Abstract
16S rRNA gene molecular analysis elucidated the spatiotemporal distribution of bacterial biofilm communities along a water quality gradient. Multivariate statistics indicated that terrestrial runoff, in particular dissolved organic carbon and chlorophyll a concentrations, induced shifts of specific bacterial communities between locations and seasons, suggesting microbial biofilms could be suitable bioindicators for water quality.
TEXT
Tropical coral reefs are facing global climate change (e.g., warming sea surface temperatures), and local-scale disturbances at an increasing rate (13, 21, 50). However, local-scale natural (e.g., monsoonal rainfalls and cyclones) and anthropogenic (e.g., land-based activities) impacts result in terrestrial runoff and deteriorate water quality, putting coastal regions at greater risk (18). The Proserpine River catchment in the Whitsunday Island area, Great Barrier Reef (GBR), Australia, is considered a priority coastal management region due to land clearing and agriculture (i.e., sugarcane farming) (17). In particular during the summer wet season, terrestrial runoff inshore leads to higher concentrations of nutrients, chlorophyll a (Chl a), and suspended sediments, forming cross-shelf water quality gradients (5, 7, 11, 17, 44). Changes in water quality show significant alterations in coral reef dynamics (14, 30, 44). Alterations also include associated microorganisms that predominately exist as surface-attached biofilms (32), contributing importantly to ecosystem productivity, biogeochemical fluxes (4, 27), and invertebrate larval settlement (46, 49). Bacterial communities are highly responsive indicators of changing environmental conditions (35, 40), as demonstrated in rivers (3), estuaries (33, 34), and polar (45) and temperate (8) marine waters. Biofilms may therefore also find application as a biomonitoring tool of transient spatiotemporal variability and more persistent ecosystem change due to large-scale GBR river catchment management decisions (controlling farming activities) or global climate change. Despite the suitability of bacterial biofilm communities as environmental bioindicators, biofilms in tropical coral reefs remain poorly explored.
This study therefore aimed to explore the spatiotemporal variation and response of bacterial biofilm communities in coastal coral reefs to seasonal terrestrial runoff disturbances at pristine and human-impacted sites. Biofilms were established during repeated summer wet and winter dry seasons at 5 nearshore fringing reefs along a water quality gradient in the Whitsunday Islands. A molecular approach (terminal restriction fragment length polymorphism [T-RFLP] and 16S rRNA gene clone libraries) and a multivariate statistical approach (permutational multivariate analysis of variance [PERMANOVA], principal component analysis [PCA], and distance-based redundancy analysis [dbRDA]) were used to identify the effects of location and season and the most decisive water quality parameters to determine the main controls driving microbial community shifts. Finally, the utility of bacterial biofilms as bioindicators for water quality in tropical coral reefs was evaluated.
The water quality gradient included permanent sites of the long-term Reef Plan Marine Monitoring Program comprising 3 inner nearshore islands (Pine, Daydream, and Double Cone, located <10 km from the coast and subjected to higher concentrations of nutrients and suspended sediments) and 2 outer nearshore islands (Edward and Deloraine, located >30 km from the coast and which are less exposed to terrestrial runoff) in the Whitsunday Islands, GBR (see Table S1 in the supplemental material) (7, 24, 41, 43, 44). Water quality samples (for determination of the variables Chl a concentration, total suspended solids, Secchi depth, dissolved inorganic nitrogen [DIN; the sum of NH4, NO2, and NO3], dissolved organic carbon [DOC], temperature, and salinity) were obtained monthly between 2008 and 2010 and analyzed as described in detail in references 7 and 37.
By scuba diving, three replicate glass slides were deployed vertically (6-m water depth) at 2 replicate sites (25 m apart) at each reef (52). Biofilms were developed repeatedly for ∼48 days during two seasonal cycles (2008 to 2009 and 2009 to 2010; dry season, 22°C; wet season, 29°C). Biofilms (6 replicates per island and 60 replicates per season, for a total of 120 samples) were scraped off the substrate using scalpel blades, immediately frozen in liquid nitrogen, and stored at −80°C. Total DNA was extracted from a 0.5-g biofilm (wet weight) sample using the MoBio UltraClean soil kit (MoBio Laboratories, Solana Beach, CA). Eight 16S rRNA gene clone libraries (one per island [excluding Double Cone] and season [2008 to 2009]) were constructed (Table 1). Samples were purified using the MinELUTE PCR clean-up kit (Qiagen) and cloned using a TOPO-TA cloning kit (Invitrogen). Clones (n = 655; ∼80 per library) were purified and sequenced using an ABI3730 XL automatic DNA sequencer at the Australian Genome Research Facility, Ltd. (Brisbane, Australia). Retrieved sequences were edited using Chromas Lite 2.33 (Technelysium Pty., Ltd., Australia). The Greengenes database and tools (greengenes.lbl.gov) were used for sequence alignment (10), chimera check (Bellerophon version 3 [22]), and classification (NCBI taxonomy) (31).
Table 1.
Relative proportions of the indicated phylogenetic groups in the total 16S rRNA gene clone library sequences
| Phylogenetic group | Proportion (%) of group found by location and seasona |
|||||||
|---|---|---|---|---|---|---|---|---|
| Inner nearshore |
Outer nearshore |
|||||||
| Pine Island |
Daydream Island |
Deloraine Island |
Edward Island |
|||||
| Dry (n = 81) | Wet (n = 78) | Dry (n = 84) | Wet (n = 86) | Dry (n = 83) | Wet (n = 80) | Dry (n = 83) | Wet (n = 81) | |
| Bacteroidetes | 32.14 | 20.51 | 25.41 | 31.4 | 13.1 | 23.17 | 14.12 | 18.50 |
| Cyanobacteria | 2.38 | 6.41 | 0 | 1.60 | 2.38 | 8.54 | 3.53 | 19.75 |
| Firmicutes | 0 | 0 | 1.176 | 2.30 | 3.57 | 0 | 2.35 | 0 |
| Alphaproteobacteria | 13.10 | 10.26 | 18.8 | 25.60 | 64.3 | 20.73 | 15.29 | 9.80 |
| Betaproteobacteria | 0 | 0 | 0 | 1.60 | 0 | 0 | 0 | 0 |
| Deltaproteobacteria | 4.76 | 0 | 5.88 | 0 | 1.19 | 0 | 1.17 | 1.23 |
| Gammaproteobacteria | 14.29 | 34.62 | 20 | 23.30 | 10.7 | 20.73 | 24.71 | 24.69 |
| Acidobacteria | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.23 |
| Actinobacteria | 2.38 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Oceaniserpentilla | 0 | 1.28 | 0 | 0 | 0 | 0 | 0 | 0 |
| Nitrospirae | 0 | 0 | 1.176 | 0 | 0 | 0 | 0 | 0 |
| Unclassified bacteria | 7.14 | 3.85 | 4.70 | 10.77 | 1.19 | 17.07 | 10.59 | 11.04 |
| Diatom plastids | 23.81 | 23.07 | 17.65 | 3.49 | 3.57 | 9.76 | 28.24 | 13.76 |
The values shown indicate the relative proportions of the indicated phylogenetic groups as percentages of the total 16S rRNA gene clone library sequences (where n is the number of clones sequenced).
For terminal restriction fragment polymorphism (T-RFLP), bacterial 16S rRNA genes were amplified by PCR using universal fluorescently labeled 5′-Cy5 63F forward (5′-CAGGCCTAACACATGCAAGTC-3′) and unlabeled 1389R reverse (5′-ACGGGCGGTGTGTACAAG-3′) primers (Sigma-Proligo, The Woodlands, TX) (28) as described in reference 51. Terminal restriction fragments (T-RFs) were resolved and visualized using the CEQ 8800 genetic analysis system (Beckman-Coulter, Fullerton, CA) and analyzed using the software T-Align (39). Purified DNA from individual clones was used for verification and identification of the taxonomic identity of T-RFs.
To determine location and season differences in bacterial assemblages, multivariate statistics were performed on T-RFLP data (which were square root transformed and standardized). The multivariate statistics included principal component analysis (PCA), pairwise t tests, and two-way permutational multivariate analysis of variance (PERMANOVA) based on 9,999 permutations using the Bray-Curtis distance measure and P values derived from Monte-Carlo simulations [P(MC)]) The relative fluorescence intensity of the T-RF peak area was a measure for relative abundance. Taxon contributions to the total dissimilarities were analyzed using the similarity percentage (SIMPER) routine. To determine the relationship between water quality parameters (z-transformed, where average = 0 and standard deviation [SD] = 1) and bacterial communities (16S rRNA T-RFLP data) were determined by a distance-based redundancy analysis (dbRDA) using multivariate multiple regression model (distance linear-based model [DistLM]) marginal tests with a Bray-Curtis distance matrix based on 9,999 permutations. Multivariate statistics (6, 29) were performed using the Primer 6.0 statistical software (6). Two-way analysis of variance (ANOVA) was performed to determine significant differences between relative abundances (peak area) of contributing T-RFs, using the Number Cruncher Statistical System 2007 statistical software (NCSS, Kaysville, UT) (20) (for method details, see the supplemental material).
Generally, our water quality data followed previously recognized spatial and seasonal patterns from previous years (7, 37, 38) with higher concentrations of most parameters (Chl a, suspended solids, and nutrient species) at inner compared to outer nearshore sites and higher concentrations during the wet than the dry season (see Table S2 in the supplemental material).
16S rRNA gene clone library analysis revealed that independent of location and season, the phyla Alphaproteobacteria, Gammaproteobacteria, and Bacteroidetes were consistently most frequent in bacterial biofilm communities. Bacteroidetes-affiliated sequences were consistently more frequent at inner than outer nearshore sites (Table 1), while Gammaproteobacteria were equally abundant at both locations. In contrast, Cyanobacteria were more abundant at outer than inner nearshore reefs. In contrast to the aforementioned sequences, Alphaproteobacteria-affiliated sequences were more frequent in the dry than the wet season at each location, with Daydream Island as an exception (Table 1).
Terminal restriction fragment polymorphism of 16S rRNA genes revealed a total of 60 T-RFs, of which 80% could be assigned to clones from the clone libraries, apart from 12 T-RFs that could not be confirmed by any of the clones (see Table S3 in the supplemental material). Permutational multivariate analysis of variance (PERMANOVA) of the overall community data using the factors season (wet/dry) and location (inner/outer nearshore islands, excluding Double Cone Island), revealed that at inner and outer nearshore locations bacterial biofilm community compositions were significantly different between season and island, with a significant interaction term (P = < 0.05, 2-way PERMANOVA) (see Table S4A in the supplemental material). Interaction post hoc tests revealed significant seasonal differences in communities at both locations [P(MC) < 0.05], explained by the high community data variability from the wet season (see Tables S4B and S4C). Principal component analysis (PCA) showed overlap of microbial assemblages between locations, but segregation between seasons (Fig. 1). Furthermore, seasons formed distinct assemblages within each location, which may explain the significant interaction term of the PERMANOVA (Fig. 1). Similarity percentage (SIMPER) analysis revealed an overall average dissimilarity of 79.2% between locations and seasons, with Alteromonadaceae (Gammaproteobacteria) (contribution, 7.1%), Cyanobacteria (6.1%), Flavobacteriaceae (5.0%), diatom plastids (4.8%), and Roseobacter (Alphaproteobacteria) (4.0%) contributing the most to the community composition dissimilarities. These phylogenetic groups were reconfirmed by the length of the vectors in the PCA biplot. A significantly higher relative abundance of Roseobacter (16.9% higher at outer than inner locations) and Cyanobacteria (19.3%) at outer than inner nearshore locations was detected (1-way ANOVA, F = 4.04 and P = 0.0473 and F = 6.12 and P = 0.0403, respectively). The relative abundance of Flavobacteriaceae was significantly higher (by 24%) during the dry season compared to the wet season (ANOVA, F = 7.07 and P = 0.0092), while Alteromonadaceae increased (by 300%) in the wet season (ANOVA, F = 34.45 and P < 0.0001).
Fig 1.
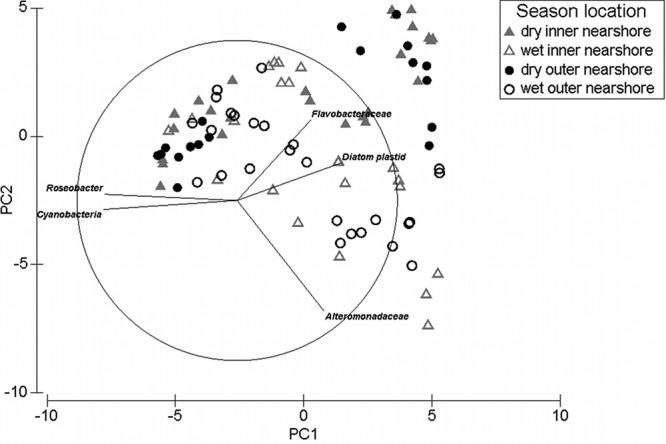
Principal component analysis (PCA) incorporating relative abundances of T-RFs (using the relative fluorescence peak intensity matrix) showing bacterial assemblages at different locations and seasons. Solid triangles, dry season, inner nearshore; open triangles, wet season, inner nearshore; solid circles, dry season, outer nearshore; open circles, wet season, outer nearshore. PC1, 13.8%; PC2, 27.8%. Vectors of the most importantly contributing T-RFs (bp), as revealed by a similarity percentage (SIMPER) analysis, are shown in the biplot and labeled with the corresponding taxons.
In a distance-based redundancy analysis (dbRDA), seven environmental variables (total suspended solids [TSS], salinity, dissolved inorganic nitrogen [DIN], dissolved organic carbon [DOC], chlorophyll a [Chl a], temperature, and Secchi depth) explained a significant amount of the variation in the microbial community data (DistLM marginal test; see Table S5 in the supplemental material), of which DOC (6.69% of the explained variance) and Chl a (6.57% variance) contributed the most to microbial community differences (Fig. 2; see Table S5). dbRDA illustrated that Alteromonadaceae and diatom plastids were correlated with high Chl a concentrations, while Cyanobacteria and Roseobacter were correlated with low Chl a concentrations (Fig. 2). To some extent, Flavobacteriaceae were correlated with small amounts of DOC. Our findings suggest that the parameters Chl a and DOC, both found at high concentrations inshore during the wet season, were the main drivers in spatiotemporal bacterial community shifts. This is in concordance with previous studies on biofilms from local aquaria studies (53) and midshelf reefs from the GBR (46) and temperate marine bacterioplankton (16, 19, 23).
Fig 2.
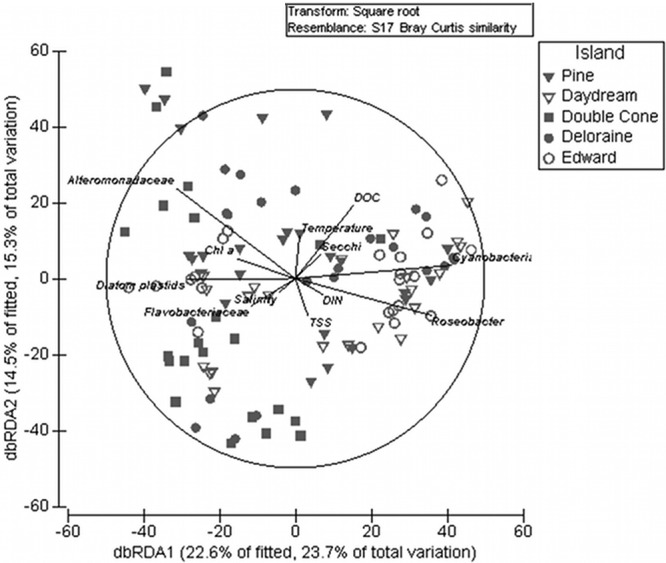
Relationship between measured coastal coral reef water quality parameters and changes in bacterial biofilm community profiles from replicate glass substrata deployed within the reef during both seasons (wet and dry). The plot represents a distance-based redundancy analysis (dbRDA) ordination of 16S rRNA T-RFLP trace data derived from biofilms generated from a Bray-Curtis distance matrix and with water quality variables chosen by significances of distance linear-based model (DisTLM) marginal tests.
16S rRNA gene sequencing and fingerprinting data (Table 2) identified five dominant, opportunistic bacterial groups correlated with changing water quality along a tropical cross-shelf gradient, growing to population sizes that can be measured by culture-independent techniques. The distributions of Gammaproteobacteria, diatom plastids, and Bacteroidetes at inner nearshore biofilm communities and Cyanobacteria and Alphaproteobacteria as the primary bacterial groups in outer nearshore biofilm communities have previously been detected in sediments (42) and biofilms (24, 52) at inshore GBR sites, the Atlantic coast (8), and bacterioplankton from a temperate river plume to the coastal water gradient (15). Seasonal distributions were in concordance with previous observations: e.g., higher abundances of Flavobacteriaceae linked to phytoplankton bloom decays (36) and Altermonadaceae in response to organic carbon loads after disturbances (1, 2, 48).
Table 2.
Bacterial diversity estimate of biofilm communities from different locations and seasons derived from 16S rRNA gene fingerprinting and clone librariesa
| Location and season | Total no. of: |
Result byb: |
|||||
|---|---|---|---|---|---|---|---|
| T-RFs | Clones sequenced | OTU | Chao1 | ACE | H′ | 1-D | |
| Overall | 60 | 655 | 463 | ||||
| Pine Island | |||||||
| Dry | 27 | 81 | 52 | 87.1 | 113 | 3.59 | 0.02 |
| Wet | 27 | 78 | 50 | 88.2 | 119 | 3.71 | 0.02 |
| Daydream Island | |||||||
| Dry | 27 | 84 | 67 | 238 | 534 | 4.07 | 0.01 |
| Wet | 28 | 86 | 71 | 253 | 246 | 4.17 | 0.01 |
| Deloraine Island | |||||||
| Dry | 28 | 83 | 62 | 73.8 | 112 | 2.86 | 0.09 |
| Wet | 35 | 80 | 63 | 221 | 509 | 3.97 | 0.01 |
| Edward Island | |||||||
| Dry | 22 | 83 | 51 | 215 | 546 | 3.58 | 0.03 |
| Wet | 33 | 81 | 47 | 97.8 | 92.6 | 3.63 | 0.02 |
Operational taxonomic units (OTU), average richness estimators, and diversity indices were calculated for each clone library.
Chao1, Chao1 richness estimator; ACE, ACE richness estimator; H′, Shannon-Weaver index of diversity; and 1-D, Simpson's index of diversity. Values for 3% unique distance similarity difference (97% species level) were generated with MOTHUR.
Several microbial bioindicator studies repeatedly reconfirmed the aforementioned phylogenetic groups with similar responses (24, 51–53). Of particular relevance were the decreasing relative abundance of Alphaproteobacteria (mostly Roseobacter) and the concomitant increase in Bacteroidetes (mostly Flavobacteriaceae) detected in biofilms in response to increased water temperature (47, 53), partial CO2 pressure (pCO2) (51), and nitrate (53). Potentially valuable as bioindicator groups to be integrated into long-term coastal health monitoring programs, these phyla should be studied in more depth (53). In contrast, phyla commonly found in soils and freshwater (Verrucomicrobia, Acidobacteria, Actinobacteria, and Deltaproteobacteria), expected to arrive with runoff waters, were minor or absent groups, as was also observed in sediments (42) and biofilms (24) from the GBR. However, PCR bias, affecting relative abundance measurements, and the limitations of T-RFLP, only capturing dominant bacteria, may be reasons for the underrepresentation of rare bacterial groups. However, fingerprinting techniques (i.e., automated rRNA intergenic spacer analysis [ARISA] and T-RFLP) have been shown to be capable of detecting significant bacterial variation in biofilms across spatial and temporal scales, representing a useful tool for water quality biomonitoring in streams (25, 26). Therefore, we propose that T-RFLP may provide a cost-effective and rapid approach to monitor land use impacts on coastal water quality in tropical coral reefs (52).
Our findings contribute to new insights into the distribution of surface-attached coastal coral reef bacterial communities. Bacteria within a biofilm respond to environmental stress over short periods (weeks) in advance of other components of the ecosystem and therefore serve as bioindicators to detect early warning responses to changes in water quality and consequent changes in ecosystem integrity. This is important in coastal coral reefs close to estuaries, where the dynamics of the system often preclude adequate characterization from single spatial and temporal point grab water quality samples. It is, however, unlikely that biofilms can be developed as absolute indicators of a condition; they are perhaps rather best employed as a relative indicator of differences across spatial and temporal scales. As nutrient-poor coral reefs display highly efficient nutrient cycling, these ecosystems are considered particularly vulnerable to excess nutrient input. For example, Chl a concentrations increased by 100% (average ± SD, 0.415 ± 0.07 μg liter−1) at outer shore locations and by 120% (average ± SD 0.86 ± 0.04 μg liter−1) at inner nearshore locations during the wet season compared to the dry season. The chlorophyll a concentration, highly correlated with particulate nitrogen, phosphorous, and suspended solids (12), is commonly used as a proxy for nutrient status and a measure of water column productivity (5). Mean annual values of Chl a concentrations for the inshore GBR are <0.45 μg liter−1 and have been suggested as water quality thresholds for coastal management (9). Inner dry season inshore GBR N concentrations lie well below this threshold, while inner wet season values show a 2-fold increase, herewith exceeding the aforementioned threshold levels. Therefore, it is suggested that Chl a and the DOC of the water quality parameters are best for indicating conditions that alter microbial composition in these tropical coastal coral reef ecosystems. Further work should therefore include investigation of the physiological and metabolic variability accompanying bacterial community shifts to understand the effects of deteriorating water quality on coral reef ecosystem productivity and functioning in order to improve coastal management.
Nucleotide sequence accession numbers.
All sequences determined in this study have been deposited in GenBank under accession no. JQ726882 to JQ727208 and JF261700 to JF262029.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Humphrey, C. Reymond, J. van Dam, and the crew on board the RV Cape Ferguson for assistance in the field. We are grateful to I. Zagorskis for providing the water quality data. We also thank C. Reymond for helpful comments on the manuscript.
The water quality data were collected as part of the Reef Plan Marine Monitoring Program, which is supported by the Great Barrier Reef Marine Park Authority (GBRMPA) through funding from the Australian Government's Caring for our Country and by the Australian Institute of Marine Science (AIMS). This project (project 3.7.1) was funded by the Australian Government Marine and Tropical Sciences Research Facility (MTSRF).
Footnotes
Published ahead of print 17 August 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Allers E, et al. 2007. Response of Alteromonadaceae and Rhodobacteriaceae to glucose and phosphorus manipulation in marine mesocosms. Environ. Microbiol. 10:2417–2429 [DOI] [PubMed] [Google Scholar]
- 2. Alonso-Saez L, Gasol JM. 2007. Seasonal variations in the contributions of different bacterial groups to the uptake of low-molecular-weight compounds in northwestern Mediterranean coastal waters. Appl. Environ. Microbiol. 73:3528–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Araya R, Tani K, Takagi T, Yamaguchi N, Nasu M. 2003. Bacterial activity and community composition in stream water and biofilm from an urban river determined by fluorescent in situ hybridization and DGGE analysis. FEMS Microbiol. Ecol. 43:111–119 [DOI] [PubMed] [Google Scholar]
- 4. Battin TJ, Kaplan LA, Denis Newbold J, Hansen CM. 2003. Contributions of microbial biofilms to ecosystem processes in stream mesocosms. Nature 426:439–442 [DOI] [PubMed] [Google Scholar]
- 5. Brodie J, De'ath G, Devlin M, Furnas M, Wright M. 2007. Spatial and temporal patterns of near-surface chlorophyll a in the Great Barrier Reef lagoon. Mar. Freshw. Res. 58:342–353 [Google Scholar]
- 6. Clarke K, Gorley RN. 2006. PRIMER v6: user manual/tutorial. PRIMER-E, Lutton, Ivybridge, United Kingdom [Google Scholar]
- 7. Cooper TF, Uthicke S, Humphrey C, Fabricius KE. 2007. Gradients in water column nutrients, sediment parameters, irradiance and coral reef development in the Whitsunday Region, central Great Barrier Reef. Estuar. Coast. Shelf Sci. 74:458–470 [Google Scholar]
- 8. Dang HY, Li TG, Chen MN, Huang GQ. 2008. Cross-ocean distribution of Rhodobacterales bacteria as primary surface colonizers in temperate coastal marine waters. Appl. Environ. Microbiol. 74:52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. De'ath G, Fabricius K. 2010. Water quality as a regional driver of coral biodiversity and macroalgae on the Great Barrier Reef. Ecol. Appl. 20:840–850 [DOI] [PubMed] [Google Scholar]
- 10. DeSantis TZ, et al. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:W394–W399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fabricius KE. 2005. Effects of terrestrial runoff on the ecology of corals and coral reefs: review and synthesis. Mar. Pollut. Bull. 50:125–146 [DOI] [PubMed] [Google Scholar]
- 12. Fabricius KE, De'ath G. 2004. Identifying ecological change and its causes: a case study on coral reefs. Ecol. Appl. 14:1448–1465 [Google Scholar]
- 13. Fabricius KE, Golbuu Y, Victor S. 2007. Selective mortality in coastal reef organisms from an acute sedimentation event. Coral Reefs 26:69 [Google Scholar]
- 14. Fabricius KE, Wild C, Wolanski E, Abele D. 2003. Effects of transparent exopolymer particles and muddy terrigenous sediments on the survival of hard coral recruits. Estuar. Coast. Shelf Sci. 57:613–621 [Google Scholar]
- 15. Fortunato CS, Herfort L, Zuber P, Baptista AM, Crump BC. 2012. Spatial variability overwhelms seasonal patterns in bacterioplankton communities across a river to ocean gradient. ISME J. 6:554–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fuhrman JA, et al. 2006. Annually reoccurring bacterial communities are predictable from ocean conditions. Proc. Natl. Acad. Sci. U. S. A. 103:13104–13109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Furnas M. 2003. Catchments and corals: terrestrial runoff to the Great Barrier Reef. Australian Institute of Marine Science, Townsville, Queensland, Australia [Google Scholar]
- 18. GESAMP 2001. Protecting the oceans from land-based activities. Land-based sources and activities affecting the quality and uses of the marine, coastal and associated freshwater environment. Joint Group of Experts on the Scientific Aspects of Marine Environmental Protection, United Nations Environment Program, International Maritime Organization, London, United Kingdom [Google Scholar]
- 19. Gilbert JA, et al. 2012. Defining seasonal marine microbial community dynamics. ISME J. 6:298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hintze J. 2007. NCSS 2007. NCSS, LLC, Kaysville, UT [Google Scholar]
- 21. Hoegh-Guldberg O, Mumby PJ, Hooten AJ. 2007. Coral reefs under rapid climate change and ocean acidification. Science 318:1737–1742 [DOI] [PubMed] [Google Scholar]
- 22. Huber T, Faulkner G, Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319 [DOI] [PubMed] [Google Scholar]
- 23. Jamieson RE, Rogers AD, Billett DSM, Smale DA, Pearce DA. 19 March 2012. Patterns of marine bacterioplankton biodiversity in the surface waters of the Scotia Arc, Southern Ocean. FEMS Microbiol. Ecol. [Epub ahead of print.] doi:10.1111/j.1574-6941.2012.01313.x [DOI] [PubMed] [Google Scholar]
- 24. Kriwy P, Uthicke S. 2011. Microbial diversity in marine biofilms along a water quality gradient on the Great Barrier Reef. Syst. Appl. Microbiol. 34:116–126 [DOI] [PubMed] [Google Scholar]
- 25. Lear G, Anderson MJ, Smith JP, Boxen K, Lewis GD. 2008. Spatial and temporal heterogeneity of the bacterial communities in stream epilithic biofilms. FEMS Microbiol. Ecol. 65:463–473 [DOI] [PubMed] [Google Scholar]
- 26. Lear G, Lewis GD. 2009. Impact of catchment land use on bacterial communities within stream biofilms. Ecol. Indicators 9:848–855 [Google Scholar]
- 27. Lock MA, Wallace RR, Costerton JW, Ventullo RM, Charlton SE. 1984. River epilithon: toward a structural-functional model. Oikos 42:10–22 [Google Scholar]
- 28. Marchesi JR, et al. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64:795–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McArdle BH, Anderson MJ. 2001. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82:290–297 [Google Scholar]
- 30. McCook LJ. 2001. Competition between corals and algal turfs along a gradient of terrestrial influence in the nearshore central Great Barrier Reef. Coral Reefs 19:419–425 [Google Scholar]
- 31. McDonald D, et al. 1 December 2011. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. [Epub ahead of print.] doi:10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mihm JW, Banta WC, Loeb GI. 1981. Effects of adsorbed organic and primary fouling films on bryozoan settlement. J. Exp. Mar. Biol. Ecol. 54:167–169 [Google Scholar]
- 33. Moss JA, Nocker A, Lepo JE, Snyder RA. 2006. Stability and change in estuarine biofilm bacterial community diversity. Appl. Environ. Microbiol. 72:5679–5688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nocker A, Lepo JE, Martin LL, Snyder RA. 2007. Response of estuarine biofilm microbial community development to changes in dissolved oxygen and nutrient concentrations. Microb. Ecol. 54:532–542 [DOI] [PubMed] [Google Scholar]
- 35. Paerl HW, Pinckney JL. 1996. A mini-review of microbial consortia: their roles in aquatic production and biogeochemical cycling. Microb. Ecol. 31:225–247 [DOI] [PubMed] [Google Scholar]
- 36. Riemann L, Steward GF, Azam F. 2000. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66:578–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schaffelke B, et al. 2010. Reef Rescue Marine Monitoring Program. Final report of AIMS activities—inshore water quality monitoring 2009/10. Reef and Rainforest Research Centre, Australian Institute of Marine Science, Townsville, Queensland, Australia [Google Scholar]
- 38. Schaffelke B, Carleton J, Skuza M, Zagorskis I, Furnas MJ. 2012. Water quality in the inshore Great Barrier Reef lagoon: implications for long-term monitoring and management. Mar. Pollut. Bull. 65:249–260 [DOI] [PubMed] [Google Scholar]
- 39. Smith CJ, et al. 2005. T-Align, a web-based tool for comparison of multiple terminal restriction fragment length polymorphism profiles. FEMS Microbiol. Ecol. 54:375–380 [DOI] [PubMed] [Google Scholar]
- 40. Tolker-Nielsen T, Molin S. 2000. Spatial organization of microbial biofilm communities. Microb. Ecol. 40:75–84 [DOI] [PubMed] [Google Scholar]
- 41. Uthicke S, Altenrath C. 2010. Water column nutrients control growth and C:N ratios of symbiont-bearing benthic foraminifera on the Great Barrier Reef, Australia. Limnol. Oceanogr. 55:1681–1696 [Google Scholar]
- 42. Uthicke S, McGuire K. 2007. Bacterial communities in Great Barrier Reef calcareous sediments: contrasting 16S rDNA libraries from nearshore and outer shelf reefs. Estuar. Coast. Shelf Sci. 72:188–200 [Google Scholar]
- 43. Uthicke S, Nobes K. 2008. Benthic Foraminifera as ecological indicators for water quality on the Great Barrier Reef. Estuar. Coast. Shelf Sci. 78:763–773 [Google Scholar]
- 44. van Woesik R, Tomascik T, Blake S. 1999. Coral assemblages and physico-chemical characteristics of the Whitsunday Islands: evidence of recent community changes. Mar. Freshw. Res. 50:427–440 [Google Scholar]
- 45. Webster NS, Negri AP. 2006. Site-specific variation in Antarctic marine biofilms established on artificial surfaces. Environ. Microbiol. 8:1177–1190 [DOI] [PubMed] [Google Scholar]
- 46. Webster NS, et al. 2004. Metamorphosis of a scleractinian coral in response to microbial biofilms. Appl. Environ. Microbiol. 70:1213–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Webster NS, Soo R, Cobb R, Negri AP. 2011. Elevated seawater temperature causes a microbial shift on crustose coralline algae with implications for the recruitment of coral larvae. ISME J. 5:759–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Weinbauer MG, et al. 2010. Bacterial community composition and potential controlling mechanisms along a trophic gradient in a barrier reef system. Aquat. Microb. Ecol. 60:15–28 [Google Scholar]
- 49. Wieczorek SK, Todd CD. 1998. Inhibition and facilitation of settlement of epifaunal marine invertebrate larvae by microbial biofilm cues. Biofouling 12:81–118 [Google Scholar]
- 50. Wilkinson C. 2004. Status of coral reefs of the world: 2004 summary. Australian Institute of Marine Science, Townsville, Queensland, Australia [Google Scholar]
- 51. Witt V, Wild C, Anthony KRN, Diaz-Pulido G, Uthicke S. 2011. Effect of ocean acidification on microbial community composition of, and oxygen fluxes through, biofilms from the Great Barrier Reef. Environ. Microbiol. 13:2976–2989 [DOI] [PubMed] [Google Scholar]
- 52. Witt V, Wild C, Uthicke S. 2011. Effect of substrate type on bacterial community composition in biofilms from the Great Barrier Reef. FEMS Microbiol. Lett. 323:188–195 [DOI] [PubMed] [Google Scholar]
- 53. Witt V, Wild C, Uthicke S. 2012. Interactive climate change and runoff effects alter O2 fluxes and bacterial community composition of coastal biofilms from the Great Barrier Reef. Aquat. Microb. Ecol. 66:117–131 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


