Abstract
Candida albicans grows within a wide range of host niches, and this adaptability enhances its success as a commensal and as a pathogen. The telomere-associated TLO gene family underwent a recent expansion from one or two copies in other CUG clade members to 14 expressed copies in C. albicans. This correlates with increased virulence and clinical prevalence relative to those of other Candida clade species. The 14 expressed TLO gene family members have a conserved Med2 domain at the N terminus, suggesting a role in general transcription. The C-terminal half is more divergent, distinguishing three clades: clade α and clade β have no introns and encode proteins that localize primarily to the nucleus; clade γ sometimes undergoes splicing, and the gene products localize within the mitochondria as well as the nuclei. Additionally, TLOα genes are generally expressed at much higher levels than are TLOγ genes. We propose that expansion of the TLO gene family and the predicted role of Tlo proteins in transcription regulation provide C. albicans with the ability to adapt rapidly to the broad range of different environmental niches within the human host.
INTRODUCTION
The ability to respond to stress is critical for survival, especially in organisms that reside in a dynamic environment such as the varied niches within a host organism. Candida species are the most prevalent fungal pathogens of humans, causing mucosal infections of the mouth, genitourinary tract, and skin, as well as life-threatening bloodstream infections. Candida albicans resides as a harmless commensal in the human gastrointestinal tract, yet it causes >50% of all systemic fungal infections. A number of traits, including the ability to switch to hyphal growth and to undergo phenotypic switches, likely contribute to the higher virulence of C. albicans than of other Candida species (35).
Genetic responses to growth in new, stressful environments include changes in gene copy number, which provide a rapid mechanism to adapt available genetic material to cope with altered conditions (7, 10, 15, 33, 42). Telomeric regions of the genome exhibit the most variation, and variation accumulates rapidly in these regions (7, 9, 12). For example, in Saccharomyces cerevisiae the SUC, MAL, and MEL families have expanded to different extents in strains bred to ferment different carbon sources (sucrose, maltose, and melibiose, respectively) (3, 7, 10, 43); the subtelomeric family of FLO genes, which encode the ability to adhere to different cellular and abiotic surfaces, have expanded in some fermentation and clinical isolates (22, 43, 45).
The telomere-associated (TLO) gene family in C. albicans is a remarkable example of gene family expansion near the telomeres. The TLO gene family is the gene family that has expanded most in C. albicans relative to the less pathogenic Candida species (6). C. albicans has 14 annotated TLO genes, compared to two TLO genes in the closely related oral pathogen Candida dubliniensis and a single TLO gene in most other Candida species (6, 44).
All but one of the TLO genes are located within 12 kb of a telomere and are often the most terminal predicted open reading frame (ORF) of each chromosome arm. A single TLO is found at an internal locus on chromosome 1 (Chr1), although whether this TLO is expressed is not known (44). In S. cerevisiae, the most terminal ORFs contain the subtelomeric gene families Y′ and X, which encode RNA helicases and transcriptional silencers, respectively (26, 27), and are actively transcribed (47).
The first TLO gene to be identified was named CTA2 and was isolated in an S. cerevisiae one-hybrid screen for C. albicans transactivating proteins (24). This implies that Tlo proteins bind (directly or indirectly) to DNA and have the potential to regulate transcription. Indeed, the predicted Tlo proteins all include a domain with high similarity to Med2, a component of the Mediator complex, which regulates the transcription of class II genes by bridging general transcriptional activators and RNA polymerase II (PolII) (20, 24, 34). A recent study (48) revealed that some Tlo proteins function as Med2-like components of the Mediator complex.
Here, we characterize the structure and expression patterns of the C. albicans TLO gene family. Phylogenetic analysis indicates that there are three clades of expressed TLO genes, α, β, and γ, all of which include a predicted Med2 domain. They differ primarily by the presence of long terminal repeat (LTR) insertions that alter the coding sequences. In addition, we identified a 15th TLO gene, organized in a head-to-tail arrangement with a TLO pseudogene copy that lacks the Med2 domain. We found that members of the TLOγ clade produce both spliced and unspliced transcripts and that the splice junctions are different in different TLOγ genes. Tlo proteins encoded by all three clades are detected in the nucleus, and the Tloγ proteins also localize to mitochondria. TLOα genes are expressed at the highest levels, with TLOγ clade transcripts and proteins expressed at much lower levels under a range of physiologically relevant growth conditions. This broad range of Tlo expression levels and different localization patterns is predicted to result in a similarly broad range of Mediator complex subunit compositions, perhaps facilitating adaptation to the broad range of host niches that C. albicans occupies.
MATERIALS AND METHODS
Growth conditions used.
Standard growth conditions were rich medium (YPAD at 30°C) (39). Assays were performed by inoculating cells in YPAD and growing them at 30°C overnight. Cultures were then diluted 1:100 in fresh YPAD and grown at 30°C for 4 h.
Bioinformatic characterization of the TLO gene family.
TLO sequences were aligned using the Multiple Sequence Comparison by Log-Expectation (MUSCLE) algorithm at http://www.ebi.ac.uk/Tools/muscle/index.html (11). The protein sequence was investigated for functional motifs using InterPro Scan (http://www.ebi.ac.uk/InterProScan/) and Pfam HMM (http://pfam.sanger.ac.uk/). Homology to other known genes was determined using BLAST methods at http://www.ncbi.nlm.nih.gov/blast/Blast.cgi/ and http://www.yeastgenome.org/. The domain architecture of Med2 was investigated using the Conserved Doman Database (CDD) (http://www.ncbi.nlm.nih.gov/sites/entrez?db=cdd). Nuclear localization signal (NLS) predictions were made with PredictNLS (http://www.predictprotein.org/).
Characterization of TLO coding sequences and splicing.
SC5314 was inoculated in YPAD and grown at 30°C overnight. Cultures were then diluted 1:100 in fresh YPAD and grown at 30°C for 3 to 4 h. RNA was harvested using the Masterpure yeast RNA extraction kit (Epicentre, Madison, WI) and reverse transcribed using primer 5′ CCAGTGAGCAGAGTGACGAGGACTCGAGCTCAAGCTTTTTTTTTTTTTTTTT 3′ with the Protoscript Moloney murine leukemia virus (M-MuLV) first-strand cDNA synthesis kit (New England Biotech, Ipswich, MA). The purity of the cDNA was verified using large-subunit ribosome protein 6 (RPL6) primers flanking an intron to ensure the absence of genomic DNA. The coding sequences of TLOα3, TLOα12, and TLOβ2 were determined using primer 5′ CCAGTGAGCAGAGTGACG 3′, homologous to the poly(A) tail anchor primer, and primers 5′ TCAACGGCAATACCAACGAC 3′, 5′ CATCAGGATACATATTAGAGG 3′, and 5′ TGGCAACAACACCAACCACGTA 3′, respectively. The TLOγ5, TLOγ7, and TLOγ13 coding sequences were amplified using primer 5′ CCAGTGAGCAGAGTGACG 3′, homologous to the poly(A) tail anchor primer, and primers 5′ TCAAGAAAAAGGCAGAGGAAGCG 3′, 5′ GGCCAAAAAAAAGGAAGAAGAGGC 3′, and 5′ GCAATGTGATTACTAGCCCC 3′, respectively. Two rounds of PCR were performed to identify the TLOγ16 coding sequences. The first round used primer 5′ CCAGTGAGCAGAGTGACG 3′, homologous to the anchor primer, and genomic primer 5′ GACTACATAACTCACTCGACG 3′, and the second round of PCR used primers 5′ GAGGACTCGAGCTCAAGC 3′, homologous to the anchor primer, and 5′ GGGGCAAAGAAAAAGGAAGA 3′, unique to TLOγ16. All products were cloned into TOPO-TA (Invitrogen, Carlsbad, CA) and sequenced.
Identification of TLOγ5 and TLOγ13 splicing was performed by amplifying the splice junction with primers 5′ CTCTGCCTTCTCTTCTTCCT 3′ and 5′ ATGCCAGAAAACCTCCAAAC 3′. Resulting amplicons were TOPO cloned and sequenced.
Assignment of TLO genes to chromosome arms.
SC5314 DNA was collected as previously described (17). PCR was performed using arm-specific primers together with a pan-TLO primer, which are all listed in Table S1 in the supplemental material. The arm-specific primers were designed against the region of unique sequence closest to the TLO.
Construction of the right arm of Chr1 was performed by amplifying an ∼4-kb fragment using the pan-TLO primer and the Chr1 right-arm-specific primer listed in Table S1 in the supplemental material. Sequential sequencing of the amplicons was used to construct the full sequence. The start codon of TLOγ4 was confirmed using primers 5′ ATGCCAGAAAACCTCCAAAC 3′ and 5′ CACACATCAGGTGATGACAG 3′, which correspond to the well-conserved TLO start codon and a TLOγ4-specific primer, respectively.
Strain construction.
Strains are listed in Table S2 in the supplemental material. Transformations were performed using lithium acetate (36). Integration of the construct at the expected locus was confirmed by PCR and sequencing. C-terminal green fluorescent protein (GFP)-tagged TLO strains were constructed by PCR amplification from plasmid p1602 (13), which contains GFP and URA3, using primers with at least 70 bp of homology to the target TLO (see Table S3 in the supplemental material). The resulting tagged TLO was identified by amplifying a fragment with a GFP-specific primer and the pan-TLO primer and subsequently sequenced. To detect the chromosome into which the construct integrated, chromosomes were separated on contour-clamped homogeneous electric field (CHEF) karyotype gels and Southern blotting (38) was used to detect the URA3 insertion. The URA3 probe was amplified by PCR using digoxigenin (DIG)-11-dUTP nucleotides (Roche, Mannheim, Germany) with primers 5′ AGACCTATAGTGAGAGAGCA 3′ and 5′ CAAACAATCCTCTACCAACA 3′ according to the manufacturer's instructions. We used only strains in which insertion was detected on only one chromosome and with one, unambiguous PCR fragment from a single chromosome arm.
Fluorescence microscopy.
Strains were inoculated in YPAD, grown at 30°C overnight, diluted 1:100 in fresh synthetic complete dropout (SDC) medium, and grown at 30°C for 3 to 4 h. 4′,6-Diamidino-2-phenylindole (DAPI) (Sigma, St. Louis, MO) diluted 1:1,000 and Mito-Tracker (Invitrogen, Carlsbad, CA), which labels mitochondria by reacting with accessible thiol groups found in the mitochondrial matrix and inner membrane, diluted 1:1,000 were added, and cultures were incubated for 25 min. Cells were washed twice in fresh SDC medium and imaged using differential interference contrast (DIC) and epifluorescence microscopy with a Nikon Eclipse E600 photomicroscope (Chroma Technology Corp., Brattleboro, VT). Digital images were collected using a CoolSnap HQ camera (Photometrics, Tucson, AZ) and MetaMorph software, version 6.2r5 (Universal Imaging Corp., Downingtown, PA). A total of 12 fields with 8 to 15 fluorescent images along the z axis, in 1-μm increments, were collected for each cell to ensure that any signal present was captured throughout the diameter of the cell body. The z series stack was then collapsed into a single image by using the stack arithmetic/maximum function of MetaMorph for analysis and image construction for presentation.
Quantitative reverse transcription-PCR (qRT-PCR) of TLO transcription.
RNA was collected using the Masterpure yeast RNA extraction kit (Epicentre, Madison, WI), and cDNA was synthesized using the Protoscript M-MuLV first-strand cDNA synthesis kit according to the manufacturer's instructions using d(T)23VN (New England Biotech, Ipswich, MA). PCR of cDNA using intron-spanning primers to RPL6 confirmed a lack of contaminating genomic DNA. Transcript abundance was quantified by SYBR green incorporation using a Lightcycler 480 II qPCR machine (Roche, Mannheim, Germany) and analyzed with the Lightcycler 480 software package v1.5.0 (Syni Corporation, Los Angeles, CA). Absolute quantification of SYBR fluorescence using the second derivative maximum value was used to calculate the threshold cycle (ΔCT) value for each TLO gene by using ACT1 and TEF1 as controls. Briefly, the ΔCT value of each TLO gene was calculated as the difference in cycle threshold between TEF1 and each TLO gene (the ratio of TEF1/ACT1 was consistent across experiments). Results represent at least three independent experiments with standard deviations.
Oligonucleotide sequences are provided in Table S4 in the supplemental material. Due to the high levels of DNA sequence homology between TLO family members, TLO-specific primer sets were designed such that one oligonucleotide hybridized to a limited number of TLO sequences (1 to 4 instead of all 14 TLO sequences) and the second oligonucleotide was specific to a single TLO transcript by including unique single nucleotide polymorphism (SNPs). TLO-specific PCR amplicons were verified by DNA sequencing of cDNA.
Splicing abundance assay.
SC5314 cDNA was used to PCR amplify both spliced and unspliced TLOγ16 transcripts using the primers described to identify both RNA isoforms above. Amplified products were run by gel electrophoresis on the same 1% gel, and band intensities were quantified using Fiji/ImageJ v1.46 (NIH, Bethesda, MD). Three independent experiments were run for each strain tested.
Western blotting assay of Tlo abundance.
Protein lysates were collected as previously described (14). Western blot assays were performed with mouse anti-GFP (Roche, Penzberg, Germany) and goat anti-Cdc28 (Santa Cruz Biotechnology, Santa Cruz, CA).
RESULTS
Identification and mapping of TLO clades.
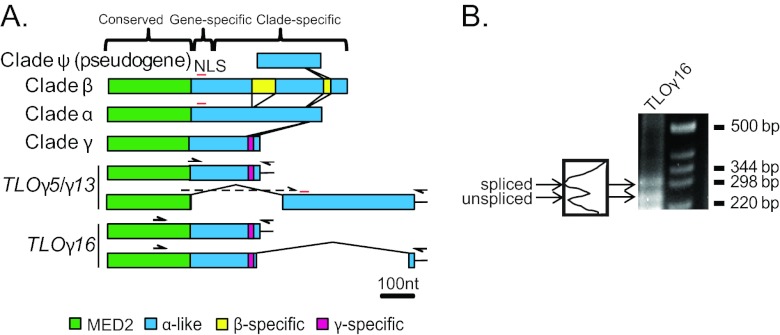
Fourteen telomere-associated (TLO) genes were previously defined and mapped to the assembled C. albicans genome sequence, primarily by using bioinformatics criteria (44). To compare the relative similarities of members of this gene family, we aligned the nucleotide sequences using MUSCLE (11). Based on sequence similarity, the TLO genes are organized into three major regions: the 5′ region (nucleotides [nt] 1 to 315), which has homology to the Med2 domain (Fig. 1A, green regions), as discussed below and in reference 48; a middle region of variable length (up to 140 nt) that contains indels with gene-specific adenosine-rich stretches; and the 3′ half, which is organized in a clade-specific manner (Fig. 1A) interspersed with clade-specific unique sequences (Fig. 1A, yellow and magenta). To distinguish these differences, we renamed the TLO genes by adding the clade letter (α, β, or γ) between “TLO” and the notation of van het Hoog et al. (44), which numbers the TLO genes based upon their chromosomal arm positions, e.g., TLOβ2.
Fig 1.
The C. albicans TLO genes form three separate clades. (A) Alignment of the TLO gene family members in the SC5314 genome and distinctions between the α, β, and γ clades. The TLO DNA sequence is roughly divided into three regions: a conserved region with the Med2 domain, highlighted in green; a gene-specific region consisting of highly variable adenosine-rich sequences; and a clade-specific region, which in the single-exon form is dissimilar between clades and highly conserved among genes within clades. A predicted nuclear localization signal (NLS) is indicated by a red line. A series of indels differentiates the TLOα/β and single-exon TLOγ sequences. Insertions of clade-specific sequences are connected by lines to indicate their positions relative to one another and color coded as indicated. Two differentially spliced transcripts were detected for TLOγ5 and TLOγ13 using a primer spanning the splicing event and against the poly(A) tail, indicated as half arrows. Sequencing confirmed splicing at nt 310 in TLOγ8 and at nt 333 in TLOγ13, in both cases joined to a second exon of ∼372 bp starting at nt 647 and 639, respectively. TLOγ16 splicing occurred 30 bp upstream of the stop codon in the unspliced transcript and spliced into a second, 45-bp exon from nt 356 to 401 downstream of the unspliced stop codon. (B) 3′ RACE of SC5314 cDNA amplified both spliced and unspliced transcripts of TLOγ16 of approximately 300 bp and 250 bp, respectively. Each transcript is expressed at roughly equivalent amounts as determined by densitometry.
The TLOα clade has 6 members, each contained by a single exon. TLOβ2, the single member of the TLOβ clade, contains two unique sequences in the 3′ half of the ORF, resulting in a single exon that is 240 bp longer than in TLOα genes (Fig. 1A). The most 5′ TLOβ-specific sequence encodes a predicted EF-hand calcium-binding domain (P < 1E−5). Each of the 7 TLOγ clade members has been predicted to encode transcripts with two possible RNA isoforms, either a single exon or a spliced transcript (44) that would produce proteins differing in length by 8 to 43 amino acids. Unspliced TLOγ clade transcripts are similar to TLOα clade members but contain several short deletions through the clade-specific region. We detected full-length (unspliced) transcripts for TLOγ5, TLOγ7, TLOγ13, and TLOγ16 (Fig. 1A). Furthermore, TLO-specific primers for either TLOγ5 or TLOγ13 detected both unspliced and spliced products (Fig. 1A).
Unexpectedly, the splice junctions previously predicted for TLOγ clade members (44) could not be detected using primers bridging the splice junction at nt 243 (data not shown). Instead, we identified a putative splice junction at nt 333 of TLOγ5 and TLOγ13, immediately upstream of the adenosine-rich, gene-specific region, using GeneScan to analyze the TLOγ clade sequences (5). GeneScan predicted a 314-nt intron, which includes a 5′ splice site at nt 333, a lariat branch point at nt 531 (UUCUUAC), and a 3′ splice junction at nt 647. The second 372-nt exon spans nt 647 to 1019 and carries 102 predicted codons. A PCR primer designed to span the 5′ splice junction at nt 333 (Fig. 1A) produced a product whose sequence confirmed that the splice junction conformed to the GeneScan prediction. The TLOγ5 and TLOγ13 spliced transcripts were sequenced, and their ORFs predict proteins with a Med2 domain most similar to the TLOγ clade and genes with a 3′ region similar to TLOα coding sequences.
Because the predicted splice donor sequence in TLOγ5 and TLOγ13 was not detected in the other 5 TLOγ clade members, it was unclear if these genes are spliced in the same manner. Indeed, analysis of TLOγ16 by 3′ rapid amplification of cDNA ends (RACE) identified two transcripts: the predicted single exon (528 nt) and a spliced form with 5′ and 3′ junction sites very different from those in TLOγ5 and TLOγ13. The 5′ splice site at nt 498 is only 30 nt upstream of the stop codon in the single-exon form of the gene. The second exon is short, only 45 bp long, and carries a TLOα-like sequence (Fig. 1A). TLOγ16 is likely to be unique in having this particular spliced isoform, because no other TLO sequence contains the necessary GT splice donor site at this position. The proportions of spliced-to-unspliced TLOγ16 mRNA were roughly equivalent (Fig. 1B). Interestingly, multiple TLOγ transcripts (e.g., TLOγ16 and TLOγ13) ended immediately following the translational termination codon followed by a poly(A) tail, suggesting that some TLO transcripts completely lack a 3′ untranslated region (UTR) (data not shown). Transcripts lacking a 3′ UTR have not been detected for other C. albicans genes and are likely to affect transcript stability and translation efficiency (23).
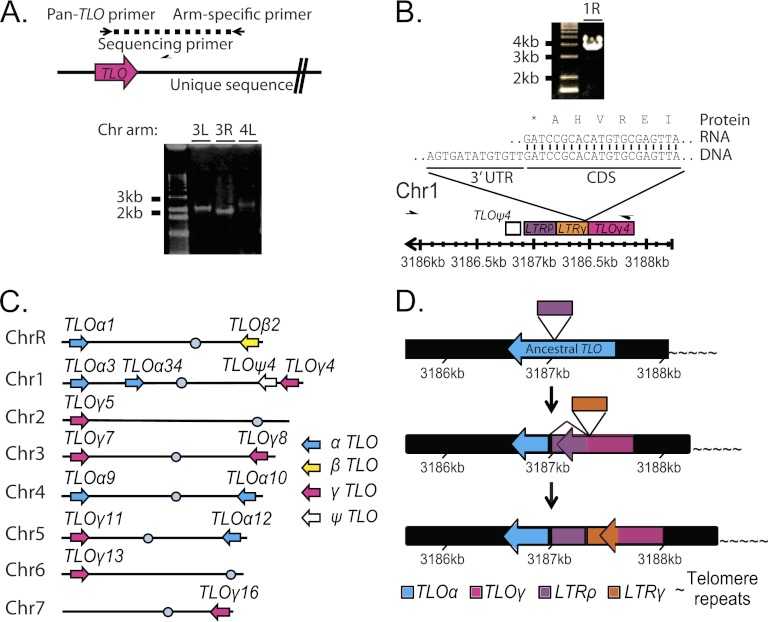
High sequence similarity between TLO gene family members and complex sequence structure in the subtelomeres could have complicated the bioinformatic assignment of specific TLO genes to chromosome arms. To experimentally test the genome position of each TLO sequence, we amplified the TLO from each chromosome arm by PCR with a single arm-specific primer anchored in a unique sequence, together with a single pan-TLO primer with homology to the highly conserved Med2 domain (Fig. 2A). In addition to the expected TLO amplicons in strain SC5314 (Fig. 2C; Table 1), the sequenced amplicons identified two tandem TLO ORFs on the right arm of Chr1 (Fig. 2B). The centromere-proximal one of these two TLO genes, previously named TLO4 (44), contains only a partial TLO sequence lacking the Med2 domain and the conserved TLO promoter sequence. qRT-PCR and 3′ RACE studies did not detect a TLO4 transcript (data not shown). Thus, we named this gene TLOψ4, to indicate that it is a pseudogene. The second TLO ORF on Chr1R was telomere proximal to TLOψ4 (at coordinates Chr1:3187887 to Chr1:3187464). This locus, now named TLOγ4, carries a TLOγ clade ORF that is disrupted by an LTR in the 3′ half of the gene. TLOγ4 is expressed and is transcribed toward the centromere, like all other TLO genes. The TLOγ4 transcript terminates within the LTR sequence, producing a protein that contains the Med2 domain and a short (36-amino-acid [aa]) C-terminal tail that is homologous to TLOγ clade members. Thus, SC5314 contains 14 transcribed TLO genes and one pseudogene.
Fig 2.
Organization of TLO genes in C. albicans SC5314. (A) TLO genes were amplified from each chromosome arm as indicated in the cartoon. Primers are represented as half-arrows. A representative gel shows amplification of both arms of Chr3 (3L and 3R) and the left arm of Chr4 (4L). (B) Arm-specific PCR as in panel A produced a large PCR product that, when sequenced, confirmed the presence of an unannotated TLO gene, TLOγ4, on the right arm of chromosome 1. The end of the transcript terminated in the gamma LTR sequence shortly after the Med2 domain. This cDNA was cloned three times independently, and no alternate RNA isoforms were identified. (C) TLO assignment to chromosome arms in strain SC5314 is depicted and reflects PCR and sequencing information obtained as in panel A. TLO genes are color coded by clade as indicated. (D) The ancestral C. albicans TLO was most similar to the TLOα clade. A rho LTR disrupted an ancestral TLO, producing a novel 3′ end. The disrupted TLO could splice out the LTR sequence and recreate the TLOα-like sequence. This disruption is the progenitor of the TLOγ clade. Further evolution has occurred on Chr1 where a second LTR insertion has disrupted the TLOγ clade gene and produced TLOγ4, which is truncated, terminating in the more recent LTR insertion, compared to other TLOγ clade members. The numbers indicate the position of the TLO gene on chromosome 1.
Table 1.
List of TLO genes in SC5314 and important features
| Namea | Phylogenetic nameb | Alternate splicingc | Length (aa)d | Functional motif(s) (aa)e | NLSf |
|---|---|---|---|---|---|
| TLO1 | TLOα1 | No | 258 | Med2 (1–109) | Yes |
| TLO2 | TLOβ2 | No | 273 | Med2 (1–119), EF-hand (196–208) | Yes |
| TLO3 | TLOα3 | No | 255 | Med2 (1–115) | Yes |
| TLO34 | TLOα34 | No | 331 | Med2 (75–183) | Yes |
| TLO4 | TLOψ4 | No | NAg | NA | NA |
| TLOγ4 | No | 140 | Med2 (1–104) | No | |
| TLO5 | TLOγ5 | Yes | 176 | Med2 (1–108) | No, yes |
| TLO7 | TLOγ7 | ? | 169, 206 | Med2 (1–107) | No |
| TLO8 | TLOγ8 | ? | 169 | Med2 (1–108) | No |
| TLO9 | TLOα9 | No | 225 | Med2 (1–108) | Yes |
| TLO10 | TLOα10 | No | 219 | Med2 (1–108) | Yes |
| TLO11 | TLOγ11 | ? | 169 | Med2 (1–108) | No |
| TLO12 | TLOα12 | No | 252 | Med2 (1–108) | Yes |
| TLO13 | TLOγ13 | Yes | 176, 213 | Med2 (1–108) | No, yes |
| TLO16 | TLOγ16 | Yes | 179, 184 | Med2 (1–108) | No |
Based on the work of van het Hoog et al. (44).
Based on this study.
Based on RT-PCR analysis (Fig. 1).
Predicted protein length (unspliced, spliced).
Predicted functional motifs with Pfam/Prosite scores above P < 1 × 10−5.
Presence of predicted NLS based on PredictNLS with score above P < 1 × 10−5 (unspliced, spliced).
NA, not applicable.
Localization of Tlo proteins.
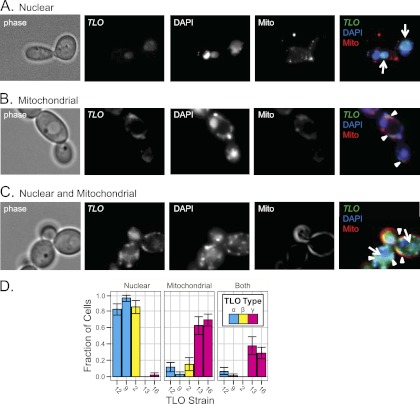
The presence of a Mediator complex subunit 2 (Med2) domain (P = 7.3E−24) at the 5′ end of predicted TLO ORFs suggests that the TLO genes are C. albicans Med2 homologs. The Mediator complex is a large, multisubunit transcriptional coactivator for polymerase II (PolII)-transcribed genes (8) expected to localize to the nucleus, and the identification of TLO genes as having DNA binding activity (21) bolsters the prediction. Furthermore, nuclear localization signals (NLSs) are predicted in TLOα and TLOβ sequences just 3′ of the Med2 domain (Fig. 1A). Thus, we performed fluorescence microscopy on TLO gene products that were tagged with green fluorescent protein (GFP) at the C terminus of their single-exon forms. Sequence containing the highest level of polymorphism within 150 bp 3′ of the TLO coding sequence provided the basis for targeting GFP to specific TLO genes. To identify the TLO gene tagged with GFP, we sequenced an amplified fragment from the GFP tag into the TLO Med2 domain. To ensure that this gene was on the expected chromosome end, we also analyzed chromosomes separated on a CHEF gel by Southern blotting with a probe against URA3, the selectable marker used to introduce the GFP tag. Tloα9-GFP, Tloα12-GFP, and Tloβ2-GFP colocalized with DAPI staining of nuclear DNA in a single large focus per cell in >80% of cells imaged (Fig. 3A and D). In contrast, Tloγ16-GFP and Tloγ13-GFP exhibited a more complex pattern: some of the GFP signal colocalized with DAPI in the nucleus and some colocalized with DAPI to regions outside the nucleus at the cell periphery (Fig. 3B and C). This peripheral Tloγ16-GFP and Tloγ13-GFP colocalized with Mito-Tracker stain, which detects mitochondrial inner membrane and matrix proteins. Tloγ16 and Tloγ13 localized to mitochondria and the nucleus in ∼60% of cells, to mitochondria in only ∼38% of cells, and exclusively to nuclei in only ∼2% of cells imaged (Fig. 3D). Spliced Tloγ5/Tloγ13 proteins had primarily nuclear localization like that seen for Tloα and Tloβ proteins, likely due to the NLS encoded in the second exon (data not shown). Thus, unlike Tloα and Tloβ proteins, the Tloγ proteins produced from unspliced transcripts localized both to the mitochondria and to the nucleus.
Fig 3.
TLO clades localize to different cellular compartments. Localization of Tloα, Tloβ, and Tloγ clade members GFP tagged at the C terminus was determined by fluorescence microscopy. Colocalization with DAPI to stain nuclear and mitochondrial DNA and Mito-Tracker to stain the mitochondria determined localization of Tlo proteins in live cells. (A) A representative Tlo localized to the nucleus coincident with DAPI. (B) A representative Tlo localized to the mitochondria coincident with Mito-Tracker. (C) A representative Tlo localized to the nucleus and mitochondria coincident with DAPI and Mito-Tracker. (D) The fraction of cells with Tlo-GFP localized to various cellular compartments across 10 fields is displayed alongside each clade. Two Tloα and Tloγ clade members were localized to ensure clade reproducibility. Tloα and Tloβ clade members localized primarily to the nucleus, and Tloγ clade members localized to the mitochondria or mitochondria and nucleus.
TLO gene transcription levels.
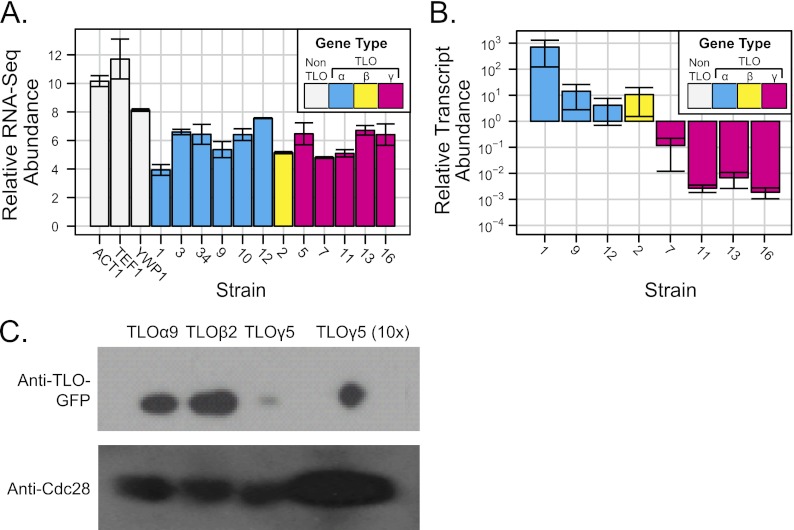
Individual members of expanded subtelomeric gene families are often expressed at different levels (25, 40, 47). To ask if all TLO genes are transcribed, we analyzed deep sequencing (RNA-Seq) data reported for C. albicans strain SC5314 by Bruno et al. (4). The abundances of most TLO transcripts appeared to be similar under standard growth conditions, suggesting that TLO genes from all clades were expressed at similar levels (Fig. 4A).
Fig 4.
Expression of C. albicans TLO genes. (A) Transcript abundance from RNA-Seq data (4) indicates similar midrange expression levels of all TLO genes. The TLO clade is indicated as follows: cyan for TLOα, yellow for TLOβ, and magenta for TLOγ. (B) qRT-PCR measured TLO transcript abundance for eight TLO genes representing all three clades in SC5314 under standard growth conditions. Transcript abundance of TLOα and TLOβ clade members was ∼10- to 100-fold greater than TLOγ clade members. TEF1 was used to normalize across replicates. The graph represents three independent experiments with technical triplicates and the standard deviations of the cycle threshold. (C) Protein abundance of Tlos was measured by Western blotting assay using Cdc28 as a loading control. Tloα and Tloβ clade members were in greater abundance than Tloγ clade members.
A concern with RNA-Seq analysis is ambiguity in the mapping of sequencing reads to specific genes that have high levels of homology to each other. For the C. albicans RNA-Seq data, each ambiguous read was resolved independently by uniform assignment to one, and only one, of the matching sequences. Given two differentially expressed genes with similar coding sequences, ambiguous mappings would be expected to make the transcript levels appear more similar than the biological reality.
To measure expression from individual TLO genes, we used quantitative reverse transcription-PCR (qRT-PCR) (31) together with gene-specific primers designed to align with single-exon sequences that were most distinctive for each TLO gene (described in detail in Materials and Methods). All assayed TLO family members, including the internal TLOα34, produced detectable transcripts, with the exception of pseudogene TLOψ4. However, in stark contrast to the predictions from RNA-Seq, steady-state levels of TLOα and TLOβ transcripts were 100- to 1,000-fold more abundant than the steady-state levels of TLOγ transcripts (Fig. 4B; note log scale on y axis). Consistent with this, much higher levels of the corresponding proteins were detected by Western blotting (Fig. 4C) (48). Therefore, we conclude that TLOα and TLOβ expression is significantly higher than TLOγ expression.
DISCUSSION
Telomere-associated genes tend to expand when they provide an adaptive advantage for growth under a new environmental condition (7, 37). The C. albicans TLO gene family has expanded dramatically since the divergence of C. albicans from C. dubliniensis (20 million years ago [MYA], 2 copies) and the other CUG clade members (50 to 200 MYA, 1 copy) (30). While the mechanism by which it provides an advantage, presumably to life as a commensal and/or pathogen within the human host, is not yet clear, here we begin to characterize the nature of the expansion. We found that TLO genes have diversified into three clades from a single ancestral gene (19), that the TLOγ clade is regulated differently at the level of mRNA splicing, and that the proteins encoded by the different clades also localize, to some degree, to different cellular compartments.
The 14 TLO genes (plus one TLO pseudogene) of SC5314 likely arose from a single ancestral TLOα member that expanded and produced the current TLO diversity (19). The close association of LTRs with TLO ORFs, as well as with the tandem TLO gene and pseudogene on Chr1R, suggests that LTR retrotransposition likely drove TLO diversification (Fig. 2D) (29). We speculate that an initial LTR retrotransposition event disrupted a TLOα-like ancestral gene, producing the TLOγ clade, with a different 3′ end in the single-exon form. Splicing of the resulting TLOγ gene, as in the case of TLOγ13 and TLOγ5, would excise the LTR and produce a transcript with a 3′ end that resembles that of TLOα clade members and that might retain a closely related function. Interestingly, the localization of the single-exon Tloγ proteins to the mitochondria portends a function that may differ from that of Tloα and Tloβ proteins (49). Distinguishing the function of mitochondrial Tlo proteins will require biochemical and further genetic analysis.
The mechanism that facilitated TLO expansion is unclear, but clues exist in the structure of the subtelomeres. A kappa LTR sequence resides telomere proximal to all TLO genes, and the Tloγ proteins produced from unspliced mRNAs are predicted to include C-terminal sequences translated from the rho LTR (44). A subsequent LTR disruption of TLOγ4 on Chr1R suggests that these elements have continued to play a role in TLO diversification. Interestingly, the one internal TLO gene, TLOα34, also is flanked by kappa LTR and rho LTR sequences, indicating that TLO genes associate with LTRs irrespective of their chromosome location and LTRs may have facilitated TLO expansion and clade differentiation (29, 46). Additionally, long tracts of highly homologous sequence that reside centromere proximal to the TLO genes may have facilitated recombination and TLO expansion.
The conserved N-terminal Mediator 2 (MED2) homology of all transcribed TLO genes strongly suggests that they encode functional components of the Mediator complex, which participates in transcription regulation (16, 24), an idea recently confirmed biochemically (48). The ratio of nonsynonymous to synonymous evolutionary changes (dN/dS ratio) of the Med2 domain across all TLO family members (0.625) suggests purifying selection for the Med2 sequence, similar to what is seen for MED2 homologs from other yeast species (2). The C termini of Med2 proteins are highly variable and species specific, presumably conferring specificity for different transcription factors in different species (32).
It is quite unusual that multiple genes encode a single Mediator subunit, but this is not completely unprecedented: plants and some other yeast species encode multiple paralogs of a single Mediator subunit (28, 41). The large number of Med2 homologs encoded by the TLO genes and the splicing variants of TLOγ clade members suggest that Mediator is not monolithic. Rather, Mediator is likely to be a related set of different variant complexes that, in C. albicans, differ by the Med2 component (18). In metazoans, Mediator complexes differ by interchangeable use of duplicated kinase module subunits and the incorporation of additional novel subunits in the core modules (2). It is tempting to speculate that different Mediator variants have different affinities for a range of transcription factors and that they ultimately impact the expression of different sets of target genes (1). Thus, the TLO gene family expansion may have greatly increased the repertoire of possible transcriptional responses by generating a broad variety of options for transcriptional outputs.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for the gift of additional SC5314 genome sequence reads from Gavin Sherlock and suggestions from Peter Tiffin in visualizing TLO relatedness. We thank Laura Burrack, Benjamin Harrison, Meleah Hickman, and P. T. Magee for valuable comments on the manuscript and all members of the Berman lab for many helpful discussions and suggestions.
This work was supported by a grant from the National Institute of Allergy and Infectious Diseases (AI075096-03S1) to J.B. and a Research Supplement to Promote Diversity in Health-Related Research award to M.Z.A.
Footnotes
Published ahead of print 24 August 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Beve J, et al. 2005. The structural and functional role of Med5 in the yeast Mediator tail module. J. Biol. Chem. 280: 41366– 41372 [DOI] [PubMed] [Google Scholar]
- 2. Bourbon HM. 2008. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 36: 3993– 4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown CA, Murray AW, Verstrepen KJ. 2010. Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr. Biol. 20: 895– 903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bruno VM, et al. 2010. Comprehensive annotation of the transcriptome of the human fungal pathogen Candida albicans using RNA-seq. Genome Res. 20: 1451– 1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burge C, Karlin S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78– 94 [DOI] [PubMed] [Google Scholar]
- 6. Butler G, et al. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459: 657– 662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carreto L, et al. 2008. Comparative genomics of wild type yeast strains unveils important genome diversity. BMC Genomics 9: 524 doi:10.1186/1471-2164-9-524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Conaway RC, Conaway JW. 2011. Function and regulation of the Mediator complex. Curr. Opin. Genet. Dev. 21: 225– 230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dreszer TR, Wall GD, Haussler D, Pollard KS. 2007. Biased clustered substitutions in the human genome: the footprints of male-driven biased gene conversion. Genome Res. 17: 1420– 1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dujon B, et al. 2004. Genome evolution in yeasts. Nature 430: 35– 44 [DOI] [PubMed] [Google Scholar]
- 11. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792– 1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Farman ML, Kim YS. 2005. Telomere hypervariability in Magnaporthe oryzae. Mol. Plant Pathol. 6: 287– 298 [DOI] [PubMed] [Google Scholar]
- 13. Gerami-Nejad M, Berman J, Gale CA. 2001. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast 18: 859– 864 [DOI] [PubMed] [Google Scholar]
- 14. Greenbaum D, Colangelo C, Williams K, Gerstein M. 2003. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 4: 117 doi:10.1186/gb-2003-4-9-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gresham D, et al. 2008. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 4:e1000303 doi:10.1371/journal.pgen.1000303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gustafsson CM, Samuelsson T. 2001. Mediator—a universal complex in transcriptional regulation. Mol. Microbiol. 41:1– 8 [DOI] [PubMed] [Google Scholar]
- 17. Hoffman CS, Winston F. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57: 267– 272 [DOI] [PubMed] [Google Scholar]
- 18. Huh WK, et al. 2003. Global analysis of protein localization in budding yeast. Nature 425: 686– 691 [DOI] [PubMed] [Google Scholar]
- 19. Jackson AP, et al. 2009. Comparative genomics of the fungal pathogens Candida dubliniensis and Candida albicans. Genome Res. 19: 2231– 2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kagey MH, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430– 435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaiser B, Munder T, Saluz HP, Kunkel W, Eck R. 1999. Identification of a gene encoding the pyruvate decarboxylase gene regulator CaPdc2p from Candida albicans. Yeast 15: 585– 591 [DOI] [PubMed] [Google Scholar]
- 22. Kellis M, Birren BW, Lander ES. 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428: 617– 624 [DOI] [PubMed] [Google Scholar]
- 23. Kondoh H, Mizutani T. 1998. Expression of the glutathione peroxidase gene lacking its 3′ untranslated region. Mol. Biol. Rep. 25: 121– 125 [DOI] [PubMed] [Google Scholar]
- 24. Kornberg RD. 2005. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30: 235– 239 [DOI] [PubMed] [Google Scholar]
- 25. Kyes SA, Kraemer SM, Smith JD. 2007. Antigenic variation in Plasmodium falciparum: gene organization and regulation of the var multigene family. Eukaryot. Cell 6: 1511– 1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lebrun E, et al. 2001. Protosilencers in Saccharomyces cerevisiae subtelomeric regions. Genetics 158: 167– 176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Louis EJ, Haber JE. 1992. The structure and evolution of subtelomeric Y' repeats in Saccharomyces cerevisiae. Genetics 131: 559– 574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mathur S, Vyas S, Kapoor S, Tyagi AK. 2011. The Mediator complex in plants: structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress. Plant Physiol. 157: 1609– 1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maxwell PH, Burhans WC, Curcio MJ. 2011. Retrotransposition is associated with genome instability during chronological aging. Proc. Natl. Acad. Sci. U. S. A. 108: 20376– 20381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mishra PK, Baum M, Carbon J. 2007. Centromere size and position in Candida albicans are evolutionarily conserved independent of DNA sequence heterogeneity. Mol. Genet. Genomics 278: 455– 465 [DOI] [PubMed] [Google Scholar]
- 31. Nolan T, Hands RE, Bustin SA. 2006. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 1: 1559– 1582 doi:10.1038/nprot.2006.236 [DOI] [PubMed] [Google Scholar]
- 32. Novatchkova M, Eisenhaber F. 2004. Linking transcriptional mediators via the GACKIX domain super family. Curr. Biol. 14: R54– R55 [DOI] [PubMed] [Google Scholar]
- 33. Pavelka N, Rancati G, Li R. 2010. Dr Jekyll and Mr Hyde: role of aneuploidy in cellular adaptation and cancer. Curr. Opin. Cell Biol. 22: 809– 815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Peng J, Zhou JQ. 2012. The tail-module of yeast Mediator complex is required for telomere heterochromatin maintenance. Nucleic Acids Res. 40: 581– 593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ramirez-Zavala B, Reuss O, Park YN, Ohlsen K, Morschhauser J. 2008. Environmental induction of white-opaque switching in Candida albicans. PLoS Pathog. 4: e1000089 doi:10.1371/journal.ppat.1000089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sanglard D, Ischer F, Monod M, Bille J. 1996. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob. Agents Chemother. 40:2300– 2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schuller D, et al. 2012. Genetic diversity and population structure of Saccharomyces cerevisiae strains isolated from different grape varieties and winemaking regions. PLoS One 7: e32507 doi:10.1371/journal.pone.0032507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Selmecki A, Bergmann S, Berman J. 2005. Comparative genome hybridization reveals widespread aneuploidy in Candida albicans laboratory strains. Mol. Microbiol. 55:1553– 1565 [DOI] [PubMed] [Google Scholar]
- 39. Sherman F. 1991. Getting started with yeast. Methods Enzymol. 194: 3– 21 [DOI] [PubMed] [Google Scholar]
- 40. Taylor JE, Rudenko G. 2006. Switching trypanosome coats: what's in the wardrobe? Trends Genet. 22: 614– 620 [DOI] [PubMed] [Google Scholar]
- 41. Thakur JK, et al. 2009. Mediator subunit Gal11p/MED15 is required for fatty acid-dependent gene activation by yeast transcription factor Oaf1p. J. Biol. Chem. 284: 4422– 4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Torres EM, et al. 2010. Identification of aneuploidy-tolerating mutations. Cell 143: 71– 83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Turakainen H, Naumov G, Naumova E, Korhola M. 1993. Physical mapping of the MEL gene family in Saccharomyces cerevisiae. Curr. Genet. 24: 461– 464 [DOI] [PubMed] [Google Scholar]
- 44. van het Hoog M, et al. 2007. Assembly of the Candida albicans genome into sixteen supercontigs aligned on the eight chromosomes. Genome Biol. 8: R52 doi:10.1186/gb-2007-8-4-r52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Verstrepen KJ, Klis FM. 2006. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 60:5– 15 [DOI] [PubMed] [Google Scholar]
- 46. Xiao H, Jiang N, Schaffner E, Stockinger EJ, van der Knaap E. 2008. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319: 1527– 1530 [DOI] [PubMed] [Google Scholar]
- 47. Yamada M, Hayatsu N, Matsuura A, Ishikawa F. 1998. Y'-Help1, a DNA helicase encoded by the yeast subtelomeric Y' element, is induced in survivors defective for telomerase. J. Biol. Chem. 273: 33360– 33366 [DOI] [PubMed] [Google Scholar]
- 48. Zhang A, et al. 2012. The Tlo proteins are stoichiometric components of Candida albicans Mediator anchored via the Med3 subunit. Eukaryot. Cell 11: 874– 884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang F, Sumibcay L, Hinnebusch AG, Swanson MJ. 2004. A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4p. Mol. Cell. Biol. 24: 6871– 6886 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.