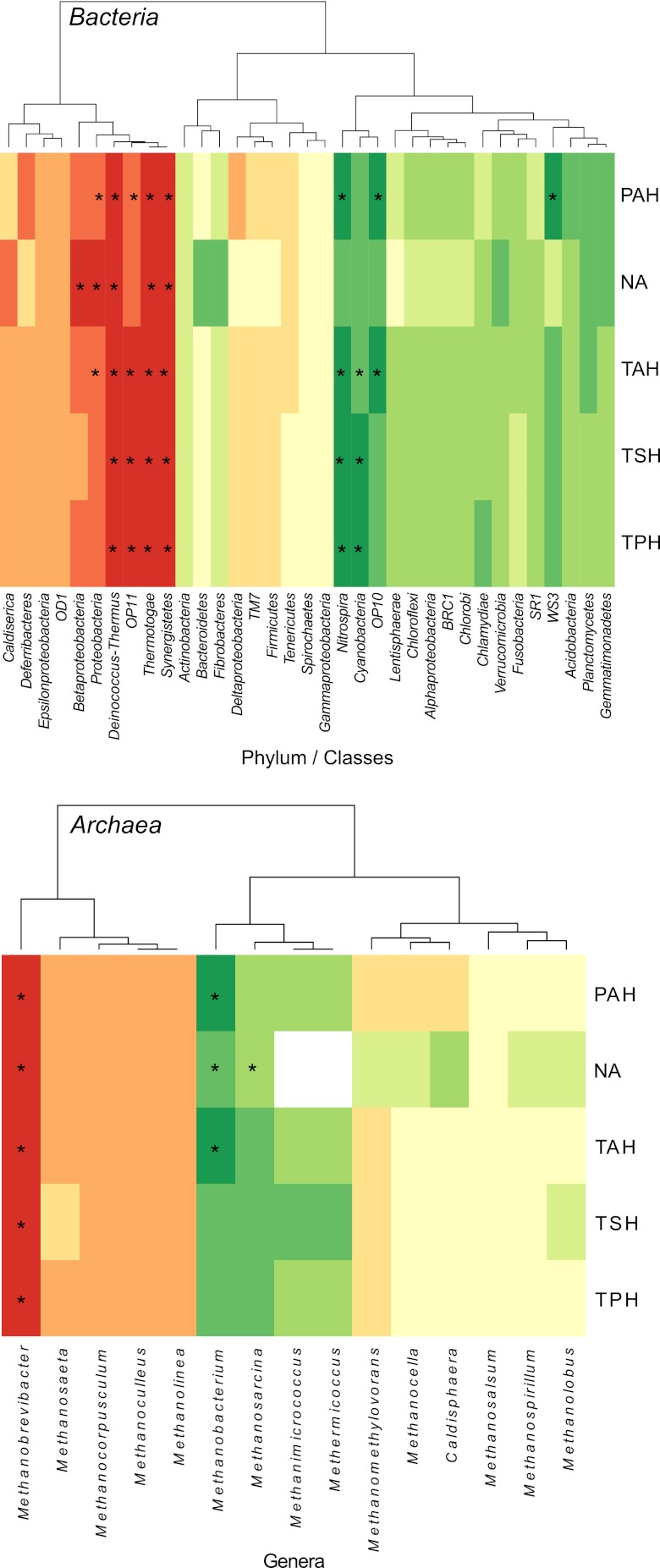
In order to highlight individual bacterial phyla/classes and archaeal genera that could be used as bioindicators, we carried out correlation analyses with sediment chemical data. Spearman (monotonous) correlations were carried out because the large differences in most physicochemical parameters between the tailings ponds and the river sediments would have produced spurious Pearson (linear) correlations. Among the dominant genera, the relative abundances of Proteobacteria and Betaproteobacteria were positively correlated with the concentration of naphthenic acids, while the abundance of Proteobacteria was also positively correlated to the concentrations of TAHs and EPA-PAHs (Fig. 5). The relative abundance of Cyanobacteria was negatively correlated with the concentrations of TPHs, TSHs, and TAHs. The relative abundance of Betaproteobacteria was negatively correlated with distance from the nearest tailings pond, with Betaproteobacteria being more abundant in or near tailings ponds (rs = −0.509, P = 0.037). In contrast, the relative abundance of Acidobacteria was positively correlated to the same distance, as described above (rs = 0.565, P = 0.018).
Fig 5.
Heat map based on Spearman correlations between the relative abundance of bacterial phylum/classes (derived from 454 sequencing) and Archaea genera (derived from Ion Torrent sequencing). Red, strong positive correlation; yellow, weak correlation; green, strong negative correlation; *, significant correlations (at P < 0.05).
Archaeal genera were mostly negatively correlated to chemical data, with the exception of Methanobrevibacter, which was positively correlated to the concentrations of TPHs, TSHs, TAHs, EPA-PAHs, and NAs (Fig. 5). Significant negative correlations included Methanobacterium to TAHs, NAs, and EPA-PAHs and Methanosarcina to NAs (Fig. 5). The relative abundance of Methanobacterium was positively correlated to the distance from the nearest tailings pond (rs = 0.680, P = 0.0027), being more abundant far from the tailings ponds.
Several bacterial genera (see Table S3 in the supplemental material) were significantly correlated with sediment chemical data, and we present the 10 strongest positive and negative correlations in Table 3. Six genera showed strong positive correlations with TPHs, TSHs, TAHs, EPA-PAHs, or NAs: Schumannella (Actinobacteria), Hydrogenophaga (Betaproteobacteria), Azonexus (Betaproteobacteria), Salinimicrobium (Bacteroidetes), Achromobacter (Betaproteobacteria), and Gillisia (Bacteroidetes) (Table 3). Four genera showed strong negative correlations with TPHs, TSHs, TAHs, NAs, or EPA-PAHs: Sorangium (Deltaproteobacteria), Hyalangium (Deltaproteobacteria), Rhodopila (Alphaproteobacteria), and Mesorhizobium (Alphaproteobacteria) (Table 3).
Table 3.
The 10 strongest positive and negative correlations between bacterial genera (derived from 454 sequencing) and hydrocarbon concentrationsa
| Correlation | r | P value | Genus | Chemical |
|---|---|---|---|---|
| Largest | ||||
| 1 | 0.827 | 0.0001 | Schumannella | TSHs |
| 2 | 0.819 | 0.0001 | Schumannella | TPHs |
| 3 | 0.794 | 0.0002 | Hydrogenophaga | EPA-PAHs |
| 4 | 0.787 | 0.0002 | Schumannella | TAHs |
| 5 | 0.783 | 0.0006 | Azonexus | NAs |
| 6 | 0.778 | 0.0002 | Schumannella | EPA-PAHs |
| 7 | 0.764 | 0.0004 | Salinimicrobium | EPA-PAHs |
| 8 | 0.763 | 0.0009 | Achromobacter | NAs |
| 9 | 0.742 | 0.0007 | Gillisia | EPA-PAHs |
| 10 | 0.731 | 0.0009 | Azonexus | EPA-PAHs |
| Smallest | ||||
| 1 | −0.821 | 0.0001 | Sorangium | TAHs |
| 2 | −0.815 | 0.0001 | Hyalangium | EPA-PAHs |
| 3 | −0.810 | 0.0001 | Hyalangium | TAHs |
| 4 | −0.805 | 0.0001 | Hyalangium | TSHs |
| 5 | −0.800 | 0.0001 | Hyalangium | TPHs |
| 6 | −0.784 | 0.0002 | Rhodopila | EPA-PAHs |
| 7 | −0.779 | 0.0003 | Sorangium | EPA-PAHs |
| 8 | −0.775 | 0.0004 | Sorangium | TPHs |
| 9 | −0.765 | 0.0005 | Sorangium | TSHs |
| 10 | −0.758 | 0.0004 | Mesorhizobium | EPA-PAHs |
TPHs, total petroleum hydrocarbons; TSHs, total straight-chain hydrocarbons; TAHs, total aromatic hydrocarbons; NAs, naphthenic acids; EPA-PAHs, sum of the U.S. Environmental Protection Agency 16 priority polycyclic aromatic hydrocarbons.
Ion Torrent sequencing.
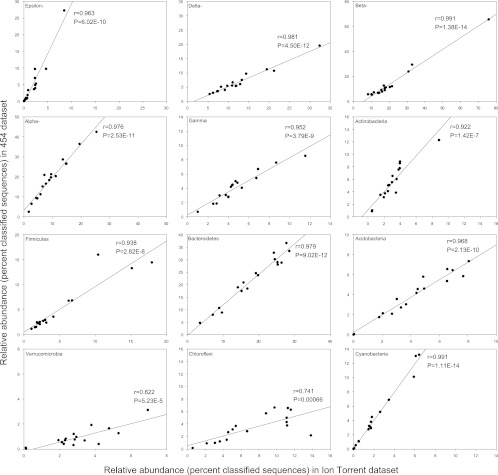
In order to confirm the suitability of Ion Torrent sequencing for microbial ecology studies, we generated two bacterial 16S rRNA gene data sets, one using 454 sequencing and one using Ion Torrent sequencing. The two regions amplified overlapped, and we analyzed these two data sets separately and compared the results generated from an ecological perspective. The relative abundances of the most dominant phyla and Proteobacteria classes derived from the two data sets were strongly correlated (Fig. 6). However, the slope of the curves differed, indicating some preferential amplification, most probably due to the different forward primer used. Unifrac-based PCoA ordinations revealed almost identical patterns in community structure derived from the 454 and Ion Torrent data, with the samples from the Syncrude tailings ponds clustering away from all other samples (see Fig. S1 in the supplemental material). The significant correlations found between taxa derived from the 454 data set and environmental variables were also found in a majority of cases for the data derived from the Ion Torrent data set (Table 4).
Fig 6.
Pearson correlation between the relative abundance of dominant phylum and Proteobacteria classes in 454 and Ion Torrent data sets. Epsilon-, Delta-, Beta-, Alpha-, and Gamma-, Epsilonproteobacteria, Deltaproteobacteria, Betaproteobacteria, Alphaproteobacteria, and Gammaproteobacteria, respectively.
Table 4.
Comparison of correlation between abundant phylum/classes and hydrocarbon concentrations for 454 and Ion Torrent data setsa
| Phylum/class | Chemical | 454 |
Ion Torrent |
||
|---|---|---|---|---|---|
| r | P | r | P | ||
| Betaproteobacteria | NAs | 0.680 | 0.005 | 0.599 | 0.018 |
| Proteobacteria | TAHs | 0.488 | 0.049 | 0.328 | 0.198 |
| Proteobacteria | NAs | 0.599 | 0.018 | 0.570 | 0.027 |
| Proteobacteria | EPA-PAHs | 0.490 | 0.048 | 0.221 | 0.393 |
| Cyanobacteria | TPHs | −0.522 | 0.034 | −0.517 | 0.036 |
| Cyanobacteria | TSHs | −0.556 | 0.022 | −0.542 | 0.027 |
| Cyanobacteria | TAHs | −0.490 | 0.048 | −0.502 | 0.042 |
Discordant values are in boldface. TPHs, total petroleum hydrocarbons; TSHs, total straight-chain hydrocarbons; TAHs, total aromatic hydrocarbons; NAs, naphthenic acids; EPA-PAHs, sum of the U.S. Environmental Protection Agency 16 priority polycyclic aromatic hydrocarbons.
To evaluate the reliability of the classification of 16S rRNA sequences generated by the Ion Torrent, the 16S rRNA gene of known archaeal and bacterial strains was amplified with fusion primers, pooled with environmental samples, sequenced, and analyzed in an identical manner as the environmental samples. For Archaea, from the 128 sequences retrieved, 93.8% (n = 120) were classified to the right genus (Methanobrevibacter), and the remaining sequences were not classified to the genus level. No misclassification occurred at the genus level. For Bacteria, from the 17,874 sequences retrieved, 17,412 (97.4%) were classified to the right genus (Alkanindiges) and 17,822 (99.7%) were classified to the right family (Moraxellaceae), with 408 (2.3%) of these Moraxellaceae sequences being unclassified at the genus level. Twelve sequences (0.07%) were wrongly classified in genera within the Proteobacteria but also in other phyla.
To evaluate the accuracy of Ion Torrent sequencing, we also carried out a resequencing of E. coli DH10B. This resulted in 713,430 sequences of 106 bp on average, for a total of 75.65 Mbp and a mean coverage depth of 16.14 times. Among these, 64.22 Mbp had phred-like quality scores of 20 or more. When taking the first 50 bp into account (653,330 mapped reads), 493,323 reads (76.9%) had perfect matches with the reference sequence, 108,005 reads (16.8%) had one mismatch with the reference sequence, and 40,061 reads (6.2%) had two or more mismatches with the reference sequence, resulting in a per base accuracy of 99.41%. When taking the first 100 bp into account (554,053 mapped reads), 320,534 reads (60.6%) had a perfect match with the reference sequence, 130,477 reads (24.7%) had one mismatch with the reference sequence, and 77,689 reads (14.7%) had two or more mismatches with the reference sequence, resulting in a per base accuracy of 99.47%.
DISCUSSION
One of the concerns regarding aquatic environments in the region are the tailings ponds that contain toxic compounds such as PAHs and NAs. Although the impact of these toxic substances on aquatic microbial life is not known, such knowledge could lead to the identification of bioindicator taxa, improved monitoring, and effective remediation strategies.
Ecological effects of oil sands mining operations on microorganisms.
The Bacteria and Archaea inhabiting the studied tailings ponds were significantly different (P < 0.05) from those found in river sediments. This observation is in line with previous PCR-denaturing gradient gel electrophoresis studies that reported a clustering of bacterial communities from affected ponds that were distinct from the communities in off-site wetlands (11). Furthermore, we showed here that bacterial and archaeal communities located close to the tailings ponds were more similar to each other and to those from the tailings ponds, suggesting that tailings might influence microbial communities in their direct vicinity. This effect was not observed when looking at samples taken further away from the ponds and is corroborated by the significant correlations found between distance to the closest pond and aromatic hydrocarbons. Thus, both chemical and microbiological data indicate that proximity to tailings ponds is influencing microbial communities in river sediments. This influence may be due to leaching or seepage of tailings water into the neighboring aquatic environments, since there is no deliberate discharge of tailings pond material to the environment. There are some indications of a potential hydraulic connectivity between the ponds (which are often above grade, setting up a hydraulic head) and the at-grade or subsurface natural water bodies (see reference 17), which could explain the results observed here. The correlation to tailings ponds is not just due to the presence of natural bitumen in the riverbeds or due to proximity to the Athabasca River, because all sites cut through oil sands formations and the sites very near the Athabasca River were also distinct from the sites nearest the ponds.
The bacterial and archaeal phyla and genera observed here in oil sands tailings sediments were similar to those previously observed in other direct 16S rRNA gene sequencing studies of Alberta oil sands tailings ponds. These included the genera Pseudomonas, Thauera, Rhodoferax, Acidovorax, Thiobacillus, and Brachymonas (29, 33, 37). However, as expected, bacterial diversity was lower in tailings ponds sediments where the concentration of toxic compounds was the greatest. It is also likely that the lower microbial diversity reflects the limited variety of carbon sources available, with mainly hydrocarbons available in the ponds but a potentially higher diversity of carbon sources available in natural environments. Bacterial diversity was also generally lower in sediments from Steepbank Creek, which is more heavily disturbed by mining operations than Ells River and Firebag Creek. This significant reduction in bacterial diversity may have repercussions on ecosystem processes and on the speed with which tailings pond reclamation takes place. Higher diversity means that a given process can be carried out under a wider range of environmental conditions, which often results in higher and more stable process rates in a fluctuating environment (36, 41). It has previously been suggested that a diverse microbial community would have to be present in oil sands tailings for remediation to take place (14).
Even though they were obtained from tailing ponds receiving the same type of material, the three pond sediment samples harbored distinct microbial communities. The Suncor pond was considered full by 2006 and is in the process of being transformed into a fine tailings drying site. This would indicate that the sediments from this pond would have a higher bulk density than those obtained from the active Syncrude sites. In fact, sediment size (clay percentage) had a significant effect on the microbial community structure, indicating that differences in bulk density might drive the differences observed between the pond samples by changing oxygen availability. The water depth in the Suncor pond is also shallow relative to the Syncrude site, which might again change oxygen availability in the sediments. Large differences within the Syncrude sediments were also observed, which is not surprising since sediment size within tailing ponds can vary substantially due to differences in originating material through time. These results indicate that there is a large within- and between-pond variability and that pond remediation strategies will probably need to be adapted to the specific pond conditions (e.g., aerobic versus anaerobic degraders).
Indicator taxa.
It is very difficult to chemically distinguish the heavy hydrocarbons produced during mining and the extracted hydrocarbons produced through upgrading activities from those that are naturally occurring in the Athabasca River ecosystem, since they originate from the same source. Naphthenic acids are seen as the most promising chemical indicators of potential downstream effects after release (10), and recent work suggests that it may be possible to identify NAs specific to a source (35). Microorganisms, however, are sensitive to low pollutant concentrations, and it is conceivable that they could serve as more accurate indicators for oil sands mining impacts. This study served as a stepping stone to identifying microbial bioindicators related to oil sands mining activities. Several of the measured chemical compounds appeared to have a negative influence on specific microbial taxa, probably linked to their toxicity, although nutrient-like effects cannot be ruled out. For instance, Cyanobacteria were negatively correlated to sediment hydrocarbon content, being relatively less abundant in tailings ponds and in river sediments with higher hydrocarbon content. In other studies, Cyanobacteria have been reported to be sensitive to low concentrations of pharmaceutical pollutants (47). As well, NAs have been shown to influence phytoplankton community composition (26). In aquatic ecosystems, Cyanobacteria are involved in major processes (C and N fixation) that are at the base of ecosystem productivity. A decrease in primary production resulting from a decreased presence of Cyanobacteria could also have a cascade effect on microbial communities.
In contrast, some taxa were positively correlated to hydrocarbon compounds and naphthenic acids, which could indicate either that these taxa were less affected by the toxicity of PAHs and NAs or that they might be able to use some of these compounds as carbon sources. Additional physicochemical factors might also play a role beyond hydrocarbons and NAs: it would seem unlikely that a strict aerobe would be found in all samples or that a strict anaerobe would be found in shallow (oxic) river sediments. For instance, Azonexus, a nitrogen-fixing bacterium, was positively correlated to sediment PAH concentrations, suggesting that this genus was more abundant in disturbed environments. In fact, strains from the closely related Azoarcus genus were first isolated from petroleum refinery oily sludge (21) and were shown to degrade toluene anaerobically (48). Another genus positively correlated with NA concentration was Achromobacter, which is a strict aerobe. This observation is in line with observations from other studies showing that species from this genus are capable of NA degradation (2, 3, 19). The strongest positive correlation observed for Archaea, however, was between the methanogen genus (strict anaerobe) Methanobrevibacter (Methanobacteriales) and NAs, TPHs, TSHs, and TAHs. Methane generation from Mildred Lake alone (where samples SY1A and SY1B were collected), estimated to be as high as 43,000 m3 day−1 (18), is another environmental issue associated with oil sands mining. More work is necessary, however, to confirm that the microbial taxa highlighted above are indeed effective bioindicators of pollution caused by oil sands mining operations. It should be ascertained that correlations reported here are related to real relationships and not spurious correlations caused by a common response to another (environmental) factor, such as oxygen. Indeed, several of the most abundant taxa found in the tailings ponds are strict or facultative anaerobes (e.g., Rhodoferax, Smithella, Thiobacillus) (see Table S3 in the supplemental material), and their presence might not be indicative of a preference for contaminated conditions but, rather, might be indicative of the limited oxygen available in the pond sediments due to the sampling depth or the higher clay content of the sediments. However, the presence of methanogens in all the samples indicates that some anaerobic niches were potentially available even in the shallower river sediments.
16S rRNA gene amplicon sequencing with the Ion Torrent PGM.
The present study is one of the first to use Ion Torrent's PGM to sequence 16S rRNA amplicons from environmental samples. As with any new sequencing technology, however, many believe that high error rates might blur the microbial community picture generated. We therefore carried out work for extensive validation of the technique in the context of microbial ecology studies. The Ion Torrent sequencing approach was first evaluated by sequencing the same bacterial 16S rRNA region using 454 and Ion Torrent. The 454 sequencing approach is widely accepted as accurate and has been used in numerous studies. The Ion Torrent generated a data set that was highly similar to the one generated by 454. Some differences in the relative abundance at the phylum level were observed and were most probably related to a primer bias, as the forward primer was not the same for the two sequencing runs because of the current technical limitation to the amplicon size that can enter in the emulsion PCR procedure during Ion Torrent library preparation. Having said that, the 454 and Ion Torrent data sets were almost interchangeable, and both would have yielded the exact same ecological conclusions. We used the 454 data set in our ecological analyses simply because of the longer read length.
The second validation analysis was to sequence known Bacteria and Archaea strains and to carry the sequencing results through our analysis pipeline. Using this approach, we determined that no misclassification had occurred for Archaea and that less than 0.1% of the Bacteria sequences were misclassified. The small amount of misclassified bacterial sequences could be due to several different factors: (i) errors in the main part of the sequence, (ii) errors in the MID, which caused migration from one bin to the other, or (iii) carryover during the initial PCR or the emulsion PCR procedure. Sequencing errors for Ion Torrent mainly occur at long homopolymer stretches (28). However, the longest homopolymer stretch in the 16S rRNA gene region for Alkanindiges is four, which is normally not problematic. The majority of the sequencing errors are thus expected to occur randomly along the sequence. Following this, it is highly unlikely that an Alkanindiges sequence could randomly mutate and be classified in a genus from another phylum with high confidence. Similarly, it is highly unlikely that with random sequencing errors a MID would end up with the sequence of another MID, since they were designed by Ion Torrent to be resistant to multiple sequencing errors (see Table S1 in the supplemental material) and the beginning of the sequence is where the quality is highest. One possibility is contamination during the initial PCR, which is surprising since all our no-template controls yielded no visible product. However, with the very low percentage observed (<0.1% of reads), it is possible that a very slight, undetectable contamination occurred during the amplification. Another possibility is carryover of template during emulsion PCR on the OneTouch instrument. Incomplete washing of the machine and the use of similar MIDs in two subsequent runs could have resulted in a small percentage of carryover. Overall, these data provide confidence that despite some very low level of misclassification, the Ion Torrent data are valid and representative of the environmental community in our samples.
The last test that we performed was to resequence an E. coli strain, which revealed highly accurate base calls. The per base accuracy was in the range of previously published values for Ion Torrent, which was reported to be comparable to other next-generation sequencing platforms (28, 34). Altogether, these validation tests give us confidence in the quality, accuracy, and reliability of the sequence data produced by Ion Torrent. The Ion Torrent PGM is ideally suited for environmental microbiology studies because of its rapid turnaround time (run time, about 2 h) and low sequencing run costs (∼US $500) and we expect that with the improvements in read lengths (currently at 250 bp; 400 bp is expected by the end of 2012), it will gain wide acceptance and use.
Conclusions.
This study revealed that river sediments in close proximity to oil sands tailings ponds were chemically and microbiologically more similar to each other and to those from the tailings ponds than to samples obtained from further away. Bacterial and archaeal taxa that were strongly correlated to contaminant presence were also identified. Importantly, these taxa have potential as bioindicators. Additionally, we demonstrated the reliability of Ion Torrent sequencing in the context of environmental microbiology studies. In conclusion, the indicator species identified in this study will help monitor and mitigate oil sands mining impacts on the Athabasca watershed ecosystem.
Supplementary Material
ACKNOWLEDGMENTS
We are thankful to Terry Bell for developing the Ion Torrent archaeal 16S sequencing method and to Christine Maynard for Ion Torrent sequencing. M. Hewitt, M. McMaster, V. Tumber, and D. Turcotte are acknowledged for organization and execution of water and sediment sampling.
This project was undertaken with the financial support of Environment Canada's Strategic Technology Applications of Genomics in the Environment (STAGE) Program.
Footnotes
Published ahead of print 24 August 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Bell TH, et al. 2011. Identification of nitrogen-incorporating bacteria in petroleum-contaminated Arctic soils by using [15N]DNA-based stable isotope probing and pyrosequencing. Appl. Environ. Microbiol. 77:4163–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blakley ER. 1978. The microbial degradation of cyclohexanecarboxylic acid by a β-oxidation pathway with simultaneous induction to the utilization of benzoate. Can. J. Microbiol. 24:847–855 [DOI] [PubMed] [Google Scholar]
- 3. Blakley ER. 1974. The microbial degradation of cyclohexanecarboxylic acid: a pathway involving aromatization to form p-hydroxybenzoic acid. Can. J. Microbiol. 20:1297–1306 [Google Scholar]
- 4. Claesson MJ, et al. 2009. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4:e6669 doi:10.1371/journal.pone.0006669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Crowe AU, Plant AL, Kermode AR. 2002. Effects of an industrial effluent on plant colonization and on the germination and post-germinative growth of seeds of terrestrial and aquatic plant species. Environ. Pollut. 117:179–189 [DOI] [PubMed] [Google Scholar]
- 6. Del Rio LF, Hadwin AKM, Pinto LJ, MacKinnon MD, Moore MM. 2006. Degradation of naphthenic acids by sediment micro-organisms. J. Appl. Microbiol. 101:1049–1061 [DOI] [PubMed] [Google Scholar]
- 7. Duncan GA, LaHaie GG. 1979. Size analysis procedures used in the sedimentology laboratory. National Water Research Institute, Canada Centre for Inland Waters, Hydraulics Division, Burlington, Ontario, Canada [Google Scholar]
- 8. Galand PE, Casamayor EO, Kirchman DL, Potvin M, Lovejoy C. 2009. Unique archaeal assemblages in the Arctic Ocean unveiled by massively parallel tag sequencing. ISME J. 3:860–869 [DOI] [PubMed] [Google Scholar]
- 9. Gentes M-L, Waldner C, Papp Z, Smits JEG. 2006. Effects of oil sands tailings compounds and harsh weather on mortality rates, growth and detoxification efforts in nestling tree swallows (Tachycineta bicolor). Environ. Pollut. 142:24–33 [DOI] [PubMed] [Google Scholar]
- 10. Giesy JP, Anderson JC, Wiseman SB. 2010. Alberta oil sands development. Proc. Natl. Acad. Sci. U. S. A. 107:951–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hadwin AKM, et al. 2006. Microbial communities in wetlands of the Athabasca oil sands: genetic and metabolic characterization. FEMS Microbiol. Ecol. 55:68–78 [DOI] [PubMed] [Google Scholar]
- 12. Hamady M, Lozupone C, Knight R. 2010. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 4:17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Headley JV, Akre C, Conly FM, Peru KM, Dickson LC. 2001. Preliminary characterization and source assessment of PAHs in tributary sediments of the Athabasca River, Canada. Environ. Forensics 2:335–345 [Google Scholar]
- 14. Headley JV, McMartin DW. 2004. A review of the occurrence and fate of naphthenic acids in aquatic environments. J. Environ. Sci. Health A Tox. Hazard Subst. Environ. Eng. 39:1989–2010 [DOI] [PubMed] [Google Scholar]
- 15. Headley JV, et al. 2009. Aquatic plant-derived changes in oil sands naphthenic acid signatures determined by low-, high- and ultrahigh-resolution mass spectrometry. Rapid Commun. Mass Spectrom. 23:515–522 [DOI] [PubMed] [Google Scholar]
- 16. Herman DC, Fedorak PM, MacKinnon MD, Costerton JW. 1994. Biodegradation of naphthenic acids by microbial populations indigenous to oil sands tailings. Can. J. Microbiol. 40:467–477 [DOI] [PubMed] [Google Scholar]
- 17. Holden AA, Donahue RB, Ulrich AC. 2011. Geochemical interactions between process-affected water from oil sands tailings ponds and north Alberta surficial sediments. J. Contam. Hydrol. 119:55–68 [DOI] [PubMed] [Google Scholar]
- 18. Holowenko FM, MacKinnon MD, Fedorak PM. 2000. Methanogens and sulfate-reducing bacteria in oil sands fine tailings waste. Can. J. Microbiol. 46:927–937 [PubMed] [Google Scholar]
- 19. Johnson RJ, et al. 2011. Microbial biodegradation of aromatic alkanoic naphthenic acids is affected by the degree of alkyl side chain branching. ISME J. 5:486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kelly EN, et al. 2010. Oil sands development contributes elements toxic at low concentrations to the Athabasca River and its tributaries. Proc. Natl. Acad. Sci. U. S. A. 107:16178–16183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a. Kelly EN, et al. 2009. Oil sands development contributes polycyclic aromatic compounds to the Athabasca River and its tributaries. Proc. Natl. Acad. Sci. U. S. A. 106:22346–22351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laguerre G, Bossand B, Bardin R. 1987. Free-living dinitrogen-fixing bacteria isolated from petroleum refinery oily sludge. Appl. Environ. Microbiol. 53:1674–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lawrence JR, Swerhone GDW, Wassenaar LI, Neu TR. 2005. Effects of selected pharmaceuticals on riverine biofilm communities. Can. J. Microbiol. 51:655–669 [DOI] [PubMed] [Google Scholar]
- 23. Lawrence JR, et al. 2012. Molecular and microscopic assessment of the effects of caffeine, acetaminophen, diclofenac and their mixtures on river biofilm communities. Environ. Toxicol. Chem. 31:508–517 [DOI] [PubMed] [Google Scholar]
- 24. Lawrence JR, et al. 2009. Comparative microscale analysis of the effects of triclosan and triclocarban on the structure and function of river biofilm communities. Sci. Total Environ. 407:3307–3316 [DOI] [PubMed] [Google Scholar]
- 25. Lawrence JR, et al. 2008. Community-level assessment of the effects of the broad-spectrum antimicrobial chlorhexidine on the outcome of river microbial biofilm development. Appl. Environ. Microbiol. 74:3541–3550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leung SS, MacKinnon MD, Smith REH. 2003. The ecological effects of naphthenic acids and salts on phytoplankton from the Athabasca oil sands region. Aquat. Toxicol. 62:11–26 [DOI] [PubMed] [Google Scholar]
- 27. Lister A, Nero V, Farwell A, Dixon DG, Van Der Kraak G. 2008. Reproductive and stress hormone levels in goldfish (Carassius auratus) exposed to oil sands process-affected water. Aquat. Toxicol. 87:170–177 [DOI] [PubMed] [Google Scholar]
- 28. Loman NJ, et al. 2012. Performance comparison of benchtop high-throughput sequencing platforms. Nat. Biotech. 30:434–439 [DOI] [PubMed] [Google Scholar]
- 29. Penner TJ, Foght JM. 2010. Mature fine tailings from oil sands processing harbour diverse methanogenic communities. Can. J. Microbiol. 56:459–470 [DOI] [PubMed] [Google Scholar]
- 30. Phillips LA, Armstrong SA, Headley JV, Greer CW, Germida JJ. 2010. Shifts in root-associated microbial communities of Typha latifolia growing in naphthenic acids and relationship to plant health. Int. J. Phytoremediation 12:745–760 [DOI] [PubMed] [Google Scholar]
- 31. Pollet I, Bendell-Young LI. 2000. Amphibians as indicators of wetland quality in wetlands formed from oil sands effluent. Environ. Toxicol. Chem. 19:2589–2597 [Google Scholar]
- 32. Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. 2011. Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12:38 doi:10.1186/1471-2105-12-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ramos-Padron E, et al. 2011. Carbon and sulfur cycling by microbial communities in a gypsum-treated oil sands tailings pond. Environ. Sci. Technol. 45:439–446 [DOI] [PubMed] [Google Scholar]
- 34. Rothberg JM, et al. 2011. An integrated semiconductor device enabling non-optical genome sequencing. Nature 475:348–352 [DOI] [PubMed] [Google Scholar]
- 35. Rowland SJ, Scarlett AG, Jones D, West CE, Frank RA. 2011. Diamonds in the rough: identification of individual naphthenic acids in oil sands process water. Environ. Sci. Technol. 45:3154–3159 [DOI] [PubMed] [Google Scholar]
- 36. Schwartz MW, et al. 2000. Linking biodiversity to ecosystem function: implications for conservation ecology. Oecologia 122:297–305 [DOI] [PubMed] [Google Scholar]
- 37. Siddique T, Fedorak PM, Foght JM. 2006. Biodegradation of short-chain n-alkanes in oil sands tailings under methanogenic conditions. Environ. Sci. Technol. 40:5459–5464 [DOI] [PubMed] [Google Scholar]
- 38. Wang Z, Fingas M, Li K. 1994. Fractionation of a light crude oil and identification and quantitation of aliphatic, aromatic, and biomarker compounds by GC-FID and GC-MS, part I. J. Chromatogr. Sci. 32:361–366 [Google Scholar]
- 39. Wang Z, Fingas M, Li K. 1994. Fractionation of a light crude oil and identification and quantitation of aliphatic, aromatic, and biomarker compounds by GC-FID and GC-MS, part II. J. Chromatogr. Sci. 32:367–382 [Google Scholar]
- 40. Wang Z, Fingas M, Sergy G. 1994. Study of 22-year-old Arrow oil samples using biomarker compounds by GC/MS. Environ. Sci. Technol. 28:1733–1746 [DOI] [PubMed] [Google Scholar]
- 41. Wardle DA. 2002. Communities and ecosystems: linking the aboveground and belowground components. Princeton University Press, Princeton, NJ [Google Scholar]
- 42. Wilmotte A, Van der Auwera G, De Wachter R. 1993. Structure of the 16 S ribosomal RNA of the thermophilic cyanobacterium Chlorogloeopsis HTF (‘Mastigocladus laminosus HTF') strain PCC7518, and phylogenetic analysis. FEBS Lett. 317:96–100 [DOI] [PubMed] [Google Scholar]
- 43. Wyndham RC, Costerton JW. 1981. Heterotrophic potentials and hydrocarbon biodegradation potentials of sediment microorganisms within the Athabasca oil sands deposit. Appl. Environ. Microbiol. 41:783–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wyndham RC, Costerton JW. 1981. In vitro microbial degradation of bituminous hydrocarbons and in situ colonization of bitumen surfaces within the Athabasca oil sands deposit. Appl. Environ. Microbiol. 41:791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yergeau E, et al. 2012. Shifts in soil microorganisms in response to warming are consistent across a range of Antarctic environments. ISME J. 6:692–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yergeau E, Lawrence JR, Korber DR, Waiser MJ, Greer CW. 2010. Meta-transcriptomic analysis of the response of river biofilms to pharmaceutical products using anonymous DNA microarrays. Appl. Environ. Microbiol. 76:5432–5439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yergeau E, Sanschagrin S, Waiser MJ, Lawrence JR, Greer CW. 2012. Sub-inhibitory concentrations of different pharmaceutical products affect the meta-transcriptome of river biofilm communities cultivated in rotating annular reactors. Environ. Microbiol. Rep. 4:350–359 [DOI] [PubMed] [Google Scholar]
- 48. Zhou J, Fries MR, Chee-Sanford JC, Tiedje JM. 1995. Phylogenetic analyses of a new group of denitrifiers capable of anaerobic growth on toluene and description of Azoarcus tolulyticus sp. nov. Int. J. Syst. Bacteriol. 45:500–506 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.