Abstract
Extremely thermoacidophilic microbes, such as Sulfolobus solfataricus, are strict chemoheterotrophs despite their geologic niche. To clarify their ecophysiology, the overlapping roles of endoglucanases and carbohydrate transporters were examined during growth on soluble cellodextrins as the sole carbon and energy source. Strain-specific differences in genome structure implied a unique role for one of three endogenous endoglucanases. Plasmid-based endoglucanase expression promoted the consumption of oligosaccharides, including cellohexaose (G6) through cellonanaose (G9). Protein transporters required for cellodextrin uptake were identified through mutagenesis and complementation of an ABC transporter cassette, including a putative oligosaccharide binding protein. In addition, ablation of the binding protein compromised growth on glucose and alpha-linked oligosaccharides while inactivation of a previously described glucose transporter had no apparent impact. These data demonstrate that S. solfataricus employs a redundant mechanism for soluble cellodextrin catabolism having both substrate uptake and extracytoplasmic hydrolytic components.
INTRODUCTION
Sulfolobus solfataricus is an extremely thermophilic and acidophilic organism that thrives in hot acidic habitats, growing optimally between pH values of 3 and 4 at 70 to 90°C (7, 12). S. solfataricus belongs to the archaeal domain and the phylum Crenarchaeota (36). It is an obligate aerobe and chemoheterotroph that grows on a variety of reduced carbon sources, including simple sugars and more complex carbohydrates, such as starch (12, 17) and cellulose (38). Even though its native habitat excludes plants, nearby vegetation may be introduced by natural events. Resident conditions are likely to promote chemical transformation of this lignocellulosic material, providing carbon for growth.
Glycosyl hydrolase enzymes are classified into 128 families (GHF) and 14 clans. Those derived from extremophiles have gained considerable attention because of their inherent thermostability and resistance to extremes of pH (5, 6, 10, 15, 19–23, 27, 31, 32). Three homologous open reading frames (ORFs) have been identified in the genome of S. solfataricus strain P2 (DSM1617) as putative extracellular cellulases (35). These genes encode endoglucanases belonging to GHF 12 and lack a cellulose binding domain (CBD). However, an N-terminal signal peptide ranging from 21 to 24 amino acids is present that could guide membrane targeting and/or secretion. The Sso2534 gene product, identified in the growth medium of S. solfataricus strain MT4, exhibited optimal endoglucanase activity on carboxymethyl cellulose (CMC) at pH 5.8 and 65°C (23). A recombinant form of Sso1949, cloned from S. solfataricus strain P2 (DSM1617) and expressed in Escherichia coli, had optimum activity at pH 1.8 and 80°C. The enzyme utilized cellotetraose as the smallest substrate and generated cellobiose and cellotriose as the primary products, indicating it was an endoglucanase (19). Both endoglucanase and xylanase activities were identified for the native form of Sso1354 (9, 27). The enzyme was purified from S. solfataricus strain Oα and hydrolyzed CMC, producing cellobiose and cellooligomers under optimal conditions of pH 3.5 and 90°C (27).
The presence of these endoglucanases implies the existence of oligosaccharide transporters that would enable substrate uptake and subsequent catabolism. Binding-protein-dependent ABC transporters are a common transport system in S. solfataricus (2). Sugar transporters in these organisms are assigned to two categories, oligodipeptide transporters and carbohydrate uptake transporters (2, 3). The oligodipeptide transporters possess a signal sequence followed by a catalytic domain, a serine/threonine linker, and a hydrophobic domain, while sugar transporters contain a type III signal sequence followed by a hydrophobic domain, a serine/threonine linker, and a catalytic domain (2, 35). While these glucose transporter genes were expressed constitutively, those involved in arabinose uptake were inducible (24). Sso3053 (annotated as MalE) was previously reported as a maltose binding protein due to the transcription of the gene in the presence of maltose, while Sso2669 (CbtA) bound cellobiose (13). Interestingly, a recently described oligodipeptide transporter (16) also provides a receptor for virus-host interaction (14). To obtain an understanding of the potential overlap between sugar transporters and oligosaccharide hydrolysis, in this study, a genetics-based approach was used to examine their impacts on cellodextrin utilization.
MATERIALS AND METHODS
Archaeal strains and construction.
The strains used in this study are listed in Table 1. The strains were grown in modified Allen's medium in batch culture at 80°C and pH 3.0 with aeration, as described previously (4, 7). Five-milliliter cultures were cultivated in glass screw-cap test tubes placed in rotary tube-drum agitators mounted in forced-air incubators with external DC motors. A mixture of soluble cellodextrins prepared by dilute acid hydrolysis of Sigmacell (Sigma) was added to modified Allen's medium at a concentration of 1:4 (vol/vol). Other sugars were added as indicated at 0.2% (wt/vol). For a complex medium, tryptone was added at 0.2% (wt/vol). Growth was monitored by absorption at 540 nm. The differences between biological replicates are shown in the figures as error bars. Spent culture medium was obtained by successive passages of specific cell lines with cell removal by sterile filtration. A solid medium was prepared as described previously (17).
Table 1.
Strain list
| Strain | Description | Reference(s) or source |
|---|---|---|
| PBL2000 | S. solfataricus strain 98/2 (wild type) | 30 |
| PBL2025 | PBL2000; deletion of Sso3004 to Sso3050 | 33 |
| DSM1617 | S. solfataricus P2 (wild type) | DSMZ |
| PBL2028 | PBL2000; deletion of Sso3017 to Sso3050; IS1::Sso3053 | 18, 33 |
| PBL2091 | PBL2025; deletion of Sso0615 to Sso0616 (pyrEF) by markerless exchange | This work |
| PBL2092 | PBL2091; Sso1949::lacS, Sso2534::pyrEF by linear recombination | This work |
| PBL2074 | PBL2025; Sso1949::lacS by linear recombination | This work |
| PBL2105 | PBL2025; deletion of Sso3052 to Sso3060 using plasmid pBN1202 (pUC19; Sso3052::lacS::Sso3060) by consecutive single crossover | This work |
| PBL2099 | PBL2091/pBN1226 (pKlacS; Sso1354::C-His6) | This work |
| PBL2106 | PBL2025; deletion of Sso2833 to Sso2850 using plasmid pBN1204 (pUC19; Sso2833::lacS::Sso2850) by linear recombination | This work |
| PBL2113 | PBL2028/pBN1239 (pKlacS; Sso3053) | This work |
S. solfataricus strain 98/2 (PBL2000 [30]) was from laboratory stocks, while S. solfataricus strain P2 (DSM 1617) was obtained from DSMZ. S. solfataricus strain PBL2025, derived from PBL2000 (33), incurred a 58-kbp spontaneous deletion in the chromosome, encompassing ORFs Sso3004 to Sso3050 as annotated in the genome (35). The deletion included Sso3019, which encodes the sole cytoplasmic β-glycosidase (lacS). The deletion of lacS allowed the gene to be used as a functional marker for targeted disruption of other genes (37). Sso1949 was disrupted in PBL2025 by insertion of lacS to create strain PBL2074. Simultaneous inactivation of Sso1949 and Sso2534 used strain PBL2091, a Lac− Pyr− double mutant, by disruption of Sso1949 by lacS insertion and disruption of Sso2534 by pyrEF insertion, resulting in strain PBL2092. To assess the role of Sso1354, strain PBL2099 was constructed by transformation of strain PBL2091 with plasmid pBN1226. pBN1226 encodes an expression copy of Sso1354 using its native promoter fused to a C-terminal polyhistidine tag and cloned in plasmid pKlacS at the SphI site (genome coordinates 1192003 to 1193148). pKlacS is a derivative of pJ (pBN1078) with addition of lacS, including 150 nucleotides of flanking DNA encoding its native promoter and terminator (26). In addition, pKlacS has a replacement of the S. solfataricus pyrEF genes and promoter with those of Sulfolobus acidocaldarius derived from chromosomal positions 1361371 to 1362890 at XmaI and KpnI sites. PBL2091 is a derivative of PBL2025 in which chromosomal positions 535869 to 537242, DNA spanning the pyrEF genes, were deleted by markerless exchange (34).
Sso3053 was inactivated by deletion using consecutive single crossovers, creating strain PBL2105. Sequences spanning coordinates 2805424 to 2805923 cloned in pUC57 at SacI/BamHI sites and 2815262 to 2815761 at PstI/SphI sites, respectively, were derived from either end of the region spanning ORFs Sso3052 to Sso3060. lacS was inserted at a SalI site in pUC57. Integration of this circular nonreplicating construct into strain PBL2025 using a lacS-based selection (37) followed by successive single crossovers deleted Sso3052 through Sso3060. The spontaneous deletion in strain PBL2028 was mapped as described previously (33) by PCR and DNA sequencing throughout the tested region. Complementation of the Sso3053 disruption mutation in strain PBL2028 used plasmid pBN1239. pBN1239 is a pKlacS derivative encoding Sso3053 with its native promoter amplified from coordinates 2806121 to 2808446 and inserted at an SphI site. PBL2106, a strain lacking Sso2847 and its flanking genes, was produced by deletion of the region including ORFs Sso2833 through Sso2850 and spanning coordinates 2595288 to 2611082. This mutation employed linear DNA recombination, as described previously (25), using a linear PCR amplicon generated from a pUC57 template including 500 bp from Sso2833 inserted at the SacI/BamHI site and a 500-bp region from Sso2850 inserted at a the PstI/SphI site, with the selective marker lacS inserted between these regions at the SalI site.
Genomic analysis of S. solfataricus strains.
NCBI BLASTp was used to analyze the duplicated region centered around Sso1354 in S. solfataricus strain P2 (DSM1617). The corresponding but deleted region in strain 98/2 (PBL2000) (30) was identified by PCR analysis with primers flanking the deletion region, followed by sequencing and NCBI BLASTn analysis.
Mixed-acid hydrolysis of cellulose.
Generation of mixed soluble cellodextrins from hydrolysis of microcrystalline cellulose was as described previously (39) with modifications. Sigmacell in 2-g amounts was gradually added to 16 ml of ice-cold concentrated hydrochloric acid while stirring. Then, 4 ml of ice-cold concentrated sulfuric acid was gradually added, and the flask was capped with a rubber stopper and stirred gently for 4 h at room temperature. The hydrolysate was then poured into 180 ml of −20°C acetone, chilled at −20°C for 2 h, and then filtered through a Millipore AP40 glass fiber filter. The filter cake of cellodextrins was washed four times with 20 ml −20°C acetone. The filter cake was resuspended with 60 ml of deionized water and stirred overnight in an open container to evaporate residual acetone. The next day, the pH for the hydrolysate was adjusted to 3.0 using potassium hydroxide, and the mixture was centrifuged at 7,000 × g for 10 min to pellet the insoluble material. The supernatant was collected and stored at 4°C.
TLC of cellodextrins.
Thin-layer chromatography (TLC) was performed with ventilation on K6 (4860-820; Whatman) 20- by 20-cm glass plates coated with 60 Å silica to a thickness of 250 μm. The TLC plates were heated to 80°C for 30 min for silica activation and prior to sample application. One microliter of the sample was loaded 1.5 cm away from the bottom of the plate. The 150-ml mobile phase consisted of a mixture of 2:1:1 (vol/vol/vol) ethyl acetate-acetic acid-water. The samples were resolved three times for 3 h each time, combined with air drying for 15 min. After drying, color was developed by spraying a solution of 1.5% (wt/vol) thymol in 95:5 (vol/vol) ethanol-sulfuric acid over the plate and heating over a hotplate at 120°C until color development was complete.
HPLC analysis of cellodextrins.
Spent culture supernatant samples were diluted 1:8 (vol/vol) with high-performance liquid chromatography (HPLC) grade water and filtered through Phenomenex RC membrane 0.45-μm filters. The diluted samples were neutralized where necessary prior to liquid chromatography (LC) injection. Five-microliter volumes were characterized in triplicate using an Agilent 1200 Series Rapid Resolution LC system (Agilent Technologies, Santa Clara, CA) with two Phenomenex Rezex RSO-Oligosaccharide Ag+ columns connected in series. A compatible guard column charged with silver ions was employed. The columns were heated to 40°C using an external column heater, and solvent was applied at a binary pump flow rate of 0.25 ml/minute. Water was used as the mobile phase, and an Agilent 1200 series Refractive Index Detector (RID) was used to detect the eluting sugars. The RID was set to positive mode and the optical unit temperature to 40°C. Data acquisition and integration were carried out using Agilent Chemstation software to quantify the various cellodextrins in each sample.
RESULTS
Strain-specific genome variation and endoglucanase content.
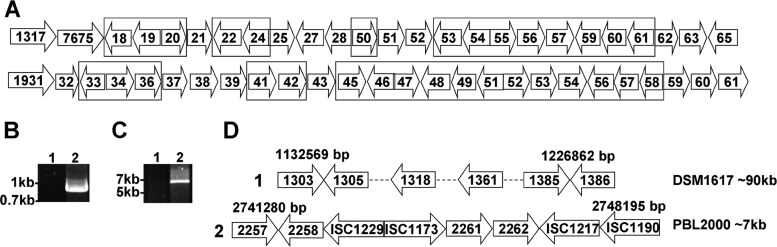
It has been noted previously that the genome of S. solfataricus strain P2 (DSM1617) includes a chromosomal duplication of 14 genes centered at position 1192998 in a region predicted to be involved in carbohydrate metabolism (8). Upon closer examination, it was apparent that the duplicated region in strain P2 (DSM1617) was comprised of 20 genes spanning ORFs Sso1318 through Sso1361 and again between Sso1933 and Sso1958, with several intervening transposons (Fig. 1A and Table 2). PCR amplification of endoglucanase Sso1354 using strain 98/2 (PBL2000) genomic DNA was not successful, suggesting the gene was absent (Fig. 1B). PCR using primer pairs for genes Sso1305 through Sso1385, which are present in strain P2 (DSM1617), was also not successful in amplifying cognate sequences from strain 98/2 (PBL2000). Ultimately, the boundaries of a larger region that was absent from the strain 98/2 (PBL2000) genome were determined by PCR using a forward primer for Sso1303 and a reverse primer for Sso1388, resulting in a 6.875-kb amplicon (Fig. 1C) rather than the nearly 90-kb region present in strain P2 (DSM1617). In strain P2 (DSM1617), this region closely resembles one that is present between Sso1933 and Sso1958 and presumably arose by duplication of one of the two segments. In strain 98/2 (PBL2000), two genes, Sso_2261 and Ssol_2262, were flanked by transposons ISC1229, ISC1173, and ISC1217. Ssol_2258 is a partial gene present in strain 98/2 (PBL2000) and shares 79% identity with Sso1305. A schematic contrasting these regions within their respective genomes is shown in Fig. 1D. The identities of the genes in the duplicated region range from 75 to 90% (Table 2). Though strain 98/2 lacked Sso1354, it contained both Sso1949 and Sso2534. The percent amino acid sequence identity between Sso1949 and Sso1354 (from DSM1617) is 83%, and only 27% between Sso1949 and Sso2534. The identity between Sso1949 and Sso2534 orthologs in the two strains was 100%. Based on this information and the lack of BLAST hits using GHF 5, 6, 7, 8, 9, 10, 12, 18, 19, 26, 44, 45, 48, 51, 61, 74, and 124 domains as queries, strain P2 (DSM1617) has three endoglucanases while strain 98/2 (PBL2000) has only two.
Fig 1.
Duplication and deletion in S. solfataricus strains 98/2 (PBL2000) and P2 (DSM1617). (A) Flanking regions of Sso1354 and Sso1949 in P2. Regions that are duplicated within the strain P2 genome flanking the Sso1354 gene are indicated (boxes). (B) PCR analysis of Sso1354 (P17 and P18 [Table 3]). Lanes: 1, strain 98/2 (PBL2000); 2, strain P2 (DSM1617). (C) PCR analysis of the site of duplication from Sso1303 (primer P3) to Sso1388 (P4) (Table 3). Lanes: 1, strain P2; 2, strain 98/2. (D) Analysis of the DNA sequence of a PCR amplicon spanning Sso1303 to Sso1388 in strains P2 (D1) and 98/2 (D2). Genome coordinates are indicated above the ORFs. Amplicon sizes are indicated on the right. The dashed lines indicate regions not shown.
Table 2.
ORFs in the P2 genome duplication
| Duplicated ORFs | Annotation | Reference(s) | Identity (%) |
|---|---|---|---|
| Sso1933/1320 | Hypothetical | 83 | |
| Sso1934/1319 | Putative multidrug ABC transporters | 92 | |
| Sso1936/1318 | Putative ABC transporters | 94 | |
| Sso1941/1324 | Putative thiamine biosynthesis | 91 | |
| Sso1942/1322 | Ketol-acid reductoisomerase ilvC | 90 | |
| Sso1946/1951 | Insertion elements; ISC1290/ISC1190 | - | |
| Sso1945/1350 | Hypothetical; 1945 is the N-terminal half of Sso1350 | 89 | |
| Sso1947/1350 | Hypothetical; 1947 is the C-terminal half of Sso1350 | 87 | |
| Sso1948/1353 | Beta glycosidases | 11 | 86 |
| Sso1949/1354 | Endoglucanases | 19, 27 | 83 |
| Sso1952/1355 | Putative carboxypeptidases | 97 | |
| Sso1953/1356 | Hypothetical | 82 | |
| Sso1954/1357 | Putative dolichyl-phosphate mannose synthases | 88 | |
| Sso1956/1359 | Hypothetical | 83 | |
| Sso1957/1360 | Hypothetical | 76 | |
| Sso1958/1361 | Putative major facilitator superfamily | 90 |
Table 3.
Primers
| Primer | Name | Sequence |
|---|---|---|
| P1 | Sso1354 with native promoter SphI Forward | TAGATAGTGCATGCGAATTATCCAGTCCTCTATTTCTCTTACG |
| P2 | Sso1354 C-terminal 6×His tag SphI Reverse | ATCTAGCTGCATGCTCAGTGGTGGTGGTGGTGGTGGAGGAGAGTTTCAGAAAAGTTGGA |
| P3 | Sso1303 Sso1388 Forward | TTGGCTTAAAGTATAGCCTTTA |
| P4 | Sso1303 Sso1388 Reverse | CTGCTCTGCTTTGAAGATTCT |
| P5 | Sso3052 SacI Forward | CATCAGAGCTCCAAACAACGCTCCTTTAGGG |
| P6 | Sso3052 BamHI Reverse | ATCCAGGATCCTTATTCTGGTGCGTAGACGTTAGG |
| P7 | Sso3060 PstI Forward | CATCACTGCAGCTATCTATAATCTAGGATAACCCTTATAAC |
| P8 | Sso 3060 SphI Reverse | ATCCAGCATGCTATCCCATTTAGGTGATGTTGTTAG |
| P9 | Sso2833 SacI Forward | CATCAGAGCTCAGCACAAGGGTTTTCCCTTT |
| P10 | Sso2833 BamHI Reverse | ATCCAGGATCCTCATACCTTAAAGTTTAATATCGTCCA |
| P11 | Sso2850 PstI Forward | CATCACTGCAGTTAGCAATTTAGATGCCAGGATG |
| P12 | Sso2850 SphI Reverse | ATCCAGCATGCAGAATGTATCGGATGATCAGAATACG |
| P13 | Sso3053 internal Forward | AACAATACGCTAATTTAACGAATCC |
| P14 | Sso3053 internal Reverse | ATTCCTTTAGCTAGAAGGCCATATG |
| P15 | Sso3053 SphI Forward | AGTCAGCATGCCCATAAATATAAAAGTTTCGCCATTT |
| P16 | Sso3053 SphI Reverse | GTACAGCATGCTTATCTTCTTCTTACCACTACATAAGCTACT |
| P17 | Sso1354 internal Forward | ATCATTTACTTGCATCAACA |
| P18 | Sso1354 internal Reverse | TTAGAGGAGAGTTTCAGAAAAGTTGG |
| P19 | Sso2847 internal Forward | TACCCAGCAAAAGGATGACC |
| P20 | Sso2847 internal Reverse | CCTCCGTTGCCTCAATACAT |
In vivo function of Sso1354 endoglucanase.
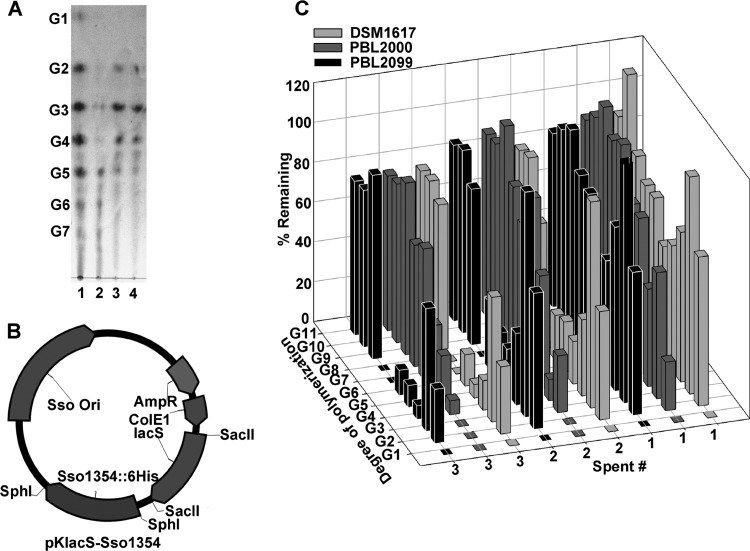
To determine the in vivo role of endoglucanase Sso1354, residual soluble cellodextrins present after repeated passage of strains 98/2 (PBL2000) and P2 (DSM1617) in a spent defined medium containing soluble cellodextrins as the sole carbon and energy source were analyzed by TLC (Fig. 2A). Strain DSM1617 exhibited preferential depletion of longer polymers relative to strain PBL2000 (Fig. 2A, lanes 2 and 3). There was a significant difference in the consumption of higher oligosaccharides, especially after the second and third passages of spent media. To clarify the specific identity of the cellodextrin consumption pattern, an LC method was developed that could fractionate these polymers through G10 (DP10; cellodecaose) based on the use of two Phenomenex RSO columns in tandem. In addition, since one of the differences in the endoglucanase contents of these strains was the unique presence of Sso1354 in strain P2, a derivative of strain 98/2, called PBL2099, having a plasmid-borne copy of Sso1354 expressed via its native promoter was constructed (Fig. 2B). Analysis of residual cellodextrins remaining in spent medium after one cycle of growth produced a pattern on TLC that resembled that of strain DSM1617 rather than the strain 98/2 derivative (Fig. 2A, lane 4). LC analysis of the passaged medium after each of three successive cycles of growth of the three strains (Fig. 2C) indicated the strain 98/2 derivative (30) preferentially depleted the medium of shorter cellodextrins, primarily from G2 through G5, particularly after one and two cycles of growth. There was also a decrease in the amount of the longer cellodextrins up to G8 after the third cycle of growth. In contrast, strain DSM1617 and the strain 98/2 derivative having Sso1354 (PBL2099) exhibited increased consumption of longer cellodextrins, including G4 through G8, particularly after the second and third cycles of growth. Unlike the unmodified wild-type strain 98/2 (PBL2000), strains P2 and PBL2099 also exhibited larger amounts of residual G2 and G3. In all cases, glucose was not present in the spent culture supernatants, indicating rapid and or preferential consumption. The effect of prolonged exposure of added cellodextrins to dilute acid at the cultivation temperature of 80°C in the absence of cells was also examined. No change was observed in the relative distribution of cellodextrins after the three cycles of incubation (data not shown), indicating there was little if any chemical hydrolysis of the soluble cellodextrins.
Fig 2.
Effect of Sso1354 on cellodextrin consumption by strain 98/2. (A) TLC analysis of cellodextrin consumption. Lanes: 1, cellodextrin mixture (no cells); 2, strain 98/2 (PBL2000) spent medium; 3, strain P2 (DSM1617) spent medium; 4, PBL2099 (PBL2025 with pBN1226 [pKlacS-Sso1354]) spent medium. (B) pBN1226 (pKlacS-Sso1354). Shown are the pRN1 plasmid origin (Sso ori) and selectable marker (lacS), E. coli shuttle vector components (AmpR and ColE1), and transgene C-terminally His-tagged Sso1354 plus the natural promoter. (C) HPLC analysis of spent medium after 3 cycles of growth of strain 98/2 (PBL2000) and PBL2099 and strain P2 (DSM1617). The samples were analyzed in triplicate, and the standard deviations were less than 10%.
Characterization of endoglucanases.
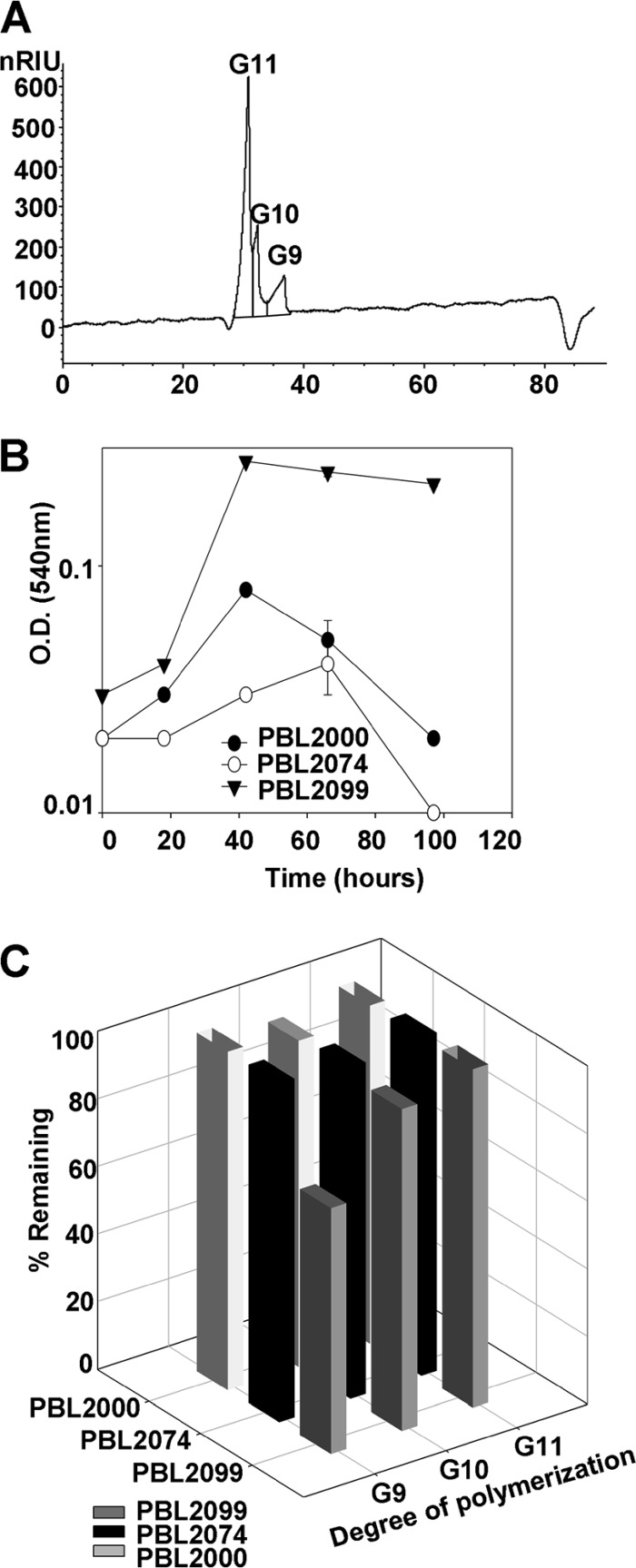
To better understand the roles of the other two endogenous endoglucanases (Sso1949 and Sso2534) present in both strains 98/2 (PBL2000) and P2 (DSM1617), a derivative of 98/2 lacking them was constructed and evaluated. Strain PBL2092 carried a pyrEF disruption of Sso2534 and a lacS disruption of Sso1949. This strain was passaged repeatedly in the cellodextrin medium, and the residual contents of the spent medium were examined by LC (Fig. 3A). Since this strain lacks detectable endoglucanases, it was likely that cellodextrin consumption during passage arose through the combined action of a cellodextrin transporter(s) and the cytoplasmic enzyme LacS, a beta-glycosidase (29). LacS has been shown previously to hydrolyze G5 in vitro (28). To test the importance of LacS for utilization of cellodextrins, growth of the lacS mutant PBL2025 was tested, and no growth was observed. This verified a role for LacS in cellodextrin consumption and implied the existence of a cellodextrin transporter. Since the residual cellodextrin content resulting from passage of strain PBL2092 resulted in an enrichment of longer cellodextrins, this medium was used to test the specific in vivo roles of Sso1354, Sso1949, and Sso2534. Strain 98/2 (PBL2000) and mutant derivatives lacking Sso1354 (PBL2099) or Sso1949 (PBL2074) were grown in the spent medium produced by strain PBL2092. Strain PBL2099 grew better than either of the other two strains (Fig. 3B). LC analysis of the spent medium of each of the strains showed specific depletion of G9 (cellononaose) of about 30% relative to the starting amount (Fig. 3C). This demonstrates that Sso1354 has activity toward longer cellodextrins in vivo, whereas the other two endoglucanases do not show any significant role in hydrolysis of longer cellodextrins, while their actions toward shorter cellodextrins may be obscured due to the actions of other proteins (see below).
Fig 3.
Cellodextrin consumption patterns. (A) HPLC chromatogram of spent medium produced by Sso2092 (third cycle) showing enrichment of G9 to G11. nRIU, nano-refractive index units. (B) Growth of strains in spent medium (third cycle) of PBL2092 (strain 98/2 derivative lacking endoglucanases). The growth analyses were conducted in replicates, and the error bars indicate the differences between the biological replicates. (C) HPLC analysis of spent media of strains 98/2 (PBL2000), PBL2074, and PBL2099 in PBL2092 spent medium (third cycle). The samples were analyzed in triplicate, and the standard deviations were less than 10%.
Characterization and overlap of cellodextrin transporters.
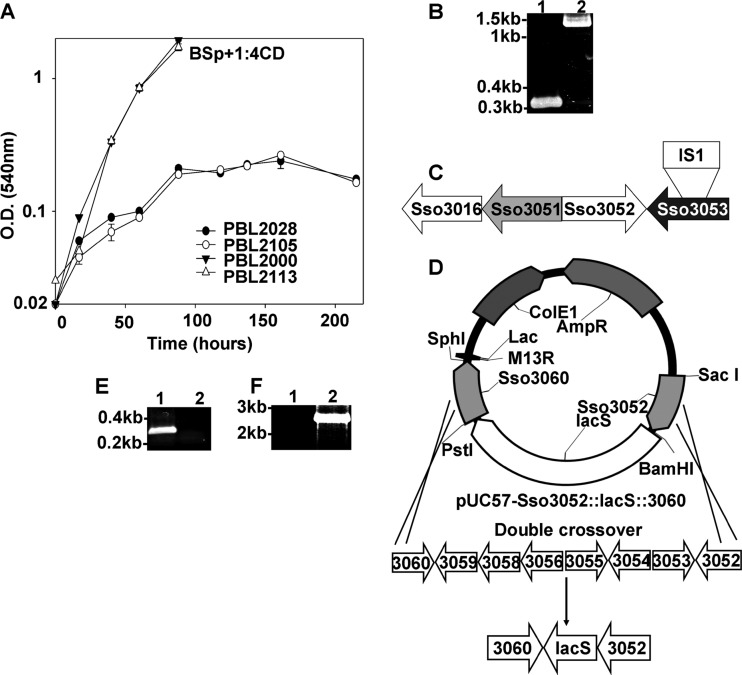
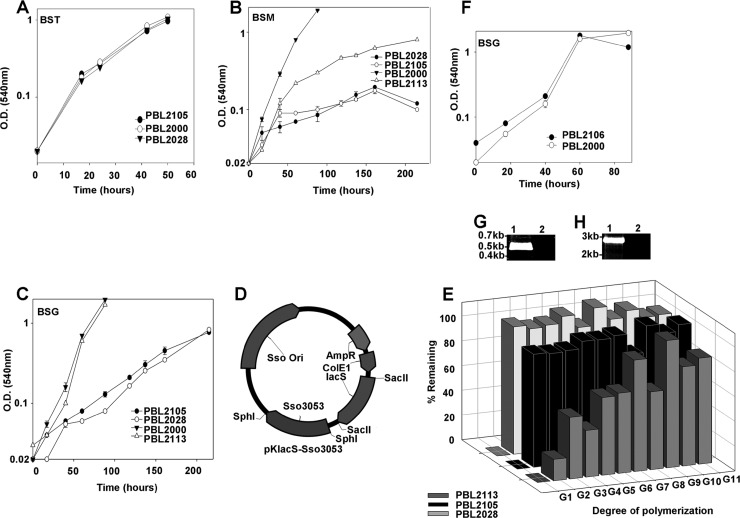
Since LacS was shown to be required for cellodextrin consumption in the absence of endoglucanases, a cellodextrin transporter must exist to enable substrate uptake. The occurrence of a cellodextrin transporter was implied by the phenotype of strain PBL2028, a spontaneous strain 98/2 derivative with a large chromosomal deletion that was unable to grow on soluble cellodextrins (Fig. 4A). PBL2028 arose from a screen for lacS mutants (18). PCR analysis of the mutation mapped the deletion endpoints to Sso3050 and immediately adjacent to a putative ABC transporter (Fig. 4B and C). In addition, PBL2028 encoded an IS element (IS1) inserted within Sso3053 at nucleotide 723. As Sso3053 exhibited homology to an oligosaccharide transporter (TM1223) from Thermotoga maritima, it was possible this mutation was responsible for the inability of strain PBL2028 to utilize cellodextrins as a sole carbon and energy source. To test this possibility, a strain 98/2 derivative (PBL2105) was constructed lacking Sso3053 and its flanking putative ABC transporter components (Fig. 4D, E, and F). Strain PBL2105 grew normally in complex medium (Fig. 5A); however, the strain grew poorly using maltose or glucose as the carbon source (Fig. 5B and C). As prior studies have suggested that glucose is transported via a glucose binding facilitator (Sso2847) (1), a strain was constructed lacking Sso2847 and its flanking ABC transporter components and then tested for growth using glucose as the carbon source. This strain grew normally relative to the wild-type strain 98/2 (PBL2000), indicating Sso2847 was not required for glucose uptake (Fig. 5F, G, and H).
Fig 4.
Sso3053 deletion and insertional inactivation strains, (A) Growth analyses of various strains in a defined soluble cellodextrin medium. The analyses were done in replicates, and the error bars indicate the differences between the biological replicates. (B) PCR analysis of Sso3053 disruption in strain PBL2028 using primers P13 and P14 (Table 3). Lanes: 1, strain 98/2 (PBL2000); 2, PBL2028. (C) Schematic of the PBL2028 strain genotype showing deletion of genes from Sso3017 through Sso3050 and IS1 insertion in Sso3053. (D) Deletion of the Sso3053 ABC transporter cluster using plasmid pBN1202, resulting in strain PBL2105. (E) PCR analysis of Sso3053 deletion (using P13 and P14 [Table 3]). Lanes: 1, strain 98/2 (PBL2000); 2, PBL2105. (F) PCR reconfirmation analysis of Sso3053 deletion (using P5 and P8 flanking the deletion region [Table 3]). Lanes: 1, 98/2; 2, PBL2105.
Fig 5.
Growth of strains and cellodextrin analysis. (A) Complex (BST) medium. (B) Maltose (BSM) medium. (C) Glucose (BSG) medium. (D) pBN1239 (pKlacS plus Sso3053). All growth analyses were done in replicates, and the error bars indicate the differences between the biological replicates. (E) HPLC analysis of spent cellodextrin media of PBL2113, PBL2105, and PBL2028. The samples were analyzed in triplicate, and the standard deviations were less than 10%. (F) Growth patterns for strain 98/2 (PBL2000) and PBL2106 in glucose medium (BSG). (G) PCR analysis of the Sso2847 deletion (P19 and P20) (Table 3). Lanes: 1, strain 98/2 (PBL2000); 2, PBL2106. (H) PCR analysis of the Sso2847 deletion using primers amplifying the flanking regions (P9 and P12). Lanes: 1, PBL2106; 2, PBL2000.
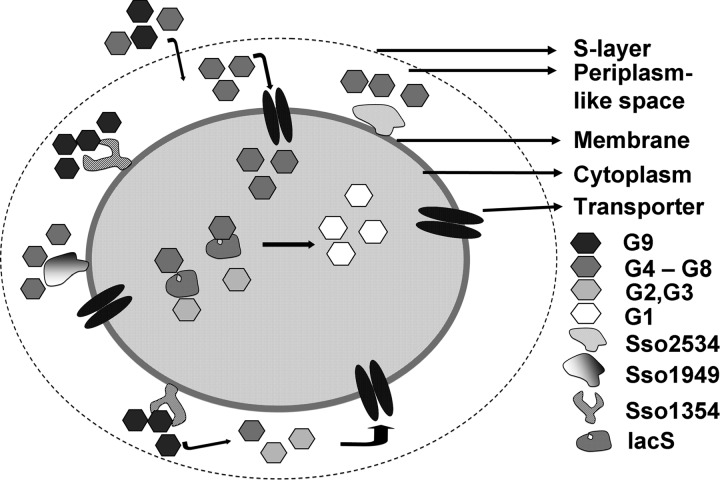
The importance of Sso3053 was evaluated by complementation using strain PBL2028, which carried an IS element insertion in the gene (Fig. 4C). PCR analysis confirmed the presence of Sso3051 and Sso3054 in PBL2028. The remaining components of the transporter cluster are Sso3055 (ATP binding), Sso3058 (permease), and Sso3059 (permease). Strain PBL2028 was transformed with either pKlacS (encoding Sso3053 under the control of its endogenous promoter) or pBN1239, and the transformed cells were tested for the ability to grow using lactose as the carbon source. Only PBL2028 transformed with pBN1239 (strain PBL2113) gained the ability to grow using lactose. While growth using glucose was fully restored, PBL2113 regained only partial ability to use maltose (Fig. 5B and C). The effect of Sso3053 manipulation on consumption of cellodextrins was then evaluated (Fig. 4A). Strain PBL2028 lacking both lacS and Sso3053 exhibited slight growth on the soluble cellodextrin mixture due to the presence of residual glucose, while it showed normal growth in complex medium (Fig. 5E). Similar results were obtained with the strain with Sso3053 deleted despite the presence of lacS (Fig. 5A). In contrast, complementation of Sso3053 restored the ability to use cellodextrins (Fig. 4A). The HPLC analysis of cellodextrins in spent media of the strains showed the gain of ability to consume the cellodextrin in PBL2113 (Fig. 5E). Taken together, these data indicate there are two overlapping systems for the catabolism of cellodextrins in S. solfataricus, and a model describing these systems is presented (Fig. 6). Membrane-targeted endoglucanases hydrolyze polymers prior to import, and a cellodextrin transporter translocates oligomers into the cytoplasm for subsequent hydrolysis by LacS, a beta-glycosidase. The combined activities of these systems ensure rapid and efficient consumption of cellodextrins.
Fig 6.
Model illustrating the hydrolysis and transport of cellodextrins in S. solfataricus.
DISCUSSION
This study shows the occurrence of two overlapping systems for cellodextrin consumption in S. solfataricus. One system is comprised of multiple extracytoplasmic endoglucanases, and the other system includes a broad-substrate ABC transporter combined with an intracellular beta-glycosidase (LacS). While three types of endoglucanases have been described (9, 19, 23, 27), this study shows that S. solfataricus strains vary in their contents of these enzymes and that they confer on the host organism different abilities to utilize cellodextrins. The locations of these endoglucanases is also not yet clear. While they appear to be membrane targeted, there have been reports that at least one is fully secreted (23) while the other two could remain membrane associated. Action toward soluble cellodextrins would arise after such substrates penetrate the S layer through its pores.
In vivo analysis demonstrated that strain 98/2 (PBL2000) had a reduced ability to hydrolyze and consume oligosaccharides ranging from G6 to G9 relative to strain P2 (DSM1617). Though strain 98/2 encodes two of the three endoglucanases present in strain P2, the specific addition of Sso1354 to strain 98/2 improved its ability to catabolize longer cellodextrins, G6 through G8, and conferred the ability to catabolize G9. These data suggest Sso1354 specifically targets hydrolysis of longer oligosaccharides. In addition, if Sso1354 arose by gene duplication in strain P2, it may have been in response to the availability of long cellodextrins as carbon and energy sources.
Unexpectedly, analysis of the pattern of cellodextrin consumption by the strain 98/2 derivative lacking both endogenous endoglucanases Sso1949 and Sso2534 (PBL2092) showed there was continued consumption of cellodextrins through G8. This pattern of consumption depended on the presence of both LacS and the cellodextrin transporter component Sso3053. Prior reports are consistent with the role of LacS in this process because of its reported in vitro hydrolytic activity toward cellodextrins up to G5 (cellopentaose) (28). The ability of transporter Sso3053 to translocate cellodextrins from G2 through G8, as well as maltose and to a limited extent glucose, reveals a novel broad substrate preference ranging from monosaccharides to polysaccharides and from alpha-linked to beta-linked sugars. Together, LacS and the Sso3053 ABC transporter comprise a system for cellodextrin consumption that is independent of any of the endogenous endoglucanases. As strains P2 (DSM1617) and 98/2 (PBL2000) originate from different continents (30, 40), the presence of both systems for cellodextrin consumption indicates these systems provided distinct evolutionary benefits that led to their maintenance in separate lineages.
Deletion of the Sso3053 transporter system in strain PBL2028 compromised glucose catabolism. This defect was overcome by Sso3053 complementation. However, the reduced growth rate of the complemented strain indicated maltose catabolism was only partially restored. This may be a result of the absence of a different gene cluster missing in strain PBL2028 encoded by Sso3043 to Sso3048. The specific role of these genes in the transport of oligosaccharides remains to be determined. Since the Sso3053 disruption (in strain PBL2028) reduced the growth rate in a glucose minimal medium and this defect was restored by complementation, it is reasonable to conclude that Sso3053 is at least partially responsible for the uptake of glucose. Strain PBL2028 was able to grow normally in complex medium. Interestingly, inactivation of a previously described glucose transporter system involving Sso2847 (1) had no effect on the rate of growth using glucose. These results could be reconciled by proposing that the two transporters operate at different glucose concentrations or that there exist additional transport systems for glucose in strain 98/2 relative to strain P2. Combining mutations in both transport systems in a single cell line could provide insight into their relative contributions to the uptake of glucose.
ACKNOWLEDGMENTS
The efforts of Katie Starostka, Xiuyun Zhao, Amanda Daugherty, and Yukari Maezato are acknowledged.
This research was supported by funds from the Department of Energy (DE-FG36-08GO88055).
Footnotes
Published ahead of print 31 August 2012
REFERENCES
- 1. Albers SV, et al. 1999. Glucose transport in the extremely thermoacidophilic Sulfolobus solfataricus involves a high-affinity membrane-integrated binding protein. J. Bacteriol. 181: 4285– 4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Albers SV, Koning SM, Konings WN, Driessen AJ. 2004. Insights into ABC transport in archaea. J. Bioenerg. Biomembr. 36: 5– 15 [DOI] [PubMed] [Google Scholar]
- 3. Albers SV, Van de Vossenberg JL, Driessen AJ, Konings WN. 2001. Bioenergetics and solute uptake under extreme conditions. Extremophiles 5: 285– 294 [DOI] [PubMed] [Google Scholar]
- 4. Allen MB. 1959. Studies with Cyanidium caldarium, an anomalously pigmented chlorophyte. Arch. Mikrobiol. 32: 270– 277 [DOI] [PubMed] [Google Scholar]
- 5. Bok JD, Yernool DA, Eveleigh DE. 1998. Purification, characterization, and molecular analysis of thermostable cellulases CelA and CelB from Thermotoga neapolitana. Appl. Environ. Microbiol. 64: 4774– 4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Breves R, et al. 1997. Genes encoding two different beta-glucosidases of Thermoanaerobacter brockii are clustered in a common operon. Appl. Environ. Microbiol. 63: 3902– 3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brock TD, Brock KM, Belly RT, Weiss RL. 1972. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 84: 54– 68 [DOI] [PubMed] [Google Scholar]
- 8. Brouns SJJ, et al. 2005. The hyperthermophilic archaeon Sulfolobus: from exploration to exploitation, p 261–276 In Inskeep WP, McDermott TR. (ed), Geothermal biology and geochemistry in Yellowstone National Park. Thermal Biology Institute, Montana State University, Bozeman, MT [Google Scholar]
- 9. Cannio R, Di Prizito N, Rossi M, Morana A. 2004. A xylan-degrading strain of Sulfolobus solfataricus: isolation and characterization of the xylanase activity. Extremophiles 8: 117– 124 [DOI] [PubMed] [Google Scholar]
- 10. Chhabra SR, Kelly RM. 2002. Biochemical characterization of Thermotoga maritima endoglucanase Cel74 with and without a carbohydrate binding module (CBM). FEBS Lett. 531: 375– 380 [DOI] [PubMed] [Google Scholar]
- 11. Cobucci-Ponzano B, et al. 2010. A new archaeal beta-glycosidase from Sulfolobus solfataricus: seeding a novel retaining beta-glycan-specific glycoside hydrolase family along with the human non-lysosomal glucosylceramidase GBA2. J. Biol. Chem. 285: 20691– 20703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Rosa M, Gambacorta A, Bu'lock JD. 1975. Extremely thermophilic acidophilic bacteria convergent with Sulfolobus acidocaldarius. J. Gen. Microbiol. 86: 156– 164 [DOI] [PubMed] [Google Scholar]
- 13. Elferink MG, Albers SV, Konings WN, Driessen AJ. 2001. Sugar transport in Sulfolobus solfataricus is mediated by two families of binding protein-dependent ABC transporters. Mol. Microbiol. 39: 1494– 1503 [DOI] [PubMed] [Google Scholar]
- 14. Erdmann S, Scheele U, Garrett RA. 2011. AAA ATPase p529 of Acidianus two-tailed virus ATV and host receptor recognition. Virology 421: 61– 66 [DOI] [PubMed] [Google Scholar]
- 15. Gibbs MD, et al. 2000. Multidomain and multifunctional glycosyl hydrolases from the extreme thermophile Caldicellulosiruptor isolate Tok7B.1. Curr. Microbiol. 40: 333– 340 [DOI] [PubMed] [Google Scholar]
- 16. Gogliettino M, et al. 2010. A highly selective oligopeptide binding protein from the archaeon Sulfolobus solfataricus. J. Bacteriol. 192: 3123– 3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grogan DW. 1989. Phenotypic characterization of the archaebacterial genus Sulfolobus: comparison of five wild-type strains. J. Bacteriol. 171: 6710– 6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoang V, Bini E, Dixit V, Drozda M, Blum P. 2004. The role of cis-acting sequences governing catabolite repression control of lacS expression in the archaeon Sulfolobus solfataricus. Genetics 167: 1563– 1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang Y, Krauss G, Cottaz S, Driguez H, Lipps G. 2005. A highly acid-stable and thermostable endo-beta-glucanase from the thermoacidophilic archaeon Sulfolobus solfataricus. Biochem. J. 385: 581– 588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kengen SW, Luesink EJ, Stams AJ, Zehnder AJ. 1993. Purification and characterization of an extremely thermostable beta-glucosidase from the hyperthermophilic archaeon Pyrococcus furiosus. Eur. J. Biochem. 213: 305– 312 [DOI] [PubMed] [Google Scholar]
- 21. Liebl W. 2001. Cellulolytic enzymes from Thermotoga species. Methods Enzymol. 330: 290– 300 [DOI] [PubMed] [Google Scholar]
- 22. Liebl W, et al. 1996. Analysis of a Thermotoga maritima DNA fragment encoding two similar thermostable cellulases, CelA and CelB, and characterization of the recombinant enzymes. Microbiology 142: 2533– 2542 [DOI] [PubMed] [Google Scholar]
- 23. Limauro D, Cannio R, Fiorentino G, Rossi M, Bartolucci S. 2001. Identification and molecular characterization of an endoglucanase gene, celS, from the extremely thermophilic archaeon Sulfolobus solfataricus. Extremophiles 5: 213– 219 [DOI] [PubMed] [Google Scholar]
- 24. Lubelska JM, Jonuscheit M, Schleper C, Albers SV, Driessen AJ. 2006. Regulation of expression of the arabinose and glucose transporter genes in the thermophilic archaeon Sulfolobus solfataricus. Extremophiles 10: 383– 391 [DOI] [PubMed] [Google Scholar]
- 25. Maezato Y, Dana K, Blum P. 2011. Engineering thermoacidophilic archaea using linear DNA recombination. Methods Mol. Biol. 765: 435– 445 [DOI] [PubMed] [Google Scholar]
- 26. Maezato Y, et al. 2011. VapC6, a ribonucleolytic toxin regulates thermophilicity in the crenarchaeote Sulfolobus solfataricus. RNA 17: 1381– 1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maurelli L, et al. 2008. Evidence that the xylanase activity from Sulfolobus solfataricus Oα is encoded by the endoglucanase precursor gene (sso1354) and characterization of the associated cellulase activity. Extremophiles 12: 689– 700 [DOI] [PubMed] [Google Scholar]
- 28. Nucci R, Moracci M, Vaccaro C, Vespa N, Rossi M. 1993. Exo-glucosidase activity and substrate specificity of the beta-glycosidase isolated from the extreme thermophile Sulfolobus solfataricus. Biotechnol. Appl. Biochem. 17: 239– 250 [PubMed] [Google Scholar]
- 29. Pisani FM, et al. 1990. Thermostable beta-galactosidase from the archaebacterium Sulfolobus solfataricus. Purification and properties. Eur. J. Biochem. 187: 321– 328 [DOI] [PubMed] [Google Scholar]
- 30. Rolfsmeier M, Blum P. 1995. Purification and characterization of a maltase from the extremely thermophilic crenarchaeote Sulfolobus solfataricus. J. Bacteriol. 177: 482– 485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sakon J, Adney WS, Himmel ME, Thomas SR, Karplus PA. 1996. Crystal structure of thermostable family 5 endocellulase E1 from Acidothermus cellulolyticus in complex with cellotetraose. Biochemistry 35: 10648– 10660 [DOI] [PubMed] [Google Scholar]
- 32. Saul DJ, et al. 1990. celB, a gene coding for a bifunctional cellulase from the extreme thermophile “Caldocellum saccharolyticum.” Appl. Environ. Microbiol. 56: 3117– 3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schelert J, et al. 2004. Occurrence and characterization of mercury resistance in the hyperthermophilic archaeon Sulfolobus solfataricus by use of gene disruption. J. Bacteriol. 186: 427– 437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schelert J, Drozda M, Dixit V, Dillman A, Blum P. 2006. Regulation of mercury resistance in the crenarchaeote Sulfolobus solfataricus. J. Bacteriol. 188: 7141– 7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. She Q, et al. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. U. S. A. 98: 7835– 7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Woese CR, Kandler O, Wheelis ML. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. U. S. A. 87: 4576– 4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Worthington P, Hoang V, Perez-Pomares F, Blum P. 2003. Targeted disruption of the alpha-amylase gene in the hyperthermophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 185: 482– 488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wurtzel O, et al. 2010. A single-base resolution map of an archaeal transcriptome. Genome Res. 20: 133– 141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang YH, Lynd LR. 2003. Cellodextrin preparation by mixed-acid hydrolysis and chromatographic separation. Anal. Biochem. 322: 225– 232 [DOI] [PubMed] [Google Scholar]
- 40. Zillig W, et al. 1980. The Sulfolobus-“Caldariella” group: taxonomy on the basis of the structure of DNA-dependent RNA polymerases. Arch. Microbiol. 125: 259– 269 [Google Scholar]