Abstract
In a previous study, system level analysis of adaptively evolved yeast mutants showing improved galactose utilization revealed relevant mutations. The governing mutations were suggested to be in the Ras/PKA signaling pathway and ergosterol metabolism. Here, site-directed mutants having one of the mutations RAS2Lys77, RAS2Tyr112, and ERG5Pro370 were constructed and evaluated. The mutants were also combined with overexpression of PGM2, earlier proved as a beneficial target for galactose utilization. The constructed strains were analyzed for their gross phenotype, transcriptome and targeted metabolites, and the results were compared to those obtained from reference strains and the evolved strains. The RAS2Lys77 mutation resulted in the highest specific galactose uptake rate among all of the strains with an increased maximum specific growth rate on galactose. The RAS2Tyr112 mutation also improved the specific galactose uptake rate and also resulted in many transcriptional changes, including ergosterol metabolism. The ERG5Pro370 mutation only showed a small improvement, but when it was combined with PGM2 overexpression, the phenotype was almost the same as that of the evolved mutants. Combination of the RAS2 mutations with PGM2 overexpression also led to a complete recovery of the adaptive phenotype in galactose utilization. Recovery of the gross phenotype by the reconstructed mutants was achieved with much fewer changes in the genome and transcriptome than for the evolved mutants. Our study demonstrates how the identification of specific mutations by systems biology can direct new metabolic engineering strategies for improving galactose utilization by yeast.
INTRODUCTION
Microbe-based production of fuels and chemicals has been extensively investigated since it may contribute to the establishment of a more sustainable society. In this context, the development of microorganisms with efficient substrate utilization and product formation is a requirement. Evolutionary engineering has traditionally been used for this kind of improvement in industry, since it can result in strategies that are not predicted and hence cannot be obtained through rational design (23). Evolutionary strategies have gained renewed interest with the progress of tools in systems biology since this has allowed for the identification of governing mutations that can subsequently be implemented through site-directed mutagenesis using the concept of inverse metabolic engineering (1, 15). Furthermore, understanding the evolution process is useful for the identification of novel metabolic engineering strategies (2, 5). One of the typical patterns during adaptive evolution is the saturation of fitness with a proportional increase of mutations with the number of generations (2, 10). This phenomenon seems to be partially explained by the accumulation of deleterious mutations. Therefore, introduction of beneficial mutations into a parental strain to remove negative mutations is normally the last step when evolutionary engineering is used for strain development (25). Another finding from adaptive evolution studies is that the decline of fitness is due to negative epistasis among beneficial mutations, which means that the impact of combining beneficial mutations is less than the sum of individual mutations (4, 20). This finding suggests that different combinations of beneficial genetic changes may result in more advanced phenotype, and this opens the way for multiple inverse metabolic engineering strategies.
In a previous study we characterized evolved mutants of yeast that showed improved galactose utilization compared to a reference strain (17). Three isolated mutants from different populations derived from identical adaptive evolution processes were analyzed by using systems biology tools, and the evolved mutants were also compared to metabolically engineered strains. No mutations were detected in known galactose pathway and regulatory pathways, whereas all three evolved mutants had mutations in the Ras/PKA signaling pathway, and application of one of these mutations showed an improved specific galactose uptake rate. One unique mutation that may link with specific changes of transcripts and metabolites in the ergosterol pathway was also identified, namely, a mutation in ERG5. In addition to the above-mentioned mutations, PGM2 encoding phosphoglucomutase was validated as a metabolic engineering target since it was found to be overexpressed in all of the evolved mutants, which was consistent with the earlier finding that overexpression of this enzyme resulted in improved galactose uptake (3, 13, 17), whereas overexpression of GAL genes either individually or in combination does not result in increased galactose uptake (7). We therefore constructed site-directed mutants that have each of the identified mutations and combined them with PGM2 overexpression to evaluate how much the phenotypes of the adaptively evolved mutants could be recovered by these defined genetic changes. The site-directed mutants were constructed by introducing three point mutations, RAS2Lys77(from evolved mutant 62A), RAS2Tyr112 (from evolved mutant 62B), and ERG5Pro370 (from evolved mutant 62B) to a reference strain, and the mutants were further engineered by transformation of a high-copy-number plasmid with constitutive promoter for overexpression of PGM2. We present the results of a comparative analysis of the mutants with only site-directed mutations and mutants with a combination of site-directed mutations and PGM2 overexpression. All of the engineered strains were compared to a reference strain, a previously constructed strain, and the two evolved strains. The comparative analysis involved quantitative fermentation physiology and measurement of the transcriptome and targeted intracellular metabolites. Based on our analysis of the constructed mutants, additional strategies for improving galactose utilization are discussed together with the use of adaptive evolution for improving strain performance.
MATERIALS AND METHODS
Yeast strains and plasmids.
The Saccharomyces cerevisiae strains used in the present study are summarized in Table 1. S. cerevisiae CEN.PK113-5D was used to construct site-directed mutants and combined mutants due to the availability of the URA3 marker gene with the same genotype background as strain 7D except for that particular gene. Reference strains, evolved mutants and site-directed mutants, were preserved on a synthetic minimal (SM) medium containing yeast nitrogen base without amino acids (6.9 g/liter; Formedium, Hun-stanton, United Kingdom), complete supplement mixture (0.77 g/liter; MP Biomedicals, Solon, OH), glucose (20 g/liter), and agar (20 g/liter). Other strains carrying plasmids were maintained on the same SM medium, except for the complete supplement mixture without uracil (0.77 g/liter). To avoid uracil supplement-related effects, an empty plasmid with the URA3 gene as a marker, pSP-GM2, was used for transformation into the CEN.PK113-5D strain and the site-directed mutants (22). To overexpress the PGM2 gene, pPGM2 was used, which is a high-copy-number plasmid containing the constitutive promoter of the PMA1 gene and URA3 as a marker (3).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Group | Source or referencea |
|---|---|---|---|
| 7D | MATa SUC2 MAL2-8C (CEN.PK113-7D) | Reference strain | SR&D |
| 62A | 7D; total 21 SNPs, including RAS2Lys 77 | Evolved mutants | 17 |
| 62B | 7D; total of 104 SNPs, including RAS2Tyr112 and ERG5Pro370 | Evolved mutants | 17 |
| 5D | MATa SUC2 MAL2-8C ura3-52 (CEN.PK113-5D) | Reference strain | SR&D |
| 5DU | 5D, pSP-GM2(URA3) | Reference strain | This study |
| RAU | 5D, pSP-GM2(URA3); RAS2Lys77 (from 62A) | Site-directed mutants | This study |
| RBU | 5D, pSP-GM2(URA3); RAS2Tyr112 (from 62B) | Site-directed mutants | 17b |
| EBU | 5D, pSP-GM2(URA3); ERG5Pro370 (from 62B) | Site-directed mutants | This study |
| PGM2 | 5D, pPGM2(URA3, PPMA1-PGM2) | Engineered mutant | 3 |
| RAP | 5D, pPGM2(URA3, PPMA1-PGM2); RAS2Lys77 | Combined mutants | This study |
| RBP | 5D, pPGM2(URA3, PPMA1-PGM2); RAS2Tyr112 | Combined mutants | This study |
| EBP | 5D, pPGM2(URA3, PPMA1-PGM2); ERG5Pro370 | Combined mutants | This study |
SR&D, Scientific Research & Development GmbH, Oberursel, Germany.
The plasmid was added in the present study.
Strain construction and cultivation.
Site-directed mutants were constructed by a previously described method (17). Three point mutations—RAS2Lys77(identified in evolved mutant 62A), RAS2Tyr112 (identified in evolved mutant 62B), and ERG5Pro370 (identified in evolved mutant 62B)—were introduced into strain CEN.PK113-5D separately. The oligomers used in the present study are summarized in Table 2. Approximately 1 kb of sequence, including the mutation in the center region and part of URA3 gene (ca. 20 to 22 mer) in the end, was amplified by PCR. The marker gene URA3 was amplified into two parts. The 5′-end (716 bp) and the 3′ end (1,028 bp) of the URA3 gene were amplified separately. Fusion PCR between the 1-kb sequence and each partial URA3 was implemented, and then two cassettes were prepared. The cassettes were transformed into S. cerevisiae CEN.PK113-5D, and the sequence that had the mutation in the target gene together with the URA3 marker was integrated into the chromosome by homologous recombination. The strains that had URA3 could be screened out in selection medium, which was SD medium without uracil. These strains were streaked onto the same medium one more time to purify single colonies. The single isolate clones were streaked onto 5-fluoorotic (5-FOA) acid medium, which consisted of SD medium supplemented with 30 mg of uracil and 750 mg of 5-fluoroortic acid monohydrate (Formedium)/liter, to loop out the URA3 marker. In this process, two types of clones were obtained: one containing the point mutation in the target gene and the other with the wild-type sequence of the target gene. The site-directed mutants were selected through colony PCR by comparing different concentration of PCR product when the primer that had the mutation at the 3′ end was used. To check the selected clones, the genes were sequenced, and presence of the site-directed mutation was confirmed. Finally, prototrophic site-directed mutants (strains RAU, RBU, and EBU) were constructed by transformation with the plasmid, pSP-GM2 containing the URA3 gene. The combined mutants RAP, RBP, and EBP were constructed by transformation of the plasmid pPGM2 into the site-directed mutants. The culture medium and condition used in the present study were as described earlier (17). The fermentation physiology data for S. cerevisiae strains CEN.PK113-7D, PGM2, 62A, and 62B were generated in a previous study (17).
Table 2.
Oligomers used in this study
| Target | Oligomer | Sequence (5′–3′)a | Use |
|---|---|---|---|
| URA3 | URA3-5P1 | TTCGGCTTCATGGCAATTCC | Half of URA3 (5′ URA3) |
| URA3-3P1 | GAGCAATGAACCCAATAACGAAATC | Half of URA3 (5′ URA3) | |
| URA3-5P2 | CTTGACGTTCGTTCGACTGATGAGC | Half of URA3 (3′ URA3) | |
| URA3-3P2 | ATCGATAAGCTTGATATCGAATTCC | Half of URA3 (3′ URA3) | |
| RAS2 | RAS2-5P1 | CCACTCTTTATCTGACTCTTCTGC | Fusion with 5′ URA3 |
| RAS2-3P1 | GGAATTGCCATGAAGCCGAATTCACATTTTTACCGTTGGCAGC | Fusion with 5′ URA3 | |
| RAS2-5P2 | GGAATTCGATATCAAGCTTATCGATCCACTCTTTATCTGACTCTTCTGC | Fusion with 3′ URA3 | |
| RAS2-3P2 | TTCACATTTTTACCGTTGGCAGC | Fusion with 3′ URA3 | |
| RAS2-4AW | GCCGTTGCGCATGTATTG | Selection of wild-type, RAS2 | |
| RAS2-4AM | GCCGTTGCGCATGTATTT | Selection of mutants, RAS2Lys77 | |
| RAS2-4BW | AATTGGAACATAGTCGGTATC | Selection of wild-type, RAS2 | |
| RAS2-4BM | AATTGGAACATAGTCGGTATA | Selection of mutants, RAS2Tyr112 | |
| RAS2-5P | GTTTTAGCCGTTGTCTTCTCTT | Sequencing of RAS2 | |
| RAS2-3P | GTTCTTTTCGTCTTAGCGTTTC | Sequencing of RAS2 | |
| ERG5 | ERG5-5P1 | CGCCTTATCATTGAACTCTT | Fusion with 5′ URA3 |
| ERG5-3P1 | GGAATTGCCATGAAGCCGAAGGGAAAATTGTAGCGAAAAC | Fusion with 5′ URA3 | |
| ERG5-5P2 | GGAATTCGATATCAAGCTTATCGATCGCCTTATCATTGAACTCTT | Fusion with 3′ URA3 | |
| ERG5-3P2 | GGGAAAATTGTAGCGAAAAC | Fusion with 3′ URA3 | |
| ERG5-4BW | GTAGACATGTCATTGTTAC | Selection of wild-type, ERG5 | |
| ERG5-4BM | GTAGACATGTCATTGTTAG | Selection of mutants, ERG5Pro370 | |
| ERG5-5P | GAGAATAACTACGAGCCCCAG | Sequencing of ERG5 | |
| ERG5-3P | GGTCTCTCTTTTTGAAAGTCAG | Sequencing of ERG5 |
Sequences used for making fusion regions with URA3 are marked in boldface. Different sequences used toseparate wild-type and mutant are underlined.
Transcriptome analysis.
The transcriptome was measured by using Affymetrix Yeast Genome 2.0 arrays. The set of differentially expressed genes (the Limma test [Bioconductor] obtained using R Language version 2.13.2 was applied [adjusted P value of <0.01]) were analyzed by comparing the site-directed mutants, the combined mutants, and the PGM2 strain after background subtraction of the transcriptome of the 5DU strain. A second set of differentially expressed genes (adjusted P value of <0.01) was identified by observing the contrast among the combined mutants and the evolved mutants. For the evolved mutants, the parental strain is 7D, and background subtraction was therefore based on the 7D strain. The transcriptome data for S. cerevisiae strains CEN.PK113-7D, PGM2, 62A, and 62B were as published in a previous study (17). Functional gene enrichment was done by using the updated g:Profiler (24). The heat maps of specific pathways were visualized by using MultiExperiment Viewer software (Dana-Farber Cancer Institute, Boston, MA).
Gene expression data.
Gene expression data were deposited in the Gene Expression Omnibus database under accession number GSE36118.
Analysis of carbohydrates and sterols.
Harvested cells were directly used for the extraction of carbohydrates and sterols at 10 and 100 mg (cell dry weight), respectively. The quantification of carbohydrates (trehalose and glycogen) and sterols (ergosterol and dihydroergosterol) used in the present study was described earlier (17). The metabolite concentration data of the CEN.PK113-7D, PGM2, 62A, and 62B strains were measured in a previous study (17) and used for combined analysis here.
RESULTS
Fermentation physiology of reconstructed mutants.
The S. cerevisiae strains used in the present study are summarized in Table 1. Three point mutations—RAS2Lys77(identified in evolved mutant 62A), RAS2Tyr112 (identified in evolved mutant 62B), and ERG5Pro370 (identified in evolved mutant 62B)—were introduced into the CEN.PK113-5D strain separately. To avoid uracil supplement-related effects, an empty plasmid having the URA3 gene as a marker, pSP-GM2, was transformed into the CEN.PK113-5D strain and the strains carrying the site-directed mutations. The resulting prototrophic strains were designated 5DU (reference strain), RAU (carrying RAS2Lys77), RBU (carrying RAS2Tyr112), and EBU (carrying ERG5Pro370). Based on a comparison of these four strains, it was found that each of the single point mutations resulted in an improvement of both the maximum specific growth rate and the specific galactose uptake rate (Fig. 1). The largest effect was observed for RAU, and this single mutation resulted in a 42% increase in the maximum specific growth rate and a 57% increase in the specific galactose uptake rate compared to the reference strain 5DU. This improvement was much greater than what that obtained in the evolved mutants 62A and 62B derived from the prototrophic CEN.PK113-7D strain, namely, a 23 to 24% increase in the maximum specific growth rate and a 18 to 26% increase in the maximum specific galactose uptake rate. Although that for the RBU strain that carries another point mutation in the same gene (RAS2), the improvement of galactose utilization was much less. The EBU strain showed the smallest increase of galactose utilization among the three site-directed mutants.
Fig 1.
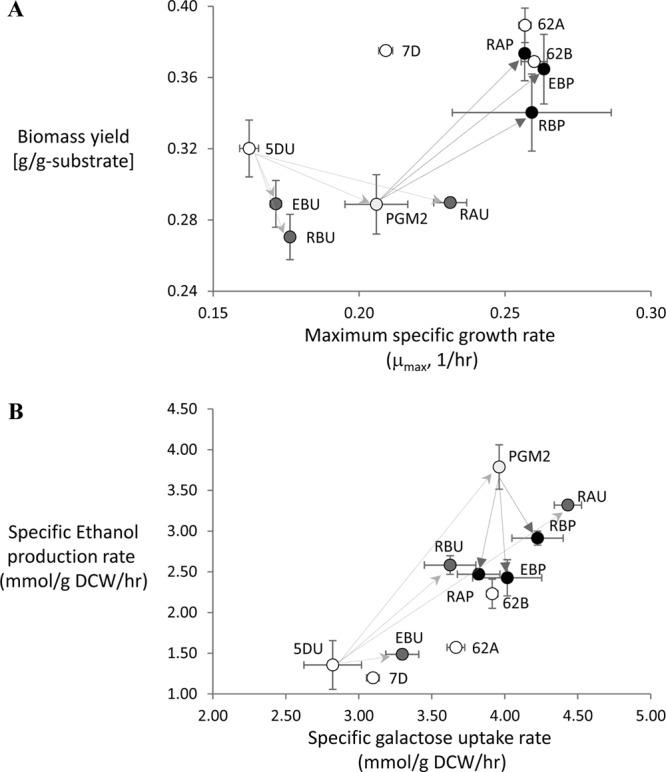
Data for the overall fermentation physiology of the site-directed mutants (RAU, RBU, and EBU) and the combined mutants (RAP, RBP, and EBP) compared to the reference strains 5DU and 7D, the engineered strain PGM2, and the corresponding evolved mutants 62A and 62B. (A) Correlation between maximum specific growth rate and biomass yield. (B) Correlation between specific galactose uptake rate and specific ethanol production rate. The arrows indicate the relation between reference strains and mutants. Error bars represent the standard deviations from biological duplicates.
Combination of the beneficial point mutations with overexpression of PGM2 was evaluated by transformation of pPGM2, including the URA3 marker instead of the empty plasmid pSP-GM2 into the site-directed mutants and the reference strain CEN.PK113-5D, and the resulting strains were named RAP, RBP, EBP, and PGM2 (Table 1). The PGM2 strain exhibited a 27% increase of the maximum specific growth rate and a 40% increase of the specific galactose uptake rate compared to 5DU. These improvements were better than those observed for the RBU and EBU strains (Fig. 1) but consistent with what we have reported earlier (3). Combination of the single point mutations from the evolved mutants and PGM2 overexpression resulted in strains that have gross phenotypes that are almost completely identical to those of the evolved mutants, i.e., in terms of the maximum specific growth rate on galactose, the specific galactose uptake rate and the specific ethanol production rate, they were very similar (Fig. 1; see Table S1 in the supplemental material). The two mutations in RAS2, as well as the ERG5Pro370 mutation, combined with PGM2 overexpression, resulted in a ca. 60% increase in the maximum specific growth rate and a 35 to 50% increase in the specific galactose uptake rate. This is remarkable since the two reference strains 5DU and 7D showed some differences in terms of the maximum specific growth rate, but RAP, RBP, and EBP still reached the same level of galactose utilization as the evolved mutants. We performed an evaluation of the overall carbon fluxes, and this shows that the addition of PGM2 overexpression in the single point mutations resulted in increased respiration and reduced fermentation even with the increase in the maximum specific growth rate (Fig. 2; see Table S2 in the supplemental material). The biomass yield for the combined mutants (RAP, RBP, and EBP) showed a range similar to those for the evolved mutants.
Fig 2.
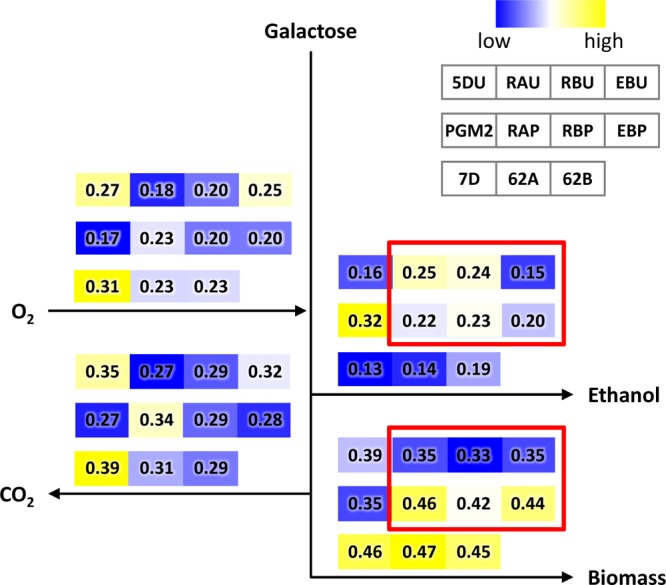
Comparison of overall carbon fluxes for all of the strains used in the present study. The boxes represent yield values expressed as C mol/C mol of galactose (mol/C mol of galactose for oxygen). Colors indicate the relative sizes of values for each line of strains. Red boxes indicate an additional effect of PGM2 overexpression in the site-directed mutants.
Molecular differences among the reconstructed mutants.
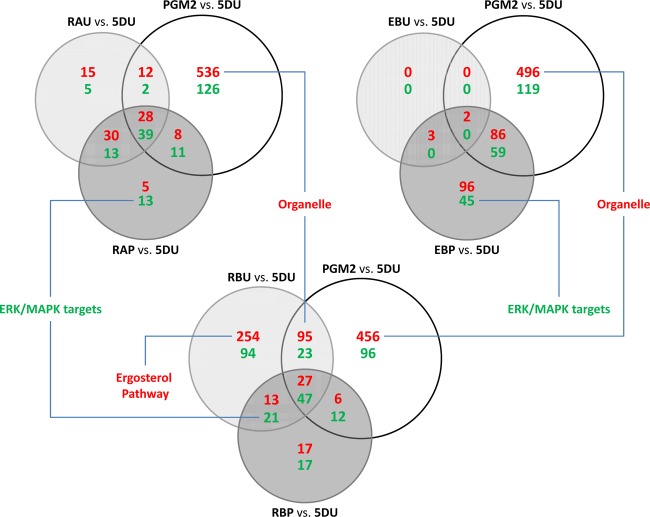
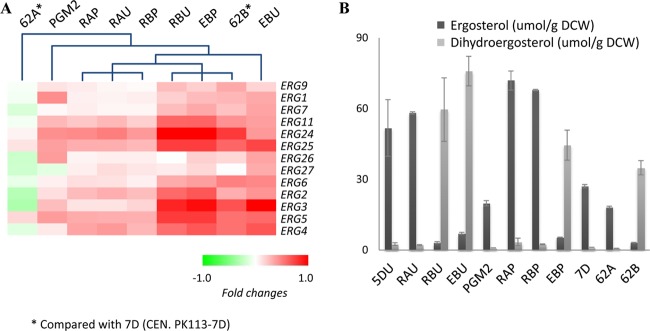
The site-directed mutants (RAU, RBU, and EBU), the PGM2 strain, and the combined mutants (RAP, RBP, and EBP) were compared to find common and unique changes in their transcriptomes (Fig. 3). The transcriptome in each strain was compared to the reference strain (CEN.PK113-5D containing an empty plasmid), and genes with significant changes in its transcription were identified (correcting for multiple testing). Conserved transcriptional changes in the different mutants were evaluated using Venn diagrams as illustrated in Fig. 3. The PGM2 strain showed the largest number of transcriptional alterations among all of the reconstructed mutants (762 significantly changed genes), whereas the combined mutants, even though they also have PGM2 overexpression, had much lower number of genes with significant changed transcription, i.e., around 150 to 300 genes. Functional enrichment based on the GO term database for the transcriptional changes in the PGM2 strain was mostly in organelle-related genes (Fig. 3; see also data set S1 in the supplemental material). A functional category of genes with significant changes in transcription in the combined mutants RAP and EBP was ERK/MAPK target related genes based on the REACTOME database. The RBU strain also showed many more transcriptional changes (389 upregulated and 185 downregulated) than RAU (85 upregulated and 59 downregulated). Especially, one of the gene enrichment categories in the uniquely upregulated genes of RBU (254 genes) was ergosterol metabolism. Transcripts and metabolites of the ergosterol pathway were therefore analyzed in more detail (Fig. 4). All strains that have mutation of ERG5Pro370 showed overall upregulation of ERG genes and a higher ratio of dihydroergosterol to ergosterol. The mutation of RAS2Tyr112 also showed this pattern, while the combination of this mutation with PGM2 overexpression results in a loss of this pattern (RBP strain). The mutation of ERG5Pro370 had no significant changes in its transcriptome except for five genes in the ergosterol pathway, which are significantly upregulated. The combination with PGM2 overexpression (EBP strain), however, induced more transcriptional changes, but when this strain is compared to the PGM2 strain, the change in the transcriptome is much smaller.
Fig 3.
Effect of mutations (RAS2Lys77, RAS2Tyr112, and ERG5Pro 370) and the combination with PGM2 overexpression compared to the engineered strain PGM2 by differentially expressed genes. Differentially expressed genes (P < 0.01) are categorized as Venn diagrams. The functional enrichment of genes in each part was analyzed by hypergeometric distribution based on the KEGG, Reactome, and GO term databases. Red text indicates upregulation, and green text indicates downregulation.
Fig 4.
Changes in ergosterol metabolism in the reconstructed strains illustrated as fold changes in gene transcription and the concentrations of ergosterol and dihydroergosterol compared to the other strains. (A) Hierarchical clustering of all strains is computed using fold changes of genes in ergosterol metabolism. (B) Concentrations of ergosterol and dihydroergosterol. Error bars represent the standard deviations from biological duplicates.
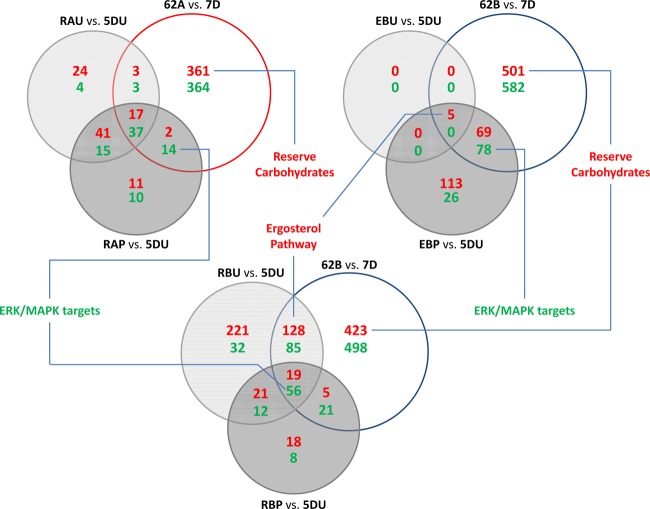
Comparison to evolved mutants.
We next compared the reconstructed strains with the evolved mutants at the transcriptome level to analyze mutual and exclusive changes in their transcriptome (Fig. 5). The transcriptome of the evolved strains, 62A and 62B, was analyzed in our earlier study (17) and was found to display a much larger number of transcriptional changes than all of the reconstructed mutants, i.e., around 700 to 1,100 genes had significant changes in their transcription. The result of functional enrichments for uniquely changed genes in the evolved mutants included reserve carbohydrate metabolism (Fig. 5; see also data set S2 in the supplemental material). The reconstructed strains RAU and RAP covered only 7.5 and 8.7% of the transcriptional changes of the 62A evolved mutant, respectively. The RBU strain showed 23.3% of coverage of the transcriptional changes of the 62B evolved mutant, whereas the combined mutant, RBP covered only 8.2% of the transcriptional changes. In case of the ERG5Pro370 mutation derived from the evolved mutant 62B (EBU strain), only 0.4% of the transcriptional changes of the 62B strain were covered. However, when PGM2 overexpression was introduced together with the ERG5Pro370 mutation (EBP strain), almost 12.3% of the transcriptional changes in 62B were covered. The common change in all of the strains that showed improved galactose availability was upregulation of PGM2. The other common change in the combined mutants and the evolved mutants was the functional category of ERK/MPPK targets. Genes involved in the ergosterol pathway were detected to have significantly changed expression in both RBU and 62B, whereas this is not the case for the RBP strain. Also, the common change among the EBU, EBP, and 62B strains was genes of the ergosterol pathway based on the KEGG database.
Fig 5.
Comparison of the reconstructed strains to the evolved strains 62A and 62B through differentially expressed genes. Differentially expressed genes (P < 0.01) are categorized as Venn diagrams. The functional enrichment of genes in each part was analyzed by the hypergeometric distribution based on the KEGG, Reactome, and GO term databases. Red text indicates upregulation, and green text indicates downregulation.
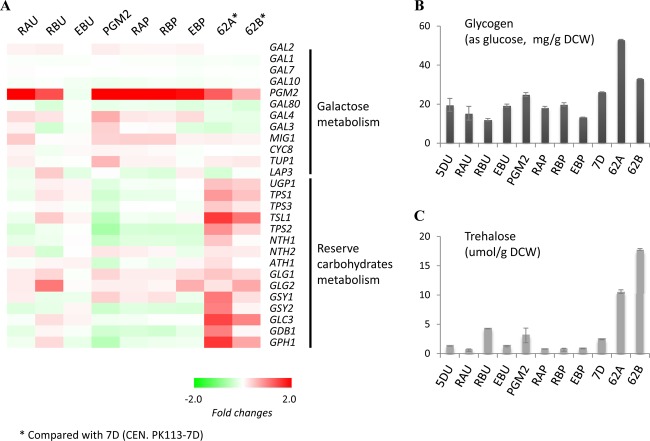
Changes in galactose and reserve carbohydrate metabolism.
The effects of the mutations in RAS2 for regulation of PGM2 and reserve carbohydrate metabolism were analyzed in terms of transcripts and metabolites. The mutations, RAS2Lys77 and RAS2Tyr112, induced upregulation of PGM2 but not reserve carbohydrate metabolism (Fig. 6). The level of PGM2 upregulation by the mutations in RAS2 was similar to what is found in the PGM2 strain that have overexpression of PGM2 through constitutive expression using a high-copy-number plasmid. Transcripts and metabolites in reserve carbohydrate metabolism showed no comparable changes for any of the reconstructed mutants as found for the evolved mutants 62A and 62B.
Fig 6.
Changes in galactose and reserve carbohydrate metabolism in reconstructed strains are shown as fold changes in the transcriptome and the concentrations of the carbohydrates. (A) Fold changes among all genes involved in galactose and reserve carbohydrate metabolism are compared to the reference strains. (B) Concentrations of glycogen. (C) Concentrations of trehalose. Error bars represent standard deviations from biological duplicates.
DISCUSSION
Few genetic and transcriptional changes are required to reach adaptive phenotypes.
During adaptive evolution, there is the possibility of having negative or neutral mutations, and these may be inherited for many generations, since their effect could be compensated for by other beneficial mutations (10). Identification of beneficial mutations and removing negative mutations is clearly one of the important points in strain development (25). Negative epistasis among beneficial mutations is also possible (20). In Escherichia coli undergoing adaptive evolution it has been found that spontaneous mutations appear proportionally with the number of cell generations; however, the increase of fitness to a specific environment showed a saturation pattern (2). There are therefore clear discordances between the mutation generation rate and the adaptation rate. Recently, this observation has been explained by the concept of diminishing return epistasis among beneficial mutations (4, 20). E. coli generated and accumulated beneficial mutations; however, the improvement of fitness resulting from their combination was less than that obtained as the sum of the individual mutations. Therefore, interaction among beneficial mutations could be negative for phenotypic progress. In other words, expressed phenotypes may be changeable depending on the combination of beneficial changes. This phenomenon should be very carefully considered in metabolic engineering as the reconstruction of strains based on a combination of beneficial targets identified in evolved mutants may result in improved strains since it not only results in removing negative or neutral mutations but also removes negative epistasis effects. Evolved mutant 62A achieved improved galactose availability with 21 point mutations, including 6 mutations in the coding regions (17), whereas the site-directed mutant RAU showed an even higher specific galactose uptake rate with only one point mutation RAS2Lys77 identified in 62A with a much reduced overall transcriptional alteration. Strain 62A showed a slightly higher maximum specific growth rate than strain RAU; however, based on comparison to the corresponding reference strains 5DU and 7D, respectively, the improvement in specific growth rate was much higher for RAU. It is unclear why the two reference strains showed differences in terms of the maximum specific growth rate and biomass yield. Their transcriptional differences indicate the changes in cell wall and membrane composition (see data set S3 in the supplemental material), and this may be generated by different Ura3p activity because of different copy numbers. The combined mutants RAP, RBP, and EBP, even though they underwent far fewer genetic and molecular changes than the evolved mutants, also achieved almost the same galactose utilization as the evolved ones. These results indicate that fewer genetic changes leading to less transcriptional alteration resulted in better performance of the strain compared to the cumulative mutations obtained from adaptive evolution. The changes in reserve carbohydrate metabolism were considered critical for galactose utilization because of not only common changes in the three evolved mutants but also close linkage with galactose metabolism by sharing metabolites (17). However, the reconstructed mutants obtained the same gross phenotype as the evolved mutants without any changes in reserve carbohydrate metabolism. This finding indicates that all changes occurring in connection with adaptive evolution, even those present in different evolved mutants, may be at least nonessential for improving the phenotype. There could, however, be a beneficial effect of the change in reserve carbohydrate metabolism observed in all of the evolved mutants, and it is possible that this effect was not detected here. However, it seems that many changes that are not related to galactose utilization appeared during the adaptive evolution.
RAS2Lys77 (RAU) and RAS2Tyr112 (RBU) induced the upregulation of PGM2 but not reserve carbohydrate metabolism, and RAS2Tyr112 (RBU) and ERG5Pro370 (EBU) upregulate the ergosterol pathway.
In our previous study we inferred that the identified mutations in the Ras/PKA signaling pathway may have triggered the upregulation of PGM2 and reserve carbohydrates metabolism because not only were the mutations identified in all of the evolved mutants but also on the promoter region of PGM2, and genes involved in reserve carbohydrate metabolism contained STER elements involved in Ras activation (27). The upregulation of PGM2 in the evolved strains could increase galactose utilization as proven in earlier studies (3, 13, 17, 21). In the present study, the mutations RAS2Lys77 (RAU) and RAS2Tyr112 (RBU) showed a certain relation to the upregulation of PGM2 but not to any changes in reserve carbohydrate metabolism. The Ras/PKA signaling pathway is involved in glucose sensing, stress response, and many other cellular functions (27). The Ras2 protein is one of the regulatory components that control the activity of protein kinase A (PKA). Ras2 interacts with several molecules such as GTP, guanine exchange factor (Cdc25), GTPase, and adenylate cyclase (Cyr1), and protein structure studies to elucidate binding site with those molecules have been implemented using site-directed mutations (14, 26). One of the mutations RAS2Val19 in yeast has been well investigated by evaluating its effect on PKA activity and cellular metabolism (16, 18). Incorporation of that mutation resulted in constitutive activation of PKA by strong GTP binding through inhibition of GTPase (26). The effects of the mutation were the transcriptional downregulation of genes in galactose metabolism and reduction of glycogen accumulation (18). The mutations RAS2Lys77 (RAU) and RAS2Tyr112 (RBU) in the present study seem to have more complex roles than the change in PKA activity. Their positions in RAS2 are not directly involved in known binding sites of molecules (26). No changes in galactose metabolic genes (GAL1, GAL7, and GAL10) were detected (Fig. 6A). In addition to these different effects compared to mutations in other studies such as RAS2Val19 mutation, the RAS2Lys77 (RAU) and RAS2Tyr112 (RBU) mutations showed their unique features in terms of differences in the extent of galactose utilization and the induction of transcriptional changes. One of the unique changes in RAS2Tyr112 (RBU) was a change in the expression of genes involved in the ergosterol pathway (Fig. 4). The transcriptional level of ERG genes and the ratio of ergosterol and dihydroergosterol in this mutant were very similar to those for the strain containing the ERG5Pro370 (EBU) mutation. There have been no reports of a direct relationship between Ras2 and ergosterol metabolism in yeast. This phenomenon seems to be a distinctive feature of the RAS2Tyr112 (RBU) mutation.
Concerning the effect of the ERG5 mutation on galactose metabolism, we speculate that the higher amount of dihydroergosterol instead of ergosterol may loosen the cell membrane rigidity, which may result in the induction of trehalose biosynthesis for stress protection. Increased production of trehalose may increase galactose uptake by consuming glucose-1-phosphate, which is a feed-forward inhibitor of Pgm2 (17). However, in the present study, only a negligible effect of mutation in the ergosterol pathway on galactose metabolism was detected. This result means that the ERG5Pro370 (EBU) mutation has no direct effect on improving galactose utilization, and this may be the reason why EBU only showed a very small improvement in galactose utilization (Fig. 1). It is, however, interesting that the unique changes in the ergosterol pathway observed in the evolved mutant 62B may be derived from not only the ERG5Pro370 (EBU) mutation but also the RAS2Tyr112 (RBU) mutation.
Combination of PGM2 overexpression and mutations in RAS2 and ERG5 recovers adaptive phenotypes in galactose utilization.
The combination of ERG5Pro370 mutation with PGM2 overexpression (EBP strain) showed a pattern similar to those of the other combined strains, which not only indicate a role for ERG5Pro370 in ergosterol metabolism but also suggest that it has a positive interaction with PGM2 overexpression for improving galactose utilization. PGM2 overexpression by constitutive promoter in a high-copy-number plasmid (PGM2 strain) mainly increased fermentation, as seen in the higher specific ethanol production rate and upregulation of organelle related genes (Fig. 1 and 3) (8, 19). When the three point mutations were combined with PGM2 overexpression, the metabolism changed to have increased respiration, resulting in a higher maximum specific growth rate and thereby also a higher biomass yield on galactose with downregulation of stress related signaling ERK/MAPK targets, which are related to stress response of osmolarity and starvation. PGM2 overexpression in RAU and RBU strains, which already showed upregulation of this gene, may somehow result in different Pgm2p functions that could not only be dedicated to galactose metabolism but also to the cellular state. The main function of PGM2 (phosphoglucomutase) is the conversion of glucose-1-phosphate to glucose-6-phosphate, but other functions such as involvement in the synthesis of cell wall and cellular calcium ion homeostasis have been reported (6, 12). Moreover, conditional posttranscriptional modification of PGM2 has been reported (9, 11). This modification is controlled by culture conditions such as heat shock and carbon sources.
In conclusion, we found that only a few mutations identified in adaptively evolved yeast strains are necessary to confer the same gross phenotype. Furthermore, these mutations may be additive in the sense that the overexpression of a single gene may result in a large genome-wide transcriptional response, but when this overexpression is combined with point mutations, many of these transcriptional changes disappear. This is most likely due to an effect of regulation of cellular stress response in the mutations of the Ras/PKA signaling pathway. Thus, it is clear that the use of adaptive evolution with the identification of beneficial changes is a powerful strategy in metabolic engineering since it allows for the identification of strategies that involves modulation of the cells regulatory system.
Supplementary Material
ACKNOWLEDGMENTS
We thank the CJ CheilJedang Doctoral Fellowship for financial support (to K.-K.H.). We acknowledge financial contributions from the Chalmers Foundation, The Knut and Alice Wallenberg Foundation, the European Union-funded project UNICELLSYS (contract 201142), the European Research Council (grant 247013), and the Novo Nordisk Foundation.
Footnotes
Published ahead of print 17 August 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Atsumi S, et al. 2010. Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol. Syst. Biol. 6:449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barrick JE, et al. 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461:1243–1247 [DOI] [PubMed] [Google Scholar]
- 3. Bro C, Knudsen S, Regenberg B, Olsson L, Nielsen J. 2005. Improvement of galactose uptake in Saccharomyces cerevisiae through overexpression of phosphoglucomutase: example of transcript analysis as a tool in inverse metabolic engineering. Appl. Environ. Microbiol. 71:6465–6472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chou HH, Chiu HC, Delaney NF, Segre D, Marx CJ. 2011. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 332:1190–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Conrad TM, Lewis NE, Palsson BO. 2011. Microbial laboratory evolution in the era of genome-scale science. Mol. Syst. Biol. 7:509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Daran JM, Bell W, Francois J. 1997. Physiological and morphological effects of genetic alterations leading to a reduced synthesis of UDP-glucose in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 153:89–96 [DOI] [PubMed] [Google Scholar]
- 7. de Jongh WA, et al. 2008. The roles of galactitol, galactose-1-phosphate, and phosphoglucomutase in galactose-induced toxicity in Saccharomyces cerevisiae. Biotechnol. Bioeng. 101:317–326 [DOI] [PubMed] [Google Scholar]
- 8. DeRisi JL, Iyer VR, Brown PO. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680–686 [DOI] [PubMed] [Google Scholar]
- 9. Dey NB, Bounelis P, Fritz TA, Bedwell DM, Marchase RB. 1994. The glycosylation of phosphoglucomutase is modulated by carbon source and heat shock in Saccharomyces cerevisiae. J. Biol. Chem. 269:27143–27148 [PubMed] [Google Scholar]
- 10. Elena SF, Lenski RE. 2003. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet. 4:457–469 [DOI] [PubMed] [Google Scholar]
- 11. Fu L, et al. 1995. The posttranslational modification of phosphoglucomutase is regulated by galactose induction and glucose repression in Saccharomyces cerevisiae. J. Bacteriol. 177:3087–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fu L, Miseta A, Hunton D, Marchase RB, Bedwell DM. 2000. Loss of the major isoform of phosphoglucomutase results in altered calcium homeostasis in Saccharomyces cerevisiae. J. Biol. Chem. 275:5431–5440 [DOI] [PubMed] [Google Scholar]
- 13. Garcia Sanchez R, Hahn-Hagerdal B, Gorwa-Grauslund MF. 2010. PGM2 overexpression improves anaerobic galactose fermentation in Saccharomyces cerevisiae. Microb. Cell Fact 9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hermann-Le Denmat S, Jacquet M. 1997. Yeast RAS2 mutations modulating the ras-guanine exchange factor interaction. FEBS Lett. 403:95–99 [DOI] [PubMed] [Google Scholar]
- 15. Herring CD, et al. 2006. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat. Genet. 38:1406–1412 [DOI] [PubMed] [Google Scholar]
- 16. Hlavata L, Aguilaniu H, Pichova A, Nystrom T. 2003. The oncogenic RAS2(val19) mutation locks respiration, independently of PKA, in a mode prone to generate ROS. EMBO J. 22:3337–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hong KK, Vongsangnak W, Vemuri GN, Nielsen J. 2011. Unraveling evolutionary strategies of yeast for improving galactose utilization through integrated systems level analysis. Proc. Natl. Acad. Sci. U. S. A. 108:12179–12184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Howard SC, Deminoff SJ, Herman PK. 2006. Increased phosphoglucomutase activity suppresses the galactose growth defect associated with elevated levels of Ras signaling in Saccharomyces cerevisiae. Curr. Genet. 49:1–6 [DOI] [PubMed] [Google Scholar]
- 19. James TC, Campbell S, Donnelly D, Bond U. 2003. Transcription profile of brewery yeast under fermentation conditions. J. Appl. Microbiol. 94:432–448 [DOI] [PubMed] [Google Scholar]
- 20. Khan AI, Dinh DM, Schneider D, Lenski RE, Cooper TF. 2011. Negative epistasis between beneficial mutations in an evolving bacterial population. Science 332:1193–1196 [DOI] [PubMed] [Google Scholar]
- 21. Lee KS, et al. 2011. Improved galactose fermentation of Saccharomyces cerevisiae through inverse metabolic engineering. Biotechnol. Bioeng. 108:621–631 [DOI] [PubMed] [Google Scholar]
- 22. Partow S, Siewers V, Bjorn S, Nielsen J, Maury J. 2010. Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae. Yeast 27:955–964 [DOI] [PubMed] [Google Scholar]
- 23. Patnaik R. 2008. Engineering complex phenotypes in industrial strains. Biotechnol. Prog. 24:38–47 [DOI] [PubMed] [Google Scholar]
- 24. Reimand J, Arak T, Vilo J. 2011. g:Profiler—a web server for functional interpretation of gene lists (2011 update). Nucleic Acids Res. 39:W307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Warner JR, Patnaik R, Gill RT. 2009. Genomics enabled approaches in strain engineering. Curr. Opin. Microbiol. 12:223–230 [DOI] [PubMed] [Google Scholar]
- 26. Wood DR, et al. 1994. Biochemical characterization of yeast RAS2 mutants reveals a new region of ras protein involved in the interaction with GTPase-activating proteins. J. Biol. Chem. 269:5322–5327 [PubMed] [Google Scholar]
- 27. Zaman S, Lippman SI, Zhao X, Broach JR. 2008. How Saccharomyces responds to nutrients. Annu. Rev. Genet. 42:27–81 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.