Abstract
The function of a number of genes in the gliotoxin biosynthetic cluster (gli) in Aspergillus fumigatus remains unknown. Here, we demonstrate that gliK deletion from two strains of A. fumigatus completely abolished gliotoxin biosynthesis. Furthermore, exogenous H2O2 (1 mM), but not gliotoxin, significantly induced A. fumigatus gliK expression (P = 0.0101). While both mutants exhibited significant sensitivity to both exogenous gliotoxin (P < 0.001) and H2O2 (P < 0.01), unexpectedly, exogenous gliotoxin relieved H2O2-induced growth inhibition in a dose-dependent manner (0 to 10 μg/ml). Gliotoxin-containing organic extracts derived from A. fumigatus ATCC 26933 significantly inhibited (P < 0.05) the growth of the ΔgliK26933 deletion mutant. The A. fumigatus ΔgliK26933 mutant secreted metabolites, devoid of disulfide linkages or free thiols, that were detectable by reverse-phase high-performance liquid chromatography and liquid chromatography-mass spectrometry with m/z 394 to 396. These metabolites (m/z 394 to 396) were present at significantly higher levels in the culture supernatants of the A. fumigatus ΔgliK26933 mutant than in those of the wild type (P = 0.0024 [fold difference, 24] and P = 0.0003 [fold difference, 9.6], respectively) and were absent from A. fumigatus ΔgliG. Significantly elevated levels of ergothioneine were present in aqueous mycelial extracts of the A. fumigatus ΔgliK26933 mutant compared to the wild type (P < 0.001). Determination of the gliotoxin uptake rate revealed a significant difference (P = 0.0045) between that of A. fumigatus ATCC 46645 (9.3 pg/mg mycelium/min) and the ΔgliK46645 mutant (31.4 pg/mg mycelium/min), strongly suggesting that gliK absence and the presence of elevated ergothioneine levels impede exogenously added gliotoxin efflux. Our results confirm a role for gliK in gliotoxin biosynthesis and reveal new insights into gliotoxin functionality in A. fumigatus.
INTRODUCTION
Gliotoxin was discovered in 1936, and its structure was elucidated in 1943 (19, 38). Much initial interest in the molecule was directed toward exploiting its antifungal activities; however, after observations that gliotoxin exhibited growth-inhibitory effects against animal cells, it was posited as a putative anti-cancer or organ transplant rejection agent, and a large number of subsequent studies investigated the mechanism of action of gliotoxin against mammalian cell types (3, 24, 37).
Until recently, the mechanism of gliotoxin biosynthesis had proven to be especially refractory to analysis, despite multiple studies between 1960 and 1985 involving radiolabeled precursor feeding and analysis of proteins produced during gliotoxin biosynthesis (2, 18). Since the discovery of the gliotoxin gene cluster gli (14), now known to contain 13 genes, the function of a number of cluster genes has been established by a combination of targeted gene deletion and phenotypic and biochemical analyses (4, 10, 11, 13, 22, 31–33, 34, 36). gliZ [a Zn(II)2-Cys(6) binuclear cluster domain transcription factor] deletion resulted in attenuation of gli cluster expression and the absence of gliotoxin production (4). The nonribosomal peptide synthetase GliP has been shown to be essential for gliotoxin biosynthesis and to catalyze cyclo-l-phenylalanyl-l-seryl formation from the cognate precursor amino acids (1). Glutathione (GSH) has been implicated as the immediate thiol source for both sulfur atoms in gliotoxin, and a reactive acyl imine intermediate has been proposed to act as an acceptor for glutathionylation catalyzed by the glutathione S-transferase GliG (11, 31). The gliotoxin oxidoreductase, GliT, a key self-protection gene against gliotoxin, is regulated independently of gliZ and catalyzes disulfide bridge closure (32, 33). The involvement of GliC has also been investigated (31), whereby GliC-mediated oxygenation of a gliotoxin biosynthetic intermediate was essential for subsequent thiolation catalyzed by GliG. Additionally, Schrettl et al. (33) demonstrated that gliH was also essential for gliotoxin biosynthesis, but that it did not play a role in self-protection against gliotoxin. Comparative metabolomic analysis (13), using two-dimensional nuclear magnetic resonance, has revealed the presence of 19 gli-dependent metabolites (biosynthetic intermediates or shunt metabolites) in wild-type Aspergillus fumigatus which are absent from the gliZ deletion strain (4). Interestingly, this work also demonstrated that gliP and gliI deletion mutant metabolomes did not contain any gliZ-dependent compounds. While this might be expected for GliP absence, which normally catalyzes the initiation step in gliotoxin biosynthesis (1), it is unclear why gliI (a putative aminotransferase) deletion resulted in the complete absence of gliZ-dependent metabolites, although the absence of gliZ and gliP expression in A. fumigatus ΔgliI has been noted (13).
Probably one of the most cryptic genes in the gli cluster is gliK (CADRE identifier AFUA_6G09700; http://www.cadre-genomes.org.uk [23]). However, the role of the protein encoded by gliK in A. fumigatus has not been characterized to date. Interestingly, fungi which contain the major facilitator superfamily (MFS), rather than an ABC transporter associated with gliotoxin production, also possess an adjacent gliK-type gene (28), whereas organisms that contain the ABC transporter do not contain a gliK gene within the relevant gene cluster. Conceivably, therefore, it is possible that the protein encoded by gliK is involved in epipolythiodioxopiperazine efflux with GliA, an MFS transporter in the gli cluster. In any case, given the effective annotation of GliK as a protein of unknown function and the presence of orthologs in a range of fungal species (n = 56 with >30% identity), functional annotation of the gene is essential to further our knowledge of epipolythiodioxopiperazine (ETP) biosynthesis.
Here, we demonstrate that deletion of gliK in two A. fumigatus strains results in acquisition of significant gliotoxin and hydrogen peroxide sensitivity, disrupted gliotoxin biosynthesis, metabolome remodelling, and impaired gliotoxin uptake. Unexpectedly, we further demonstrate that gliotoxin presence protects A. fumigatus against hydrogen peroxide-induced oxidative stress.
MATERIALS AND METHODS
Strains, growth conditions, and oligonucleotides.
A. fumigatus strains (see Table S1 in the supplemental material) were grown at 37°C in Aspergillus minimal medium (AMM). AMM contained 1% (wt/vol) glucose as the carbon source, 5 mM ammonium tartrate as the nitrogen source, and trace elements according to Pontecorvo et al. (29). Liquid cultures were performed with 200 ml AMM or Czapek-Dox in 500-ml Erlenmeyer flasks inoculated with 108 conidia. For growth assays, 103 conidia of the respective strains were point inoculated on AMM or Czapek-Dox plates containing the relevant supplements and incubated for 72 h at 37°C. Oligonucleotides used are listed in Table S2 in the supplemental material.
Generation of A. fumigatus mutant strains.
For the generation of both ΔgliK strains, the bipartite marker technique was used (25). Briefly, A. fumigatus strains ATCC 46645 and ATCC 26933 were cotransformed with two DNA constructs, each containing an incomplete fragment of a pyrithiamine resistance gene (ptrA) (21) fused to 1 and 1.1 kb of gliK flanking sequences, respectively. These marker fragments shared a 557-bp overlap in the ptrA cassette, which served as a potential recombination site during transformation. During transformation, homologous integration of each fragment into the genome flanking gliK allows recombination of the ptrA fragments and generation of the intact resistance gene at the site of recombination. Two rounds of PCR generated each fragment. First, each flanking region was amplified from ATCC 46645 genomic DNA using primers gliKP1 and gliKP4 for flanking region A (1.25 kb) and gliKP2 and gliKP3 for flanking region B (1.2 kb). Subsequent to gel purification, the fragments were digested with SpeI and HindIII, respectively. The ptrA selection marker was released from plasmid pSK275 (a kind gift from Sven Krappmann, Goettingen, Germany) by digestion with SpeI and HindIII and ligated with the two flanking regions A and B described above. For generation of the ΔgliK mutants, two overlapping fragments were amplified from the ligation products using primers gliKP5 and optrA2 for fragment C (2.6 kb) and primers gliKP6 and optrA1 for fragment D (2.2 kb). Subsequently, ATCC 46645 and ATCC 26933 were transformed simultaneously with the overlapping fragments C and D. Complementation of gliK was undertaken in the A. fumigatus ΔgliK46645 strain using selection on phleomycin and confirmed by Southern analysis (see Fig. S1 in the supplemental material).
RNA isolation and real-time PCR.
Fungal RNA was isolated and purified from A. fumigatus hyphae crushed in liquid nitrogen using the RNeasy plant minikit (Qiagen). RNA was treated with DNase 1 (Invitrogen), and cDNA synthesis from mRNA (500 ng) was performed using a first-strand transcription cDNA synthesis kit (Roche) with oligo(dT) primers. The gene encoding calmodulin (AFUA_4G10050), which is constitutively expressed in A. fumigatus, served as a control in reverse transcription-PCR (RT-PCR) experiments (6). Real-time PCR was performed using the LightCycler 480 Sybr green 1 master mix (Roche) on a LightCycler 480 real-time PCR system as previously described (27). For real-time PCRs, a 1/10 dilution of cDNA from each sample was used as a template and each reaction was performed in triplicate.
Phenotypic assays.
A. fumigatus wild-type and mutant strains were incubated at 37°C for up to 72 h in the presence of either gliotoxin (0 to 20 μg/ml), H2O2 (0 to 5 mM), gliotoxin and H2O2 (H2O2, 0 to 1 mM; gliotoxin, 0 to 10 μg/ml), or voriconazole (0 to 0.25 μg/ml).
Analysis of gliotoxin and related metabolite production.
To analyze gliotoxin and related metabolite production, A. fumigatus wild-type and mutant strains were grown at 37°C for 48 h in AMM or Czapek-Dox medium. Supernatants were chloroform extracted overnight, and fractions were dried to completion under vacuum. Extracts were resolubilized in methanol and analyzed using reverse-phase high-performance liquid chromatography (RP-HPLC) and LC-mass spectrometry (LC-MS) as previously described (30, 33). Reduction and alkylation (using 5′-iodoacetamidofluorescein; 5′-IAF) of organic extracts was performed as described previously (11, 12). For intracellular metabolite analysis, mycelia were lysed as described previously (7, 17) prior to reduction and alkylation (11, 12). Alternatively, total mycelial thiol quantitation was undertaken using aldrithiol-4 (11 mg/ml in ethanol) analysis (35).
Determination of gliotoxin uptake rate.
A. fumigatus ATCC 46645 (n = 3 biological replicates) and ΔgliK46645 (n = 3) strains were grown at 37°C in minimal medium (MM), which contained 1% (wt/vol) glucose as the carbon source, 20 mM l-glutamine as the nitrogen source, and trace elements according to Pontecorvo et al. (29) for 24 h. At 24 h, gliotoxin (5 μg/ml final) was added to the cultures, and 2-ml aliquots of supernatant were removed at 0, 15, 30, and 45 min. After chloroform extraction, all specimens were resuspended in methanol (55 μl) followed by RP-HPLC analysis (30, 33) for quantitation of residual gliotoxin. Corresponding mycelia from replicate specimens were lyophilized and weighted.
Statistical analysis.
All data were analyzed using built-in GraphPad Prism (version 5.01) functions as specified. The level of significance was set at P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***) unless otherwise stated. Post hoc comparisons between groups were performed using the Bonferroni multiple-comparison test unless otherwise stated. All graphs were compiled using Graphpad Prism (version 5.01) unless otherwise stated.
RESULTS
Deletion of gliK from A. fumigatus.
A. fumigatus ΔgliK mutants were generated by transformation of A. fumigatus strains ATCC 46645 and ATCC 26933, respectively, as described in Materials and Methods, using the bipartite marker technique and pyrithiamine selection, with modifications (21, 25). Targeted gliK deletions in both ATCC 46645 and 26933 genetic backgrounds were obtained at a frequency of 1/113 and 1/58 colonies screened, respectively (see Fig. S1 in the supplemental material). These two strains were chosen because A. fumigatus ATCC 46645 exhibits extremely low-level gliotoxin secretion using MM, whereas the ATCC 26933 strain is a potent gliotoxin producer (see below) (33).
Gene expression analysis.
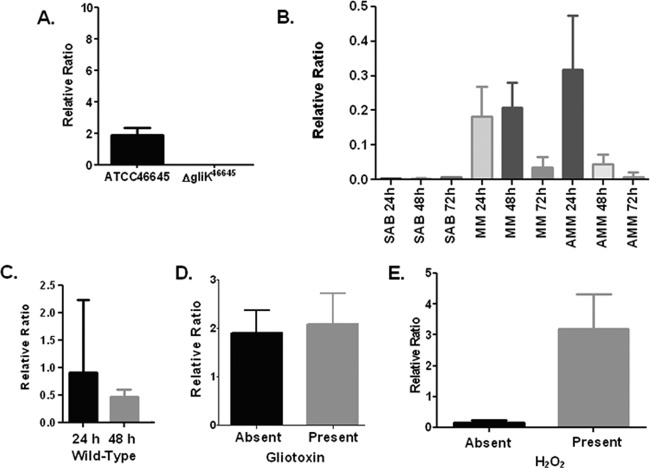
gliK mRNA abundance was assessed in A. fumigatus ATCC 46645 in different liquid medium types, namely, Sabouraud medium, MM, and AMM. The cultures were grown for 24, 48, and 72 h, respectively (Fig. 1). mRNA abundance of gliK never exceeded a ratio of 1, indicating that it is at a much lower level than the calmodulin reference gene (6). gliK mRNA abundance was highest in AMM at 24 h yet was absent from Sabouraud media at all time points. The mRNA abundance of gliK in MM was equivalent at 24 and 48 h, whereas the abundance level dropped dramatically at 72 h (P < 0.1). In AMM cultures, the highest mRNA abundance of gliK was at 24 h, which was reduced considerably at 48 h (P < 0.01) and was barely detectable at 72 h (P < 0.001). This indicates that gliK mRNA abundance is highest in MM and at early time points during culture. Since gliK mRNA abundance was highest in AMM, this medium was chosen to confirm the absence of gliK mRNA in the A. fumigatus ΔgliK46645 mutant (Fig. 1). gliK was expressed in A. fumigatus ATCC 46645 at 24 and 48 h, whereas the ΔgliK46645 strain did not express gliK, thereby confirming the deletion of gliK from the genome. gliK expression was restored following complementation (see Fig. S2 in the supplemental material). Liquid cultures of A. fumigatus ATCC 46645 in AMM were spiked with gliotoxin (5 μg/ml final) at 21 h and the cultures harvested at 24 h (33), and it was observed that gliK mRNA abundance was not significantly increased in the presence of gliotoxin (Fig. 1), indicating that gliK expression is not controlled by the presence of gliotoxin. AMM cultures of A. fumigatus ATCC 46645 were also subjected to H2O2-induced oxidative stress (1 mM) at 21 h, and the cultures were harvested at 24 h. The abundance of gliK mRNA was significantly upregulated, from a relative ratio of 0.1665 to 3.193 (P = 0.0101), in the presence of H2O2 (Fig. 1). A. fumigatus gliA is adjacent to gliK in the genome; however, deletion of gliK did not prevent gliA expression in the mutant strains (see Fig. S2).
Fig 1.
(A) qRT-PCR analysis of gliK expression in A. fumigatus ATCC 46645 and the ΔgliK46645 mutant in AMM at 24 h at 37°C. (B) Quantitative RT-PCR (qRT-PCR) analysis of gliK expression at 24, 48, and 72 h in A. fumigatus ATCC 46645 cultured in various media at 37°C. (C) qRT-PCR analysis of gliK expression in A. fumigatus ATCC 46645 and the ΔgliK46645 mutant, respectively, in AMM at selected time points at 37°C. (D) qRT-PCR analysis of gliK expression in A. fumigatus ATCC 46645 in AMM at 24 h at 37°C, with and without exogenous gliotoxin spiked into the media. Column 1, gliK expression in A. fumigatus ATCC 46645; column 2, gliK expression in A. fumigatus ATCC 46645 spiked with gliotoxin (5 μg/ml final concentration). (E) qRT-PCR analysis of gliK expression in A. fumigatus ATCC 46645 in AMM at 24 h at 37°C, with and without hydrogen peroxide spiked into the media. Column 1, gliK expression in A. fumigatus ATCC 46645; column 2, gliK expression in A. fumigatus ATCC 46645 spiked with 1 mM hydrogen peroxide (final concentration). SAB, Sabouraud medium.
Gliotoxin significantly inhibits growth of A. fumigatus ΔgliK.
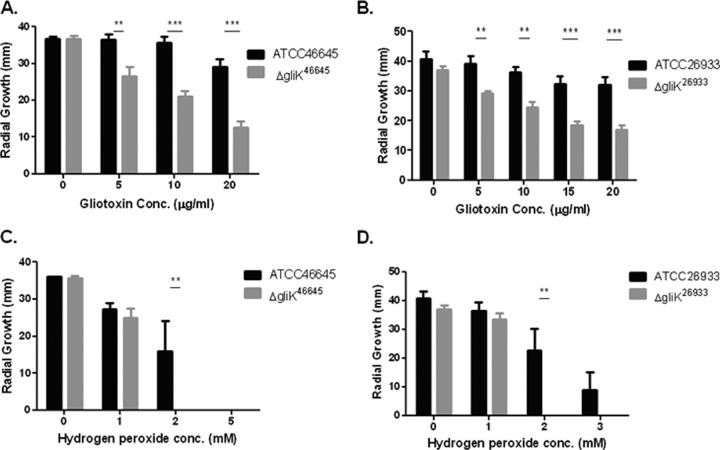
It has been demonstrated that A. fumigatus is capable of self protection against gliotoxin, and that deletion of gliT significantly increased strain sensitivity to exogenous gliotoxin (32, 33). Both ΔgliK mutants were also significantly more sensitive to gliotoxin (P < 0.001), and a reduced growth rate was seen at all time points, with the greatest difference in growth seen at 72 h (Fig. 2). After 72 h of growth on AMM plates, there was a significant decrease in the radial growth of the A. fumigatus ΔgliK46645 strain compared to the wild-type strain when gliotoxin was present in the media (Fig. 2 and 3). The difference was highly significant at gliotoxin concentrations of 10 and 20 μg/ml (P < 0.001). A. fumigatus ATCC 46645 also exhibited a decreased radial growth (∼20%) at the highest gliotoxin concentration used (20 μg/ml) compared to that in media without gliotoxin, indicating that the A. fumigatus wild type is sensitive to high levels of gliotoxin. The decrease in radial growth due to the presence of exogenous gliotoxin was also observed in the A. fumigatus ΔgliK26933 strain at all gliotoxin concentrations tested (Fig. 2 and 3) and was highly significant at 10 and 20 μg/ml gliotoxin, respectively (P < 0.001). As seen for A. fumigatus ATCC 46645, ATCC 26933 showed a decrease in growth ability in gliotoxin (20 μg/ml), where there was a reduction in radial growth of 21% compared to that in media without gliotoxin. The abundance of gliT mRNA was determined in A. fumigatus ΔgliK26933 and ΔgliK46645 strains and was not found to be affected, indicating that the sensitivity to gliotoxin observed in A. fumigatus ΔgliK26933 and ΔgliK46645 strains was not due to attenuated A. fumigatus gliT expression (see Fig. S3 in the supplemental material).
Fig 2.
A. fumigatus ΔgliK is sensitive to gliotoxin- and H2O2-mediated inhibition. Effect of gliotoxin on the growth of A. fumigatus ATCC 46645 and the ΔgliK46645 mutant (A) and ATCC 26933 and the ΔgliK26933 mutant (B). Significant growth inhibition of A. fumigatus ΔgliK46645 and ΔgliK26933 mutants is evident at 72 h. Effect of hydrogen peroxide on the growth of A. fumigatus ATCC 46645 and the ΔgliK46645 mutant (C) and A. fumigatus ATCC 26933 and the ΔgliK26933 mutant (D). Significant growth inhibition of the A. fumigatus ΔgliK26933 mutant is evident at 72 h.
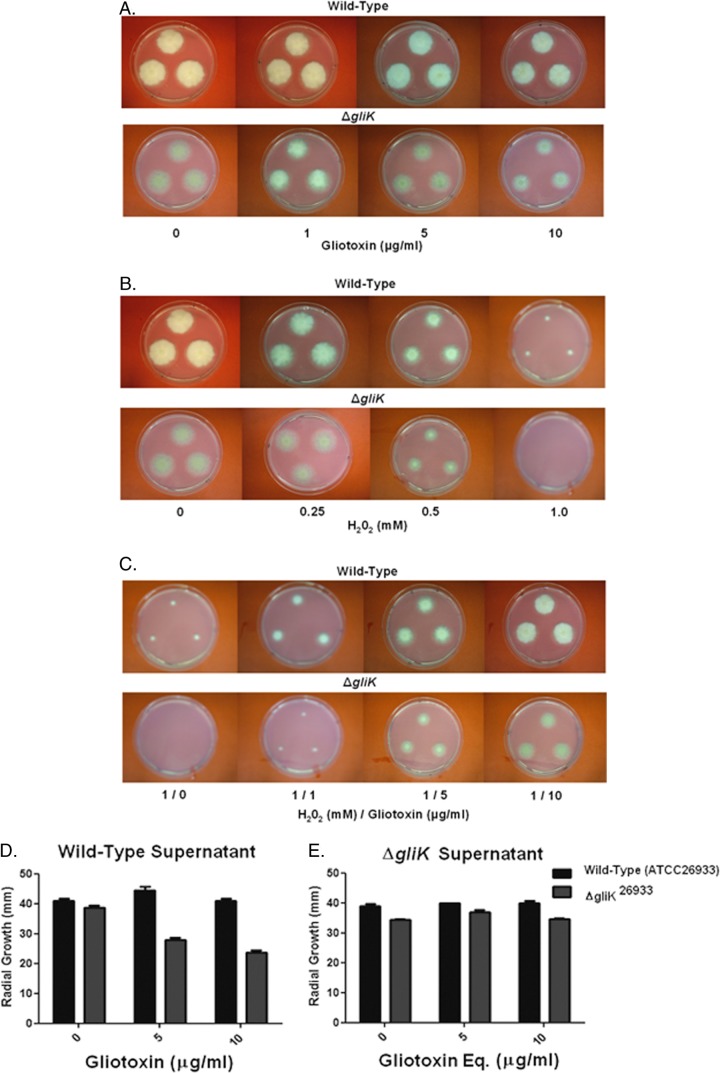
Fig 3.
Individual and combined effects of gliotoxin (0 to 10 μg/ml) and H2O2 on wild-type A. fumigatus and the ΔgliK mutant on Czapek-Dox medium. (A) Gliotoxin (10 μg/ml) significantly inhibits ΔgliK mutant growth. (B) H2O2 (1 mM) completely inhibits the growth of the A. fumigatus ΔgliK mutant compared to that of the wild type. (C) Gliotoxin (1 to 10 μg/ml) significantly relieves H2O2-mediated growth inhibition in a dose-dependent manner. (D) Organic extracts from A. fumigatus ATCC 26933 culture supernatants (containing gliotoxin at 0 to 10 μg/ml) exhibit significant growth inhibition of the ΔgliK mutant compared to the wild type. (E) Organic extracts from A. fumigatus ΔgliK culture supernatants (gliotoxin was absent, but equivalent [Eq] volumes of organic extract were used) do not affect wild-type or ΔgliK mutant growth.
H2O2 significantly inhibits growth of the A. fumigatus ΔgliK mutant.
After 72 h of growth on AMM plates (containing 2 mM H2O2), a significant difference in the radial growth of the A. fumigatus ΔgliK46645 mutant compared to the wild type was observed (P < 0.01) (Fig. 2). In fact, the A. fumigatus ΔgliK mutant was unable to grow at 2 mM H2O2. At 1 mM H2O2, both strains exhibited equivalent radial growth. The A. fumigatus ΔgliK26933 strain showed the same pattern of growth on H2O2 as the ΔgliK46645 strain (Fig. 2), and there was significantly different radial growth at 2 mM H2O2 between the A. fumigatus ATCC 26933 and ΔgliK26933 strains (P < 0.01), as the mutant strain was incapable of any growth at this concentration. This inability of the mutant strain to grow was also observed at 3 mM H2O2, but this was not found to be significant as the wild-type growth was also severely diminished. Conversely, no significant growth difference between A. fumigatus ATCC 46645 and ΔgliK46645 strains was observed at 72 h on AMM plates in the presence of voriconazole (0 to 0.25 μg/ml). Interestingly, at a low voriconazole concentration (0.05 μg/ml), radial growth of the ΔgliK strains was greater than that of the wild type; however, this difference was not significant (data not shown).
Exogenous gliotoxin relieves H2O2 inhibition of A. fumigatus growth.
Figure 3 illustrates the inhibitory effects of gliotoxin and H2O2, respectively, against A. fumigatus wild-type and ΔgliK strains on Czapek-Dox plates. Here, it can be seen that A. fumigatus ΔgliK mutant growth is completely inhibited at 1 mM H2O2; however, gliotoxin addition (1 to 10 μg/ml) significantly relieves this inhibition (P < 0.05) in a dose-dependent manner. Moreover, wild-type growth is similarly recovered in the presence of gliotoxin (P < 0.05). Clearly, gliotoxin presence acts to attenuate the H2O2-induced oxidative stress independently of the gliK status by either a direct or indirect mechanism (Fig. 3). Organic extracts from A. fumigatus ATCC 26933 exert significant growth-inhibitory effects against the ΔgliK26933 mutant (Fig. 3D), while in reverse experimentation no inhibitory effect was detectable in extracts from ΔgliK strains (Fig. 3E). Thus, we conclude that gliotoxin is primarily responsible for the growth inhibition observed in Fig. 3D.
Deletion of gliK abolishes gliotoxin biosynthesis and secretion.
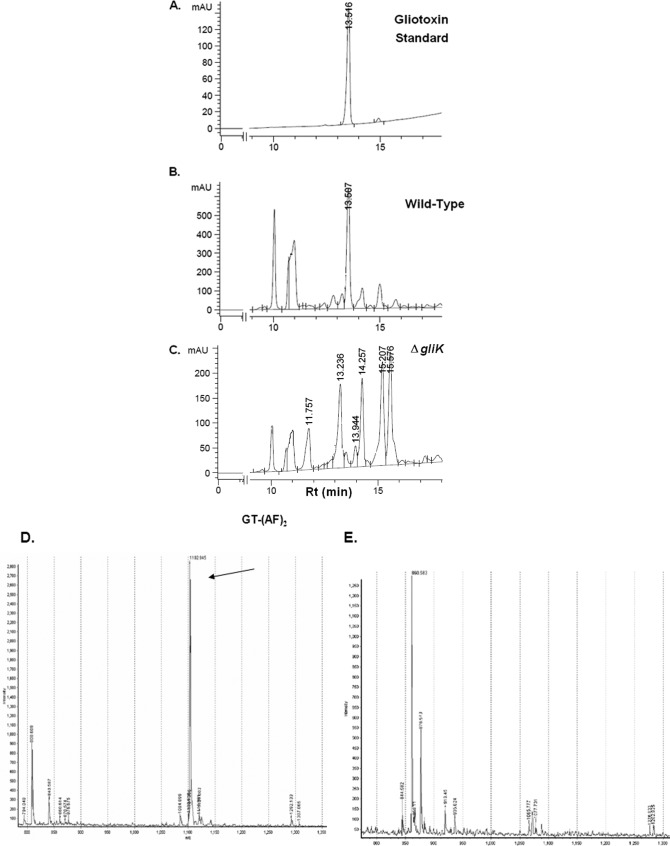
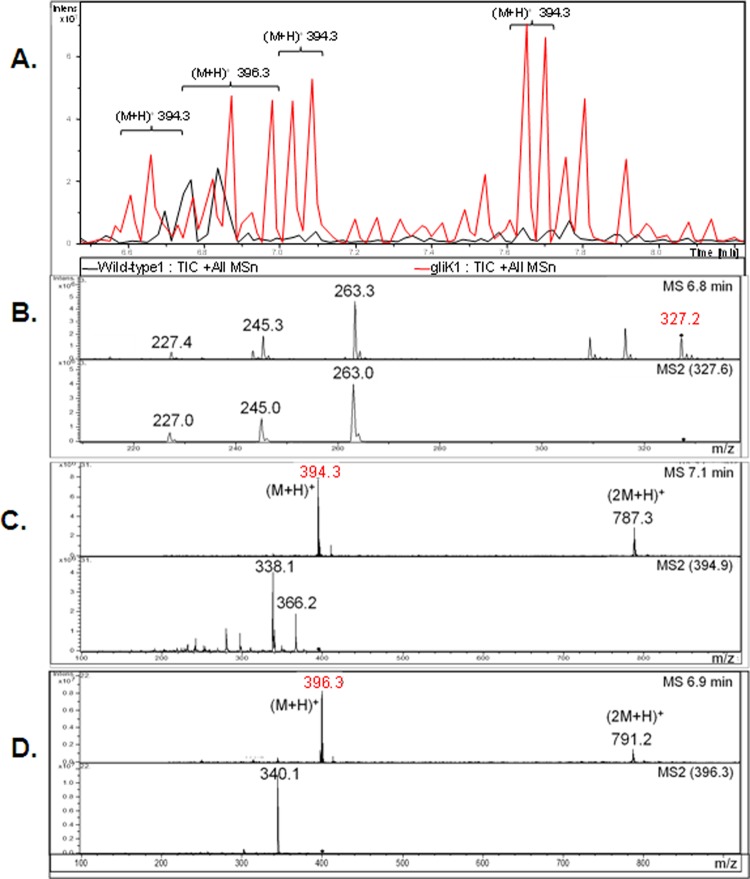
A. fumigatus ATCC 46645 in AMM did not produce significant amounts of gliotoxin, therefore it was difficult to determine if gliK deletion had an effect on gliotoxin production (data not shown). RP-HPLC analysis of A. fumigatus ATCC 26933 showed clear gliotoxin production (in AMM or Czapek-Dox) at a retention time (Rt) of 13.507 min, which is comparable to the standard gliotoxin peak (13.516 min) (Fig. 4). A number of mutant-specific metabolites with Rt of 11.757, 13.236, 14.257, 15.207, and 15.576 min were evident in cultures from the A. fumigatus ΔgliK26933 strain (Fig. 4C). RP-HPLC analysis of NaBH4-reduced organic extracts, with and without subsequent 5′-IAF alkylation (11, 12), from wild-type and mutant cultures indicated the absence of gliotoxin production by the deletion mutant (data not shown). Matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) MS analysis of reduced and alkylated organic extracts from the wild type confirmed the presence of the gliotoxin [i.e., the presence of diacetamidofluorescein-gliotoxin, or GT-(AF)2], while labeled gliotoxin was undetectable in organic extracts from the ΔgliK26933 mutant (Fig. 4D and E). LC-MS analysis confirmed the absence of gliotoxin production and revealed the presence of metabolites [(M + H)+ = 394 and 396] that are significantly upregulated in culture supernatants of the A. fumigatus ΔgliK26933 mutant compared to the wild type (P = 0.0024 [fold difference = 24.1] and P = 0.0003 [fold difference = 9.6], respectively) (Fig. 5). These appear to be related compounds that differ by 2 Da. This relationship is supported by the identical difference between their relative daughter ion base peaks [(M + H) + = 338 and 340, respectively]. The presence of dimers of these compounds is also noted [(2 M+H)+ = 787 and 791, respectively]. These ions are all singly charged as deduced by interrogation of the isotopic peaks of each compound. Importantly, the absence of these secreted metabolites from A. fumigatus ΔgliG supernatants (see Fig. S4 in the supplemental material) confirms the occurrence of a GliG-mediated reaction prior to that catalyzed by GliK. LC-MS analysis of organic extracts of A. fumigatus wild-type supernatants yielded spectra with dominant peaks between retention times 6.7 and 6.9 min which were attributable to the elution of gliotoxin and confirmed by the presence of the parent ion [(M + H)+ = 327.2] and characteristic daughter ions [(M + H)+ = 263, 245, 227] (Fig. 5).
Fig 4.
RP-HPLC confirms gliotoxin absence from A. fumigatus ΔgliK. Standard gliotoxin with a retention time of 13.51 min (A), ATCC 26933 with gliotoxin at an Rt of 13.50 min (B), and the ΔgliK26933 mutant lacking a corresponding gliotoxin peak (C). Detection was at 254 nm. (D) MALDI-TOF analysis reveals diacetamidofluorescein-gliotoxin [GT-(AF)2] (m/z 1102.94) (12) following reduction and alkylation of organic extracts from A. fumigatus ATCC 26933. (E) GT-(AF)2 is not present in organic extracts of the A. fumigatus ΔgliK26933 mutant.
Fig 5.
(A) Total ion chromatograph (TIC; overlaid) of organic extracts from wild-type (black) and ΔgliK26933 mutant (red) culture supernatants. (B) Gliotoxin (m/z 327.2) was identified in the mass spectra from the wild-type culture supernatant but was not found in the ΔgliK26933 mutant culture supernatant. Comparative profiling identified compounds (m/z 394.3 and 396.3) that were present at higher concentrations in the ΔgliK26933 mutant than in wild-type culture supernatants. Mass spectra from the molecular species, m/z 394.3 (C) and m/z 396.3 (D), are shown. Selected parent ions from MS are indicated (◆), with resultant MS/MS spectra included directly below.
Elevated levels of EGT in A. fumigatus ΔgliK.
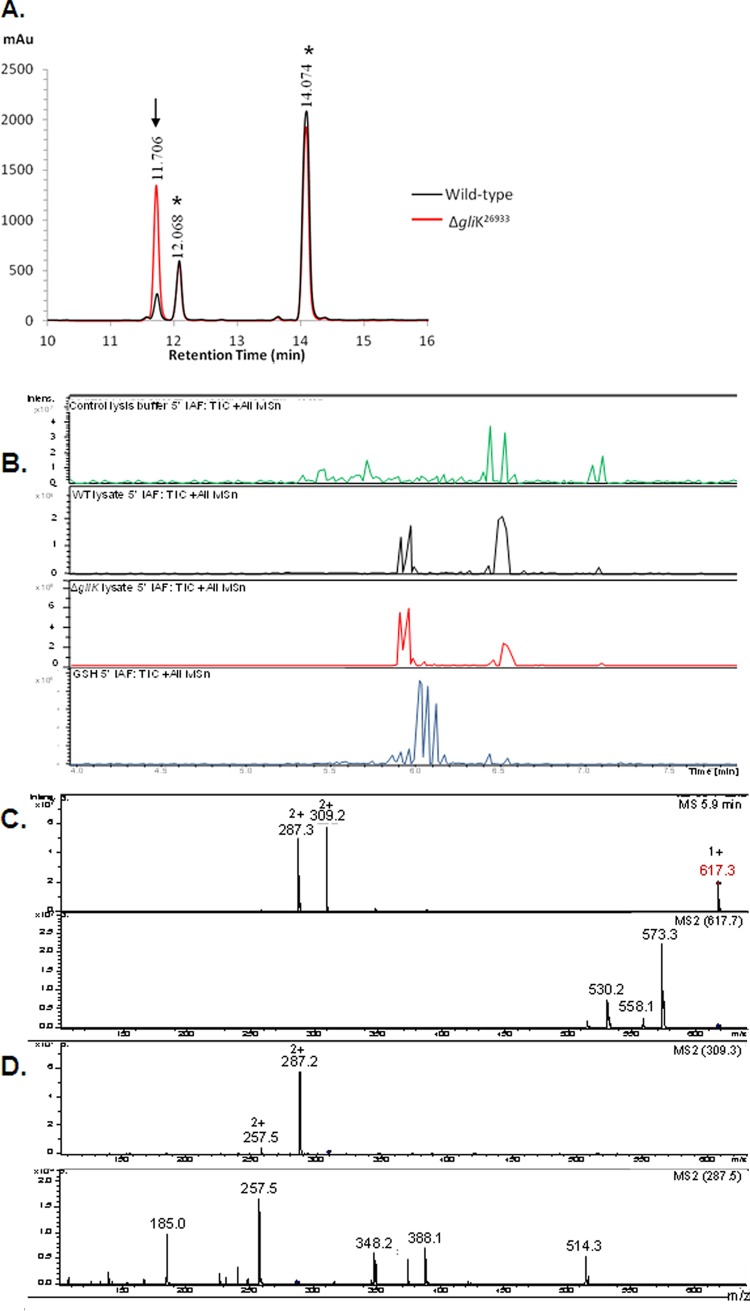
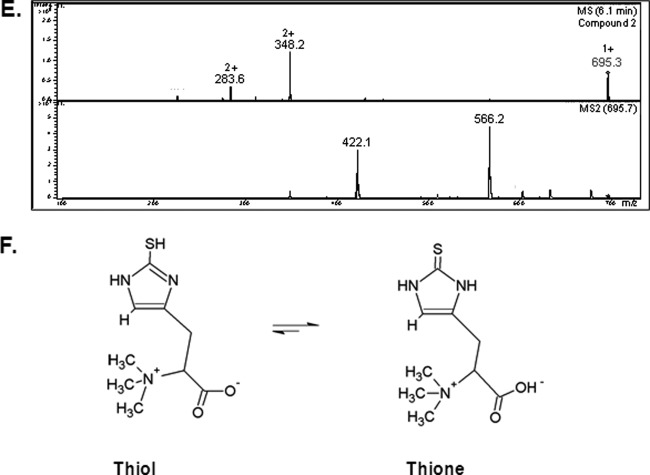
Interestingly, aldrithiol-4 titration revealed significantly more free thiol groups per mg protein in the ΔgliK26933 mutant than in wild-type mycelial lysates (P = 0.0028; n = 3; fold difference, 2.2). This suggested the presence of elevated amounts of free thiols, possibly in low-molecular-mass metabolite(s) related to disruption of gliotoxin biosynthesis, in the absence of gliK. However, no unique intracellular metabolite was evident by RP-HPLC analysis of A. fumigatus ΔgliK mycelial lysates (data not shown). RP-HPLC analysis of 5′-IAF-treated mycelial lysates, either unreduced or subjected to prior NaBH4-mediated reduction, indicated the presence of an alkylated molecular species at an Rt of 11.7 min (Fig. 6A). Following purification, total ion chromatograph (TIC) analysis of this alkylated compound from A. fumigatus wild-type and ΔgliK26933 strains exhibited retention times between 5.9 and 6.0 min, attributable to a compound with (M + H)+ = 617. 3 (Fig. 6B), and the labeled metabolite was present at significantly higher levels in the ΔgliK26933 mutant than in the wild-type lysates (P < 0.001; n = 6; fold difference, 4.96). No equivalent metabolite was detected in the control samples. A number of doubly charged molecular species were also detected [(M + 2H)2+ = 309.2, 287.2], formed as a result of double protonation of the molecular ion [(M + H)+ = 617. 3] or of daughter ions [(M + H)+ = 573.3], respectively (Fig. 6C and D). The mass of the intracellular unlabeled metabolite, present at elevated levels in the A. fumigatus ΔgliK mutant, was 229 Da, which was deduced by subtracting the acetamidofluorescein mass from that of the molecular ion of the labeled compound (617.3 − 388.3 Da = 229 Da). Importantly, alkylation of pure GSH, followed by LC-MS analysis, revealed the expected mass of acetamidofluorescein-glutathione (m/z = 695) (Fig. 6E) at a different Rt (6.1 min) compared to that of the monothiol metabolite from A. fumigatus (Fig. 6C). Moreover, MS/MS fragmentation indicated a neutral loss of 129 Da, corresponding to the loss of the γ-glutamyl moiety from GSH (40). This was not detectable following LC-tandem MS (MS/MS) analyses of the fungal monothiol species, thereby eliminating the possibility that it is GSH. This compound was subsequently identified, for the first time in A. fumigatus, as ergothioneine (EGT; 229 Da; C9H15N3O2S), which exists as a tautomer of a thiol and thione form (Fig. 6F) (2a). The fragmentation pattern observed included a neutral loss of 44 (617.3 to 573.3) corresponding to the loss of COO−, a neutral loss of 59 (573.3 to 514.3) corresponding to N(CH3)3+, and a neutral loss of 126.2 (514.3 to 388.1) corresponding to C5N2H6S. This MS/MS analysis correlated with the fragmentation pattern observed for the selenium analog of ergothioneine, selenoneine (39). Consequently, this analysis confirmed the presence of elevated levels of ergothioneine in A. fumigatus ATCC 26933 following deletion of the gliK gene. Ergothioneine levels in the gliKC strain returned to those found in the wild-type (ATCC 46645) (see Fig. S5 in the supplemental material), thereby confirming the reconstitution of gliK in the A. fumigatus ΔgliK46645 mutant.
Fig 6.
RP-HPLC analysis of A. fumigatus ATCC 26933 wild-type (black) and ΔgliK26933 mutant (red) mycelial lysates with no reduction prior to 5′-IAF labeling (NR plus 5′-IAF). (A) Absorbance detection at 254 nm is shown for all samples. 5′-IAF was detected at Rt of 12.068 and 14.074 min (*). A 5′-IAF-labeled metabolite (AF11.7) eluted at an Rt of 11.706 min (indicated by arrow). (B) Total ion chromatograph (TIC) of the 11- to 12-min fraction from RP-HPLC analyses of 5′ IAF-labeled mycelial lysates of A. fumigatus ATCC 26933 wild-type (black) and ΔgliK26933 mutant (red) strains. A control sample, comprising the equivalent fraction from 5′-IAF labeling of lysate buffer, is also shown (green). MSn, multiple-stage mass spectrometry. (C) Mass spectra of the compounds observed at retention times of 5.9 to 6.0 min from each of the A. fumigatus samples revealed a singly charged molecule with (M + H)+ = 617.3 along with two doubly charged molecules with m/z 309.2 and 287.3, respectively. (D) Compounds were not found in the mass spectra of the control. MS/MS (MS2) analyses of each of these three ions was carried out. (E) Mass spectra of the compound observed at retention times of 6.0 to 6.2 min in 5′-IAF-labeled GSH revealed a singly charged molecule with (M + H)+ = 695.3, corresponding to acetamidofluorescein-glutathione, along with two doubly charged molecules with m/z = 348.2 and 283.6, respectively. (F) Ergothioneine.
gliK deletion significantly impedes gliotoxin efflux from A. fumigatus.
RP-HPLC analysis confirmed that although A. fumigatus ATCC 46645 produced a residual amount of gliotoxin, 0.3 ± 0.13 (means ± standard deviations) ng/mg mycelium, the A. fumigatus ΔgliK46645 mutant did not produce gliotoxin (Fig. 7A). Thus, the uptake of exogenously added gliotoxin (5 μg/ml), by both wild-type and ΔgliK46645 strains in liquid cultures over 0 to 45 min, was determined by quantifying the residual gliotoxin concentration in supernatants. Significantly less gliotoxin was present in culture supernatants of the A. fumigatus ΔgliK46645 mutant than in that of ATCC 46645 after gliotoxin exposure at 15 min (P = 0.0102) and 30 min (P = 0.0045) (Fig. 7B). This implied that gliotoxin was either effluxing more slowly from the mycelia of A. fumigatus ΔgliK46645 after uptake or is removed more rapidly from the supernatant. After 45 min, the level of gliotoxin increased in the ΔgliK culture supernatants, possibly because alternative efflux mechanisms had been activated. The uptake rate of gliotoxin by the wild-type and mutant strains was calculated at 15-min intervals. At 15 min, the A. fumigatus ΔgliK46645 mutant exhibited a gliotoxin uptake rate of 31.443 pg/mg mycelia/min, more than three times that of A. fumigatus ATCC 46645 (9.337 pg/mg mycelia/min) (P = 0.0039). The wild-type strain exhibited a relatively constant level of gliotoxin for 45 min in the supernatant, indicating that the wild-type strain displayed a balance between uptake and efflux of gliotoxin (Fig. 7B and C). In the A. fumigatus ΔgliK46645 mutant, gliotoxin levels began to stabilize in the supernatant after the initial uptake in the first 15 min of exposure. Thus, it appears that deletion of gliK has disrupted a gliotoxin efflux mechanism in A. fumigatus, because elevated (or constant) gliotoxin levels in A. fumigatus ΔgliK46645 mutant culture supernatants would have been predicted if gliK encoded a component of a gliotoxin uptake mechanism.
Fig 7.
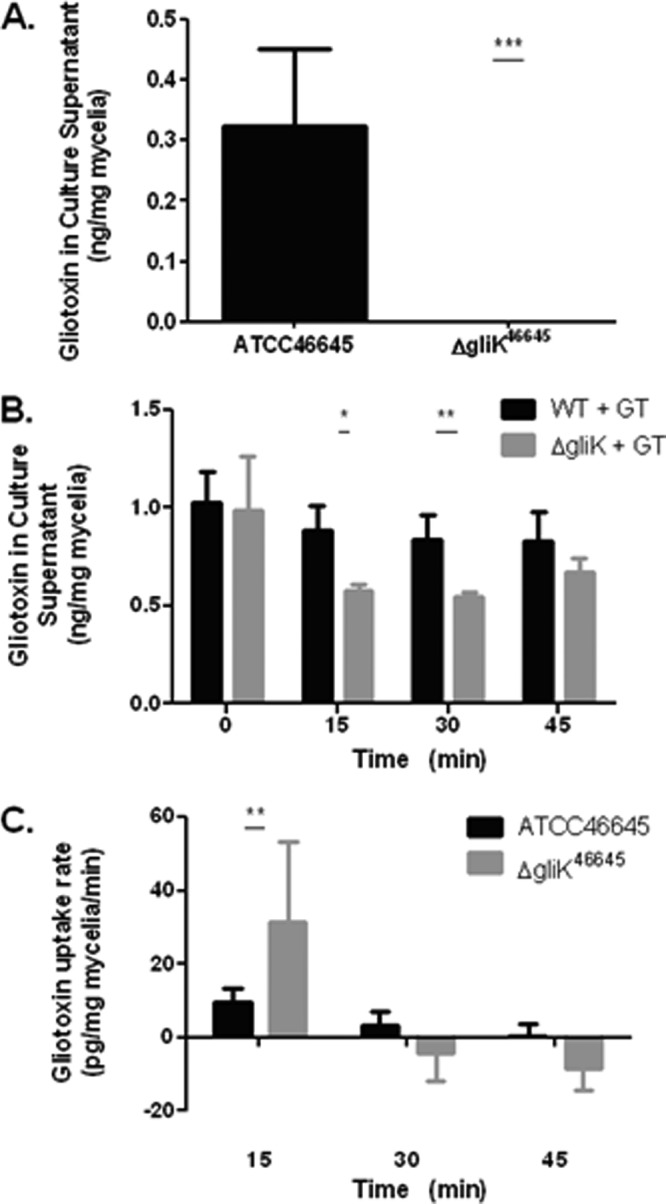
(A) Gliotoxin (GT) production in AMM liquid cultures from ATCC 46645 and ΔgliK46645 strains. (B) The amount of gliotoxin remaining in culture supernatants at time intervals after exposure of A. fumigatus ATCC 46645 and ΔgliK46645 strains to gliotoxin (5 μg/ml) over a 0- to 45-min duration. (C) Gliotoxin uptake rate by A. fumigatus ATCC 46645 and ΔgliK46645 strains in liquid cultures at selected time points.
DISCUSSION
Although orthologs of A. fumigatus gliK exist in other fungi, bioinformatic analysis of gliK in terms of comparative sequence or conserved domain analysis has provided no definitive information as to its function (28). Here, we reveal that gliK deletion from A. fumigatus results in acquisition of gliotoxin- and H2O2-sensitive phenotypes and loss of gliotoxin production in two strains. Further, we observe that H2O2 induces gliK expression and, unexpectedly, that gliotoxin addition significantly relieves the H2O2-induced growth inhibition in both A. fumigatus wild-type and ΔgliK strains. The presence of (i) significantly elevated levels of external metabolites of m/z 394 to 396, in addition to (ii) an elevation in intracellular ergothioneine levels in A. fumigatus ΔgliK strains, were determined. Finally, a significantly reduced rate of exogenously added gliotoxin efflux from A. fumigatus ΔgliK compared to wild-type strains is observed. This finding may be due to either gliotoxin reduction by ergothioneine or to consequent gliK involvement in gliotoxin secretion.
Targeted deletion of gliK in two A. fumigatus strains was achieved, and the integrity and expression of the adjacent gene (gliA) was unaffected by the deletion strategy employed (see Fig. S2 in the supplemental material). Likewise, it has been confirmed that deletion of gliT, and consequent phenotypes observed, were gene specific and not caused by alterations in the structure or expression of the surrounding genes, gliF and gliH (33). gliK deletion resulted in the complete absence of cognate gene expression, thereby ensuring that all subsequent phenotypes were directly caused by GliK absence. The maximum yield of gliotoxin is highly influenced by growth conditions, especially the culture media (20). Consequently, assessment of gliK expression under different culture conditions was undertaken to establish optimal conditions for assessment of the relationship between gliK expression and gliotoxin production. gliK expression was found to be highest at 24 h in AMM, so this time point and medium were used in all subsequent gliK expression analyses and gliotoxin uptake studies. gliK expression was also evident in MM at 24 and 48 h, although it had decreased by 72 h, indicating that gliK is normally expressed early on and may not be required in the later stages of fungal culture. gliK expression was undetectable in Sabouraud media at any time point in wild-type and A. fumigatus ΔgliK46645 mutant cultures, thereby confirming gliK absence from the latter strain.
Genes in the gliotoxin biosynthetic cluster have been associated with protection against gliotoxin. For example, gliT, a gliotoxin oxidoreductase, has been shown to play a self-protection role against gliotoxin, as disruption of this gene renders the fungus completely sensitive to exogenous gliotoxin (5 μg/ml) (32, 33). However, no alteration in sensitivity to H2O2 was observed for the A. fumigatus ΔgliT mutant. The gliotoxin transporter, gliA, was shown to confer resistance to gliotoxin in the Leptosphaeria maculans ΔsirA mutant (15). In the present study, deletion of gliK in A. fumigatus ATCC 46645 and ATCC 26933 resulted in mutant strains that became highly sensitive to exogenous gliotoxin and H2O2 but were unaffected by exposure to the antifungal voriconazole. Although a statistically significant difference in growth rates was observed between A. fumigatus wild-type and respective ΔgliK strains, addition of gliotoxin did not completely suppress growth of ΔgliK46645 and ΔgliK26933 mutants, even at high concentrations of gliotoxin (20 μg/ml). Thus, observed gliotoxin sensitivity was less apparent than that previously encountered for A. fumigatus ΔgliT (32, 33). Hence, we hypothesized that although gliK plays a role in self protection against gliotoxin, it is more likely that any metabolite (biosynthetic intermediate or shunt metabolite) produced consequent to gliK deletion mediates sensitivity to exogenous gliotoxin. We further conclude that it is not simply the loss of gliK which sensitizes A. fumigatus to gliotoxin, since gliK is not expressed in the A. fumigatus ΔgliZ mutant, and this strain exhibits a wild-type gliotoxin phenotype (33).
Although exogenous gliotoxin (5 μg/ml) had no effect on gliK expression in A. fumigatus ATCC 46645, we found that H2O2-induced oxidative stress resulted in an almost 20-fold increase (0.1665 versus 3.193) in gliK expression (P = 0.0101). Moreover, exposure of A. fumigatus wild-type, ΔgliK26933, and ΔgliK46645 strains to hydrogen peroxide also resulted in differential growth responses. Here, increased mutant sensitivity to hydrogen peroxide (2 mM) was observed whereby the mutant strains were unable to grow, unlike both wild-type strains, which exhibited approximately 50% growth. Since gliotoxin biosynthesis is impaired in the A. fumigatus ΔgliK mutant, these observations suggest that either gliK absence, a specific biosynthetic intermediate, or ergothioneine presence augments H2O2-mediated oxidative stress. Relevantly, it has been demonstrated that the A. fumigatus ΔgliG strain does not exhibit increased sensitivity to gliotoxin or H2O2 (11). Indeed, it has also been noted that gliotoxin may play a role in regulating the redox status of A. fumigatus (7, 33). However, despite extensive analyses, no evidence of enhanced gliotoxin efflux was detectable following exposure of A. fumigatus to H2O2.
In an attempt to unravel the unique response of the A. fumigatus ΔgliK mutant to gliotoxin- and H2O2-mediated oxidative stress, we performed coaddition experiments to determine if the individual inhibitory effects of gliotoxin and H2O2 were additive. To our surprise, we found that gliotoxin actually relieved H2O2-induced growth inhibition of both wild-type and gliK deletion strains of A. fumigatus. Relevantly, it has been noted that gliotoxin catalyzes H2O2 reduction and that it was a novel thioredoxin substrate, because no H2O2-reducing activity was evident in the absence of thioredoxin reductase or thioredoxin in mammalian cells (8). These authors also demonstrated that gliotoxin inhibited the H2O2-induced angiogenesis of human umbilical vein endothelial cells. However, at present the precise mechanism for our observations in A. fumigatus remains undetermined, and work is under way to explore this phenomenon. Notably, organic extracts of wild-type culture supernatants inhibited gliK growth, yet the opposite was not observed. This result is in accordance with Coleman et al. (9), who observed that gliotoxin-deficient organic extracts of the A. fumigatus ΔgliP mutant did not exhibit inhibitory properties against Candida albicans.
Gliotoxin production by A. fumigatus ATCC 46645 was at too low a level to reliably assess if the ΔgliK mutation had any effect on gliotoxin production. This was not the case for A. fumigatus ATCC 26933, where the amount of gliotoxin produced was readily detectable prior to NaBH4-mediated reduction (11, 12), and subsequent reduction confirmed gliotoxin presence by the appearance of the dithiol form at a reduced retention time (3) in organic extracts of the wild-type culture supernatants (data not shown). A method for gliotoxin detection by alkylating the reduced form of gliotoxin with 5′-iodoacetamidofluorescein (5′-IAF) followed by RP-HPLC or MALDI-MS identification (12) provided confirmatory data for gliotoxin production by the A. fumigatus wild-type strain and deficiency in ΔgliK strains, respectively. Thus, we conclude that gliotoxin production and secretion by the ΔgliK26933 mutant was abolished. However, significantly elevated levels of two hydrophobic metabolites were detectable in organic extracts of the ΔgliK26933 mutant, which exhibited m/z 394 to 396 following LC-MS analysis. These metabolites did not contain free thiols, and as noted above they did not possess growth-inhibitory properties associated with organic extracts of wild-type organic extracts. Also, the observed m/z did not correspond to any metabolite present in the A. fumigatus wild-type intracellular metabolome (13) or secretome of the A. fumigatus ΔgliG strain (11, 31). Moreover, this mass (393 to 395 Da) is greater than that of gliotoxin, 326 Da, and strongly suggests the presence of either a shunt metabolite(s) or biosynthetic intermediate of higher mass than gliotoxin, as previously observed (31). Our analyses revealed that these metabolites were detectable following deletion of gliT but absent from the gliG deletion strain. This means that the presence of these metabolites (m/z 394 to 396) is GliG dependent, and that accumulation of these intermediates/shunt metabolites occurs in the absence of GliK. This positions GliK in the gliotoxin biosynthetic pathway as catalyzing a reaction after the diglutathionylation event effected by GliG but preceding GliT-mediated disulfide oxidation. Future purification and structural characterization will enable unambiguous identification of the nature of these extracellular metabolites.
Intriguingly, significantly elevated levels of ergothioneine were detected in aqueous extracts of A. fumigatus ΔgliK26933 mutant mycelial lysates compared to the wild type. The biosynthetic pathway of ergothioneine, in mycobacteria, involves sulfurization through incorporation of γ-glutamylcysteine (γGC) (33a). Observation of increased ergothioneine levels in ΔgliK may support a putative trans function of GliK as a γ-glutamyl cyclotransferase (GGCT), with deletion of this gene resulting in reduced catabolism of the substrate for ergothioneine biosynthesis. Interestingly, GliK appears in part to encode a gamma-glutamyl cyclotransferase-like (GGCT-like) domain that is responsible for catalyzing the formation of pyroglutamic acid (5-oxoproline) from dipeptides containing γ-glutamate (26).
Based on our observations that both A. fumigatus ΔgliK mutants were deficient in gliotoxin biosynthesis and acquired increased sensitivity to exogenous gliotoxin (5 μg/ml), the relative uptake rates of exogenous gliotoxin by A. fumigatus ATCC 46645 and ΔgliK46645 strains was undertaken to further investigate the role of gliK. Here, we reasoned that if the ΔgliK46645 mutant could take up gliotoxin normally but exhibit impaired efflux compared to the wild type, then the exogenous levels of gliotoxin should decrease more rapidly, and to a greater extent, in ΔgliK46645 cultures. The ATCC 46645 strain was selected because the low levels of endogenous gliotoxin produced (33) would not interfere with measurement of exogenously added gliotoxin. Previous work (Stephen Carberry, personal communication) had suggested that a 45-min experimental period would be optimal for assessment of gliotoxin uptake by A. fumigatus. At 15 min after gliotoxin addition, there was a significantly lower level of gliotoxin present in A. fumigatus ΔgliK46645 mutant supernatants compared to the wild type (P = 0.0102). This difference was also evident at 30 min (P = 0.0045); however, at 45 min the level of exogenous gliotoxin in supernatants of the A. fumigatus ΔgliK46645 mutant began to increase and no statistically significant difference in exogenous gliotoxin levels between mutant and wild type was apparent (wild type versus the ΔgliK46645 mutant, 0.831 ± 0.356 and 0.668 ± 0.171 ng/mg mycelium, respectively). This is possibly due to activation of the autoprotective gliT response, which has also been proposed to facilitate gliotoxin efflux from A. fumigatus (33). A differential gliotoxin uptake rate for the phytopathogens Pythium ultimum and Rhizoctania solani, expressed as μg · mg dry weight−1, has been reported (16). Interestingly, these authors noted that gliotoxin uptake was complete within 1 min, and they observed passive, adsorptive-type kinetics and that uptake was significantly lower in gliotoxin-resistant strains. Overall, we propose that exogenous gliotoxin levels drop rapidly (0 to 30 min) in the A. fumigatus ΔgliK46645 mutant due to impaired efflux, which may be due to reduction of gliotoxin by ergothionine, which prevents its release, or because gliK acts in association with gliA, the MFS transporter (15), to maintain gliotoxin homeostasis in A. fumigatus and to facilitate gliotoxin efflux under normal conditions. It should be noted that if gliK had been involved in gliotoxin uptake, one would have observed either no change or an elevation of exogenous gliotoxin levels in the A. fumigatus ΔgliK46645 mutant. This was not observed. Moreover, our data suggest that GliT (33) participates in gliotoxin secretion, possibly in conjunction with GliA functionality as an efflux pump (15), which may partly compensate for the loss of GliK, provided that the amount of gliotoxin does not exceed the levels with which the cell can cope. Sirodesmin, a related ETP toxin, is produced by L. maculans, and its biosynthesis is encoded by a multigene cluster analogous to that which encodes gliotoxin production in A. fumigatus (15). Deletion of sirA, the sirodesmin transporter gene, also led to increased sensitivity to exogenous sirodesmin and gliotoxin. Interestingly, introduction of A. fumigatus gliA recovered resistance to exogenous gliotoxin (10 μM), but not sirodesmin, in L. maculans ΔsirA::gliA. Production and secretion of sirodesmin actually increased by 39% in the L. maculans ΔsirA strain compared to the wild type, leading to speculation about the presence of alternative toxin efflux mechanisms (15). Further, Bradshaw et al. (5) suggested that the MFS transporter encoded by dotC in Dothistroma septosporum was not the major efflux pump for dothistromin and concluded that there are other factors besides DotC facilitating the efflux of the toxin. Thus, toxin secretion may be multifactorial and require not only efflux pumps but also defined intracellular conditions to enable efficient toxin-pump interaction as part of the secretion process.
In summary, targeted deletion of A. fumigatus gliK, a component of the gliotoxin biosynthetic cluster, in two fungal strains which produce low and high levels of gliotoxin, respectively, resulted in acquisition of (i) sensitivity to exogenous gliotoxin, (ii) impaired biosynthesis of gliotoxin, (iii) an altered extra- and intracellular metabolome, and (iv) increased sensitivity to H2O2. We identify gliK as a component of the gliotoxin biosynthetic system in A. fumigatus and propose that decreased efflux of exogenously added gliotoxin in the A. fumigatus ΔgliK mutant is due to significantly elevated levels of ergothioneine which may reduce gliotoxin and impede efflux. Future work will investigate these hypotheses and dissect the mechanistic action of GliK.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded in part by the Irish Health Research Board (RP/2006/043) and Science Foundation Ireland (PI/11/1188). R.A.O. was a recipient of an Irish Research Council for Science Engineering and Technology Embark Ph.D. Fellowship. M.S. was a recipient of an EU Marie Curie award (MTKD-CT-2004-014436; coordinator, Shirley O'Dea). S.K.D. was funded by a Society for General Microbiology Studentship. Quantitative PCR instrumentation and RP-HPLC and LC-MS facilities were funded by competitive awards from Science Foundation Ireland (grant no. SFI/07/RFP/GEN/F571/ECO7) and the Irish Higher Education Authority, respectively.
Footnotes
Published ahead of print 17 August 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Balibar CJ, Walsh CT. 2006. GliP, a multimodular nonribosomal peptide synthetase in Aspergillus fumigatus, makes the diketopiperazine scaffold of gliotoxin. Biochemistry 45:15029–15038 [DOI] [PubMed] [Google Scholar]
- 2.Behling RA, Fischer AG. 1980. Formation of phenylalanylserine and cyclo-phenylalanylseryl by protoplasts of Gliocladium virens. Int. J. Biochem. 11:457–458 [DOI] [PubMed] [Google Scholar]
- 2a.Bello MH, Barrera-Perez V, Morin D, Epstein L. 2012. The Neurospora crassa mutant NcΔEgt-1 identifies an ergothioneine biosynthetic gene and demonstrates that ergothioneine enhances conidial survival and protects against peroxide toxicity during conidial germination. Fungal Genet. Biol. 49:160–172 [DOI] [PubMed] [Google Scholar]
- 3.Bernardo PH, Brasch N, Chai CL, Waring P. 2003. A novel redox mechanism for the glutathione-dependent reversible uptake of a fungal toxin in cells. J. Biol. Chem. 278:46549–46555 [DOI] [PubMed] [Google Scholar]
- 4.Bok JW, et al. 2006. GliZ, a transcriptional regulator of gliotoxin biosynthesis, contributes to Aspergillus fumigatus virulence. Infect. Immun. 74:6761–6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradshaw RE, Feng Z, Schwelm A, Yang Y, Zhang S.2009. Functional analysis of a putative dothistromin toxin MFS transporter gene. Toxins 1:173–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns C, et al. 2005. Identification, cloning, and functional expression of three glutathione transferase genes from Aspergillus fumigatus. Fungal Genet. Biol. 42:319–327 [DOI] [PubMed] [Google Scholar]
- 7.Carberry S, et al. 2012. Gliotoxin effects on fungal growth: mechanisms and exploitation. Fungal Genet. Biol. 49:302–312 [DOI] [PubMed] [Google Scholar]
- 8.Choi HS, Shim JS, Kim JA, Kang SW, Kwon HJ. 2007. Discovery of gliotoxin as a new small molecule targeting thioredoxin redox system. Biochem. Biophys. Res. Commun. 359:523–528 [DOI] [PubMed] [Google Scholar]
- 9.Coleman JJ, Ghosh S, Okoli I, Mylonakis E. 2011. Antifungal activity of microbial secondary metabolites. PLoS One 6:e25321 doi:10.1371/journal.pone.0025321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cramer RA, Jr, et al. 2006. Disruption of a nonribosomal peptide synthetase in Aspergillus fumigatus eliminates gliotoxin production. Eukaryot. Cell 5:972–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis C, et al. 2011. The role of glutathione S-transferase GliG in gliotoxin biosynthesis in Aspergillus fumigatus. Chem. Biol. 18:542–552 [DOI] [PubMed] [Google Scholar]
- 12.Davis C, et al. 2011. Single-pot derivatisation strategy for enhanced gliotoxin detection by HPLC and MALDI-ToF mass spectrometry. Anal. Bioanal. Chem. 401:2519–2529 [DOI] [PubMed] [Google Scholar]
- 13.Forseth RR, et al. 2011. Identification of cryptic products of the gliotoxin gene cluster using NMR-based comparative metabolomics and a model for gliotoxin biosynthesis. J. Am. Chem. Soc. 133:9678–9681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardiner DM, Cozijnsen AJ, Wilson LM, Pedras MS, Howlett BJ. 2004. The sirodesmin biosynthetic gene cluster of the plant pathogenic fungus Leptosphaeria maculans. Mol. Microbiol. 53:1307–1318 [DOI] [PubMed] [Google Scholar]
- 15.Gardiner DM, Jarvis RS, Howlett BJ. 2005. The ABC transporter gene in the sirodesmin biosynthetic gene cluster of Leptosphaeria maculans is not essential for sirodesmin production but facilitates self-protection. Fungal Genet. Biol. 42:257–263 [DOI] [PubMed] [Google Scholar]
- 16.Hancock RW, Jones JG. 1988. Mechanism of gliotoxin action and factors mediating gliotoxin sensitivity. J. Gen. Microbiol. 134:2067–2075 [Google Scholar]
- 17.Hearn VM, Mackenzie DW. 1980. Mycelial antigens from two strains of Aspergillus fumigatus: an analysis by two-dimensional immunoelectrophoresis. Mykosen 23:549–562 [PubMed] [Google Scholar]
- 18.Johns N, Kirby GW, Bu'Lock JD, Ryles AP. 1975. Stereospecific exchange of a beta-methylene proton in phenylalanine preceding biosynthetic incorporation into gliotoxin. J. Chem. Soc. Perkin 1:383–386 [DOI] [PubMed] [Google Scholar]
- 19.Johnson JR, Bruce WF, Ditcher JD. 1943. Gliotoxin, the antibiotic principle of Gliocladium fibriatum I production, physical and biological properties. J. Am. Chem. Soc. 65:2005–2009 [Google Scholar]
- 20.Kosalec I, Pepeljnjak S, Jandrlic M. 2005. Influence of media and temperature on gliotoxin production in Aspergillus fumigatus strains. Arh. Hig. Rada Toksikol. 56:269–273 [PubMed] [Google Scholar]
- 21.Kubodera T, Yamashita N, Nishimura A. 2000. Pyrithiamine resistance gene (ptrA) of Aspergillus oryzae: cloning, characterization and application as a dominant selectable marker for transformation. Biosci. Biotechnol. Biochem. 64:1416–1421 [DOI] [PubMed] [Google Scholar]
- 22.Kupfahl C, et al. 2006. Deletion of the gliP gene of Aspergillus fumigatus results in loss of gliotoxin production but has no effect on virulence of the fungus in a low-dose mouse infection model. Mol. Microbiol. 62:292–302 [DOI] [PubMed] [Google Scholar]
- 23.Mabey JE, et al. 2004. CADRE: the Central Aspergillus Data REpository. Nucleic Acids Res. 32:D401–D405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullbacher A, Moreland AF, Waring P, Sjaarda A, Eichner RD. 1988. Prevention of graft-versus-host disease by treatment of bone marrow with gliotoxin in fully allogeneic chimeras and their cytotoxic T cell repertoire. Transplantation 46:120–125 [DOI] [PubMed] [Google Scholar]
- 25.Nielsen ML, Albertsen L, Lettier G, Nielsen JB, Mortensen UH. 2006. Efficient PCR-based gene targeting with a recyclable marker for Aspergillus nidulans. Fungal Genet. Biol. 43:54–64 [DOI] [PubMed] [Google Scholar]
- 26.Oakley AJ, et al. 2008. The identification and structural characterization of C7orf24 as gamma-glutamyl cyclotransferase. An essential enzyme in the gamma-glutamyl cycle. J. Biol. Chem. 283:22031–22042 [DOI] [PubMed] [Google Scholar]
- 27.O'Hanlon KA, et al. 2012. Non-ribosomal peptide synthetases pesL and pes1 are essential for fumigaclavine C production in Aspergillus fumigatus. Appl. Environ. Microbiol. 78:3166–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patron NJ, et al. 2007. Origin and distribution of epipolythiodioxopiperazine (ETP) gene clusters in filamentous ascomycetes. BMC Evol. Biol. 7:174 doi:10.1186/1471-2148-7-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pontecorvo G, Roper JA, Hemmons LM, Macdonald KD, Bufton AW. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141–238 [DOI] [PubMed] [Google Scholar]
- 30.Reeves EP, Messina CG, Doyle S, Kavanagh K. 2004. Correlation between gliotoxin production and virulence of Aspergillus fumigatus in Galleria mellonella. Mycopathologia 158:73–79 [DOI] [PubMed] [Google Scholar]
- 31.Scharf DH, et al. 2011. A dedicated glutathione S-transferase mediates carbon-sulfur bond formation in gliotoxin biosynthesis. J. Am. Chem. Soc. 133:12322–12325 [DOI] [PubMed] [Google Scholar]
- 32.Scharf DH, et al. 2010. Transannular disulfide formation in gliotoxin biosynthesis and its role in self-resistance of the human pathogen Aspergillus fumigatus. J. Am. Chem. Soc. 132:10136–10141 [DOI] [PubMed] [Google Scholar]
- 33.Schrettl M, et al. 2010. Self-protection against gliotoxin–a component of the gliotoxin biosynthetic cluster, GliT, completely protects Aspergillus fumigatus against exogenous gliotoxin. PLoS Pathog. 6:e1000952 doi:10.1371/journal.ppat.1000952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Seebeck F. 2010. In vitro reconstitution of Mycobacterial ergothioneine biosynthesis. J. Am. Chem. Soc. 132:6632–6633 [DOI] [PubMed] [Google Scholar]
- 34.Spikes S, et al. 2008. Gliotoxin production in Aspergillus fumigatus contributes to host-specific differences in virulence. J. Infect. Dis. 197:479–486 [DOI] [PubMed] [Google Scholar]
- 35.Stack D, Frizzell A, Tomkins K, Doyle S. 2009. Solid phase 4′-phosphopantetheinylation: fungal thiolation domains are targets for chemoenzymatic modification. Bioconjug. Chem. 20:1514–1522 [DOI] [PubMed] [Google Scholar]
- 36.Sugui JA, et al. 2007. Gliotoxin is a virulence factor of Aspergillus fumigatus: gliP deletion attenuates virulence in mice immunosuppressed with hydrocortisone. Eukaryot. Cell 6:1562–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waring P, Beaver J. 1996. Gliotoxin and related epipolythiodioxopiperazines. Gen. Pharmacol. 27:1311–1316 [DOI] [PubMed] [Google Scholar]
- 38.Weindling R, Emerson OH. 1936. The isolation of a toxic substance from a culture filtrate of Trichoderma. Phytopathology 26:1068–1070 [Google Scholar]
- 39.Yamashita Y, Yamashita M. 2010. Identification of a novel selenium-containing compound, selenoneine, as the predominant chemical form of organic selenium in the blood of bluefin tuna. J. Biol. Chem. 285:18134–18138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan ZY, Caldwell QW. 2004. Stable-isotope trapping and high-throughput screenings of reactive metabolites using the isotope MS signature. Anal. Chem. 76:6835–6847 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.