Abstract
The cell division cycle is tightly regulated by the activation and inactivation of a series of proteins that control the replication and segregation of organelles to the daughter cells. During the past decade, we have witnessed significant advances in our understanding of the cell cycle in Trypanosoma brucei and how the cycle is regulated by various regulatory proteins. However, many other regulators, especially those unique to trypanosomes, remain to be identified, and we are just beginning to delineate the signaling pathways that drive the transitions through different cell cycle stages, such as the G1/S transition, G2/M transition, and mitosis-cytokinesis transition. Trypanosomes appear to employ both evolutionarily conserved and trypanosome-specific molecules to regulate the various stages of its cell cycle, including DNA replication initiation, spindle assembly, chromosome segregation, and cytokinesis initiation and completion. Strikingly, trypanosomes lack some crucial regulators that are well conserved across evolution, such as Cdc6 and Cdt1, which are involved in DNA replication licensing, the spindle motor kinesin-5, which is required for spindle assembly, the central spindlin complex, which has been implicated in cytokinesis initiation, and the actomyosin contractile ring, which is located at the cleavage furrow. Conversely, trypanosomes possess certain regulators, such as cyclins, cyclin-dependent kinases, and mitotic centromere-associated kinesins, that are greatly expanded and likely play diverse cellular functions. Overall, trypanosomes apparently have integrated unique regulators into the evolutionarily conserved pathways to compensate for the absence of those conserved molecules and, additionally, have evolved certain cell cycle regulatory pathways that are either different from its human host or distinct between its own life cycle forms.
INTRODUCTION
The eukaryotic cell cycle represents an evolutionarily conserved process involving an ordered and tightly controlled series of molecular events. In general, the cell cycle can be considered two distinct events, DNA replication (the S phase) and mitosis (the M phase), which are separated by two gap phases (G1 and G2). These events must be regulated to ensure that they occur in the correct order and that they occur only once per cell cycle. The cell division cycle of Trypanosoma brucei, an early branched microbial eukaryote and the causative agent of human sleeping sickness, follows the typical eukaryotic cell cycle regulatory scheme, but it also possesses a number of unique features and appears to be more complicated than previously thought (41). A trypanosome cell contains a number of single-copy organelles and cytoskeletal structures, such as the nucleus, mitochondrion, kinetoplast (mitochondrial DNA network), basal body, Golgi apparatus, and flagellum, all of which need to be accurately duplicated and segregated prior to cell division. Therefore, well-coordinated regulation of organelle segregation is essential to ensure precise cell division, which occurs longitudinally from the anterior toward the posterior end of the cell.
In recent years, we have witnessed significant advances in our understanding of the trypanosome cell cycle and its regulation by factors that are either evolutionarily conserved or unique to the kinetoplastid parasites (Table 1). Here, I review the regulation of the trypanosome cell cycle by these factors, with a focus on the signaling pathways that drive the progression through different stages of the cell cycle, such as the G1/S, G2/M, and mitosis-cytokinesis transitions. Moreover, I also highlight the unique features and unusual mechanisms of trypanosome cell cycle regulation and compare them with that of its human host, as these could prove to be novel targets for chemotherapy against this dreadful human pathogen.
Table 1.
Cell cycle regulatory proteins in Trypanosoma brucei
| Cell cycle stage regulated | Gene product(s) | Human homolog(s) | Function | Reference(s) |
|---|---|---|---|---|
| G1/S transition | CYC2 | None | G1 cyclin | 43, 70 |
| CRK1 | Not known | G1 CDK | 109, 110 | |
| CRK2 | Not known | G1 CDK | 109, 110 | |
| DNA replication and S-phase progression | Orc1/Cdc6 | ORC1 | Origin recognition complex | 35 |
| Orc1b | None | Origin recognition complex | 20 | |
| Orc4 | ORC4 | Origin recognition complex | 107 | |
| Tb3120 | None | Origin recognition complex | 107 | |
| Tb7980 | None | Origin recognition complex | 107 | |
| Mcm2-7 | Mcm2-7 | CMG component | 20 | |
| Cdc45 | Cdc45 | CMG component | 20 | |
| GINS (Sld5, Psf1, Psf2, Psf3) | GINS (Sld5, Psf1, Psf2, Psf3) | CMG component | 20 | |
| TLK1 | TLK1, TLK2 | Toulsed-like kinase | 64 | |
| Asf1A, Asf1B | Asf1A, Asf1B | Histone H3 chaperone | 64 | |
| G2/M transition | CYC6 | Cyclin B | Mitotic cyclin | 42, 70 |
| CYC8 | Cyclin B | Mitotic cyclin | 70 | |
| CRK3 | Cdk1 | Mitotic CDK | 109 | |
| CRK9 | None | Mitotic CDK | 37 | |
| Mitosis | AUK1, CPC1, CPC2 | Aurora B, none, none | Chromosomal passengers | 65, 71, 108 |
| TLK1 | TLK1, TLK2 | Toulsed-like kinase | 64 | |
| KIN-A | None | Divergent kinesin | 65 | |
| KIN-B | None | Divergent kinesin | 65 | |
| Kif13-1 | MCAK | Kinesin-13 | 18, 118 | |
| APC1, Cdc27 | APC1, Cdc27 | Anaphase-promoting complex | 61 | |
| SMC3, SCC1 | SMC3, SCC1 | Cohesin | 11, 34 | |
| Separase | Separase | Protease | 11 | |
| NOP86 | None | Nucleolar protein | 14 | |
| Cytokinesis | AUK1, CPC1, CPC2 | AUK1, none, none | Chromosomal passengers | 65, 71, 108 |
| PLK | PLK1 | Polo-like kinase | 44, 62 | |
| MOB1 | MOB1 | Unknown | 45 | |
| PK50, PK53 | NDR1 | Nuclear DBF-2-related kinase | 75 | |
| TRACK | RACK1 | Rho kinase | 94 | |
| RHP | RhoA | RhoA-like GTPase | 1 | |
| CFB2 | None | F-box protein | 8 | |
| ARL2 | Arl2 | Small GTPase | 90 | |
| AIR9 | None | Microtubule-associated protein | 77 | |
| KAT80, KAT60 | p80 katanin, p60 katanin | Katanin | 9 | |
| SPA | Spastin | Spastin | 9 |
REGULATION OF THE G1/S TRANSITION
The G1 phase is one of the two gap phases in the eukaryotic cell cycle in which the cell senses environmental conditions and decides whether to proliferate, quiesce, or differentiate. One of the key regulators of the G1/S transition is the cyclin-dependent kinase (CDK), which is bound and activated by a G1 cyclin. In the fission yeast Schizosaccharomyces pombe, the cyclin Puc1 cooperates with the CDK Cdc2 to promote the G1/S transition, whereas in the budding yeast Saccharomyces cerevisiae, at least three cyclins (Cln1 to Cln3) are required for the G1/S transition through sequential activation of the CDK Cdc28 (52). Regulation of the G1/S transition in animals is more complicated, with several families of cyclins and CDKs regulating the G1/S transition. At early G1 phase, Cdk4 and/or Cdk6 are activated by D-type cyclins (D1 to D3), and at late G1 phase, Cdk2 is activated by binding to cyclin E (E1 and E2), leading to passage through the G1/S boundary (47, 96). The trypanosome genome encodes a total of 10 cyclins and 11 Cdc2-related kinases (CRKs) (41). Seven of the 10 cyclins resemble the budding yeast PHO80 cyclin, which is involved in signaling phosphate starvation, whereas the other three cyclins are homologous to the mitotic B-type cyclins found in animals and plants (41). RNA interference (RNAi)-mediated silencing of some of these cyclins demonstrated that CYC2 plays a major role in promoting G1/S transition (43, 70) and that CYC4 likely plays a minor role in G1/S transition (70). RNAi of CRK1 or CRK2 also arrests the trypanosome cells at the G1 phase, suggesting their roles in the G1/S transition (109, 110). CRK1 appears to interact with four cyclins, CYC2, CYC4, CYC5, and CYC7, whereas CRK2 only interacts with CYC2 (36). These observations suggest the involvement of multiple cyclins and two CRKs in G1/S control in trypanosomes, which resembles the animal cyclin/CDK system and appears to be more complicated than the system in fungi.
DNA REPLICATION LICENSING AND REGULATION OF S-PHASE PROGRESSION
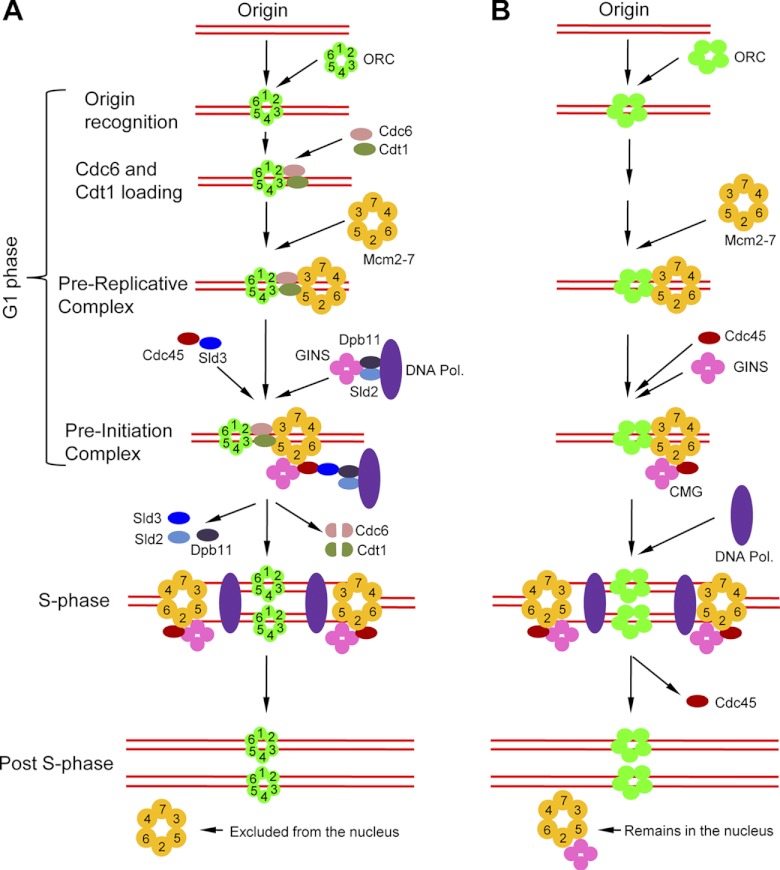
High-fidelity DNA replication is essential for maintenance of genome stability in eukaryotes. To ensure that genomic DNA is replicated only once per cell cycle, eukaryotes have evolved an intricate replication licensing system. During the G1 phase, DNA replication origins are occupied by the origin recognition complex (ORC), which consists of six related AAA+ ATPases, Orc1 to Orc6 (Fig. 1A). Subsequently, Cdc6, another AAA+ ATPase, is recruited by the ORC onto the origins, which further loads Cdt1 and the hexameric minichromosome maintenance complex, Mcm2-7, onto the origins to form a prereplicative complex (pre-RC). Assembly of the pre-RC, which is known as replication licensing, only occurs during G1 and is tightly regulated (13, 91). Initiation of DNA replication is then triggered by S-phase CDK (S-CDK) and Dbf4-dependent kinase (DDK) or Cdk7 (63). S-CDK phosphorylates two replication factors, Sld2 and Sld3 (106, 120), which generates binding sites for Dpb11, a BRCT domain-containing protein (2). Sld2 is a component of the preloading complex (pre-LC) that also includes Dpb11, the GINS (Go-Ichi-Ni-San) complex, which consists of Sld5, Psf1, Psf2, and Psf3 (55, 60, 105), and the leading-strand polymerase ϵ (Pol ε) (80). Additionally, the GINS complex also serves as an accessory factor for DNA Pol α-primase (21). Sld3 forms a complex with Cdc45, and this complex is recruited onto replication origins via Cdc45-mediated interaction with the Mcm2-7 complex (76, 99, 121). Dpb11 appears to bridge Sld3-Cdc45 with Sld2, which recruits the pre-LC to origins (63). Cdc45 also interacts with DNA Pol α-primase; therefore, it likely recruits the latter to origins (111). The DDK (Dbf4-Cdc7) or Cdk7 phosphorylates the N-terminal tails of several subunits in the Mcm2-7 complex, which promotes formation of the Cdc45-Mcm2-7 complex (100). Once the pre-LC is loaded onto origins, Cdc6 (in budding yeast) or Cdt1 (in animals) is phosphorylated and degraded by the 26S proteasome, whereas Dpb11, Sld2, and Sld3 likely dissociate from the replisome (63), thereby leaving alone DNA polymerase as well as Cdc45, Mcm2-7, and the GINS complex. The latter form, the so-called CMG (Cdc45/Mcm2-7/GINS) complex, moves along the replication forks and functions as the replicative helicase to unwind duplex DNA (15, 30, 53, 79). After DNA replication is initiated, at least three independent mechanisms are involved in preventing DNA rereplication in budding yeast: nuclear exclusion of Mcm2-7, proteolysis of Cdc6 and Cdt1, and mitotic CDK-mediated inactivation of the ORC (82). In contrast, prevention of DNA rereplication in animals occurs through nuclear exclusion of Cdc6 and degradation of Cdt1 (13).
Fig 1.
Comparison of DNA replication initiation and licensing between fungi (A) and T. brucei (B). Note that most of the replication factors found in fungi have orthologs in humans; therefore, humans and fungi share similar mechanisms of DNA replication. Currently, only five ORC proteins have been identified in trypanosomes, and it remains to be determined whether there is a sixth ORC and whether the ORC proteins form a complex in trypanosomes. Also note that only one DNA polymerase is depicted here, for the sake of simplicity.
DNA replication in eukaryotes is coupled with chromatin assembly. The fundamental repeating unit of chromatin is the nucleosome, which consists of 146 bp of DNA wrapped around an octamer of core histone proteins (H2A, H2B, H3, and H4). Histone deposition can occur by DNA replication-coupled or replication-independent mechanisms that, for the former type, is mediated by chromatin assembly factor 1 (CAF-1), which binds to histones H3 and H4 and delivers them to replicating DNA through interactions with proliferating cell nuclear antigen (PCNA), the DNA polymerase processivity protein (for a review, see reference 28). CAF-1 physically interacts with Asf1, a histone H3/H4 chaperone that is required for DNA replication (29), and the Tousled-like kinase (TLK) has been found to phosphorylate Asf1 to promote DNA replication in animals (101, 102). However, fungi appear to lack a TLK homolog (102), suggesting the lack of a TLK-mediated DNA replication pathway. Nevertheless, although there are distinctions in the regulation of DNA replication between simple and complex eukaryotic organisms, the fundamental mechanism of DNA replication is believed to be well-conserved among all eukaryotes (26).
Regulation of DNA replication in Trypanosoma brucei appears to be different from that in other eukaryotes. It was previously thought that trypanosomes expressed a single Orc1/Cdc6-like protein. This Orc1/Cdc6-like protein (TbOrc1/Cdc6) associates with the chromatin throughout the cell cycle and is essential for DNA replication in the procyclic form (35). However, a careful search of the trypanosome genome using yeast and human ORC proteins as the query identified another Orc1-like protein, TbOrc1b, which interacts with TbOrc1/Cdc6 in vitro and in vivo (20). Further, affinity purification of interacting proteins of TbOrc1/Cdc6 identified three novel proteins, among which one is distantly related to Orc4 and the other two (Tb3120 and Tb7980) have little sequence homology to known ORC proteins (107). The tight association of TbOrc1/Cdc6 with the chromatin throughout the cell cycle strongly suggests its role as a component of the ORC, rather than as the licensing factor Cdc6, which is known to be either degraded (in fungi) or exported out of the nucleus (in metazoa) after DNA replication. In total, trypanosomes express at least five ORC proteins (TbOrc1/Cdc6, TbOrc1b, TbOrc4, Tb3120, and Tb7980) (Table 1), but whether the five proteins form a complex and whether the sixth ORC protein is present require further investigation.
Despite the divergence of ORC proteins, trypanosomes apparently contain a well-conserved CMG complex, which possesses in vitro helicase activity and interacts, via Mcm3, with both TbOrc1/Cdc6 and TbOrc1b in vitro and in vivo (20). However, homologs of Cdt1, Sld2, Sld3, and the Cdc7-Dbf4 complex are not found in the trypanosome genome (10), likely because they are highly divergent. Moreover, given that TbOrc1/Cdc6 functions as a component of the ORC rather than Cdc6, it is not clear how the CMG complex is loaded onto the replication origins in the absence of the Cdc6/Cdt1 pair. Intriguingly, Mcm3 appears to interact with both TbOrc1/Cdc6 and TbOrc1b (20), but whether interactions with Mcm3 are sufficient to recruit the CMG complex to the origins remains to be investigated. Additionally, unlike in yeast, where Mcm2-7 is exported out of the nucleus after DNA replication (13, 81), Mcm2-7 in trypanosomes remains in the nucleus throughout the cell cycle (20). Instead, Cdc45 in trypanosomes appears to be exported out of the nucleus after DNA replication (20). These findings suggest that trypanosomes have evolved distinct mechanisms in promoting DNA replication initiation and preventing DNA rereplication. Based on our current understanding of DNA replication initiation in other eukaryotes (26) and the identification of the above-mentioned protein complexes in trypanosomes (20, 35, 107), we propose a model of DNA replication in trypanosomes (Fig. 1B). During the G1 phase, the replication origins are bound by a highly divergent ORC. Currently, at least five ORC proteins have been identified, but whether they form a complex remains to be examined. Subsequently, the Mcm2-7 complex is recruited onto the origins, likely via Mcm3-mediated binding to TbOrc1/Cdc6 and TbOrc1b, which leads to the formation of the prereplicative complex. This complex, however, does not possess DNA helicase activity. When cells start to enter S phase, the GINS complex and Cdc45 are loaded onto replication origins through interactions with Mcm2-7, leading to the formation of the CMG complex, which possesses DNA helicase activity and is capable of unwinding duplex DNA. When DNA polymerase is loaded and DNA replication starts, the CMG complex moves along the replication forks as part of the replisome. Finally, after DNA replication is complete, Cdc45 is excluded from the nucleus to prevent DNA rereplication (Fig. 1B). It should be noted that regulation of DNA replication is a complex process and requires the interplay of many replication factors at the origins. Indeed, homologs of TLK, Asf1, and PCNA have been identified and are found essential for DNA replication in trypanosomes (56, 64). Undoubtedly, more replication factors await our further exploration, and more work needs to be done to integrate these factors into existing pathways.
Intriguingly, a DNA replication licensing system analogous to the nuclear licensing system appears to function in controlling the replication of mitochondrial DNA in trypanosomes, which is known as the kinetoplast DNA (kDNA) (58). Replication of kDNA in trypanosomes occurs, prior to the nuclear S phase, only once per cell cycle, suggesting the presence of a tight control mechanism, but how the kDNA copy number is controlled is less understood. Recent studies demonstrated that an HslVU protease, a counterpart of the eukaryotic 26S proteasome in bacteria, regulates kDNA replication in the trypanosome mitochondrion by controlling the abundance of TbPIF2, a mitochondrial helicase that promotes kDNA replication (66, 74). These discoveries provided convincing evidence for the presence of a licensing system in kDNA replication. However, unlike the nuclear DNA replication licensing system, in which the licensing factors Cdc6 and Cdt1, but not the Mcm2-7 helicase, are degraded by the 26S proteasome, TbHslVU protease degrades the helicase TbPIF2, suggesting that the kDNA replication licensing system is different from and is likely less complicated than the nuclear replication licensing system.
REGULATION OF THE G2/M TRANSITION
The G2 phase is another gap phase in the cell cycle in which the cell assesses the state of chromosome replication and prepares to undergo mitosis and cytokinesis. Progression through the G2/M boundary in eukaryotes is known to involve B-type cyclins and their respective CDK partners. In the budding yeast, at least four B-type cyclins (Clb1 to Clb4) are required to activate Cdc28 for G2/M promotion, whereas in the fission yeast, two B-type cyclins (Cig1 and Cdc13) are responsible for the G2/M transition (52). In mammals, Cdk1 associates with several A- and B-type cyclins, and the Cdk1-cyclin B complex is the key regulator of the G2/M transition (47). Entry into mitosis is controlled through the activation of Cdk1-cyclin B, which sets the mitotic state. Cdk1 is held in an inactive state by Wee1- and Myt1-mediated phosphorylation during G2 phase, but it can be fully activated by Cdc25-mediated dephosphorylation, which promotes mitotic entry (47). Cdc25 is activated by Polo-like kinase (PLK), which is a potent regulator of M phase and is conserved from yeast to humans (3). In addition to activating Cdc25, PLK also promotes degradation of the Cdk1 inhibitor Wee1 (115). Human PLK1 is activated by Aurora A kinase in the G2 phase through phosphorylation of the conserved threonine residue in the activation loop (T-loop) of PLK1, which constitutes the most critical event to promote the G2/M transition (97). Moreover, Aurora A kinase also directly activates Cdc25 by phosphorylating the latter at a site different from that phosphorylated by PLK (25).
The G2/M transition in trypanosomes appears to be primarily controlled by the CRK3 and CYC6 pair (42, 70, 109). Intriguingly, CYC6 also forms a complex with CRK9 that is also essential for G2/M progression (37). Moreover, in addition to pairing with CYC6, CRK3 also interacts with CYC2 (36, 114), which is known as the primary G1 cyclin in trypanosomes (43, 70), indicating that CYC2 may also play a role in the G2/M transition through activating CRK3. However, it is not clear how the two cyclins cooperate with CRK3 to drive the cell cycle through the G2/M boundary. Furthermore, RNAi of CYC8 also delays the G2/M transition in the procyclic form of T. brucei (70), but the CRK partner of CYC8 remains to be identified. These studies suggest that two CRKs (CRK3 and CRK9) and multiple cyclins (CYC6, CYC2, and CYC8) are involved in promoting the G2/M transition. The trypanosome genome also encodes the homolog of Wee1 kinase (Tb927.4.3420), but close homologs of Myt1 kinase and Cdc25 phosphatase are not found in the genome. It is likely that trypanosomes express highly divergent Myt1 and Cdc25 homologs, as in the case of the divergent ORC proteins (107). Moreover, trypanosomes express an essential Polo-like kinase homolog (TbPLK), but it is excluded from the nucleus throughout the cell cycle and is not required for mitosis (22, 44, 62). Therefore, it is unlikely that TbPLK could play any role in activating CRK3 for mitotic entry. How CRK3 is activated to promote mitotic entry remains a mystery.
REGULATION OF SPINDLE ASSEMBLY AND CHROMOSOME SEGREGATION
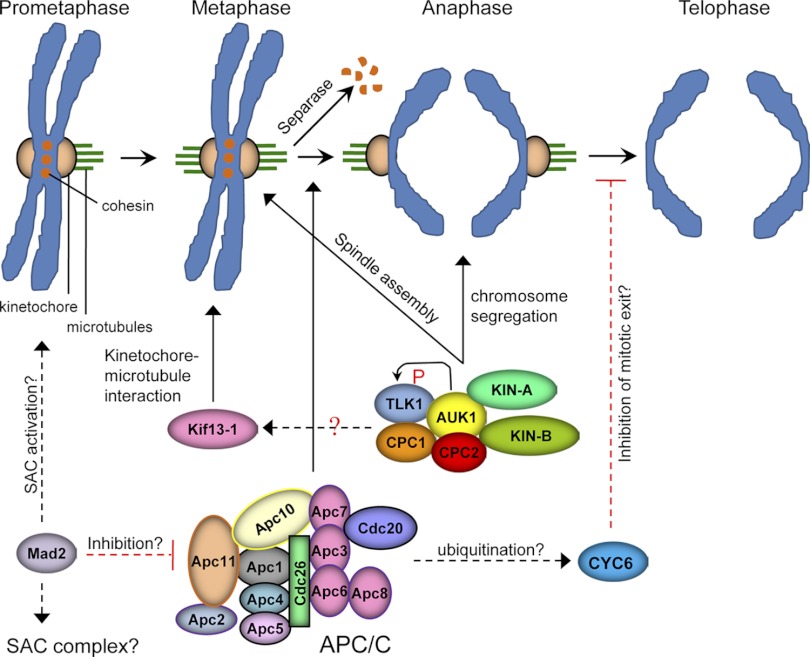
Chromosome segregation is driven by microtubule bundles, which are termed kinetochore fibers, in the bipolar mitotic spindle. In animals, spindle microtubules are nucleated either by microtubule organizing centers (MTOCs, or centrosomes) or through chromatin-mediated pathways, whereas in yeast spindle microtubules are nucleated by spindle pole bodies (SPBs) that are embedded in the nuclear envelope. In contrast to cells with centrosomes or SPBs, acentrosomal cells rely exclusively on chromatin-directed pathways in which microtubules are nucleated and stabilized near kinetochores by Ran GTPase and Aurora B kinase (23). Once a bipolar spindle is established, maintenance of the bipolar spindle requires the cooperative action of microtubule motors, such as the plus-end-directed motor kinesin-5, the minus end-directed motor kinesin-14, and the kinesin-13 microtubule depolymerase MCAK (for a review, see reference 32). Aurora A-mediated phosphorylation of kinesin-5 and MCAK and Aurora B-mediated phosphorylation of MCAK are both required for bipolar spindle assembly. In addition, Aurora B also plays a critical role in promoting chromosome biorientation by correcting microtubule-kinetochore misattachments, which are in part attributed to the regulation of MCAK activity at the kinetochores (for a review, see reference 17). Moreover, microtubule-kinetochore attachment errors are also monitored by the spindle assembly checkpoint (SAC), which delays anaphase onset by inhibiting the activity of the anaphase-promoting complex/cyclosome (APC/C) until all of the kinetochores have properly attached to the spindle microtubules. Once chromosome biorientation is established and SAC is satisfied, Securin, the inhibitor of separase, is ubiquitinated by APC/C and degraded, thus releasing active separase, which then cleaves the sister chromatid Cohesin for chromosome segregation. Moreover, APC/C activation also triggers cyclin B degradation, which inactivates Cdk1 and thereby enables exit from mitosis (for a review, see reference 86).
Trypanosomes undergo a “closed” mitosis, during which the nuclear envelope remains intact throughout the cell cycle. The parasite does not have centrosomes but possess MTOCs in the form of basal bodies that nucleate flagellum but are not involved in spindle assembly (83). The absence of centrosomes suggests that assembly of the bipolar spindle is likely mediated by chromatin-directed pathways. Indeed, proteins required for centrosome-mediated spindle assembly, such as Aurora A kinase and kinesin-5, are missing in the trypanosome genome, whereas those involved in chromatin-based pathways, such as Aurora B kinase and MCAK, are expressed and have been found essential for spindle assembly and/or spindle microtubule dynamics in trypanosomes (18, 71, 108, 118). However, the Aurora B homolog in trypanosomes, TbAUK1, forms a unique chromosomal passenger complex (CPC) with two novel proteins, TbCPC1 and TbCPC2, both of which exhibit little sequence homology to the well-conserved chromosomal passenger proteins (INCENP, Survivin, and Borealin) in fungi and animals (65, 68). This novel CPC displays a dynamic localization during mitosis and cytokinesis by migrating from chromosomes to the central spindle during the metaphase-anaphase transition, from the central spindle to the anterior tip of the new flagellum attachment zone (FAZ) during the mitosis-cytokinesis transition, and finally by traveling along the cleavage furrow between the two separating daughter cells (65, 69). It is known that the three CPC proteins in animals function as a single structural unit that targets Aurora B to various subcellular locations during the cell cycle (54), and INCENP, in addition to its roles in mediating Aurora B localization, also functions as a substrate activator of Aurora B (12). Another substrate activator of Aurora B is TLK, which initially was thought to be specific to multicellular organisms (46, 102). Surprisingly, trypanosomes express two homologs of TLK, TbTLK1 and TbTLK2, of which only TbTLK1 is required for spindle assembly and chromosome segregation and is a substrate of TbAUK1 (64) (Fig. 2), but it is not clear whether it functions as a TbAUK1 activator like its animal counterpart. Moreover, whether TbCPC1 and/or TbCPC2 functions as an activator of TbAUK1 and whether any of them is responsible for TbAUK1 targeting are also not known. Further investigation of the function of TbTLK1 and the two CPC proteins will facilitate our understanding of TbAUK1 regulation and the mechanistic roles of TbAUK1 in spindle assembly.
Fig 2.
Regulation of spindle assembly, chromosome segregation, and mitotic exit in T. brucei. Solid lines indicate confirmed regulation pathways, whereas dashed lines indicate regulation still to be verified experimentally. Black lines indicate positive regulation, whereas red lines indicate negative regulation (inhibition). Question marks indicate that the mechanism behind the regulation remains unknown. P, phosphorylation.
Despite lacking the kinesin-5 homolog in the genome, two orphan kinesins, TbKIN-A and TbKIN-B, likely cooperate with TbAUK1 to regulate spindle assembly in trypanosomes (65, 68) (Fig. 2). Both kinesins are enriched in the nucleus and the central spindle and associate with TbAUK1, but whether they are substrates of TbAUK1 and how they regulate spindle assembly remain to be investigated. Another essential mitotic kinesin found in trypanosomes is TbKif13-1, which is a member of the kinesin-13 family and possesses microtubule depolymerase activity (18, 118). RNAi of TbKif13-1 resulted in defects in spindle disassembly and chromosome segregation (18, 118), indicating the conserved roles of the kinesin-13 family in trypanosome mitosis. It is not known whether TbKif13-1 is regulated by TbAUK1 (Fig. 2); however, given the conserved roles of both proteins in trypanosomes, they likely function in the same pathway to promote kinetochore-microtubule attachment, although this remains to be examined.
Another trypanosome protein that is localized to the mitotic spindle is a nucleolar protein, TbNOP86, which appears also to be required for chromosome segregation (14). However, the biochemical function of TbNOP86 is not known, and we know little about the mechanistic role of TbNOP86 in chromosome segregation. Additionally, several other proteins have also been reported to be involved in mitosis and chromosome segregation, such as TbAGOI (24), an Argonaute protein required for RNAi, and the small ubiquitin-like modifier (SUMO) (73). The involvement of TbAGOI in mitosis indicates the importance of the RNAi pathway in heterochromatin formation at the centromeres and therefore the involvement in chromosome segregation. SUMO is known to be conjugated to a number of mitotic proteins in other eukaryotes, including all the components of the CPC (6). A recent proteomic screen for SUMO-conjugated proteins in Trypanosoma cruzi identified a number of cell cycle regulatory proteins that are potentially modified by SUMO (7). These SUMO conjugates in T. cruzi include the T. brucei homologs of CRK2 (110), protein phosphatase 2A (PP2A) (67), several DNA replication factors, such as PCNA (56), replication factor C, and DNA polymerase δ, and the structural maintenance of chromosome 3 (SMC3) protein, a subunit of the sister chromatid cohesin complex that is involved in holding together the sister chromatids (11). However, the significance of SUMO modification on these proteins and the consequence on trypanosome cell cycle regulation remain to be explored.
Trypanosomes express all the components of the APC/C, the E3 ubiquitin ligase that is essential for the transition from metaphase to anaphase (61). Surprisingly, only two APC/C subunits, APC1 and Cdc27, are essential for trypanosome mitosis (61). RNAi of APC1 or Cdc27 arrested the procyclic form in metaphase and the bloodstream form in anaphase (61), indicating that the APC/C likely functions at different cell cycle phases in the two life cycle forms. In principal, APC/C is responsible for degradation of Securin in order to activate separase; however, the Securin homolog in trypanosomes has not been identified, likely due to its high sequence divergence. Nevertheless, homologs of separase and the components of the cohesin complex (SMC1, SMC3, SCC1, and SCC3) are all found in the trypanosome genome, and functional studies on SMC3, SCC1, and separase demonstrated that they are all essential for chromosome segregation (11, 34). Trypanosomes appear to express only one subunit of the SAC complex, TbMad2 (Tb927.3.1750), with the remaining four SAC subunits (Mad2, Mad3, Bub1, and Bub3) all missing from the genome. It is not clear whether TbMad2 forms a unique SAC complex with trypanosome-specific proteins, as in the case of the CPC (65), or if Mad2 itself is sufficient to perform all the essential functions of the SAC complex, such as monitoring kinetochore-microtubule attachment errors and activating the APC/C (Fig. 2). Finally, none of the APC/C substrates has been identified or validated in trypanosomes, which significantly hinders our understanding of the regulation of the metaphase-anaphase transition. The B-type cyclin is a known APC/C substrate, and its degradation is required for mitotic exit (86). Trypanosome CYC6 appears to be a short-lived protein (unpublished data), but whether its degradation is mediated by APC/C and is required for mitotic exit require further investigation (Fig. 2).
REGULATION OF THE MITOSIS-CYTOKINESIS TRANSITION AND CYTOKINESIS
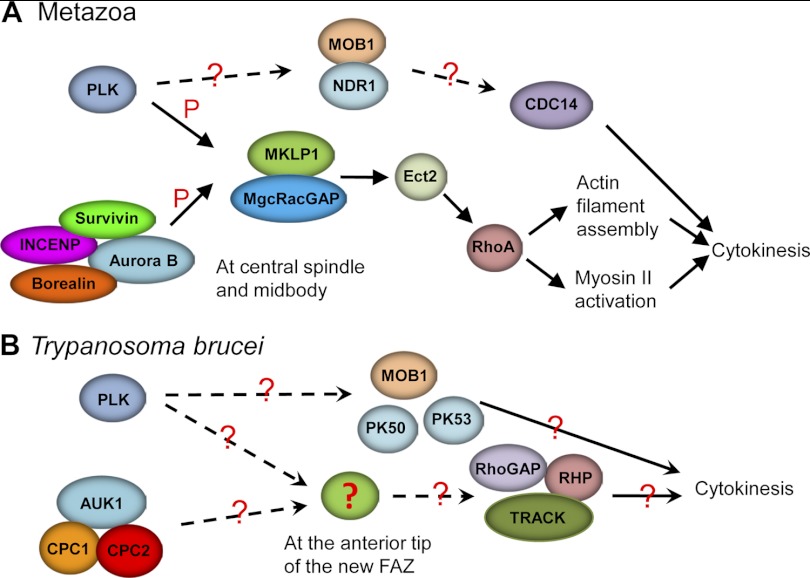
Cytokinesis is the final step of cell division and requires a complex interplay of many regulatory proteins at the cytokinesis initiation site and on the cleavage furrow. It also requires the interplay between the components of the cytoskeleton and the cell membrane (104). The mechanisms underlying cytokinesis initiation and completion in eukaryotes are still not well understood, but it is believed that the core regulatory pathways are well conserved across evolution (5, 33). The cleavage plane in fungi and animals is known to be defined by the central spindle, and the cleavage furrow in these organisms contains a contractile ring structure composed of actin and myosin II (33, 84, 88). Cytokinesis initiation in animals requires two well-conserved protein kinases, Aurora B and Polo-like kinases, both of which are concentrated on the central spindle and the midbody, where they cooperate to regulate the centraspindlin complex, which consists of a kinesin-6 family member, MKLP1, and a Rho GTPase-activating protein, MgcRacGAP. The centraspindlin complex then recruits the guanine nucleotide exchange factor Ect2 to the midbody, and the latter activates the small GTPase RhoA for further activation of the formation of the actomyosin contractile ring (16, 33). In fungi, however, the Aurora-like kinase appears to be nonessential for cytokinesis, and the Polo-like kinase constitutes an upstream component of the so-called SIN (septum initiation network) or MEN (mitotic exit network), which involves a number of proteins, such as MOB1, DBF2, Cdc14, for example, all well conserved between budding and fission yeast (39). Mammalian homologs of several SIN/MEN components have also been identified and were found to be essential for cytokinesis (Fig. 3A), but how they are regulated by Polo-like kinase has not been well defined (19).
Fig 3.
Comparison of the cytokinesis regulatory pathways mediated by Aurora B kinase and Polo-like kinase between T. brucei and its human host. Solid lines indicate confirmed regulation pathways, whereas dashed lines indicate regulation that remains to be verified. Question marks indicate that the mechanism remains unknown. In trypanosomes, TbPLK and TbAUK1 may regulate an unknown factor(s) (green oval with a red question mark) at the anterior tip of the new FAZ for cytokinesis initiation, which could be a functional ortholog of the MKLP1/MgcRacGAP complex in metazoa. P, phosphorylation.
Cytokinesis in trypanosomes, however, is drastically different from that in fungi and animals (89, 116). The cleavage plane in a dividing trypanosome cell is likely defined by the position of the new flagellum and the FAZ (59, 84, 92, 116, 117). As a consequence, cytokinesis is initiated from the anterior tip of the new FAZ, and the cleavage furrow ingresses unidirectionally along the long axis from the anterior toward the posterior end of the cell (59, 116, 117). Strikingly, there is no evidence for the existence of an actomyosin contractile ring at the cleavage furrow (31), indicating that the cleavage furrow in this early branched eukaryote may possess an unusual structure with novel components.
Despite the different mode of cytokinesis and the lack of an actomyosin-based cytokinetic apparatus in trypanosomes, both Aurora B-like kinase (TbAUK1) and Polo-like kinase (TbPLK) are implicated in cytokinesis (44, 62, 71, 108). In addition to its multiple roles in mitosis (see above), TbAUK1 apparently also regulates cytokinesis initiation, furrow ingression, and abscission, although the mechanistic roles of TbAUK1 in these processes remain to be defined. TbPLK is excluded from the nucleus throughout the cell cycle, but instead it is localized to the basal body and the bilobed structure adjacent to the Golgi apparatus during early cell cycle stages, and during late cell cycle stages it is concentrated at the anterior tip of the new FAZ, where it may promote cytokinesis initiation (22, 62, 112). Apparently, instead of being enriched on the central spindle, as in fungi and animals, TbAUK1 and TbPLK are both concentrated at the anterior tip of the new FAZ during late stages of the cell cycle (22, 65, 68, 69, 112). However, it remains unknown whether TbPLK and TbAUK1 interact with each other or colocalize at the anterior tip of the new FAZ. Trypanosomes apparently lack homologs of the partner proteins of Aurora B and Polo-like kinases identified in other eukaryotes, suggesting that trypanosomes may have evolved distinct TbAUK1- and TbPLK-mediated pathways for cytokinesis initiation. In fact, both TbAUK1 and TbPLK possess some unusual features in subcellular localization and regulation. TbAUK1 resides in a novel CPC and displays a unique trans-localization during the mitosis-cytokinesis transition (65, 68, 69), whereas TbPLK employs its kinase domain for substrate binding, but not the Polo box domain (PBD) as found for fungi and animals (119). Nevertheless, it is very likely that TbPLK and TbAUK1 cooperate, by regulating some common factor(s) at the anterior tip of the new FAZ, to promote cytokinesis initiation (Fig. 3B), and future efforts should be focused on identifying this factor(s) and understanding its role(s) in cytokinesis initiation. Intriguingly, a recent study reported the identification of a highly divergent Rho-like small GTPase, TbRHP, and its RhoGAP, TbOCRL, in trypanosomes (1). RNAi of both proteins resulted in defects in cytokinesis, in addition to a spindle formation defect (1), suggesting a key role for them in trypanosome cytokinesis. Unlike in humans, in which active RhoA is enriched at the site of cleavage furrow formation (87), TbRHP is found diffusely localized throughout the cytoplasm (1). However, it should be noted that these proteins may contain a pool of inactive TbRHP that may obscure the visualization of active TbRHP. Nevertheless, identification of the RhoA-like small GTPase and its GAP partner suggests that a RhoA-mediated cytokinesis pathway is present in trypanosomes, likely downstream of TbAUK1 and TbPLK, but whether the latter regulates the activation of TbRHP and, if it does, how the two kinases activate TbRHP to promote cytokinesis require further investigation (Fig. 3B). Despite these unknowns in the signaling pathway, it is clear that TbRHP participates in cytokinesis through interactions with TRACK, a homolog of the receptor for activated C kinase 1 (RACK1), which is known to be involved in controlling the onset and progression of cytokinesis in trypanosomes (94).
Trypanosomes also express a few SIN/MEN pathway proteins, such as homologs of MOB1 (MOB1A and MOB1B) and the homologs of yeast DBF2 kinase and human NDR1 kinase (PK50 and PK53). Intriguingly, trypanosome MOB1 appears to play different roles in the two life cycle forms. It is required for the completion of cytokinesis in the bloodstream form, but it is involved in positioning the cleavage furrow in the procyclic form (45). PK50 and PK53 are both essential for cytokinesis initiation in the bloodstream form (75). Surprisingly, neither PK50 nor PK53 interacts with MOB1 in vivo in trypanosomes (75), arguing whether they function together like their yeast and human counterparts. Moreover, it is also not known whether MOB1 and PK50/PK53 are regulated by TbPLK. Nevertheless, it appears that some of the cytokinesis regulatory pathways that are conserved in fungi and animals likely also function in trypanosomes (Fig. 3B).
Although Polo-like kinase and Aurora B kinase are key players in the signaling pathways of cytokinesis, regulation of cytokinesis initiation and completion involves more than 100 proteins in the midbody and the cleavage furrow in fungi and animals (89, 103), and many of these proteins are conserved among fungi and animals (88). However, the number of cytokinesis proteins identified in trypanosomes is still limited. Recently, a few proteins have been shown to be essential for cytokinesis in trypanosomes, such as a type III phosphatidylinositol 4-kinase (93), KMP-11 (72), the protein arginine methyltransferase (27), the small GTPase ARL2 (90), Katanin and Spastin (9), the F-box protein CFB2 (8), a microtubule-associated protein, AIR9 (77), adenylyl cyclases (95), and a kinetoplastid-specific kinesin, TbKIN-C, and its partner kinesin, TbKIN-D (50, 51), but how they are involved in cytokinesis has not been well established. Among these proteins, some are associated with basal bodies and/or the flagellum (72, 98), whereas others are localized predominantly in the cytoplasm (27, 93) or are involved in regulating microtubule dynamics (9, 50, 51, 90). Since defects in basal body replication/segregation and flagellum attachment unambiguously inhibit cytokinesis in trypanosomes, it is not possible to assign a direct role for these basal body and flagellar proteins in cytokinesis. Nevertheless, the involvement of many cytoskeletal proteins in cytokinesis suggests the requirement of the coordination between cell morphogenesis and cell division.
CONCLUSIONS AND FUTURE PERSPECTIVES
Significant advances in our understanding of the molecular mechanisms of trypanosome cell division have been made over the past decade, thanks to the availability of the genome sequence, the power of reverse genetics methods, and the fast advances in the exploration of cell cycle regulation in other model eukaryotes that have offered important references for trypanosome cell cycle studies. It is well established that many cell cycle regulatory pathways in trypanosomes have been very well preserved during evolution, although trypanosomes did evolve unusual checkpoint control mechanisms and employ a number of trypanosome-specific cell cycle regulators, some of which are likely the functional, but not the structural, orthologs of the regulators in other eukaryotes. Moreover, it is also well perceived that there are significant differences in cell cycle regulation between different life cycle stages, although the molecular mechanisms underlying these differences remain elusive.
Despite these advances, however, there are still many unanswered questions and likely many more unique cell cycle regulatory pathways awaiting our further exploration. So far, we have little information about the components of the cleavage furrow and the components of the kinetochores. We still do not know exactly how the CPC travels across the nuclear envelope to reach the anterior tip of the new FAZ and how CPC promotes cytokinesis. We also don't know how new membranes are delivered to the cleavage furrow and how the abscission machinery works to cleave the membrane and the microtubule cytoskeleton. Moreover, little is known about the cell cycle checkpoints in trypanosomes and how they differ from the checkpoints in humans and between the different life cycle stages. Our current efforts in analyzing trypanosome homologs of those conserved cell cycle regulators discovered in fungi and animals have greatly facilitated the dissection of the trypanosome cell cycle, but this approach clearly is not applicable for depiction of the unique cell cycle regulatory pathways in trypanosomes. Future endeavors should be directed toward the use of a combination of biochemistry, bioinformatics, genetics, and proteomics approaches for in-depth exploration of the trypanosome-specific regulators and integration of these regulators into the existing pathways. For example, tandem affinity purification of protein complexes provides an unparalleled method for identification of novel partners of known cell cycle regulators (40, 65). A forward genetics approach, through an RNAi screen, can also provide great promise for identification of novel cell cycle regulators (78), but an efficient mutant selection scheme is necessary to distinguish between the true cell cycle regulators and those playing indirect roles in cell cycle control. Moreover, investigation of the cell cycle-regulated transcriptome (4) and proteome offers a global view of the expression pattern of cell cycle regulatory proteins and certainly could uncover numerous cell cycle regulators. Finally, quantitative phosphoproteomics, sometimes combined with chemical genetics, have been employed to systematically identify the substrates of mitotic kinases, such as Aurora B kinase, Polo-like kinase, and CDKs in fungi and animals (38, 48, 49, 57, 85). Similar studies have not been carried out in trypanosomes, but recent establishment of the SILAC proteomics system in trypanosomes (113) provided the foundation for application of the phosphoproteomic approach to depict the signaling networks of trypanosome mitotic kinases. Undoubtedly, exploration of the unusual pathways and their novel regulators in trypanosomes not only will further our understanding of the mechanisms of cell cycle control but also could provide novel drug targets for chemotherapy.
ACKNOWLEDGMENTS
I apologize to my colleagues whose work could not be cited due to space constraints. I am very grateful to the TriTryp sequencing project, which greatly accelerated the identification of numerous cell cycle regulatory proteins. I also thank members of my laboratory for critical reading of the manuscript.
Work in my laboratory is supported by startup funds from the University of Texas Medical School at Houston and by NIH grants AI090070 and AI093897.
Footnotes
Published ahead of print 3 August 2012
REFERENCES
- 1. Abbasi K, DuBois KN, Leung KF, Dacks JB, Field MC. 2011. A novel Rho-like protein TbRHP is involved in spindle formation and mitosis in trypanosomes. PLoS One 6: e26890 doi:10.1371/journal.pone.0026890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Araki H, Leem SH, Phongdara A, Sugino A. 1995. Dpb11, which interacts with DNA polymerase II ε in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc. Natl. Acad. Sci. U. S. A. 92: 11791–11795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Archambault V, Glover DM. 2009. Polo-like kinases: conservation and divergence in their functions and regulation. Nat. Rev. Mol. Cell Biol. 10: 265–275 [DOI] [PubMed] [Google Scholar]
- 4. Archer SK, Inchaustegui D, Queiroz R, Clayton C. 2011. The cell cycle regulated transcriptome of Trypanosoma brucei. PLoS One 6: e18425 doi:10.1371/journal.pone.0018425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Balasubramanian MK, Bi E, Glotzer M. 2004. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr. Biol. 14: R806–R818 [DOI] [PubMed] [Google Scholar]
- 6. Ban R, Nishida T, Urano T. 2011. Mitotic kinase Aurora-B is regulated by SUMO-2/3 conjugation/deconjugation during mitosis. Genes Cells 16: 652–669 [DOI] [PubMed] [Google Scholar]
- 7. Bayona JC, et al. 2011. SUMOylation pathway in Trypanosoma cruzi: functional characterization and proteomic analysis of target proteins. Mol. Cell. Proteomics 10: M110.00736 doi:10.1074/mcp.M110.007369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Benz C, Clayton CE. 2007. The F-box protein CFB2 is required for cytokinesis of bloodstream-form Trypanosoma brucei. Mol. Biochem. Parasitol. 156: 217–224 [DOI] [PubMed] [Google Scholar]
- 9. Benz C, Clucas C, Mottram JC, Hammarton TC. 2012. Cytokinesis in bloodstream stage Trypanosoma brucei requires a family of katanins and spastin. PLoS One 7: e30367 doi:10.1371/journal.pone.0030367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berriman M, et al. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309: 416–422 [DOI] [PubMed] [Google Scholar]
- 11. Bessat M, Ersfeld K. 2009. Functional characterization of cohesin SMC3 and separase and their roles in the segregation of large and minichromosomes in Trypanosoma brucei. Mol. Microbiol. 71: 1371–1385 [DOI] [PubMed] [Google Scholar]
- 12. Bishop JD, Schumacher JM. 2002. Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B kinase stimulates Aurora B kinase activity. J. Biol. Chem. 277: 27577–27580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blow JJ, Dutta A. 2005. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 6: 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boucher N, Dacheux D, Giroud C, Baltz T. 2007. An essential cell cycle-regulated nucleolar protein relocates to the mitotic spindle where it is involved in mitotic progression in Trypanosoma brucei. J. Biol. Chem. 282: 13780–13790 [DOI] [PubMed] [Google Scholar]
- 15. Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K. 2005. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 19: 1905–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carmena M. 2008. Cytokinesis: the final stop for the chromosomal passengers. Biochem. Soc. Trans. 36: 367–370 [DOI] [PubMed] [Google Scholar]
- 17. Carmena M, Ruchaud S, Earnshaw WC. 2009. Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr. Opin. Cell Biol. 21: 796–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chan KY, Matthews KR, Ersfeld K. 2010. Functional characterisation and drug target validation of a mitotic kinesin-13 in Trypanosoma brucei. PLoS Pathog. 6: e1001050 doi:10.1371/journal.ppat.1001050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. D'Amours D, Amon A. 2004. At the interface between signaling and executing anaphase: Cdc14 and the FEAR network. Genes Dev. 18: 2581–2595 [DOI] [PubMed] [Google Scholar]
- 20. Dang HQ, Li Z. 2011. The Cdc45/Mcm2-7/GINS protein complex in trypanosomes regulates DNA replication and interacts with two Orc1-like proteins in the origin recognition complex. J. Biol. Chem. 286: 32424–32435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. De Falco M, et al. 2007. The human GINS complex binds to and specifically stimulates human DNA polymerase alpha-primase. EMBO Rep. 8: 99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. de Graffenried CL, Ho HH, Warren G. 2008. Polo-like kinase is required for Golgi and bilobe biogenesis in Trypanosoma brucei. J. Cell Biol. 181: 431–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dumont J, Desai A. 2012. Acentrosomal spindle assembly and chromosome segregation during oocyte meiosis. Trends Cell Biol. 22: 241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Durand-Dubief M, Bastin P. 2003. TbAGO1, an argonaute protein required for RNA interference, is involved in mitosis and chromosome segregation in Trypanosoma brucei. BMC Biol. 1: 2 doi:10.1186/1741-7007-1-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dutertre S, et al. 2004. Phosphorylation of CDC25B by Aurora-A at the centrosome contributes to the G2-M transition. J. Cell Sci. 117: 2523–2531 [DOI] [PubMed] [Google Scholar]
- 26. Errico A, Costanzo V. 2010. Differences in the DNA replication of unicellular eukaryotes and metazoans: known unknowns. EMBO Rep. 11: 270–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fisk JC, et al. 2010. TbPRMT6 is a type I protein arginine methyltransferase that contributes to cytokinesis in Trypanosoma brucei. Eukaryot. Cell 9: 866–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Franco AA, Kaufman PD. 2004. Histone deposition proteins: links between the DNA replication machinery and epigenetic gene silencing. Cold Spring Harb. Symp. Quant. Biol. 69: 201–208 [DOI] [PubMed] [Google Scholar]
- 29. Franco AA, Lam WM, Burgers PM, Kaufman PD. 2005. Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C. Genes Dev. 19: 1365–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gambus A, et al. 2006. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 8: 358–366 [DOI] [PubMed] [Google Scholar]
- 31. Garcia-Salcedo JA, et al. 2004. A differential role for actin during the life cycle of Trypanosoma brucei. EMBO J. 23: 780–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gatlin JC, Bloom K. 2010. Microtubule motors in eukaryotic spindle assembly and maintenance. Semin. Cell Dev. Biol. 21: 248–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Glotzer M. 2005. The molecular requirements for cytokinesis. Science 307: 1735–1739 [DOI] [PubMed] [Google Scholar]
- 34. Gluenz E, Sharma R, Carrington M, Gull K. 2008. Functional characterization of cohesin subunit SCC1 in Trypanosoma brucei and dissection of mutant phenotypes in two life cycle stages. Mol. Microbiol. 69: 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Godoy PD, et al. 2009. Trypanosome prereplication machinery contains a single functional orc1/cdc6 protein, which is typical of Archaea. Eukaryot. Cell 8: 1592–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gourguechon S, Savich JM, Wang CC. 2007. The multiple roles of cyclin E1 in controlling cell cycle progression and cellular morphology of Trypanosoma brucei. J. Mol. Biol. 368: 939–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gourguechon S, Wang CC. 2009. CRK9 contributes to regulation of mitosis and cytokinesis in the procyclic form of Trypanosoma brucei. BMC Cell Biol. 10: 68 doi:10.1186/1471-2121-10-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Grosstessner-Hain K, et al. 2011. Quantitative phospho-proteomics to investigate the polo-like kinase 1-dependent phospho-proteome. Mol. Cell. Proteomics 10: M111.008540 doi:10.1074/mcp.M111.008540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gruneberg U, Nigg EA. 2003. Regulation of cell division: stop the SIN! Trends Cell Biol. 13: 159–162 [DOI] [PubMed] [Google Scholar]
- 40. Gunzl A, Schimanski B. 2009. Tandem affinity purification of proteins. Curr. Protoc. Protein Sci. Chapter 19: Unit 19.19.. [DOI] [PubMed] [Google Scholar]
- 41. Hammarton TC. 2007. Cell cycle regulation in Trypanosoma brucei. Mol. Biochem. Parasitol. 153: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hammarton TC, Clark J, Douglas F, Boshart M, Mottram JC. 2003. Stage-specific differences in cell cycle control in Trypanosoma brucei revealed by RNA interference of a mitotic cyclin. J. Biol. Chem. 278: 22877–22886 [DOI] [PubMed] [Google Scholar]
- 43. Hammarton TC, Engstler M, Mottram JC. 2004. The Trypanosoma brucei cyclin, CYC2, is required for cell cycle progression through G1 phase and for maintenance of procyclic form cell morphology. J. Biol. Chem. 279: 24757–24764 [DOI] [PubMed] [Google Scholar]
- 44. Hammarton TC, Kramer S, Tetley L, Boshart M, Mottram JC. 2007. Trypanosoma brucei Polo-like kinase is essential for basal body duplication, kDNA segregation and cytokinesis. Mol. Microbiol. 65: 1229–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hammarton TC, Lillico SG, Welburn SC, Mottram JC. 2005. Trypanosoma brucei MOB1 is required for accurate and efficient cytokinesis but not for exit from mitosis. Mol. Microbiol. 56: 104–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Han Z, Riefler GM, Saam JR, Mango SE, Schumacher JM. 2005. The C. elegans Tousled-like kinase contributes to chromosome segregation as a substrate and regulator of the Aurora B kinase. Curr. Biol. 15: 894–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Harper JV, Brooks G. 2005. The mammalian cell cycle: an overview. Methods Mol. Biol. 296: 113–153 [DOI] [PubMed] [Google Scholar]
- 48. Hegemann B, et al. 2011. Systematic phosphorylation analysis of human mitotic protein complexes. Sci. Signal. 4: rs12 doi:10.1126/scisignal.2001993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Holt LJ, et al. 2009. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325: 1682–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hu H, et al. 23 May 2012, posting date. An orphan kinesin in trypanosomes cooperates with a kinetoplastid-specific kinesin to maintain cell morphology through regulating subpellicular microtubules. J. Cell Sci. [Epub ahead of print.] doi:10.1242/jcs.106534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hu L, Hu H, Li Z. 2012. A kinetoplastid-specific kinesin is required for cytokinesis and for maintenance of cell morphology in Trypanosoma brucei. Mol. Microbiol. 83: 565–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Humphrey T, Pearce A. 2005. Cell cycle molecules and mechanisms of the budding and fission yeasts. Methods Mol. Biol. 296: 3–29 [DOI] [PubMed] [Google Scholar]
- 53. Ilves I, Petojevic T, Pesavento JJ, Botchan MR. 2010. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol. Cell 37: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jeyaprakash AA, et al. 2007. Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell 131: 271–285 [DOI] [PubMed] [Google Scholar]
- 55. Kanemaki M, Sanchez-Diaz A, Gambus A, Labib K. 2003. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature 423: 720–724 [DOI] [PubMed] [Google Scholar]
- 56. Kaufmann D, Gassen A, Maiser A, Leonhardt H, Janzen CJ. 2012. Regulation and spatial organization of PCNA in Trypanosoma brucei. Biochem. Biophys. Res. Commun. 419: 698–702 [DOI] [PubMed] [Google Scholar]
- 57. Kettenbach AN, et al. 2011. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal. 4: rs5 doi:10.1126/scisignal.2001497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Klingbeil MM, Shapiro TA. 2009. Unraveling the secrets of regulating mitochondrial DNA replication. Mol. Cell 35: 398–400 [DOI] [PubMed] [Google Scholar]
- 59. Kohl L, Robinson D, Bastin P. 2003. Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. EMBO J. 22: 5336–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kubota Y, et al. 2003. A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes Dev. 17: 1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kumar P, Wang CC. 2005. Depletion of anaphase-promoting complex or cyclosome (APC/C) subunit homolog APC1 or CDC27 of Trypanosoma brucei arrests the procyclic form in metaphase but the bloodstream form in anaphase. J. Biol. Chem. 280: 31783–31791 [DOI] [PubMed] [Google Scholar]
- 62. Kumar P, Wang CC. 2006. Dissociation of cytokinesis initiation from mitotic control in a eukaryote. Eukaryot. Cell 5: 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Labib K. 2010. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 24: 1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li Z, Gourguechon S, Wang CC. 2007. Tousled-like kinase in a microbial eukaryote regulates spindle assembly and S-phase progression by interacting with Aurora kinase and chromatin assembly factors. J. Cell Sci. 120: 3883–3894 [DOI] [PubMed] [Google Scholar]
- 65. Li Z, et al. 2008. Identification of a novel chromosomal passenger complex and its unique localization during cytokinesis in Trypanosoma brucei. PLoS One 3: e2354 doi:10.1371/journal.pone.0002354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li Z, Lindsay ME, Motyka SA, Englund PT, Wang CC. 2008. Identification of a bacterial-like HslVU protease in the mitochondria of Trypanosoma brucei and its role in mitochondrial DNA replication. PLoS Pathog. 4: e1000048 doi:10.1371/journal.ppat.100048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li Z, Tu X, Wang CC. 2006. Okadaic acid overcomes the blocked cell cycle caused by depleting Cdc2-related kinases in Trypanosoma brucei. Exp. Cell Res. 312: 3504–3516 [DOI] [PubMed] [Google Scholar]
- 68. Li Z, Umeyama T, Wang CC. 2008. The chromosomal passenger complex and a mitotic kinesin interact with the Tousled-like kinase in trypanosomes to regulate mitosis and cytokinesis. PLoS One 3: e3814 doi:10.1371/journal.pone.0003814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li Z, Umeyama T, Wang CC. 2009. The Aurora kinase in Trypanosoma brucei plays distinctive roles in metaphase-anaphase transition and cytokinetic initiation. PLoS Pathog. 5: e1000575 doi:10.1371/journal.ppat.1000575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li Z, Wang CC. 2003. A PHO80-like cyclin and a B-type cyclin control the cell cycle of the procyclic form of Trypanosoma brucei. J. Biol. Chem. 278: 20652–20658 [DOI] [PubMed] [Google Scholar]
- 71. Li Z, Wang CC. 2006. Changing roles of aurora-B kinase in two life cycle stages of Trypanosoma brucei. Eukaryot. Cell 5: 1026–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Li Z, Wang CC. 2008. KMP-11, a basal body and flagellar protein, is required for cell division in Trypanosoma brucei. Eukaryot. Cell 7: 1941–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Liao S, Wang T, Fan K, Tu X. 2010. The small ubiquitin-like modifier (SUMO) is essential in cell cycle regulation in Trypanosoma brucei. Exp. Cell Res. 316: 704–715 [DOI] [PubMed] [Google Scholar]
- 74. Liu B, et al. 2009. Trypanosomes have six mitochondrial DNA helicases with one controlling kinetoplast maxicircle replication. Mol. Cell 35: 490–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ma J, et al. 2010. Nuclear DBF-2-related kinases are essential regulators of cytokinesis in bloodstream stage Trypanosoma brucei. J. Biol. Chem. 285: 15356–15368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Masai H, et al. 2006. Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J. Biol. Chem. 281: 39249–39261 [DOI] [PubMed] [Google Scholar]
- 77. May SF, et al. 2012. The Trypanosoma brucei AIR9-like protein is cytoskeleton-associated and is required for nucleus positioning and accurate cleavage furrow placement. Mol. Microbiol. 84: 77–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Monnerat S, Clucas C, Brown E, Mottram JC, Hammarton TC. 2009. Searching for novel cell cycle regulators in Trypanosoma brucei with an RNA interference screen. BMC Res. Notes 2: 46 doi:10.1186/1756-0500-2-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Moyer SE, Lewis PW, Botchan MR. 2006. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. U. S. A. 103: 10236–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Muramatsu S, Hirai K, Tak YS, Kamimura Y, Araki H. 2010. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol ε, and GINS in budding yeast. Genes Dev. 24: 602–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Nguyen VQ, Co C, Irie K, Li JJ. 2000. Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2-7. Curr. Biol. 10: 195–205 [DOI] [PubMed] [Google Scholar]
- 82. Nguyen VQ, Co C, Li JJ. 2001. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411: 1068–1073 [DOI] [PubMed] [Google Scholar]
- 83. Ogbadoyi E, Ersfeld K, Robinson D, Sherwin T, Gull K. 2000. Architecture of the Trypanosoma brucei nucleus during interphase and mitosis. Chromosoma 108: 501–513 [DOI] [PubMed] [Google Scholar]
- 84. Oliferenko S, Chew TG, Balasubramanian MK. 2009. Positioning cytokinesis. Genes Dev. 23: 660–674 [DOI] [PubMed] [Google Scholar]
- 85. Olsen JV, et al. 2010. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3: ra3 doi:10.1126/scisignal.2000475 [DOI] [PubMed] [Google Scholar]
- 86. Peters JM. 2006. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7: 644–656 [DOI] [PubMed] [Google Scholar]
- 87. Piekny A, Werner M, Glotzer M. 2005. Cytokinesis: welcome to the Rho zone. Trends Cell Biol. 15: 651–658 [DOI] [PubMed] [Google Scholar]
- 88. Pollard TD. 2010. Mechanics of cytokinesis in eukaryotes. Curr. Opin. Cell Biol. 22: 50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pollard TD, Wu JQ. 2010. Understanding cytokinesis: lessons from fission yeast. Nat. Rev. Mol. Cell Biol. 11: 149–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Price HP, Peltan A, Stark M, Smith DF. 2010. The small GTPase ARL2 is required for cytokinesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 173: 123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Remus D, Diffley JF. 2009. Eukaryotic DNA replication control: lock and load, then fire. Curr. Opin. Cell Biol. 21: 771–777 [DOI] [PubMed] [Google Scholar]
- 92. Robinson DR, Sherwin T, Ploubidou A, Byard EH, Gull K. 1995. Microtubule polarity and dynamics in the control of organelle positioning, segregation, and cytokinesis in the trypanosome cell cycle. J. Cell Biol. 128: 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Rodgers MJ, Albanesi JP, Phillips MA. 2007. Phosphatidylinositol 4-kinase III-beta is required for Golgi maintenance and cytokinesis in Trypanosoma brucei. Eukaryot. Cell 6: 1108–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Rothberg KG, et al. 2006. The RACK1 homologue from Trypanosoma brucei is required for the onset and progression of cytokinesis. J. Biol. Chem. 281: 9781–9790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Salmon D, et al. 2012. Cytokinesis of Trypanosoma brucei bloodstream forms depends on expression of adenylyl cyclases of the ESAG4 or ESAG4-like subfamily. Mol. Microbiol. 84: 225–242 [DOI] [PubMed] [Google Scholar]
- 96. Satyanarayana A, Kaldis P. 2009. Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene 28: 2925–2939 [DOI] [PubMed] [Google Scholar]
- 97. Seki A, Coppinger JA, Jang CY, Yates JR, Fang G. 2008. Bora and the kinase Aurora A cooperatively activate the kinase Plk1 and control mitotic entry. Science 320: 1655–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Selvapandiyan A, et al. 2007. Centrin1 is required for organelle segregation and cytokinesis in Trypanosoma brucei. Mol. Biol. Cell 18: 3290–3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Sheu YJ, Stillman B. 2006. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol. Cell 24: 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sheu YJ, Stillman B. 2010. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 463: 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Sillje HH, Nigg EA. 2001. Identification of human Asf1 chromatin assembly factors as substrates of Tousled-like kinases. Curr. Biol. 11: 1068–1073 [DOI] [PubMed] [Google Scholar]
- 102. Sillje HH, Takahashi K, Tanaka K, Van Houwe G, Nigg EA. 1999. Mammalian homologues of the plant Tousled gene code for cell-cycle-regulated kinases with maximal activities linked to ongoing DNA replication. EMBO J. 18: 5691–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Skop AR, Liu H, Yates J, III, Meyer BJ, Heald R. 2004. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science 305: 61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Straight AF, Field CM. 2000. Microtubules, membranes and cytokinesis. Curr. Biol. 10: R760–R770 [DOI] [PubMed] [Google Scholar]
- 105. Takayama Y, et al. 2003. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 17: 1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Tanaka S, et al. 2007. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445: 328–332 [DOI] [PubMed] [Google Scholar]
- 107. Tiengwe C, et al. 2012. Identification of ORC1/CDC6-interacting factors in Trypanosoma brucei reveals critical features of origin recognition complex architecture. PLoS One 7: e32674 doi:10.1371/journal.pone.0032674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Tu X, Kumar P, Li Z, Wang CC. 2006. An Aurora kinase homologue is involved in regulating both mitosis and cytokinesis in Trypanosoma brucei. J. Biol. Chem. 281: 9677–9687 [DOI] [PubMed] [Google Scholar]
- 109. Tu X, Wang CC. 2004. The involvement of two cdc2-related kinases (CRKs) in Trypanosoma brucei cell cycle regulation and the distinctive stage-specific phenotypes caused by CRK3 depletion. J. Biol. Chem. 279: 20519–20528 [DOI] [PubMed] [Google Scholar]
- 110. Tu X, Wang CC. 2005. Pairwise knockdowns of cdc2-related kinases (CRKs) in Trypanosoma brucei identified the CRKs for G1/S and G2/M transitions and demonstrated distinctive cytokinetic regulations between two developmental stages of the organism. Eukaryot. Cell 4: 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Uchiyama M, Griffiths D, Arai K, Masai H. 2001. Essential role of Sna41/Cdc45 in loading of DNA polymerase alpha onto minichromosome maintenance proteins in fission yeast. J. Biol. Chem. 276: 26189–26196 [DOI] [PubMed] [Google Scholar]
- 112. Umeyama T, Wang CC. 2008. Polo-like kinase is expressed in S/G2/M phase and associated with the flagellum attachment zone in both procyclic and bloodstream forms of Trypanosoma brucei. Eukaryot. Cell 7: 1582–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Urbaniak MD, Guther ML, Ferguson MA. 2012. Comparative SILAC proteomic analysis of Trypanosoma brucei bloodstream and procyclic lifecycle stages. PLoS One 7: e36619 doi:10.1371/journal.pone.0036619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Van Hellemond JJ, Neuville P, Schwarz RT, Matthews KR, Mottram JC. 2000. Isolation of Trypanosoma brucei CYC2 and CYC3 cyclin genes by rescue of a yeast G1 cyclin mutant: functional characterization of CYC2. J. Biol. Chem. 275: 8315–8323 [DOI] [PubMed] [Google Scholar]
- 115. van Vugt MA, Bras A, Medema RH. 2004. Polo-like kinase-1 controls recovery from a G2 DNA damage-induced arrest in mammalian cells. Mol. Cell 15: 799–811 [DOI] [PubMed] [Google Scholar]
- 116. Vaughan S, Gull K. 2008. The structural mechanics of cell division in Trypanosoma brucei. Biochem. Soc. Trans. 36: 421–424 [DOI] [PubMed] [Google Scholar]
- 117. Vaughan S, Kohl L, Ngai I, Wheeler RJ, Gull K. 2008. A repetitive protein essential for the flagellum attachment zone filament structure and function in Trypanosoma brucei. Protist 159: 127–136 [DOI] [PubMed] [Google Scholar]
- 118. Wickstead B, Carrington JT, Gluenz E, Gull K. 2010. The expanded Kinesin-13 repertoire of trypanosomes contains only one mitotic Kinesin indicating multiple extra-nuclear roles. PLoS One 5: e15020 doi:10.1371/journal.pone.0015020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Yu Z, Liu Y, Li Z. 2012. Structure-function relationship of the Polo-like kinase in Trypanosoma brucei. J. Cell Sci. 125: 1519–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Zegerman P, Diffley JF. 2007. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445: 281–285 [DOI] [PubMed] [Google Scholar]
- 121. Zou L, Stillman B. 2000. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol. Cell. Biol. 20: 3086–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]