Abstract
Prophages contribute to the evolution and virulence of most bacterial pathogens, but their role in Clostridium difficile is unclear. Here we describe the isolation of four Myoviridae phages, ϕMMP01, ϕMMP02, ϕMMP03, and ϕMMP04, that were recovered as free viral particles in the filter-sterilized stool supernatants of patients suffering from C. difficile infection (CDI). Furthermore, identical prophages were found in the chromosomes of C. difficile isolated from the corresponding fecal samples. We therefore provide, for the first time, evidence of in vivo prophage induction during CDI. We completely sequenced the genomes of ϕMMP02 and ϕMMP04, and bioinformatics analyses did not reveal the presence of virulence factors but underlined the unique character of ϕMMP04. We also studied the mobility of ϕMMP02 and ϕMMP04 prophages in vitro. Both prophages were spontaneously induced, with 4 to 5 log PFU/ml detected in the culture supernatants of the corresponding lysogens. When lysogens were grown in the presence of subinhibitory concentrations of ciprofloxacin, moxifloxacin, levofloxacin, or mitomycin C, the phage titers further increased, reaching 8 to 9 log PFU/ml in the case of ϕMMP04. In summary, our study highlights the extensive genetic diversity and mobility of C. difficile prophages. Moreover, antibiotics known to represent risk factors for CDI, such as quinolones, can stimulate prophage mobility in vitro and probably in vivo as well, which underscores their potential impact on phage-mediated horizontal gene transfer events and the evolution of C. difficile.
INTRODUCTION
Clostridium difficile is the leading cause of antibiotic-associated nosocomial diarrhea in developed countries (18). Highly virulent strains, such as NAP1/027, have caused severe outbreaks in North America and Europe since 2003 and are now spreading worldwide, reaching Central America, Asia, and Australia (14). C. difficile infections (CDI) are a consequence of antibiotic treatments that reduce the diversity of the intestinal microbiota (11). C. difficile is a strictly anaerobic spore-forming Gram-positive bacillus that causes a wide range of clinical symptoms varying from mild to severe diarrhea to fatal pseudomembranous colitis. The pathogenic potential of C. difficile lies mainly in the expression of two large exotoxins, TcdA and TcdB, encoded on a pathogenicity locus (PaLoc) (26, 29). Additional virulence determinants are probably important for full virulence of this pathogen, but little is known about these factors and their importance in the development of CDI (38).
The rapid change in the epidemiology of C. difficile over the last decade has raised several concerns, and the genetic basis for the evolution of this pathogen is still unclear. With the advent of next-generation sequencing, several genomes of C. difficile have been sequenced recently. The data obtained reveal that the horizontal transfer of mobile genetic elements (MGE), such as conjugative transposons and prophages, likely accounts for the great plasticity of the C. difficile genome (23, 39, 43). For instance, 11% of the genomic DNA of strain 630 is made up of MGE, including 8 conjugative transposons and 2 functional and highly similar prophages (39). Moreover, the new epidemic NAP1/027 strain R20291 was found to have acquired 5 unique DNA regions containing phage and transposon genes, two-component systems, and transcriptional regulators, compared with the historic NAP1/027 strain CD196 and strain 630 (43). This suggests that the acquisition of genetic material through horizontal gene transfer is important in modeling the genome of C. difficile.
Recent studies have highlighted the great diversity of prophages in the clinical isolates of C. difficile (17, 36, 42), but only 5 fully characterized phages with complete genomic sequences are available in public databases. Phages ϕC2 (20), ϕCD119 (21), and ϕCD27 (33) are members of the Myoviridae family, i.e., phages with long nonflexible contractile tails (1), whereas phages ϕCD6356 (24) and ϕCD38-2 (40) are members of the Siphoviridae family, i.e., phages with long and flexible noncontractile tails. It is noteworthy to mention that all known phages of C. difficile are temperate, i.e., they can adopt either a lytic or a temperate lifestyle upon the infection of their host. Prophages are well-known contributors to the evolution of most bacterial species, including important pathogens (10), but their role in the virulence and evolution of C. difficile is still highly speculative. Two recent studies have shown that C. difficile phages ϕCD119 and ϕCD38-2 can modulate toxin production, even if these phages do not encode identifiable virulence factors (22, 40).
DNA-damaging and SOS-inducing stresses are often good prophage inducers that can contribute to horizontal gene transfer in bacteria (3, 32). The phenomenon of spontaneous and antibiotic-triggered prophage induction has been described for several phage-host systems, and the consequences of this phenomenon can be significant. For example, the increase in the production of Shiga toxins was shown to be tightly linked with spontaneous and quinolone-triggered induction of prophages in Escherichia coli (45, 47). Spontaneous prophage-induced lysis has been associated with the release of extracellular genomic DNA from Streptococcus pneumoniae and increased biofilm formation (13). Prophage induction from a subpopulation of bacteria can also lead to the killing of competing species, thus increasing the fitness of noninduced lysogens (8, 10, 31, 41), and can also promote horizontal gene transfer among bacteria, thereby speeding up genomic evolution (5, 10). CDI is a consequence of antibiotic treatments that destroy the intestinal microbiota, and recent epidemic clones of C. difficile are resistant to numerous antibiotics, including most fluoroquinolones (37). Studying the impact of antibiotics, in particular fluoroquinolones, on horizontal gene transfer and prophage mobility during C. difficile infection is thus of great interest.
In this study, we report the isolation of four different C. difficile phages that were recovered as free viral particles in the feces of patients suffering from CDI. We studied spontaneous and antibiotic-triggered prophage induction in vitro to assess the mobility of these phages. Finally, the complete genomic sequence was determined for two of these phages, thus providing additional genomic data on an understudied group of phages.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All bacterial isolates used in this work are listed in Table 1 and were kindly provided by Louis Valiquette and Jacques Pépin from the Université de Sherbrooke. When required, C. difficile was isolated from the feces of patients suffering from CDI and was subjected to alcohol shock and growth on Clostridium difficile moxalactam norfloxacin (CDMN) selective agar (Oxoid) supplemented with 5% sheep blood, 0.1% taurocholate, and 1 mM glycine. The institutional review board of the Centre Hospitalier Universitaire de Sherbrooke (CHUS) approved the study protocol. Bacteria were routinely grown at 37°C in an anaerobic chamber (Coy Laboratories) in brain heart infusion broth (BHI) (BD Bioscience) or TY broth (3% tryptose, 2% yeast extract [pH 7.4]). All media were prereduced overnight under anaerobic conditions.
Table 1.
C. difficile isolates used in this study
| Isolate no. | PCR ribotypea | Comment |
|---|---|---|
| Isolates used for phage enrichment and detection | ||
| CD19 | 036 | |
| CD24 | 038 | |
| CD71 | 017 | |
| CD73 | 012 | |
| CD95 | 014 | |
| CD114 | 014 | |
| CD117 | 023 | |
| CD121 | 010 | |
| CD127 | 014 | |
| CD132 | 012 | |
| CD139 | 014 | |
| CD161 | 027 | |
| CD162 | 036 | |
| CD171 | 027 | |
| CD173 | 027 | |
| Naturally occurring lysogensb | ||
| CD343 | 023 | Clinical isolate carrying ϕMMP02b |
| CD368 | 026 | Clinical isolate carrying ϕMMP03b |
| CD380 | 006 | Clinical isolate carrying ϕMMP04b |
| Laboratory-generated lysogensc | ||
| CD407 | 036 | CD19 isolate lysogenized with ϕMMP01 |
| CD408 | 023 | CD117 isolate lysogenized with ϕMMP02 |
| CD411 | 023 | CD117 isolate lysogenized with ϕMMP03 |
| CD412 | 012 | CD73 isolate lysogenized with ϕMMP04 |
PCR ribotype 027 represents the current BI/NAP1/027 epidemic clone, and the other ribotype numbers were given arbitrarily according to our internal database.
Naturally occurring lysogens were isolated from stool samples that contained the indicated free phages.
Laboratory-generated lysogens were obtained upon stable infection with the indicated phage.
Bacterial DNA extraction and PCR ribotyping.
Total genomic DNA was purified using the Illustra bacterial genomic DNA extraction kit (GE Healthcare) as described previously (40). All C. difficile isolates were analyzed by PCR ribotyping using an Eppendorf Mastercycler with 20 ng purified genomic DNA and primers published by Bidet et al. (7), with modifications described elsewhere (17). Band patterns were analyzed with GelComparII (Applied Maths), and Pearson's correlation coefficient was used for cluster analyses.
Phage enrichment and isolation.
Sewage samples from two water treatment plants in Sherbrooke and human fecal samples from patients suffering from CDI collected over a 1-year period were screened for the presence of free phages. Raw sewage samples (400 ml) were passed through 1.5-μm Whatman filters (Schleicher & Schuell), followed by another filtration through 0.45-μm EZ-Pak filters (Millipore), and then 5-ml volumes from 3 different samples were pooled. Stool samples were homogenized in 10 ml of BHI, centrifuged at 4,000 x g for 60 min at room temperature, and then passed through 0.45-μm filter discs to remove bacteria. Five stool samples were then pooled before the phage enrichment procedure. For phage enrichment, 15 C. difficile isolates were used as hosts, which represented 8 different PCR ribotypes, including 3 isolates of ribotype 027 (Table 1). Enrichment was done by adding 1.25 ml of the pooled sewage sample to 1.5 ml of BHI containing 10 mM CaCl2 and 10 mM MgCl2 (BHIS) and a 2% (vol/vol) inoculum of an overnight C. difficile culture. For enrichment from stool samples, 2.5 ml of pooled stool supernatants was combined with 2.5 ml of BHIS and 2% of an overnight C. difficile culture. The next day, the cultures were centrifuged for 15 min at 4,000 × g and passed through 0.45-μm filter discs (Sarstedt). A second enrichment was performed in a total volume of 5 ml BHIS, using 2.5 ml of the first enrichment as the phage inoculum. Finally, a third enrichment step was done as described above using the second enrichment broth as the phage inoculum. Culture supernatants were then filter sterilized on 0.45-μm disks, and 0.1-ml samples were added to soft agar overlays inoculated with the same strains as the hosts, as described previously (40).
Phage purification and amplification.
Phages obtained after the enrichment procedure were purified from single isolated phage plaques using three successive rounds of soft agar overlays as described before (40). Phage titers of ≥109 PFU/ml were routinely obtained with this method.
Creation of lysogens and prophage induction.
C. difficile lysogens carrying ϕMMP02 or ϕMMP04 prophages were created by spreading dilutions of phage-sensitive C. difficile cultures on soft agar overlays containing phages (108 PFU/ml), as described previously (40). Five potentially phage-immune colonies were picked and restreaked 3 times on BHI agar plates without phage to purify the lysogens. The presence of the integrated prophage was confirmed by Southern blot hybridization using lysogenic bacterial DNA and a digoxigenin (DIG)-labeled whole-phage DNA probe (17). Alternatively, PCR with phage-specific primers was used. The functionality of the integrated prophage was verified by treating lysogens with UV light (302 nm) or mitomycin C (3 μg/ml), followed by phage DNA purification and restriction analyses, as described previously (40).
MIC determination.
The MICs for ciprofloxacin (CIP), moxifloxacin (MXF), and levofloxacin (LVX) were determined in 96-well plates. Briefly, antibiotics were serially diluted in 96-wells plates in a final volume of 0.1 ml. An equal volume of a bacterial culture at an optical density at 600 nm (OD600) of 0.3 was added to each well. Plates were incubated under anaerobic conditions at 37°C, and the OD600 was monitored every 10 min over 16 h using a PowerWave XS microplate reader (BioTek Instruments).
Prophage induction by quinolones.
Lysogenic bacteria were grown on BHI soft agar plates containing either ϕMMP02 or ϕMMP04 to ensure that bacteria were still phage immune and carrying the prophages. A single colony was picked and grown overnight in TY broth. Cultures were then washed by centrifugation in TY broth in order to eliminate any free-phage particle that could have induced spontaneously, and 0.1 ml of washed bacteria was used to inoculate 10 ml of fresh TY broth. When the OD600 reached 0.15, 0.5× the MIC and lower concentrations of antibiotics were added and the OD600 was monitored for a total of 8 h. Mitomycin C was used as a control for prophage induction (17). One-milliliter aliquots from each induction assay were then centrifuged at 14,000 x g for 1 min to remove bacterial cells, and the supernatants were stored at 4°C. Phage titers were determined by soft agar overlays containing the sensitive host strain as described earlier. A control without antibiotic was also run in parallel to determine the level of spontaneous prophage induction. At least three independent assays were performed, and the mean ± standard error of the mean (SEM) of the log PFU/ml was plotted. Student's t test and one-way analysis of variance (ANOVA) analyses were performed with Prism 5.04 (GraphPad) to determine whether antibiotics stimulated prophage induction compared to untreated controls. The level of statistical significance was set to a P value of <0.05.
Transmission electron microscopy.
Phage particles were washed in ammonium acetate, fixed onto 400-mesh Formvar/carbon-coated copper grids (Cederlane Laboratories), and negatively stained with 2% uranyl acetate (Cederlane Laboratories) as described previously (17). Phage particles were observed with a Hitachi H-7500 transmission electron microscope (TEM) operating at 60 kV, and pictures were taken with a 10-megapixel digital camera (Hamamatsu) controlled with the AMT software (Advanced Microscopy Techniques).
Phage DNA purification, restriction analysis, and Southern hybridization.
A rapid phenol-chloroform protocol was used for small-scale phage DNA purification from crude lysates (34), and the Lambda Maxi DNA purification kit (Qiagen) was used for large-scale preparations. Restriction enzyme analysis of whole-phage DNA was done as described elsewhere (40), and Southern hybridization was carried out using DIG-labeled whole-phage DNA probes (17).
Phage genome sequencing and bioinformatics analysis.
Complete phage genome sequencing was performed on a 454 GS FLX sequencer system (Roche) at the Génome Québec Innovation Center of McGill University (Montréal, QC, Canada). Single contigs were obtained for both phages, and additional sequencing was performed directly on purified phage DNA with specific primers on an ABI 3730xl sequencer (Applied Biosystems) at the genomic platform of the CHUL research center (Québec, QC, Canada). Additional sequence assembly was done using the Gap v4.10 application of the Staden v1.6.0 package. Some editing was also done using BioEdit v7.0.5.3 and Artemis 13.0. Putative open reading frames (ORFs) encoding ≥30 amino acids were predicted using GeneMark.hmm for Prokaryotes v2.8 (28). All predicted ORFs were translated into proteins using the standard ATG initiation codon or the alternative codons GTG and TTG, based on the presence of a suitable ribosome-binding site. The predicted proteins were compared with the BLASTp tools of the NCBI (4) and A CLAssification of Mobile genetic Elements (ACLAME) (27) databases. The identification of conserved domains was performed through searches in the Conserved Domains Database (CDD) (NCBI) and InterProScan analyses (46).
Nucleotide sequence accession number.
The complete genome sequences of phages ϕMMP02 and ϕMMP04 have been deposited in GenBank under the accession numbers JX145341 and JX145342, respectively.
RESULTS
Phage isolation.
Our initial goal was to isolate strictly lytic (“virulent”) phages in raw sewage samples and the feces from CDI patients using an enrichment protocol. A total of 30 sewage samples and 59 stool samples were processed, and only 6 stool samples contained free phage particles capable of infecting the C. difficile isolates we selected. Phage plaques were detected on isolates CD19, CD73, and CD117, which represent three different PCR ribotypes (Table 1). Six phages were isolated from independent stool samples, 3 of which had identical HindIII DNA restriction profiles. Hence, they were considered to be identical phages, and only one of them, ϕMMP02, was studied further (Fig. 1). Overall, 4 phages had unique HindIII restriction profiles and were thus considered to be different. Phage ϕMMP01 was isolated on strain CD19, ϕMMP02 and ϕMMP03 on strain CD117, and ϕMMP04 on strain CD73. Phage particles were observed under TEM, and they all had an isometric head with a diameter of 58 to 70 nm connected by a neck to a sheathed tail of 106 to 248 nm long and about 20 nm wide (Table 2). Some particles with contracted sheaths were observed in the lysates (data not shown), and based on our observations, these phages would be classified as members of the Myoviridae family of the order Caudovirales (1).
Fig 1.
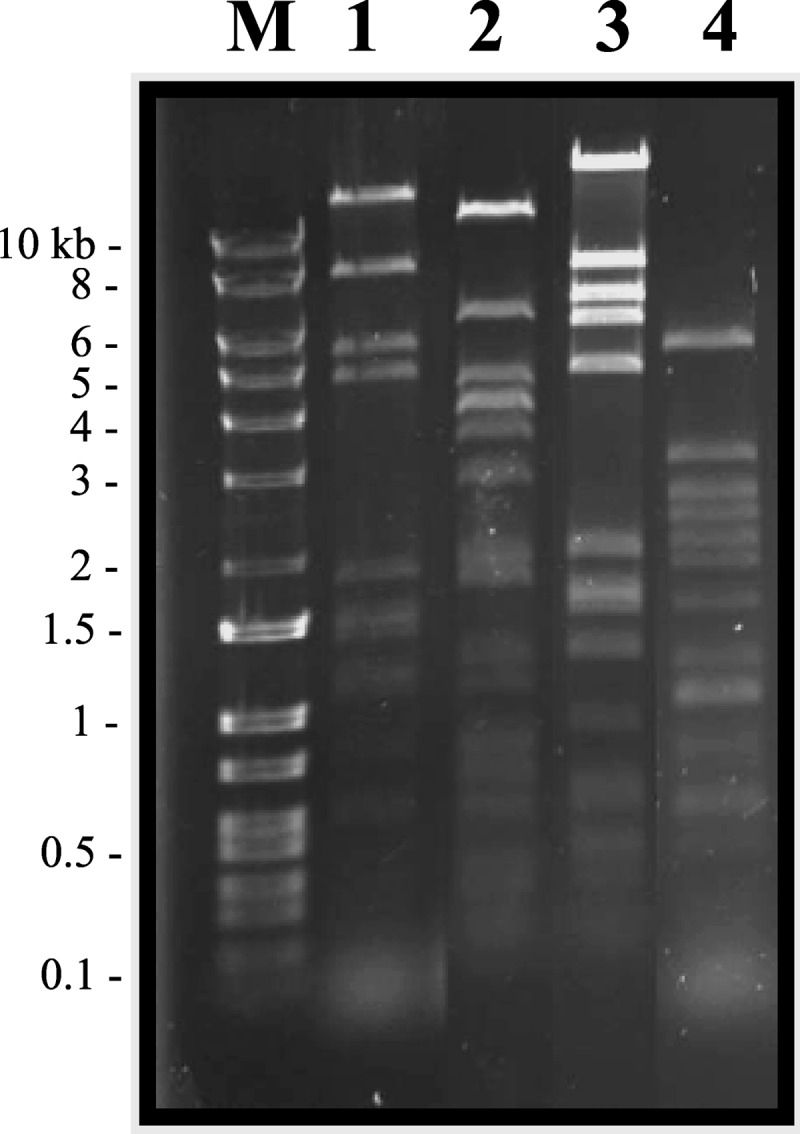
HindIII restriction profiles of isolated phages. The ethidium bromide-stained gel shows ϕMMP01 (lane 1), ϕMMP02 (lane 2), ϕMMP03 (lane 3), and ϕMMP04 (lane 4). Lane M, DNA Logic Ladder.
Table 2.
Morphological characteristics of isolated phages in the family Myoviridae
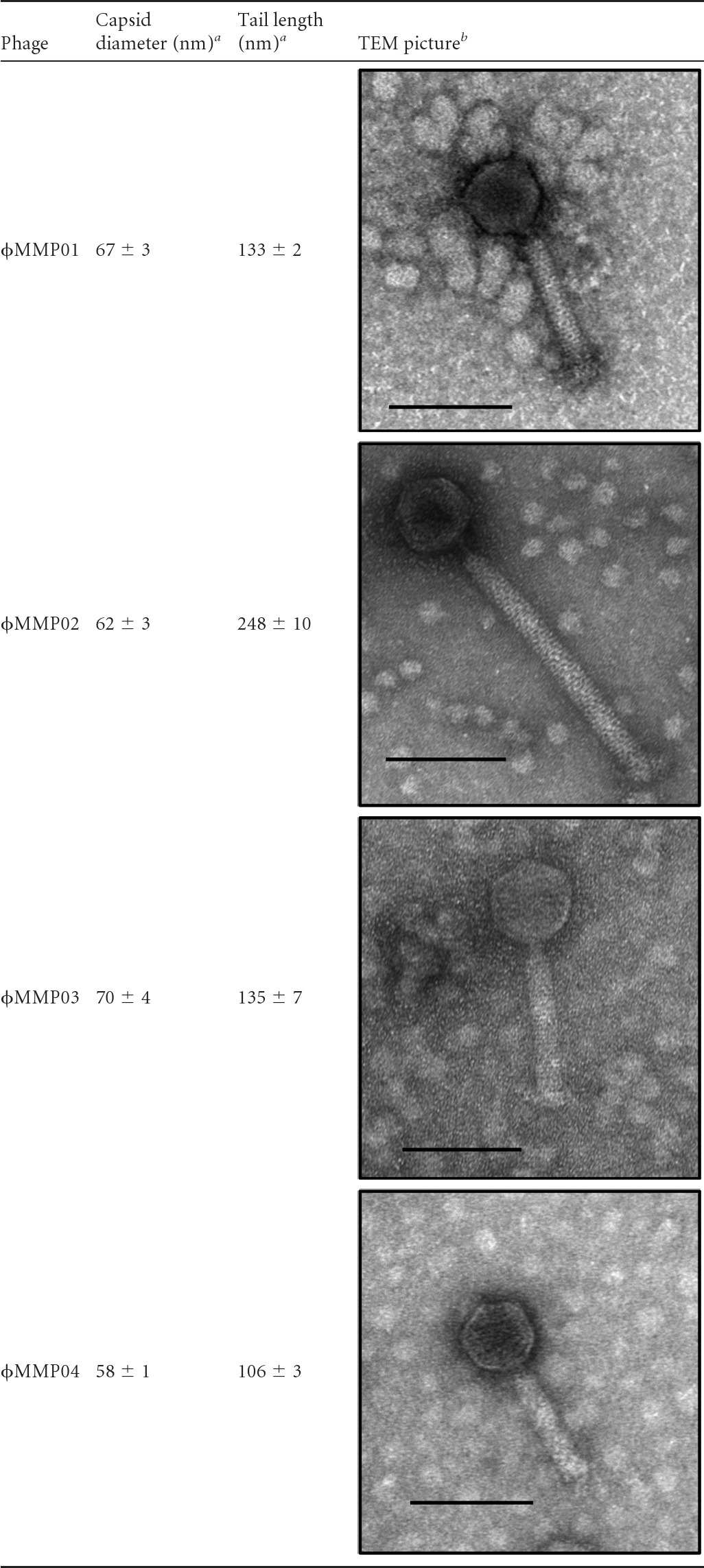
a Means of 5 measurements obtained with different viral particles.
b The black bars represent 100 nm.
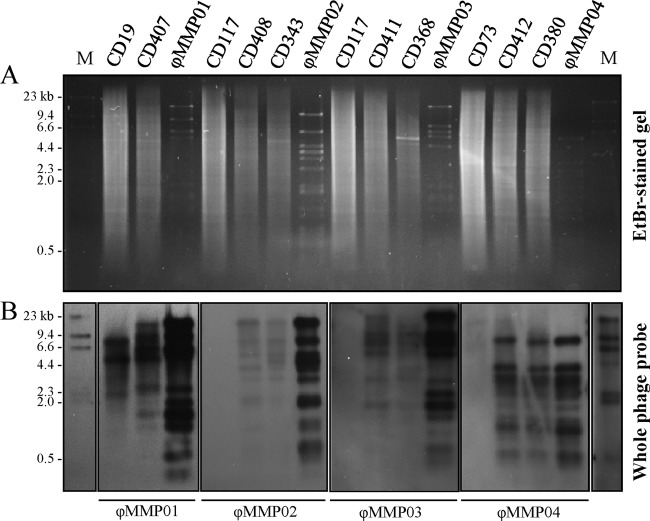
We verified whether the 15 C. difficile isolates that we used in the enrichment protocol contained endogenous prophages corresponding to those that we isolated from fecal samples. As shown in Fig. 2, Southern hybridizations with whole-phage DNA probes corresponding to ϕMMP01, ϕMMP02, ϕMMP03, and ϕMMP04 confirmed that the C. difficile-sensitive isolates CD19, CD73, and CD117 did not carry these prophages, although CD19 carried a somewhat similar prophage but with a different restriction profile. Thus, the phages that we isolated had not been induced from C. difficile isolates used in the course of our enrichment and screening protocol but were truly free phage particles present in the stool samples. We attempted to detect the phage particles by direct plating of fecal supernatants on indicator strains without prior enrichment, but the titers were below the limit of detection.
Fig 2.
Prophage detection by Southern blot hybridization with whole-phage DNA probes. DIG-labeled whole-phage DNA probes (ϕMMP01 to -04) were used to confirm the presence of corresponding prophages in wild-type strains and laboratory-generated lysogens. (A) Ethidium bromide (EtBr)-stained gel of HindIII-digested bacterial genomic DNA and purified phage DNA (ϕMMP01 to -04). (B) Southern blot hybridization of the gel shown in panel A; the phage probes used are indicated below the corresponding panels. Lane M, DIG-labeled lambda HindIII DNA marker (NEB).
Phage lifestyle.
In order to determine whether ϕMMP01, ϕMMP02, ϕMMP03, and ϕMMP04 were virulent or temperate, we infected the sensitive hosts CD19, CD73, and CD117 at a high multiplicity of infection (MOI) with the corresponding phages and screened for lysogens. Several colonies were obtained, and Southern hybridization assays were performed on the extracted genomic DNA using whole-phage DNA probes. As shown in Fig. 2, prophages with restriction profiles corresponding to each infecting phage were found in the chromosome of the CD19, CD73, and CD117 lysogens but not in the parental uninfected isolates. A few minor differences were observed between the restriction profiles of the purified phages and the lysogens, which is the consequence of integration of the phage DNA into the bacterial chromosome. Additional UV and mitomycin C treatments were done on the lysogens to confirm the functionality of the prophages (data not shown). Our results confirmed that ϕMMP01, ϕMMP02, ϕMMP03, and ϕMMP04 are all temperate phages.
Since free temperate phages were isolated directly from stool samples, we deduced that they had probably been released from indigenous C. difficile cells during infection. To demonstrate that, we isolated C. difficile from the phage-positive stool samples and looked for the presence of the corresponding prophage by Southern blot hybridization. We could not recover C. difficile from the stool sample containing ϕMMP01 due to loss of the initial sample, but as predicted, isolates CD343, CD368, and CD380 carried a prophage corresponding to ϕMMP02, ϕMMP03, and ϕMMP04, respectively (Fig. 2). This confirmed that the prophages had been induced and released by C. difficile during infection. To our knowledge, this is the first report of such in vivo prophage induction by C. difficile.
Influence of antibiotics on prophage induction.
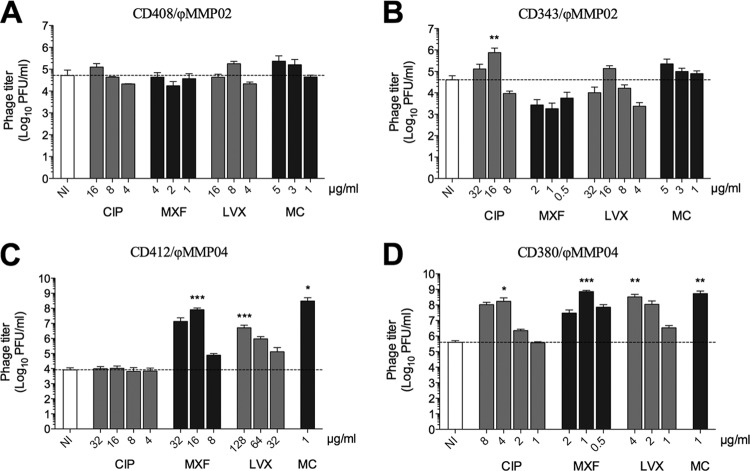
Prophage induction from lysogens was assessed in vitro in the presence of three common quinolones. We focused our analyses on ϕMMP02 and ϕMMP04 because we determined the genomic sequence of these two phages only (see below). The MICs for CIP, MXF, and LVX were determined on lysogenic isolates CD343 and CD380, which correspond to the naturally occurring clinical isolates purified from phage-positive stools and carry ϕMMP02 and ϕMMP04, respectively. The MICs were also determined on CD408 and CD412 that were obtained upon lysogenization of strains CD117 and CD73 with ϕMMP02 and ϕMMP04, respectively (Table 1). Bacteria were grown in the presence of sub-MICs (≤0.5× MIC) of antibiotics, and phage titers were determined in culture supernatants after 8 h of growth. As shown in Fig. 3, ϕMMP02 and ϕMMP04 spontaneously induced and initiated a lytic cycle, leading to the release of ∼4 to 5.5 log PFU/ml after 8 h of growth (white bars). In addition, the spontaneous induction of ϕMMP04 from the naturally occurring CD380 lysogen led to a phage titer ∼1.5 log higher than that of the laboratory-generated CD412 lysogen, suggesting a greater stability in the latter strain. In contrast, the spontaneous induction of ϕMMP02 was similar in both CD343 and CD408 lysogens, with titers of ∼5 × 104 PFU/ml (Fig. 3A and B).
Fig 3.
Effect of antibiotics on ϕMMP02 and ϕMMP04 induction. Prophage induction was assessed after 8 h of growth in the presence of various sub-MICs of ciprofloxacin (CIP), moxifloxacin (MXF), or levofloxacin (LVX). Mitomycin C (MC) was used as a positive control of induction. (A and C) Induction from laboratory-generated lysogens. (B and D) Induction from wild-type lysogens. The differences in phage titers were analyzed by one-way ANOVA followed by Dunnett's posttest using the noninduced control (NI) as the comparator. One asterisk indicates significance with a P value of <0.05, two asterisks indicate significance with a P value of <0.01, and three asterisks indicate significance with a P value of <0.001.
Treatment of C. difficile lysogens carrying ϕMMP02 with different sub-MICs of CIP, MXF, or LVX or with mitomycin C (MC) had little effect on prophage induction (Fig. 3A and B). A slight increase in the phage titers was observed at some concentrations, but these differences were not statistically significant after one-way ANOVA analyses. The only exception was observed with 16 μg/ml CIP (0.25× MIC), where a statistically significant increase in the phage titer was noted (7.5 × 105 versus 4.5 × 104 PFU/ml, P < 0.01). These results suggest that induction of the ϕMMP02 prophage by quinolones and MC is not very efficient, at least under the conditions tested.
In contrast, the ϕMMP04 prophage was more sensitive to treatment with MXF, LVX, and MC with phage titers 2 to 4 logs higher than in the untreated controls in both lysogens tested (Fig. 3C and D). For example, the highest phage titers obtained with the CD402 lysogen were 8.1 × 107, 5.1 × 106, and 3.1 × 108 PFU/ml after treatment with MXF, LVX, and MC, respectively, whereas the untreated control released 8.3 × 103 PFU/ml (Fig. 3C). For the CD380 lysogen, treatment with CIP, MXF, LVX, and MC led to the release of 1.7 × 108, 7.4 × 108, 3.3 × 108, and 5.4 × 108 PFU/ml, respectively, as opposed to a phage titer of 4 × 105 PFU/ml in the untreated control (Fig. 3D). Interestingly, treatment with CIP stimulated prophage induction in the wild-type CD380 lysogen (Fig. 3D) but not in the laboratory-generated CD402 lysogen (Fig. 3C). Taken together, our results suggest that the stability of ϕMMP02 and ϕMMP04 prophages is similar, but ϕMMP04 is significantly less stable in the presence of antibiotics.
Whole-genome comparison.
We assessed the overall genome similarity of the phages ϕMMP01, ϕMMP02, ϕMMP03, and ϕMMP04 by Southern blot hybridization with whole-phage probes (Fig. 4). We compared the four phages with each other and also with other phages from our collection (17). Hybridization with a ϕMMP02 probe revealed significant DNA similarity with ϕCD52 but limited similarity with ϕMMP01 and ϕMMP03, whereas hybridization with a ϕMMP03 probe revealed extensive similarity with ϕMMP01, ϕCD52, ϕCD630-2, and ϕCD24-2. The 4 ϕMMP phages were also found to be genetically distant from ϕCD38-2, a Siphoviridae phage that we described previously (data not shown) (40). The phage ϕMMP04 seemed to be very different from the 3 other ϕMMP phages, suggesting that this phage is genetically unique among our collection of isolates. In brief, ϕMMP01, ϕMMP02, and ϕMMP03 were similar to other known Myoviridae phages, but ϕMMP04 seemed genetically unique.
Fig 4.
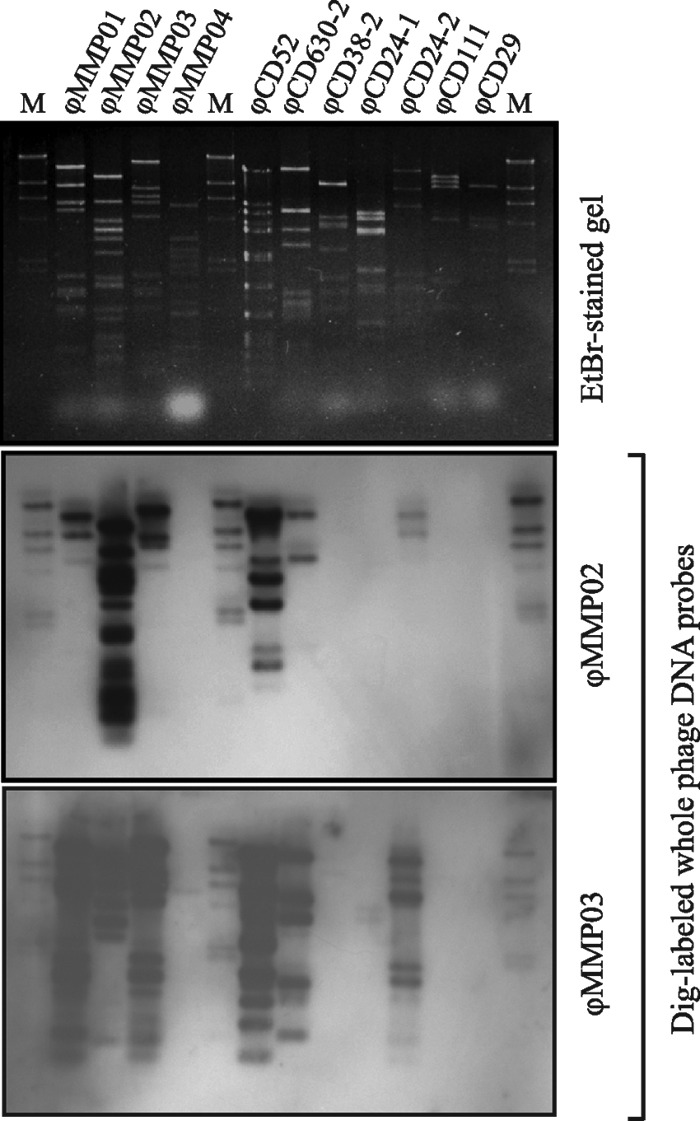
Whole-phage-genome comparison using Southern blot hybridization. Purified phage DNA from different temperate phages in our collection was compared using DIG-labeled whole-phage genomes as probes. Phages ϕCD38-2, ϕCD24-1, ϕCD111, and ϕCD29 are Siphoviridae phages, whereas all the others are Myoviridae phages. Lane M, DIG-labeled lambda HindIII DNA marker (NEB).
Genome sequencing.
The low similarity observed by Southern hybridizations between ϕMMP02, ϕMMP04, and the other phages prompted us to determine their whole genomic sequences. The complete genome of ϕMMP02 was determined after 454 sequencing and the assembly of 51,685 reads with an average length of 317 bp. The sequencing of ϕMMP04 also resulted in a single contig of 31,662 bp assembled from 23,416 reads (average length of 318 bp). For both phages, additional PCR and direct sequencing on purified phage DNA confirmed the completeness of the genomes. The phage ϕMMP02 is composed of a double-stranded DNA of 48,396 bp with an average G+C content of 29.6%, while the ϕMMP04 genome is much smaller, with 31,674 bp of double-stranded DNA and a G+C content of 30.0%. To our knowledge, this is the smallest C. difficile phage genome sequence reported so far.
Genomic organization and comparative analysis.
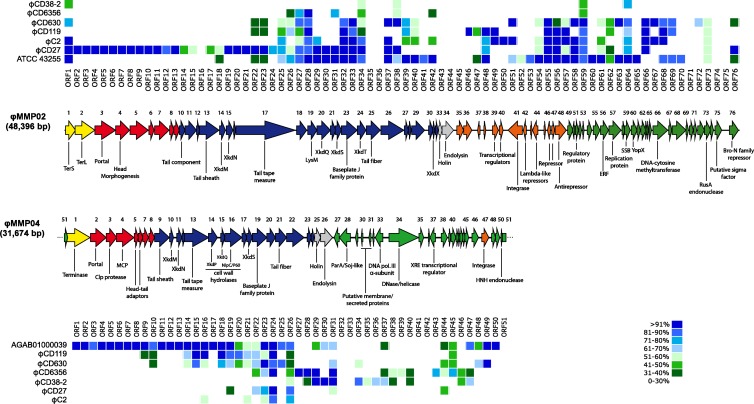
GeneMark.hmm analyses were performed on both the ϕMMP02 and ϕMMP04 genomes (28). Seventy-six and fifty-one putative open reading frames (ORFs) encoding ≥30 amino acids were found in ϕMMP02 and ϕMMP04, respectively. Comparison against the NCBI and ACLAME databases enabled us to assign a putative function to 36 of the 76 ORFs (47%) in ϕMMP02 and 24 of the 51 ORFs (47%) in ϕMMP04 (see Tables S1 and S2 in the supplemental material). The overall genomic organization of both phages appeared to be classical, with clusters of genes coding for distinct functional modules (Fig. 5). It is noteworthy to mention that virulence factors and toxin genes were not found in the two genomes using the bioinformatics approach. Protein comparison against public databases and the 5 C. difficile phage genomic sequences currently available revealed a great extent of similarity between the phage ϕMMP02 and previously characterized Myoviridae phages. For example, the whole DNA packaging, structural, and lysis modules (ORFs 1 to 34) encode proteins highly similar, and for the most part unique, to phage ϕCD27 (Fig. 5, upper panel). However, some tail proteins, as well as the holin and endolysin (ORFs 22 to 34), were also similar to phages ϕC2 and ϕCD119, prophages from C. difficile strain 630, and a putative prophage from strain ATCC 43255. In fact, besides ϕCD27, the prophage in strain ATCC 43255 was the most similar to ϕMMP02, with 30 ORFs showing >71% protein identity. Some divergence was observed in ORFs 24 to 28 between ϕMMP02 and ϕCD27, suggesting that these tail proteins are probably involved in host specificity.
Fig 5.
Genome organization of ϕMMP02 and ϕMMP04. The arrows indicate the predicted ORFs and their respective orientation. The putative functions inferred from bioinformatics analyses are indicated below the ORFs. Functional modules were assigned with regard to gene annotation and whole genomic organization, and the color code is as follows: yellow, DNA packaging; red, capsid morphogenesis; blue, tail morphogenesis; gray, lysis; orange, lysogeny; and green, DNA replication, transcription, and gene regulation. The dot matrices above and below the genomic maps show the degree of protein identity observed among ϕMMP02, ϕMMP04, and other known phages using BLASTp analysis.
In the case of ϕMMP04, little similarity was observed with previously characterized C. difficile phage genomes over the packaging, capsid, and part of the tail modules (ORFs 1 to 13) (Fig. 5, lower panel). However, similarity was observed with proteins found in Clostridium hiranonis, Clostridium cellulovorans, and Clostridium botulinum. Also, a prophage with extensive similarity to ϕMMP04 was identified in a draft genome of a C. difficile strain (UniProt accession no. AGAB01000039). The whole packaging, head and tail structural modules, as well as the lysis cassette (ORFs 1 to 26) were highly similar at the protein level (>81% identity). As for ϕMMP02, some of the tail fiber proteins (ORFs 20 to 22) diverged between ϕMMP04 and the other phages, suggesting that these proteins may also be responsible for host specificity. Some of the tail proteins (ORFs 9 to 26) were also similar to proteins from phage ϕCD119 and prophages from strain 630. We found several ORFs whose products had significant similarity with proteins from two Siphoviridae phages, ϕCD6356 and ϕCD38-2 (Fig. 5), and the similarity was concentrated downstream from the lysis cassette (ORF 26).
Lysogeny module.
The phage ϕMMP02 has a lysogeny module delimited by the endolysin gene on the left side (ORF 34) and a set of phage repressors and regulators on the right side (ORFs 42 to 48). A similar organization was also reported in other Myoviridae phages infecting C. difficile (20, 21, 33). However, a lysogeny module could not be clearly identified in ϕMMP04; an integrase gene (ORF 47) and a putative phage repressor (ORF 37) were found interspersed between other DNA replication and regulation genes (Fig. 5). This type of organization was also reported in the temperate Siphoviridae phage ϕCD38-2 (40). In brief, the similarity observed at the protein level between ϕMMP04, ϕCD38-2, and ϕCD6356 in the DNA replication/gene regulation and lysogeny modules suggests that ϕMMP04 is somewhat related to the Siphoviridae phages.
DISCUSSION
We report the isolation and characterization of four phages infecting C. difficile: ϕMMP01, ϕMMP02, ϕMMP03, and ϕMMP04. TEM observations revealed that they are morphologically similar to other Myoviridae phages in C. difficile that were recently described (17, 20, 21, 33, 36, 42). These phages were isolated as free viral particles in the feces from patients infected by C. difficile, and identical prophages were found in the chromosomes of the C. difficile strains present in the corresponding fecal samples. We demonstrated that the phages were also able to lysogenize other laboratory strains of C. difficile, confirming their temperate lifestyle. We therefore conclude that the ϕMMP phages were spontaneously induced from C. difficile in vivo. To our knowledge, this is the first report providing evidence of in vivo prophage induction during C. difficile infection.
The search for strictly lytic phages, i.e., those that can only infect and kill their host by lysis, has become very attractive in recent years because of their potential usefulness as therapeutic agents (2). Nobody has ever reported the isolation of such phages that are active against C. difficile, and all phages known to infect this species are temperate (15, 19, 21, 24, 30, 33, 35, 36, 40, 42). Using only 15 different C. difficile test strains, we were able to detect free phages in 10% of the fecal samples tested. Considering the very narrow host spectrum of C. difficile phages in general, we could probably detect more phages if additional test strains were used. Therefore, in vivo prophage induction appears to occur frequently during CDI. On the contrary, free phage particles could not be isolated in sewage samples. In fact, due to extreme oxygen sensitivity, most live cells of C. difficile are expected to be in their spore form outside the mammalian gut and are thus insensitive to phage infection. The propagation of a virulent phage under these conditions should therefore be very unlikely. Conversely, bacteria are metabolically active during infection and are thus susceptible to phage attacks. In this context, a temperate lifestyle with controlled spontaneous prophage induction from a subset of the bacterial population seems to be a better strategy for guaranteeing phage survival and dissemination than is a strictly lytic lifestyle. Such a strategy seems to be the one adopted by phages infecting Streptococcus pyogenes (9, 16).
In an effort to gain insight into the genetics of the C. difficile phages, we determined the complete genomic sequence of ϕMMP02 and ϕMMP04. No virulence factors or toxin genes could be readily inferred from bioinformatics analyses, which so far seems to be a common feature of this group of phages (20, 21, 24, 33, 39, 40). Nevertheless, recent studies have suggested that even in the absence of identifiable virulence factors or toxin genes, ϕCD119 and ϕCD38-2 prophages can influence toxin production in C. difficile (22, 40). Our comparative genomic analyses also further demonstrate the mosaic nature and the great genetic diversity of this group of phages, with ϕMMP02 and ϕMMP04 forming distinct phage families based on their head and tail structural components. Moreover, the presence of an atypical regulatory/lysogeny module and similarity with phages ϕCD6356 and ϕCD38-2 also suggest that ϕMMP04 might have arisen from a past recombination event between a Myoviridae and a Siphoviridae phage. This is the first example of such an unusual genomic organization among C. difficile phages, but only two Siphoviridae phage genomes, ϕCD6356 (24) and ϕCD38-2 (40), have been sequenced so far. As we obtain more sequences of Siphoviridae phages in the future, we might observe other examples of cross-family recombination events.
Under laboratory conditions, the ϕMMP02 and ϕMMP04 lysogens spontaneously released 4 to 5 log PFU/ml after 8 h of incubation, thus suggesting a certain degree of instability among C. difficile prophages. Similar phage titers have also been reported after growth of lysogenic staphylococcal strains under normal laboratory conditions (31). Some years ago, Mahony et al. reported the spontaneous release of two bacteriophages after in vitro screening of 94 C. difficile isolates (30). Goh et al. also noticed that 72% of their laboratory-generated lysogens carrying either ϕC2, ϕC6, or ϕC8 spontaneously lost their prophage upon storage at −70°C, while no stable lysogen of ϕC5 could be obtained (19). The biological impacts of spontaneous prophage induction on the virulence and lifestyle are well established for other bacteria, such as Staphylococcus aureus, S. pneumoniae, S. pyogenes, and E. coli (6, 12, 13, 25), but the importance of this phenomenon in C. difficile has yet to be demonstrated.
Ciprofloxacin and beta-lactams were shown to promote the transfer of pathogenicity islands (SaPIs) in S. aureus (31, 44), and norfloxacin was recently used to induce several prophages from C. difficile isolates (36, 42). We have shown that ϕMMP04 was particularly unstable compared to ϕMMP02 in the presence of ciprofloxacin, moxifloxacin, and levofloxacin, and phage titers several orders of magnitude above the titers of noninduced cultures were observed. Our in vitro results strongly suggest that antibiotics such as fluoroquinolones, known to represent risk factors for CDI (37), could also increase the frequency of prophage induction in vivo and, potentially, the frequency of phage-mediated gene transfer during CDI. Antibiotic pressure is an important driving force in bacterial evolution, and the extensive use of antibiotics that induce CDI and that stimulate prophage mobility at the same time could provide an ideal background for the rapid evolution of C. difficile and its associated phages. Further genetic and molecular studies will be needed to better grasp the impacts of prophages on the lifestyle and evolution of C. difficile.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a discovery grant from the Natural Sciences and Engineering Research Council of Canada (NSERC), by a seed grant from the Canadian Institutes of Health Research (CIHR), and by the Centre de Recherche Clinique Étienne-Le Bel. L.-C.F. is the holder of a Junior 2 research award from the Fonds de la Recherche du Québec–Santé (FRQ-S).
Footnotes
Published ahead of print 24 August 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Ackermann HW. 2009. Phage classification and characterization. Methods Mol. Biol. 501:127–140 [DOI] [PubMed] [Google Scholar]
- 2. Adhya S, Merril C. 2006. The road to phage therapy. Nature 443:754–755 [Google Scholar]
- 3. Allen HK, et al. 2011. Antibiotics in feed induce prophages in swine fecal microbiomes. mBio 2:e00260-11 doi:10.1128/mBio.00260-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 5. Asadulghani M, et al. 2009. The defective prophage pool of Escherichia coli O157: prophage-prophage interactions potentiate horizontal transfer of virulence determinants. PLoS Pathog. 5:e1000408 doi:10.1371/journal.ppat.1000408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Banks DJ, Lei B, Musser JM. 2003. Prophage induction and expression of prophage-encoded virulence factors in group A Streptococcus serotype M3 strain MGAS315. Infect. Immun. 71:7079–7086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bidet P, Barbut F, Lalande V, Burghoffer B, Petit J. 1999. Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol. Lett. 175:261–266 [DOI] [PubMed] [Google Scholar]
- 8. Bossi L, Fuentes JA, Mora G, Figueroa-Bossi N. 2003. Prophage contribution to bacterial population dynamics. J. Bacteriol. 185:6467–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Broudy TB, Fischetti VA. 2003. In vivo lysogenic conversion of Tox(−) Streptococcus pyogenes to Tox(+) with lysogenic streptococci or free phage. Infect. Immun. 71:3782–3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brussow H, Canchaya C, Hardt WD. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buffie CG, et al. 2012. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect. Immun. 80:62–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cao R, et al. 2012. Elevated enterotoxin A expression and formation in Staphylococcus aureus and its association with prophage induction. Appl. Environ. Microbiol. 78:4942–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carrolo M, Frias MJ, Pinto FR, Melo-Cristino J, Ramirez M. 2010. Prophage spontaneous activation promotes DNA release enhancing biofilm formation in Streptococcus pneumoniae. PLoS One 5:e15678 doi:10.1371/journal.pone.0015678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clements ACA, Magalhães RJS, Tatem AJ, Paterson DL, Riley TV. 2010. Clostridium difficile PCR ribotype 027: assessing the risks of further worldwide spread. Lancet Infect. Dis. 10:395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dei R. 1989. Observations on phage-typing of Clostridium difficile: preliminary evaluation of a phage panel. Eur. J. Epidemiol. 5:351–354 [DOI] [PubMed] [Google Scholar]
- 16. Fischetti V. 2007. In vivo acquisition of prophage in Streptococcus pyogenes. Trends Microbiol. 15:297–300 [DOI] [PubMed] [Google Scholar]
- 17. Fortier LC, Moineau S. 2007. Morphological and genetic diversity of temperate phages in Clostridium difficile. Appl. Environ. Microbiol. 73:7358–7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Freeman J, et al. 2010. The changing epidemiology of Clostridium difficile infections. Clin. Microbiol. Rev. 23:529–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goh S, Riley T, Chang B. 2005. Isolation and characterization of temperate bacteriophages of Clostridium difficile. Appl. Environ. Microbiol. 71:1079–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goh S, Ong PF, Song KP, Riley TV, Chang BJ. 2007. The complete genome sequence of Clostridium difficile phage phiC2 and comparisons to phiCD119 and inducible prophages of CD630. Microbiology 153:676–685 [DOI] [PubMed] [Google Scholar]
- 21. Govind R, Fralick J, Rolfe R. 2006. Genomic organization and molecular characterization of Clostridium difficile bacteriophage PhiCD119. J. Bacteriol. 188:2568–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Govind R, Vediyappan G, Rolfe RD, Dupuy B, Fralick JA. 2009. Bacteriophage-mediated toxin gene regulation in Clostridium difficile. J. Virol. 83:12037–12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. He M, et al. 2010. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc. Natl. Acad. Sci. U. S. A. 107:7527–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Horgan M, et al. 2010. Genome analysis of the Clostridium difficile phage PhiCD6356, a temperate phage of the Siphoviridae family. Gene 462:34–43 [DOI] [PubMed] [Google Scholar]
- 25. Imamovic L, Muniesa M. 2012. Characterizing RecA-independent induction of Shiga toxin2-encoding phages by EDTA treatment. PLoS One 7:e32393 doi:10.1371/journal.pone.0032393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuehne SA, et al. 2010. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467:711–713 [DOI] [PubMed] [Google Scholar]
- 27. Leplae R, Hebrant A, Wodak SJ, Toussaint A. 2004. ACLAME: a CLAssification of Mobile genetic Elements. Nucleic Acids Res. 32:D45–D49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lukashin AV, Borodovsky M. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26:1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lyras D, et al. 2009. Toxin B is essential for virulence of Clostridium difficile. Nature 458:1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mahony D, Bell P, Easterbrook K. 1985. Two bacteriophages of Clostridium difficile. J. Clin. Microbiol. 21:251–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maiques E, et al. 2006. beta-lactam antibiotics induce the SOS response and horizontal transfer of virulence factors in Staphylococcus aureus. J. Bacteriol. 188:2726–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Matsushiro A, Sato K, Miyamoto H, Yamamura T, Honda T. 1999. Induction of prophages of enterohemorrhagic Escherichia coli O157:H7 with norfloxacin. J. Bacteriol. 181:2257–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mayer MJ, Narbad A, Gasson MJ. 2008. Molecular characterization of a Clostridium difficile bacteriophage and its cloned biologically active endolysin. J. Bacteriol. 190:6734–6740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moineau S, Pandian S, Klaenhammer T. 1994. Evolution of a lytic bacteriophage via DNA acquisition from the Lactococcus lactis chromosome. Appl. Environ. Microbiol. 60:1832–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nagy E, Foldes J. 1991. Electron microscopic investigation of lysogeny of Clostridium difficile strains isolated from antibiotic-associated diarrhea cases and from healthy carriers. Acta Pathol. Microbiol. Immunol. Scand. 99:321–326 [DOI] [PubMed] [Google Scholar]
- 36. Nale JY, et al. 2012. Diverse temperate bacteriophage carriage in Clostridium difficile 027 strains. PLoS One 7:e37263 doi:10.1371/journal.pone.0037263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pepin J, et al. 2005. Emergence of fluoroquinolones as the predominant risk factor for Clostridium difficile-associated diarrhea: a cohort study during an epidemic in Quebec. Clin. Infect. Dis. 41:1254–1260 [DOI] [PubMed] [Google Scholar]
- 38. Rupnik M, Wilcox MH, Gerding DN. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7:526–536 [DOI] [PubMed] [Google Scholar]
- 39. Sebaihia M, et al. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779–786 [DOI] [PubMed] [Google Scholar]
- 40. Sekulovic O, Meessen-Pinard M, Fortier L-C. 2011. Prophage-stimulated toxin production in Clostridium difficile NAP1/027 lysogens. J. Bacteriol. 193:2726–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Selva L, et al. 2009. Killing niche competitors by remote-control bacteriophage induction. Proc. Natl. Acad. Sci. U. S. A. 106:1234–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shan J, et al. 2012. Prophage carriage and diversity within clinically relevant strains of Clostridium difficile. Appl. Environ. Microbiol. 78:6027–6034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stabler RA, et al. 2009. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 10:R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ubeda C, et al. 2005. Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Mol. Microbiol. 56:836–844 [DOI] [PubMed] [Google Scholar]
- 45. Wagner PL, et al. 2002. Bacteriophage control of Shiga toxin 1 production and release by Escherichia coli. Mol. Microbiol. 44:957–970 [DOI] [PubMed] [Google Scholar]
- 46. Zdobnov EM, Apweiler R. 2001. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848 [DOI] [PubMed] [Google Scholar]
- 47. Zhang X, et al. 2000. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181:664–670 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.