Abstract
PAK1 kinase is a crucial regulator of a variety of cellular processes, such as motility, cell division, gene transcription and apoptosis. Its deregulation is involved in several pathologies, including cancer, viral infection and neurodegenerative diseases. Due to this strong implication in human health, the complex network of signaling pathways centered on PAK1 is a subject of intensive investigations. This review summarizes the present knowledge on the multiple PAK1 intracellular localizations and on its shuttling between different compartments. The dynamics of PAK1 localization and activation are finely tuned by the cell and it is this tight control that underlies the capacity of PAK1 to participate in the regulation of many fundamental cell functions. Recently, PAK1 biosensors have been developed to visualize PAK1 activation in live cells. These new imaging tools should be of great help to better understand PAK1 biology and to conceive strategies for efficient and specific PAK1 inhibitors.
Keywords: PAK1, actin cytoskeleton, motility, Rac1, cell imaging, fluorescence resonance energy transfer (FRET), spatiotemporal dynamics, functional microscopy, biosensor
PAK1, a Tightly Regulated Kinase
PAK1 is the first discovered member1 of the mammalian PAK (p21-activated kinase) family, which comprises six proteins divided in group I (PAK1–3) and group II (PAK4–6) on the basis of their structural and functional features.2-6 We will focus on the prototype PAK1 as its implication in cancer is well established and represents an ideal target for future personalized oncology treatments. Moreover its dynamics have been extensively investigated by various cell imaging approaches.
The biological functions of PAK1 are disparate and include actin dynamics, cell motility, cell cycle progression, cell division, gene transcription, cell proliferation and apoptosis.2-6 However, it is still poorly understood how at molecular level PAK1 can perform such a variety of functions. A very reasonable assumption is that the fine control of localization and activation of PAK1 is the mechanism used by the cell to activate the right PAK1-dependent pathway according to the cell cycle status or in response to extracellular stimuli.
PAK1 is a downstream effector of the Rho-family GTPases, Rac1 and Cdc42. As all PAK proteins, PAK1 consists of a highly conserved C-terminal catalytic kinase domain and an N-terminal region with a regulatory role. The PAK1 regulatory domain contains (1) a GTPase-binding domain (GBD), (2) an auto-inhibitory switch domain (IS) and (3) several Pro-rich motifs that bind to SH3 domains of Nck and Grb2 adapters or of the PIX α/β exchange factors. Inactive PAK1 has a homodimeric trans-inhibited conformation, in which the N-terminal inhibitory IS domain of one PAK1 molecule binds and inhibits the catalytic domain of the other one in the dimer.7 When GTP-loaded Rac1 or Cdc42 (the activators) bind to the N-terminal GBD domain, a series of PAK1 conformational changes is triggered, leading to disruption of dimerization, removal of the trans-inhibitory interactions and consequent acquisition of an active state for the kinase C-terminal domain.7-9
PAK1 activation by Rac1 and Cdc42 has been well established and biochemically characterized,10 but this simple Rac1/Cdc42-PAK1 pathway explains only very partially the spatiotemporal regulation and the multiple cellular roles of PAK1. At least two levels of complexity need to be taken into account. First, there are also GTPase-independent mechanisms that regulate PAK1 kinase activity, such as phosphorylation by other kinases (including Etk,11,12 PDK113 and Cdc214,15), or interactions with other proteins; among the PAK1 partners, beside the already mentioned SH3-containing Nck, Grb2 and α/βPIX, it is worth mentioning the tumor suppressor Merlin16,17 and the integrin-binding protein Nischarin18 that both inhibit the PAK1 kinase activity. Second, PAK1 has kinase-independent functions that have been ascribed to its scaffold capacity,19-21 i.e., PAK1 in some cases acts not by phosphorylating targets but by facilitating the assembly of multi-protein signaling modules.
In the past decade, the live cell imaging came in help of classical approaches, based on biochemistry techniques and immuno-fluorescence studies. Thanks to the developments of fluorescent protein fusions and of automated video-microscopes, we start to have the right tools to decipher the dynamics of PAK1 at high resolution, both temporally and spatially. Importantly, since PAK1 is normally auto-inhibited, it is essential to know not only where/when PAK1 localizes, but also where/when PAK1 is activated.
Multiple Cellular Localizations for the Multifunctional PAK1
The functional versatility of PAK1 relies, at least in part, on its multiple cellular localizations (Fig. 1). PAK1 is a cytosolic kinase that shuttles to specific cellular locations, including plasma membrane, adhesion sites, cell-cell junctions and nucleus. The dynamics of PAK1 localization contribute to its activation/inactivation, to the efficient phosphorylation of its targets, to the selection of binding partners and, consequently, to the specificity of its biological function.
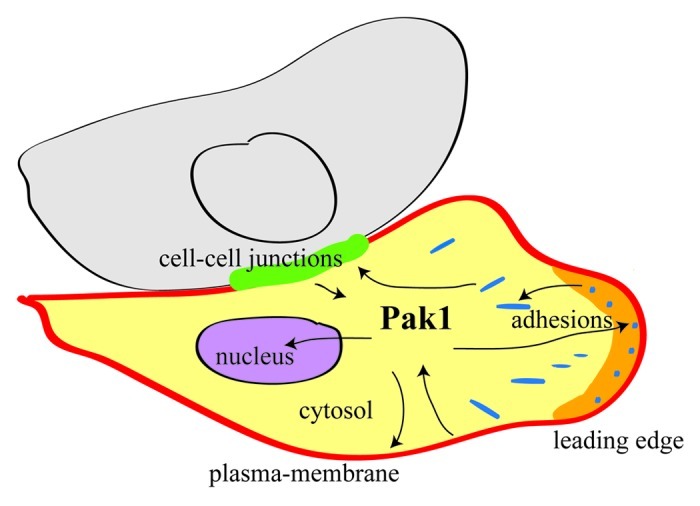
Figure 1. The multiple cellular localizations of PAK1. PAK1 shuttles between several subcellular sites. The various recruitment and activation mechanisms are all tightly controlled. At each of these localizations PAK1 performs distinct functions, by phosphorylating or interacting with specific targets.
PAK1 translocates from cytosol to membranes
Biochemical fractioning showed that PAK1 is both cytosolic and membrane-associated.22 PAK1 translocation to membranes occurs upon stimulation by growth signals and it is mediated by interaction with Nck23,24 and Grb225 SH2/SH3 adapters. Upon activation, the SH2 domains of Nck and Grb2 bind to phosphorylated Tyr residues of Tyr-kinase receptors, while their SH3 domains interact with the Pro-rich sequences in PAK1 N-terminal domain.
Recruitment of PAK1 to plasma-membrane is sufficient to partially stimulate its kinase activity, 10- to 20-fold.26,27 By using a FRET (fluorescence resonance energy transfer)-based PAK1 biosensor (Fig. 2A), it was found that membrane recruitment induces a slight conformational change of the PAK1 dimers, which shift from a closed to a semi-open state, leading to the partial PAK1 activation; full PAK1 kinase activity requires Cdc42 or Rac1 that dissociates dimers producing the open state.28
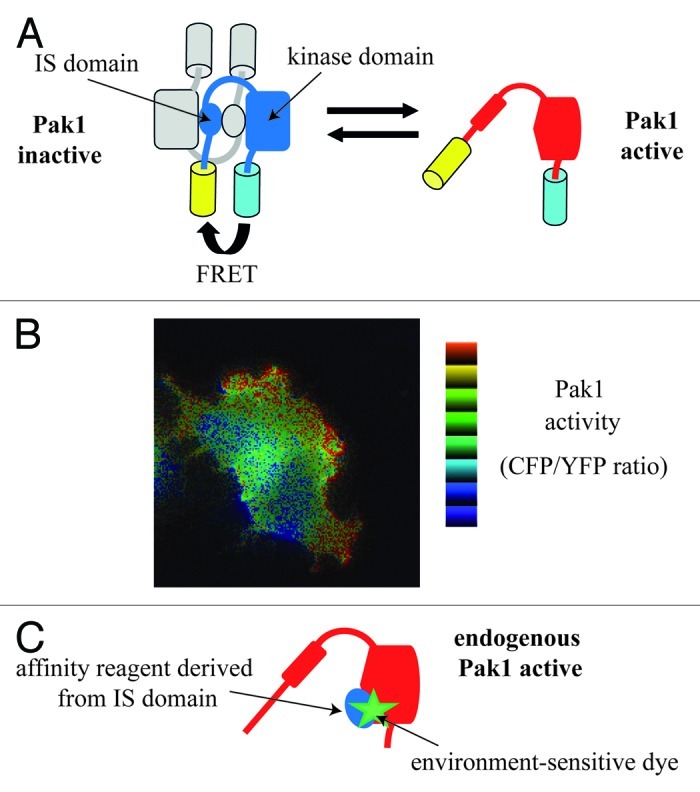
Figure 2. Biosensors for PAK1 activity. (A) Design of a FRET-based PAK1 biosensor. The Pakabi biosensor is a fusion protein comprising YFP, aa 65 to C-terminus of PAK1, and CFP. In the inactive dimeric state, FRET can occur because of the proximity between donor CFP and acceptor YFP. In the active monomeric state, CFP and YFP are moved apart, and FRET is decreased. (B) Visualization of PAK1 activity. A Cos-7 expressing Pakabix (membrane-targeted version of Pakabi) and mRFP-Cdc42wt was imaged by live FRET-microscopy during cell spreading. Note the high PAK1 activity detected at the leading edge of protrusions. (C) Strategy for an affinity-reagent PAK1 biosensor. A variant of the IS domain of PAK1 was designed in order to efficiently bind the active open state of endogenous PAK1. The conjugated environment-sensitive dye undergoes fluorescence changes upon protein-protein interaction and allows monitoring the binding of the affinity reagent to the PAK1 kinase domain.
Interestingly, membrane-localized PAK1 is hyper-responsive to GTPase stimulation,28 possibly because of its proximity with Cdc42/Rac1 molecules at the membrane or because of a higher affinity of semi-open PAK1 for Cdc42/Rac1. Moreover, other mechanisms seem to contribute to PAK1 activation at the membrane: interaction with sphingolipids,27 interaction with SH3-containing exchange factors α/βPIX28,29 and possibly also trans-phosphorylation between PAK1 molecules.28 Furthermore, the non-receptor Tyr-kinase Src, which localized on the plasma membrane, activates another non-receptor Tyr-kinase, Etk, by direct phosphorylating it at Tyr 566,30 and Etk in turn activates PAK1.11,12
These findings indicate that Nck/Grb2-mediated PAK1 translocation from cytosol to plasma-membrane is a mechanism that stimulates its kinase activity. However, it is not a simple on/off switch; multiple intermediate PAK1 activation states do exist and are dictated by the intricate balance of various incoming signals.
PAK1 at the leading edge of protrusions
Phosphorylated active PAK1 is present at the tips of protrusions of moving cells, at membrane ruffles and at the periphery of spreading cells,28,31,32 which are all sites where active actin remodeling is occurring. Indeed, PAK1 phosphorylates and/or interacts with various proteins involved in actin cytoskeleton organization, including LIM kinase33 (an inactivator of the actin-depolymerizing protein confilin), filamin34 (a cross-linker of actin filaments), p41-Arc35 (a subunit of the nucleating and branching Arp2/3 complex), MLC (myosin light chain36) and MLCK37 (myosin light-chain kinase). By the combinatory effect of phosphorylation of these targets, PAK1 regulates the dynamics of protrusions and of cell motility.38,39 In particular, a Rac1/PAK1/LIMK/cofilin pathway has been proposed as spatial organizer of the lamellipodium and lamella actin networks.40 LIM kinase is responsible for at least two distinct PAK1-dependent diseases, the metastasis/invasion of cancer cells, and neuro-cytotoxicity in Alzheimer disease.
PAK1 at adhesion sites
PAK1 has been observed at adhesion sites39,41-43 and is considered a transient component of the integrin adhesome network.44 Localization of PAK1 to adhesion sites is mediated by its binding to PIX proteins, which associate with GIT proteins (G protein coupled receptor kinase interactor) that are direct partners of paxillin, a central component of focal adhesions.19,45 Biochemical studies suggested that GIT and PIX are not simply binding partners, but rather constituent subunits of a large multi-protein complex, which can bind PAK1 to form a tri-molecular PAK-PIX-GIT signaling module.46
The use of TIRF (total internal reflection fluorescence) microscopy allows quantitative measurements on rates of adhesion assembly and disassembly. This technique revealed that PAK1 activity directly controls adhesion dynamics. Indeed, constitutively activated PAK1 accelerated both assembly and disassembly rates32; conversely, inhibition of PAK1 (by expression of the PAK auto-inhibitory domain, AID, aa 83–149 of PAK1) impaired both assembly and disassembly rates, significantly increasing adhesion lifetime.39 Moreover, adhesions of cells in which PAK1 was inhibited by AID or by RNAi silencing were longer and thinner, with an increased distribution throughout the ventral cell surface and a reduction of the zyxin protein component (a marker of mature adhesions).39 These results together indicate that PAK1 regulates assembly, maturation and turnover of adhesions.
One molecular mechanism used by PAK1 to execute its function at adhesions has been well described: it is the phosphorylation of paxillin on serine 273 (S273). PhosphoS273-paxillin efficiently binds GIT, promoting the localization of the PAK-PIX-GIT module near the leading edge of cell protrusions.32 Therefore, PAK1 acts both upstream and downstream of S273-paxillin phosphorylation in a positive feedback loop. S273-paxillin phosphorylation increases cell migration, protrusiveness and adhesion turnover.32 Since PIX is a Rac1 activator (via its GEF exchange factor activity) and PAK1 is Rac1 effector, it is possible that the phosphoS273-paxillin-PAK-PIX-GIT complex contributes both to activate Rac1 at the leading edge47,48 and to transmit its downstream signaling via PAK1.
Importantly, continuous GDP/GTP cycling of Rac1 appears to be required for adhesion turnover,32 for the organization and dynamics of protrusions, and consequently for cell migration.49 It is tempting to speculate that Rac1 cycling, PAK1 activation-inactivation cycle and adhesion turnover are dynamically connected, but the underlying molecular mechanisms still have to be discovered.
Deregulation of PAK1 localization and function at adhesions may have dramatic consequences. For example, it has been reported that certain breast cancer cell lines have endogenous hyperactive PAK1 because of its PIX-mediated mislocalization to atypical adhesions.41
PAK1 at cell-cell junctions
In epithelial cells PAK1 has been observed at regions of E-cadherin-mediated cell-cell contacts.43 More specifically, the PAK-PIX-GIT complex seems to translocate from cell-matrix adhesions to cell-cell contacts and to be involved in the establishment of contact inhibition of proliferation.43,50 In cells reaching confluence the kinetics of lateral recruitment of βPIX (presumably associated with PAK1) were accurately analyzed by confocal microscopy and turned out to be much slower than that of β-catenin, a component of adherens junction; it was necessary to wait 5–6 days after establishment of cell-cell contacts to achieve a complete lateral recruitment of βPIX-containing complexes.50 Moreover, inhibition of PAK1 (by kinase-dead PAK1 K299R or AID domain expression) did not affect the integrity of junctions,50,51 even though it impairs contact inhibition.43 These data are consistent with the conclusion that PAK1 is not necessary for establishment of cell-cell contacts, but rather for the signaling triggered by the assembly of the contacts themselves.
PAK1 kinase activity is transiently activated by junction assembly. A recently discovered target of PAK1 at this cellular location is Ajuba, an actin-binding LIM-domain protein that colocalizes with cadherins.52 Ajuba interacts with both inactive (GDP-bound) and active (GTP-bound) Rac1. T172-Ajuba phosphorylation by PAK1 increases the affinity of Ajuba for active Rac1, stabilizing the presence of Rac1-GTP at junctions,52 which is known to be important for junction maintenance in epithelia. It was found that PAK1 inhibition by AID domain or by the IPA3 inhibitor53 (1) does not perturb assembly of cell-cell contacts, but rather their maintenance, as assessed aggregation assays, and (2) impairs F-actin recruitment to cadherin complexes, as assessed by F-actin clustering experiments.52 Thus, a Rac1-PAK1-Ajuba feedback positive loop seems to participate in the stabilization of preassembled cadherin complexes and in the actin remodeling associated with junction formation. Finally, PAK1 was also shown to be required for the disruption of adherens junctions induced by constitutively active Rac1.51
PAK1 in the nucleus
Endogenous PAK1 localizes inside the nucleus of roughly 20% of the interphase cells.54 Three nuclear localization signals (NLS) were identified in the PAK1 N-terminal domain55 and the one spanning residues 243–245 is the most critical for PAK1 nuclear import; interestingly, the interaction of PAK1 with the dynein light chain LC8 protein is required for PAK1 nuclear localization.56 What does PAK1 do in the nucleus? Experiments with zebrafish showed that the presence of PAK1 in the nucleus is necessary for vertebrate development.56 Various evidences supported the notion that PAK1 is involved in regulation of gene expression, potentially by direct binding to promoters and/or transcription regulators.3,6
Again, deregulation of PAK1 localization may have dramatic consequences: a correlation has been found between PAK1 nuclear localization and tamoxifen resistance in breast cancer patients.57 In fact, inactivation of PAK1 by PAK1 blockers such as FK228, suppresses almost completely the growth of tamoxifen-resistant breast cancer xenografts in mice.58
PAK1 during mitosis
Activated PAK1 localizes to specific structures during mitosis: the chromosomes during prophase, the centrosomes in metaphase and the contraction ring during cytokinesis.54 These localizations underlie the role of PAK1 during mitosis progression.3,5,6 The PIX-GIT complex is responsible for PAK1 localization to the centrosome, where PAK1 undergoes activation by a GTPase-independent mechanism. Once activated, PAK1 dissociates from PIX-GIT complex and phosphorylates its mitotic targets.59 Among them, the Aurora kinase A and the Polo-like kinase 1 (Plk1), two key players of mitosis, are directly activated via phosphorylation by PAK1, on T288/S342 and on S49, respectively.59,60 Interestingly, both Aurora kinase A and B are essential for the growth of a variety of cancers such as breast and colon cancers, and inhibitors of these kinases such as VX-680 block their growth in vivo.61
New Tools to Study Live Dynamics of PAK1 Activity
Antibodies that recognize phosphorylated, i.e., activated, PAK1 are commercially available and can be used for immunofluorescence studies. However, such approaches on fixed cells provide only static pictures of PAK1 activity in the cell and are not very informative on temporal dynamics. Recently, a couple of strategies have been proposed to investigate PAK1 activity in living cells.
FRET-based PAK1 biosensor
FRET (fluorescence resonance energy transfer) is a non-radiative transfer of energy from a donor to an acceptor fluorophore when they are in proximity (typically less than 10 nm). This physical phenomenon is now exploited by biologists to study cellular events at molecular scale by fluorescence microscopy. In particular, FRET-based biosensors are fusion constructs, encoded by transfectable plasmid and designed to probe intracellular activities. The pioneers “camaleon” and “Raichu” biosensors allow monitoring Ca2+ production62 and activity of Ras oncogene,63 respectively. A biosensor for PAK1, called “Pakabi,” was designed by fusing two fluorophores (YFP and CFP) at the N and C termini of PAK1. In the inactive dimeric state, the YFP and CFP proximity allows FRET to occur. Upon activation, the dramatic PAK1 conformation changes move away the YFP acceptor from the CFP donor, reducing FRET (Fig. 2A). An appropriate microscope was used to acquire CFP and YFP fluorescence, the CFP/YFP ratio was calculated and used as measure of PAK1 activity. This novel PAK1 biosensor allowed the spatiotemporal visualization of PAK1 activation at the plasma membrane of nascent protrusions during cell spreading and motility28 (Fig. 2B).
The creation of Pakabi required some compromises. A first limitation is that the first 64 amino acids, including the Nck and Grb2 binding sites, had to be removed because the high flexibility of this fragment was not compatible with an efficient FRET. Second, only the artificially membrane-targeted version (Pakabix) achieved a sufficient signal-to-noise ratio for live imaging. Consequently, Pakabix can measure the activation events only if they are occurring at plasma membrane, but cannot detect activation events that are dependent on PAK1 translocation. Keeping in mind these limitations for general spatiotemporal studies, Pakabi and Pakabix remain very useful tools to detect processes and mechanisms that activate PAK1 in vivo via conformational changes.
Affinity reagent derived biosensor for PAK1
An alternative type of biosensors are designed to detect the activation state of endogenous proteins by using peptides, “affinity reagents,” that bind only to the active state of the proteins of interest; this strategy was successfully used to generate a biosensor for the GTPase Cdc42.64 Upon activation, PAK1 switches from a closed dimeric to an open monomeric conformation, in which the interaction between IS domain and kinase domains is broken. A peptide capable to interact with the IS binding site on the PAK1 kinase domain would be an ideal affinity reagent. Two different approaches have been tried to design an affinity reagent by molecular modeling. In the first approach, a structural homolog of IS domain was redesigned on the surface to have interactions with the kinase domain in the open state of PAK1.65 In the second approach, a high-affinity variant of the IS domain (aa 83–137) was engineered and inserted into a loop in a host CFP protein (Fig. 2C). This construct, named CFP-PAcKer, binds with good affinity to full-length open PAK1, but not to closed inactive PAK1. Binding can be monitored in vitro by addition of an environmentally sensitive fluorescence dye on CFP-PAcKer.66 Next step for these promising strategies will be the validation in living cells.67
Conclusion
A large amount of information has been gathered on PAK1 biology since its discovery in 19941 by using a variety of approaches. All these efforts allowed the addition of many pieces to the PAK1 puzzle. It is now clear that PAK1 activity is tightly regulated in space and time and that this tight control is necessary for PAK1 to execute various cellular roles. The future challenge is to improve the observation resolution, both at temporal and spatial scale. This challenge is taken up by the cutting-edge cell imaging technology with the development of fast-acquisition and super-resolution microscopes that paves the way to finally obtain a clear and complete description of the intricate PAK1 localization dynamics. In parallel, the generation of novel molecular imaging tools, in particular of biosensors for PAK1 activity, will be essential to study the relation between PAK1 localization and activation.
Acknowledgments
Many thanks to Carine Joffreá for critical reading of the manuscript and to Jacques Camonis for the continuous support.
Glossary
Abbreviations:
- PAK
p21-activated kinase
- SH2
Src homology 2
- SH3
Src homology 3
- GBD
GTPase-binding domain
- IS
inhibitory switch
- AID
auto-inhibitory domain
- FRET
fluorescence resonance energy transfer
- YFP
yellow fluorescent protein
- CFP
cyan fluorescent protein
- Pakabi
PAK1 activation biosensor
Footnotes
Previously published online: www.landesbioscience.com/journals/cellularlogistics/article/19817
References
- 1.Manser E, Leung T, Salihuddin H, Zhao ZS, Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–6. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 2.Bokoch GM. Biology of the p21-activated kinases. Annu Rev Biochem. 2003;72:743–81. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- 3.Kumar R, Gururaj AE, Barnes CJ. p21-activated kinases in cancer. Nat Rev Cancer. 2006;6:459–71. doi: 10.1038/nrc1892. [DOI] [PubMed] [Google Scholar]
- 4.Arias-Romero LE, Chernoff J. A tale of two Paks. Biol Cell. 2008;100:97–108. doi: 10.1042/BC20070109. [DOI] [PubMed] [Google Scholar]
- 5.Dummler B, Ohshiro K, Kumar R, Field J. Pak protein kinases and their role in cancer. Cancer Metastasis Rev. 2009;28:51–63. doi: 10.1007/s10555-008-9168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Molli PR, Li DQ, Murray BW, Rayala SK, Kumar R. PAK signaling in oncogenesis. Oncogene. 2009;28:2545–55. doi: 10.1038/onc.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parrini MC, Lei M, Harrison SC, Mayer BJ. Pak1 kinase homodimers are autoinhibited in trans and dissociated upon activation by Cdc42 and Rac1. Mol Cell. 2002;9:73–83. doi: 10.1016/S1097-2765(01)00428-2. [DOI] [PubMed] [Google Scholar]
- 8.Lei M, Lu W, Meng W, Parrini MC, Eck MJ, Mayer BJ, et al. Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell. 2000;102:387–97. doi: 10.1016/S0092-8674(00)00043-X. [DOI] [PubMed] [Google Scholar]
- 9.Lei M, Robinson MA, Harrison SC. The active conformation of the PAK1 kinase domain. Structure. 2005;13:769–78. doi: 10.1016/j.str.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Zhao ZS, Manser E. PAK and other Rho-associated kinases--effectors with surprisingly diverse mechanisms of regulation. Biochem J. 2005;386:201–14. doi: 10.1042/BJ20041638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bagheri-Yarmand R, Mandal M, Taludker AH, Wang RA, Vadlamudi RK, Kung HJ, et al. Etk/Bmx tyrosine kinase activates Pak1 and regulates tumorigenicity of breast cancer cells. J Biol Chem. 2001;276:29403–9. doi: 10.1074/jbc.M103129200. [DOI] [PubMed] [Google Scholar]
- 12.He H, Hirokawa Y, Gazit A, Yamashita Y, Mano H, Kawakami Y, et al. The Tyr-kinase inhibitor AG879, that blocks the ETK-PAK1 interaction, suppresses the RAS-induced PAK1 activation and malignant transformation. Cancer Biol Ther. 2004;3:96–101. doi: 10.4161/cbt.3.1.643. [DOI] [PubMed] [Google Scholar]
- 13.King CC, Gardiner EM, Zenke FT, Bohl BP, Newton AC, Hemmings BA, et al. p21-activated kinase (PAK1) is phosphorylated and activated by 3-phosphoinositide-dependent kinase-1 (PDK1) J Biol Chem. 2000;275:41201–9. doi: 10.1074/jbc.M006553200. [DOI] [PubMed] [Google Scholar]
- 14.Banerjee M, Worth D, Prowse DM, Nikolic M. Pak1 phosphorylation on t212 affects microtubules in cells undergoing mitosis. Curr Biol. 2002;12:1233–9. doi: 10.1016/S0960-9822(02)00956-9. [DOI] [PubMed] [Google Scholar]
- 15.Thiel DA, Reeder MK, Pfaff A, Coleman TR, Sells MA, Chernoff J. Cell cycle-regulated phosphorylation of p21-activated kinase 1. Curr Biol. 2002;12:1227–32. doi: 10.1016/S0960-9822(02)00931-4. [DOI] [PubMed] [Google Scholar]
- 16.Kissil JL, Wilker EW, Johnson KC, Eckman MS, Yaffe MB, Jacks T. Merlin, the product of the Nf2 tumor suppressor gene, is an inhibitor of the p21-activated kinase, Pak1. Mol Cell. 2003;12:841–9. doi: 10.1016/S1097-2765(03)00382-4. [DOI] [PubMed] [Google Scholar]
- 17.Hirokawa Y, Tikoo A, Huynh J, Utermark T, Hanemann CO, Giovannini M, et al. A clue to the therapy of neurofibromatosis type 2: NF2/merlin is a PAK1 inhibitor. Cancer J. 2004;10:20–6. doi: 10.1097/00130404-200401000-00006. [DOI] [PubMed] [Google Scholar]
- 18.Alahari SK, Reddig PJ, Juliano RL. The integrin-binding protein Nischarin regulates cell migration by inhibiting PAK. EMBO J. 2004;23:2777–88. doi: 10.1038/sj.emboj.7600291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown MC, West KA, Turner CE. Paxillin-dependent paxillin kinase linker and p21-activated kinase localization to focal adhesions involves a multistep activation pathway. Mol Biol Cell. 2002;13:1550–65. doi: 10.1091/mbc.02-02-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Hannigan M, Mo Z, Liu B, Lu W, Wu Y, et al. Directional sensing requires G beta gamma-mediated PAK1 and PIX alpha-dependent activation of Cdc42. Cell. 2003;114:215–27. doi: 10.1016/S0092-8674(03)00559-2. [DOI] [PubMed] [Google Scholar]
- 21.Higuchi M, Onishi K, Kikuchi C, Gotoh Y. Scaffolding function of PAK in the PDK1-Akt pathway. Nat Cell Biol. 2008;10:1356–64. doi: 10.1038/ncb1795. [DOI] [PubMed] [Google Scholar]
- 22.del Pozo MA, Price LS, Alderson NB, Ren XD, Schwartz MA. Adhesion to the extracellular matrix regulates the coupling of the small GTPase Rac to its effector PAK. EMBO J. 2000;19:2008–14. doi: 10.1093/emboj/19.9.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galisteo ML, Chernoff J, Su YC, Skolnik EY, Schlessinger J. The adaptor protein Nck links receptor tyrosine kinases with the serine-threonine kinase Pak1. J Biol Chem. 1996;271:20997–1000. doi: 10.1074/jbc.271.35.20997. [DOI] [PubMed] [Google Scholar]
- 24.Bokoch GM, Wang Y, Bohl BP, Sells MA, Quilliam LA, Knaus UG. Interaction of the Nck adapter protein with p21-activated kinase (PAK1) J Biol Chem. 1996;271:25746–9. doi: 10.1074/jbc.271.42.25746. [DOI] [PubMed] [Google Scholar]
- 25.Puto LA, Pestonjamasp K, King CC, Bokoch GM. p21-activated kinase 1 (PAK1) interacts with the Grb2 adapter protein to couple to growth factor signaling. J Biol Chem. 2003;278:9388–93. doi: 10.1074/jbc.M208414200. [DOI] [PubMed] [Google Scholar]
- 26.Lu W, Katz S, Gupta R, Mayer BJ. Activation of Pak by membrane localization mediated by an SH3 domain from the adaptor protein Nck. Curr Biol. 1997;7:85–94. doi: 10.1016/S0960-9822(06)00052-2. [DOI] [PubMed] [Google Scholar]
- 27.Bokoch GM, Reilly AM, Daniels RH, King CC, Olivera A, Spiegel S, et al. A GTPase-independent mechanism of p21-activated kinase activation. Regulation by sphingosine and other biologically active lipids. J Biol Chem. 1998;273:8137–44. doi: 10.1074/jbc.273.14.8137. [DOI] [PubMed] [Google Scholar]
- 28.Parrini MC, Camonis J, Matsuda M, de Gunzburg J. Dissecting activation of the PAK1 kinase at protrusions in living cells. J Biol Chem. 2009;284:24133–43. doi: 10.1074/jbc.M109.015271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daniels RH, Zenke FT, Bokoch GM. alphaPix stimulates p21-activated kinase activity through exchange factor-dependent and -independent mechanisms. J Biol Chem. 1999;274:6047–50. doi: 10.1074/jbc.274.10.6047. [DOI] [PubMed] [Google Scholar]
- 30.Tsai YT, Su YH, Fang SS, Huang TN, Qiu Y, Jou YS, et al. Etk, a Btk family tyrosine kinase, mediates cellular transformation by linking Src to STAT3 activation. Mol Cell Biol. 2000;20:2043–54. doi: 10.1128/MCB.20.6.2043-2054.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sells MA, Pfaff A, Chernoff J. Temporal and spatial distribution of activated Pak1 in fibroblasts. J Cell Biol. 2000;151:1449–58. doi: 10.1083/jcb.151.7.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nayal A, Webb DJ, Brown CM, Schaefer EM, Vicente-Manzanares M, Horwitz AR. Paxillin phosphorylation at Ser273 localizes a GIT1-PIX-PAK complex and regulates adhesion and protrusion dynamics. J Cell Biol. 2006;173:587–9. doi: 10.1083/jcb.200509075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edwards DC, Sanders LC, Bokoch GM, Gill GN. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat Cell Biol. 1999;1:253–9. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- 34.Vadlamudi RK, Li F, Adam L, Nguyen D, Ohta Y, Stossel TP, et al. Filamin is essential in actin cytoskeletal assembly mediated by p21-activated kinase 1. Nat Cell Biol. 2002;4:681–90. doi: 10.1038/ncb838. [DOI] [PubMed] [Google Scholar]
- 35.Vadlamudi RK, Li F, Barnes CJ, Bagheri-Yarmand R, Kumar R. p41-Arc subunit of human Arp2/3 complex is a p21-activated kinase-1-interacting substrate. EMBO Rep. 2004;5:154–60. doi: 10.1038/sj.embor.7400079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chew TL, Masaracchia RA, Goeckeler ZM, Wysolmerski RB. Phosphorylation of non-muscle myosin II regulatory light chain by p21-activated kinase (gamma-PAK) J Muscle Res Cell Motil. 1998;19:839–54. doi: 10.1023/A:1005417926585. [DOI] [PubMed] [Google Scholar]
- 37.Sanders LC, Matsumura F, Bokoch GM, de Lanerolle P. Inhibition of myosin light chain kinase by p21-activated kinase. Science. 1999;283:2083–5. doi: 10.1126/science.283.5410.2083. [DOI] [PubMed] [Google Scholar]
- 38.Smith SD, Jaffer ZM, Chernoff J, Ridley AJ. PAK1-mediated activation of ERK1/2 regulates lamellipodial dynamics. J Cell Sci. 2008;121:3729–36. doi: 10.1242/jcs.027680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delorme-Walker VD, Peterson JR, Chernoff J, Waterman CM, Danuser G, DerMardirossian C, et al. Pak1 regulates focal adhesion strength, myosin IIA distribution, and actin dynamics to optimize cell migration. J Cell Biol. 2011;193:1289–303. doi: 10.1083/jcb.201010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delorme V, Machacek M, DerMardirossian C, Anderson KL, Wittmann T, Hanein D, et al. Cofilin activity downstream of Pak1 regulates cell protrusion efficiency by organizing lamellipodium and lamella actin networks. Dev Cell. 2007;13:646–62. doi: 10.1016/j.devcel.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stofega MR, Sanders LC, Gardiner EM, Bokoch GM. Constitutive p21-activated kinase (PAK) activation in breast cancer cells as a result of mislocalization of PAK to focal adhesions. Mol Biol Cell. 2004;15:2965–77. doi: 10.1091/mbc.E03-08-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao ZS, Manser E, Loo TH, Lim L. Coupling of PAK-interacting exchange factor PIX to GIT1 promotes focal complex disassembly. Mol Cell Biol. 2000;20:6354–63. doi: 10.1128/MCB.20.17.6354-6363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zegers MMP, Forget M-A, Chernoff J, Mostov KE, ter Beest MB, Hansen SH. Pak1 and PIX regulate contact inhibition during epithelial wound healing. EMBO J. 2003;22:4155–65. doi: 10.1093/emboj/cdg398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaidel-Bar R, Geiger B. The switchable integrin adhesome. J Cell Sci. 2010;123:1385–8. doi: 10.1242/jcs.066183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manabe R, Kovalenko M, Webb DJ, Horwitz AR. GIT1 functions in a motile, multi-molecular signaling complex that regulates protrusive activity and cell migration. J Cell Sci. 2002;115:1497–510. doi: 10.1242/jcs.115.7.1497. [DOI] [PubMed] [Google Scholar]
- 46.Premont RT, Perry SJ, Schmalzigaug R, Roseman JT, Xing Y, Claing A. The GIT/PIX complex: an oligomeric assembly of GIT family ARF GTPase-activating proteins and PIX family Rac1/Cdc42 guanine nucleotide exchange factors. Cell Signal. 2004;16:1001–11. doi: 10.1016/j.cellsig.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 47.Kraynov VS, Chamberlain C, Bokoch GM, Schwartz MA, Slabaugh S, Hahn KM. Localized Rac activation dynamics visualized in living cells. Science. 2000;290:333–7. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- 48.Itoh RE, Kurokawa K, Ohba Y, Yoshizaki H, Mochizuki N, Matsuda M. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol Cell Biol. 2002;22:6582–91. doi: 10.1128/MCB.22.18.6582-6591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parrini MC, Sadou-Dubourgnoux A, Aoki K, Kunida K, Biondini M, Hatzoglou A, et al. SH3BP1, an exocyst-associated RhoGAP, inactivates Rac1 at the front to drive cell motility. Mol Cell. 2011;42:650–61. doi: 10.1016/j.molcel.2011.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu F, Jia L, Thompson-Baine A-M, Puglise JM, Ter Beest MB, Zegers MM. Cadherins and Pak1 control contact inhibition of proliferation by Pak1-betaPIX-GIT complex-dependent regulation of cell-matrix signaling. Mol Cell Biol. 2010;30:1971–83. doi: 10.1128/MCB.01247-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lozano E, Frasa MAM, Smolarczyk K, Knaus UG, Braga VMM. PAK is required for the disruption of E-cadherin adhesion by the small GTPase Rac. J Cell Sci. 2008;121:933–8. doi: 10.1242/jcs.016121. [DOI] [PubMed] [Google Scholar]
- 52.Nola S, Daigaku R, Smolarczyk K, Carstens M, Martin-Martin B, Longmore G, et al. Ajuba is required for Rac activation and maintenance of E-cadherin adhesion. J Cell Biol. 2011;195:855–71. doi: 10.1083/jcb.201107162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deacon SW, Beeser A, Fukui JA, Rennefahrt UE, Myers C, Chernoff J, et al. An isoform-selective, small-molecule inhibitor targets the autoregulatory mechanism of p21-activated kinase. Chem Biol. 2008;15:322–31. doi: 10.1016/j.chembiol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li F, Adam L, Vadlamudi RK, Zhou H, Sen S, Chernoff J, et al. p21-activated kinase 1 interacts with and phosphorylates histone H3 in breast cancer cells. EMBO Rep. 2002;3:767–73. doi: 10.1093/embo-reports/kvf157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh RR, Song C, Yang Z, Kumar R. Nuclear localization and chromatin targets of p21-activated kinase 1. J Biol Chem. 2005;280:18130–7. doi: 10.1074/jbc.M412607200. [DOI] [PubMed] [Google Scholar]
- 56.Lightcap CM, Kari G, Arias-Romero LE, Chernoff J, Rodeck U, Williams JC. Interaction with LC8 is required for Pak1 nuclear import and is indispensable for zebrafish development. PLoS One. 2009;4:e6025. doi: 10.1371/journal.pone.0006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holm C, Rayala S, Jirström K, Stål O, Kumar R, Landberg G. Association between Pak1 expression and subcellular localization and tamoxifen resistance in breast cancer patients. J Natl Cancer Inst. 2006;98:671–80. doi: 10.1093/jnci/djj185. [DOI] [PubMed] [Google Scholar]
- 58.Hirokawa Y, Arnold M, Nakajima H, Zalcberg J, Maruta H. Signal therapy of breast cancers by the HDAC inhibitor FK228 that blocks the activation of PAK1 and abrogates the tamoxifen-resistance. Cancer Biol Ther. 2005;4:956–60. doi: 10.4161/cbt.4.9.1911. [DOI] [PubMed] [Google Scholar]
- 59.Zhao Z-S, Lim JP, Ng Y-W, Lim L, Manser E. The GIT-associated kinase PAK targets to the centrosome and regulates Aurora-A. Mol Cell. 2005;20:237–49. doi: 10.1016/j.molcel.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 60.Maroto B, Ye MB, von Lohneysen K, Schnelzer A, Knaus UG. P21-activated kinase is required for mitotic progression and regulates Plk1. Oncogene. 2008;27:4900–8. doi: 10.1038/onc.2008.131. [DOI] [PubMed] [Google Scholar]
- 61.Fu J, Bian M, Jiang Q, Zhang C. Roles of Aurora kinases in mitosis and tumorigenesis. Mol Cancer Res. 2007;5:1–10. doi: 10.1158/1541-7786.MCR-06-0208. [DOI] [PubMed] [Google Scholar]
- 62.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–7. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 63.Mochizuki N, Yamashita S, Kurokawa K, Ohba Y, Nagai T, Miyawaki A, et al. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 2001;411:1065–8. doi: 10.1038/35082594. [DOI] [PubMed] [Google Scholar]
- 64.Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. Activation of endogenous Cdc42 visualized in living cells. Science. 2004;305:1615–9. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- 65.Jha RK, Leaver-Fay A, Yin S, Wu Y, Butterfoss GL, Szyperski T, et al. Computational design of a PAK1 binding protein. J Mol Biol. 2010;400:257–70. doi: 10.1016/j.jmb.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jha RK, Wu YI, Zawistowski JS, MacNevin C, Hahn KM, Kuhlman B. Redesign of the PAK1 autoinhibitory domain for enhanced stability and affinity in biosensor applications. J Mol Biol. 2011;413:513–22. doi: 10.1016/j.jmb.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jha RK, Strauss CEM. 3D structure analysis of PAKs: A Clue tot he rational design for affinity reagents and blockers. Cellular Log. 2012;2:69–78. doi: 10.4161/cl.21883. [DOI] [PMC free article] [PubMed] [Google Scholar]


