Abstract
Zoonotic pathogens in land-applied dairy wastewaters are a potential health risk. The occurrence and abundance of 10 pathogens and 3 fecal indicators were determined by quantitative real-time PCR (qPCR) in samples from 30 dairy wastewaters from southern Idaho. Samples tested positive for Campylobacter jejuni, stx1- and eaeA-positive Escherichia coli, Listeria monocytogenes, Mycobacterium avium subsp. paratuberculosis, and Salmonella enterica, with mean recoveries of genomic DNA corresponding to 102 to 104 cells ml−1 wastewater. The most predominant organisms were C. jejuni and M. avium, being detected in samples from up to 21 and 29 of 30 wastewater ponds, respectively. The qPCR detection limits for the putative pathogens in the wastewaters ranged from 16 cells ml−1 for M. avium to 1,689 oocysts ml−1 for Cryptosporidium. Cryptosporidium and Giardia spp., Yersinia pseudotuberculosis, and pathogenic Leptospira spp. were not detected by qPCR.
INTRODUCTION
In the United States, there are 9.3 million milk cows (33) producing an estimated 200 million metric tons of manure annually. Because cattle can be reservoirs of zoonotic pathogens, there is concern over the contamination of soil, water, air, and crops when the manure solids and liquids are land applied (6, 9, 44). At dairies, the solids are often removed from the manure slurries and the liquid fraction (i.e., urine, wash water) is then sent to a pond for storage or can be anaerobically digested first to produce biogas (43). During the crop-growing season, the pond wastewater is diluted with irrigation water and land applied through pressurized irrigation systems to improve the soil nutrient status. It is during spray irrigation that zoonotic pathogens could be aerosolized, increasing the risk of exposure to downwind receptors via inhalation or ingestion after deposition on fomites or food crops (8, 23). Once in the soil after manure addition, pathogens can be internalized by plants (49) and also reach recreational waters by overland flow transport during rainfall events (36, 53, 54), causing significant contamination.
Zoonotic pathogens of potential interest in cattle are Campylobacter jejuni, Escherichia coli, Leptospira spp., Listeria monocytogenes, Mycobacterium avium subsp. paratuberculosis, Salmonella spp., Yersinia spp., Giardia lamblia, and Cryptosporidium parvum (38, 39, 40). While numerous studies have measured the occurrence of these zoonotic pathogens in cattle manures and assessed their fate and transport in the environment (41, 48, 55), to our knowledge, no comprehensive studies have been conducted to quantify pathogens in dairy wastewaters. In addition, only a few studies to date have quantified a wide range of pathogens within cattle manures (20, 21, 26). Understanding the number of pathogens in any land-applied waste is particularly important when developing a quantitative microbial risk assessment (12, 17, 56). Estimation of the risk represented by pathogens in animal manures, however, has largely been based on the cultivation and enumeration of fecal indicator organisms (2, 10). It has been recently shown that molecular methods can be successfully applied with such difficult-to-analyze materials to complement culture-dependent approaches (26, 27).
In this study, we attempted to use quantitative real-time PCR (qPCR) to enumerate fecal indicator organisms and putative zoonotic pathogens in wastewaters from dairies in southern Idaho. qPCR is often used as a convenient alternative to culture-dependent techniques that has the added advantage of being able to detect viable but nonculturable (VBNC) cells of potential pathogens (7, 27, 59). While there are documented advantages and disadvantages with both molecular method- and culture-dependent approaches, it was our intent to provide the first quantitative survey of selected bacterial indicators and putative pathogens in dairy wastewaters. The bacterial indicators (enterococci, total coliforms, E. coli, and Clostridium perfringens) were also quantified using culture-dependent methodologies to gauge the effectiveness of qPCR as an enumeration technique for use with dairy wastewaters.
MATERIALS AND METHODS
Dairy operations and sample collection.
Wastewater samples were collected from 30 storage ponds at dairy operations of various levels of stocking density located in southern Idaho. Eight 500-ml near-surface samples were collected from the perimeter of each storage pond and then composited in a sterile 4-liter Nalgene container. The composite samples were transferred to our laboratory in coolers with ice packs and immediately stored in the dark under conditions of refrigeration at 5°C upon receipt. All samples were processed within 24 h of collection. Three sets of samples were collected from the ponds during the summer and fall of 2011.
Culture-dependent quantification of fecal indicators.
Prior to cultivation as described below, 10-fold serial dilutions of thoroughly mixed wastewater samples were prepared in room temperature phosphate-buffered saline (PBS). The most probable numbers (MPNs) for Escherichia coli (and total coliforms) and enterococci were determined using Colilert-18 and Enterolert detection kits, respectively, in conjunction with a Quanti-Tray 2000 (IDEXX Laboratories, Inc., Westbrook, ME). Vegetative cells and spores of C. perfringens were assayed using membrane filtration and mCP media (Neogen, Lansing, MI) with mCP selective Supplement I (Sigma-Aldrich, St. Louis, MO) as described by Armon and Payment (1). The mCP plates were anaerobically incubated at 44.5°C for 1 day, with exposure to ammonia hydroxide vapors afterward to quantify presumptive C. perfringens colonies. Use of negative controls was implemented, while positive controls consisted of E. coli (ATCC 13706), Enterococcus faecalis (ATCC 700802), and C. perfringens (ATCC 13124). All trays and plates were manually counted, and their numbers were reported as MPN and CFU per milliliter of wastewater, respectively.
Isolation of microbial DNA from the wastewater.
Three aliquots were removed from each 4-liter container of wastewater after thorough mixing. Depending on the solids content, aliquots ranging in volume from 5 to 70 ml were dispensed into 85-ml Oak Ridge tubes (Nalge Nunc International, Rochester, NY) and brought up to volume with cold PBS. After centrifugation at 10,000 × g for 10 min, the supernatant was discarded and the remaining pellet was washed twice with PBS. The pellet was then resuspended with 600 μl of PBS and then transferred to a bead-beating tube from a FastDNA Spin kit for feces (MP Biomedicals LLC, Solon, OH) and processed using a FastPrep FP120 instrument at a speed setting of 6 m s−1 for 45 s according to the manufacturer's recommendations. However, all wash and centrifugation steps were carried out twice to further reduce possible humic acid contamination. The DNA was eluted during the final step with 100 μl of N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES) buffer and then stored at −20°C until analysis by qPCR.
Quantitative PCR.
qPCR was performed with a Bio-Rad (Hercules, CA) iQ5 multicolor real-time PCR detection system using dually labeled Black Hole Quencher (BHQ) probes or iQ SYBR green Supermix (Bio-Rad). The probes were manufactured by Biosearch Technologies (Novato, CA), with FAM (6-carboxyfluorescein) and BHQ-1 as the fluorophore and quencher, respectively. Organisms enumerated in the wastewater samples are listed in Table 1, along with their target genes, individual cycling conditions, and references for primers and probes used. Quantification standards were prepared from pure cultures of C. jejuni (ATCC BAA-1062), C. perfringens (ATCC 13124), E. faecalis (ATCC 700802), E. coli O157:H7 strain 3032 (courtesy of Tom Casey, USDA-ARS), Leptospira interrogans serovar Copenhageni (ATCC BAA-1198), L. monocytogenes (ATCC BAA-679), M. avium subsp. paratuberculosis (ATCC BAA-968), Salmonella enterica serovar Typhimurium (courtesy of Pina Frantamico, USDA-ARS), and Yersinia pseudotuberculosis (ATCC 29833). Genomic DNA was extracted from the cultures using a PowerMicrobial Maxi DNA isolation kit (MO BIO Laboratories, Inc., Carlsbad, CA), while DNA for C. parvum (PRA-67D) and Giardia intestinalis (30888D) was directly purchased from the American Type Culture Collection (Manassas, VA).
Table 1.
Target genes, qPCR assays, and modified thermocycler conditions
| Target organism | Target gene | No. of targets per genome | Primers | Primer annealing temp (°C); extension time (s) | Reference(s)a |
|---|---|---|---|---|---|
| Campylobacter jejuni | VS1 | 1 | VS15-F, VS16-R | 55; 30 | 58 |
| Clostridium perfringens | Alpha toxin | 1 | cpa-F, cpa-R | 55; 30 | 15 |
| Enterococcus faecalis | 23S rRNA | 2 | ECF, ECR | 60; 30 | 18 |
| Escherichia coli | Glucuronidase | 1 | Eco-F, Eco-R | 60; 30 | 46 |
| Enterohemorrhagic E. coli | Shiga toxin | 1 | stx1-F, stx1-R | 55; 30 | 47 |
| Shiga toxin | 1 | stx2-F, stx2-R | 55; 30 | 47 | |
| Intimin | 1 | eae-F, eae-R | 55; 30 | 47 | |
| Leptospira spp. | Lipoprotein, lipL32 | 1 | 45F, 286R | 60; 30 | 51 |
| Listeria monocytogenes | Listeriolysin O | 1 | F, R | 60; 30 | 35 |
| Mycobacterium avium subsp. paratuberculosis | IS900 | 20 | F2, R2 | 65; 30 | 5, 25 |
| Salmonella enterica | Invasin | 1 | 139-F, 141-R | 65; 30 | 19 |
| Yersinia pseudotuberculosis | Invasin | 1 | inv-F, inv-R | 60; 30 | 52 |
| Cryptosporidium spp. | COWPb | 4 | P702-F, P702-R | 60; 30 | 16 |
| Giardia spp. | β-Giardin | 16 | P241-F, P241-R | 60; 30 | 16 |
Probe sequences from the associated references were used for the PCRs, except SYBR green was used for Y. pseudotuberculosis.
COWP, Cryptosporidium oocyst wall protein gene.
One microliter of nondiluted DNA template was used in a PCR with 0.3 μM (each) primer, 0.1 μM probe, 12.5 μl of iQ Supermix (Bio-Rad), and molecular analysis-grade water to achieve a final volume of 25 μl. SYBR green was used in lieu of probe only for the detection of Y. pseudotuberculosis. After an initial denaturation step (95°C for 180 s), 40 cycles of 94°C for 20 s followed by an extension time of 30 s at primer-annealing temperatures were performed as indicated in Table 1. The number of putative pathogens in the samples was calculated using the DNA concentration of the standards, genome molecular weight, and target gene frequency (26).
Pathogen recovery and detection limits.
To determine the performance of the DNA extraction technique and qPCR to detect pathogens in the dairy wastewaters, one wastewater sample was individually spiked with C. jejuni, enterohemorrhagic E. coli, L. monocytogenes, M. avium subsp. paratuberculosis, S. enterica serovar Typhimurium, and Y. pseudotuberculosis. The wastewater chosen had a pH of 7.5 and total solids of 6.7 g liter−1. In brief, an overnight culture was serially diluted in sterile PBS and then 1 ml of the appropriate dilution was dispensed into triplicate 9-ml aliquots of wastewater. The titer of the inoculum was determined by plate counting after growth of the organisms on appropriate media. For each pathogen, a total of 4 to 5 spiked dilutions were prepared, with final wastewater titer levels ranging from log10 1.5 to 8.5. After spiking, the wastewater samples were processed in a vortex apparatus for 30 s and then a 5-ml aliquot was immediately processed as previously described for the DNA isolation.
For Giardia and Cryptosporidium spp., the same spiking and DNA isolation procedures were utilized as described above, except that the cysts and oocysts were spiked into two different wastewaters to determine a possible influence of levels of solids on qPCR performance. The low and high total solids contents of the wastewaters were 3.6 g liter−1 and 12.7 g liter−1, respectively. Giardia lamblia and C. parvum at a titer of 107 cysts/oocysts ml−1 were purchased from Waterborne, Inc. (New Orleans, LA).
The percent recovery was determined for each pathogen by dividing the qPCR result by the calculated wastewater titer level, which was based on the organism density of each spiking dilution. Detection limits were determined via a linear regression analysis of the wastewater titer levels versus the qPCR cycle threshold (CT) values, where r2 > 0.96. The minimum detection limits were determined at a CT of no greater than 35, a value at which detection errors may become significant due to pipetting errors or potential traces of cross-contamination.
Statistical analysis.
Our a priori hypothesis was that there would be no statistical difference between the titers as determined by the culture-dependent techniques and qPCR for the indicator organisms. The two-sample paired t tests for the means were performed on log-transformed data using SAS statistical software version 9.2 (45). Statements of statistical significance were based on an α value of 0.05.
RESULTS AND DISCUSSION
Dairy wastewater samples and chemical properties.
Thirty freestall and open-lot dairies with <1,000 to 10,000 milk cows were targeted for this study. Most of the dairies were stocked with Holstein cows; however, in some cases, they were stocked with a combination of Holstein and Jersey cows. The freestall dairies used either a flush or a vacuum system to remove the manure from the alleys, while the open-lot dairies used a scrape system. The flushed or vacuumed manure is then commonly transferred to solid separation cells, followed, in some cases, by processing with a separator to reduce the solids content prior to discharge to the wastewater storage pond.
During the months of August and October, the pH of the wastewaters ranged from 6.6 to 8.8, with a median value of 7.7 (see Table S1 in the supplemental material). The total solids content ranged from a very low value of 0.28 g liter−1 to as high as 57.3 g liter−1, with a median value of 8.2 g liter−1. The total Kjeldahl nitrogen value ranged from 0.001 to 1.5 g liter−1, with a median value of 0.24 g liter−1. The low values, which are much lower than anticipated for dairy wastewaters, were related to the fact that a few of the wastewater ponds were flooded with irrigation water. This practice was implemented at a few of the dairies as a means to blend the wastewaters prior to land application via pressurized spray irrigation. Because at some point more irrigation water than wastewater enters the pond, the pond takes on the chemical characteristics of the irrigation water.
Culture-dependent quantification of fecal indicators.
The fecal indicator organisms were quantified in the dairy wastewaters using a MPN (enterococci, total coliforms, and E. coli) or plate-counting (C. perfringens) technique (see Table S2 in the supplemental material). Median titer values for enterococci and C. perfringens were approximately 102 MPN or CFU ml−1, respectively, throughout the sampling period. Total coliform median values were 1 to 2 orders of magnitude greater, with a maximum titer of 107 MPN ml−1 occurring in August. As expected, the titer of generic E. coli was lower than that of total coliforms in all pond samples (data not shown). On average, the organism titer levels were greater in October than in June and August, except for the titers of total coliforms, which were greatest in August. In the United Kingdom, Hutchison and coworkers (21) found that E. coli O157, Campylobacter spp., and Listeria spp. were more prevalent in fresh cattle feces during the spring months and December. Additional studies have confirmed that the prevalences and levels of various zoonotic agents within fresh cattle manures were affected in a season-dependent manner (3, 50). While too few seasonal data points were collected for a statistical analysis, our results suggest that seasonal factors such as temperature or solar irradiation could have influenced the levels of fecal indicators in the dairy wastewaters. It should be noted that changes in the levels of fecal indicators in the wastewater ponds may not be seasonal in nature and could be related to the manure aging process or nutrient status of the pond. In general, it has been reported that levels of bacterial indicators and zoonotic agents in stored livestock wastes decline over time (22, 28, 34, 57). However, regrowth of indicator bacteria to some extent, due to the nutrient-rich conditions, has also been reported (13, 28).
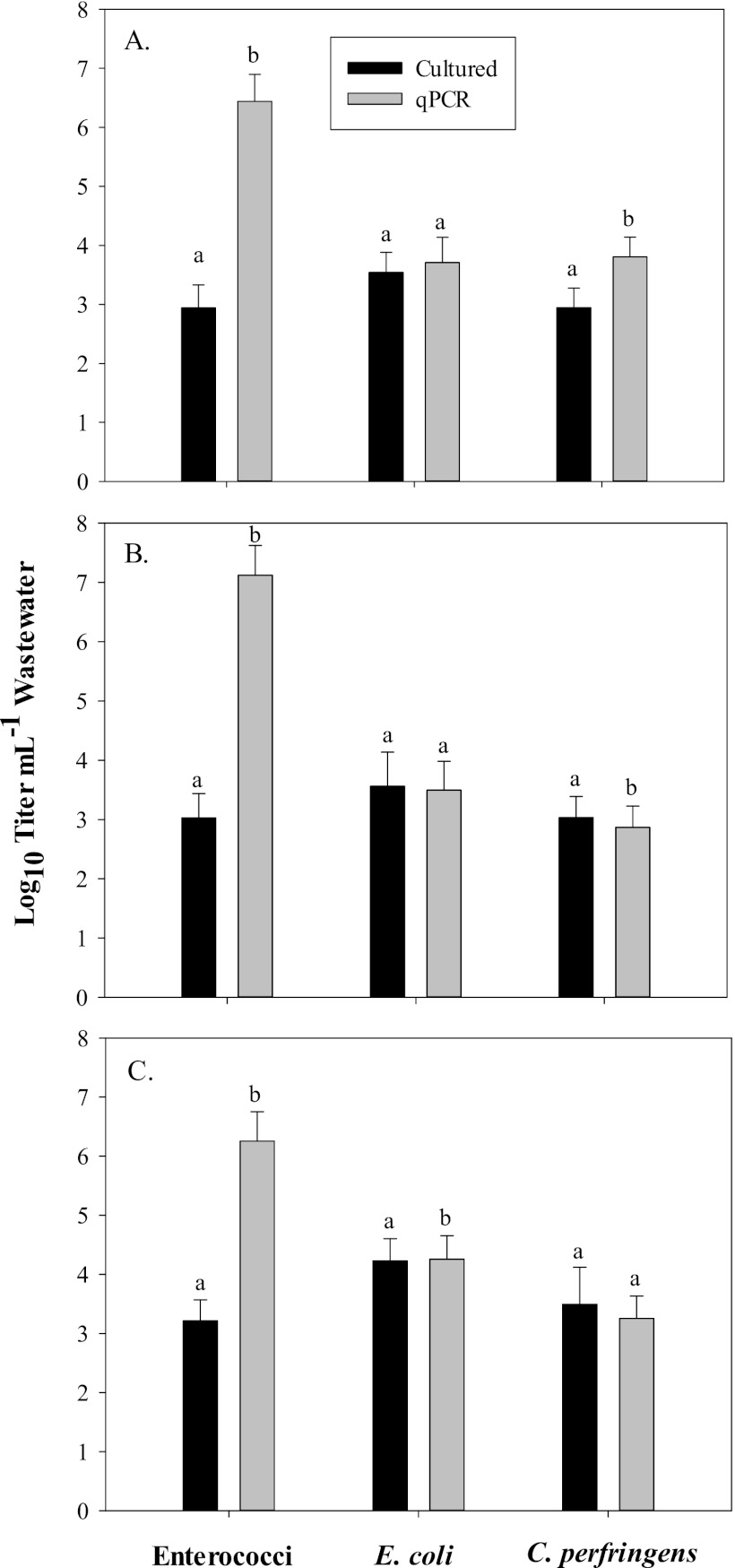
Quantification of fecal indicators by qPCR.
In addition to quantification by culture-dependent techniques, qPCR of bacterial DNA was used to calculate the titers of enterococci, E. coli, and C. perfringens in the dairy wastewaters. The qPCR results are presented in Fig. 1 along with the MPN and CFU values for comparison. During each of the 3 months in which samples were collected, the qPCR values for enterococci corresponded to 106 to 107 cells ml−1 wastewater. The qPCR method was specifically designed to detect E. faecalis, which is one of the most common enterococcus species found in the gastrointestinal tract of cattle (14, 32). qPCR values were about 3 logs greater than those determined by the MPN technique (a statistically significant difference; P < 0.0001), which indicated that the majority of enterococci might have gone into a VBNC state such as was seen in earlier studies involving bovine feces and manure (28, 30). However, since the DNA could also have been extracted from damaged or dead cells, qPCR analysis may have resulted in overestimation of actual titer values in the samples (7, 31, 37).
Fig 1.
Comparison of culture-dependent and qPCR estimates of numbers of enterococci, E. coli, and C. perfringens in the dairy wastewaters from (A) June, (B) August, and (C) October. Quantitative PCR was based on positive amplification of target genes using EC, Eco, and cpa primers for enterococci, E. coli, and C. perfringens, respectively. Columns represent means ± standard deviations (n = 30). Columns with different letters (a or b) indicate a significant difference between the two quantification techniques (P < 0.05).
In contrast to the results seen with enterococci, the average qPCR and culture-dependent values for both E. coli and C. perfringens were markedly but not statistically significantly similar in all cases (Fig. 1). In June and August, the qPCR and CFU values for C. perfringens were statistically significantly different (P < 0.0001), while the October titer values for E. coli were also different (P = 0.02). During the 3 months in which wastewater samples were collected, the titers of E. coli and C. perfringens (as determined by qPCR) ranged from about 103 to 104 cells ml−1.
Quantification of zoonotic pathogens by qPCR.
Prior to the enumeration of zoonotic bacterial pathogens in the dairy wastewaters, wastewater samples were spiked to determine the efficiency of the DNA extraction kit and precision of the qPCR technique. On average, recovery levels ranged from 56% for S. enterica to as high as 326% for C. jejuni (Table 2). Although these recovery levels are presumed to be representative of all wastewaters examined in this study, spiking was not performed on all of the wastewaters due to the logistics of conducting this task. The qPCR detection limits for the bacterial pathogens were determined to range from 16 to 1,229 cells ml−1 (Table 2). Due to the presence of multiple copies of the target gene within M. avium (i.e., 14 to 20 copies per genome), analysis performed using those sequences is more sensitive than analysis using the other targeted sequences (5), thus explaining the low detection limit of 16 cells ml−1.
Table 2.
Recovery and detection limits for bacterial pathogens in a dairy wastewater determined by qPCR after spiking
| Target organism | Wastewater titer range after spiking (log10 CFU ml−1 wastewater) | Range of recovery values (%) | Mean recovery (%)a | qPCR detection limit (no. of cells ml−1) |
|---|---|---|---|---|
| C. jejuni | 3.4–8.4 | 41–836 | 326 | 768 |
| E. coli | ||||
| stx1 | 3.2–8.2 | 38–120 | 72 | 1,229 |
| stx2 | 3.2–8.2 | 54–238 | 121 | 232 |
| eaeA | 2.2–8.2 | 41–117 | 75 | 140 |
| L. monocytogenes | 3.5–8.5 | 45–127 | 81 | 410 |
| M. avium subsp. paratuberculosis | 1.5–6.5 | 25–52 | 37 | 16 |
| S. enterica | 3.3–8.3 | 13–133 | 56 | 325 |
| Y. pseudotuberculosis | 3.2–8.2 | 126–179 | 160 | 234 |
Data represent mean values as determined by spiking triplicate wastewater samples with pathogens at several titer levels.
The putative bacterial pathogens detected in the dairy wastewater by qPCR were C. jejuni, E. coli (stx1 and eaeA positive), L. monocytogenes, M. avium, and S. enterica (Table 3). All of these organisms were detected at least twice in samples from June, but L. monocytogenes and S. enterica were not detected in the August and October samples. The organisms with the greatest number of positive detections were C. jejuni and M. avium, with 21 and 29 of 30 ponds showing the presence of those species in samples collected in June, respectively. While the greatest number of detections for all organisms occurred in that first month of analysis, the number of positive pond results generally decreased during each subsequent month. The decrease in the number of detections may have been affected by the pathogen-shedding rate of the cattle, environmental variables (e.g., solar irradiation, temperature), pond characteristics (e.g., solids content), or competition from other indigenous microorganisms leading to a reduction of pathogen survival in the ponds (29, 41, 42). The fact that Leptospira spp. were not detected in the ponds could be related to the performance of the DNA extraction, as spiking studies were not conducted with these organisms. However, sufficient recovery levels of these organisms in cattle manure have been reported from studies using similar DNA isolation techniques (26), making it more likely that they were present at levels below the detection limits. Yersinia pseudotuberculosis was also not detected, but, based on the spiking results, it can be expected that the titer in the ponds was below the detection limit of 234 cells ml−1.
Table 3.
Enumeration of pathogens in dairy wastewater samples as determined by qPCRa
| Organism | June |
August |
October |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of ponds | No. of cells (ml−1) |
No. of ponds | No. of cells (ml−1) |
No. of ponds | No. of cells (ml−1) |
|||||||
| Min | Max | Mean | Min | Max | Mean | Min | Max | Mean | ||||
| C. jejuni | 21 | <7.7 × 102 | 2.7 × 104 | 4.8 × 103 | 17 | <7.7 × 102 | 1.8 × 104 | 2.5 × 103 | 13 | <7.7 × 102 | 2.5 × 104 | 3.4 × 103 |
| E. coli | ||||||||||||
| stx1 | 13 | <1.2 × 103 | 6.3 × 103 | 2.0 × 103 | 2 | 3.6 × 103 | 9.9 × 103 | 6.8 × 103 | 5 | <1.2 × 103 | 2.7 × 103 | 1.0 × 103 |
| eaeA | 10 | 1.5 × 102 | 6.3 × 103 | 2.0 × 103 | 2 | 2.4 × 103 | 4.2 × 103 | 3.3 × 103 | 1 | 1.4 × 102 | 1.4 × 102 | 1.4 × 102 |
| L. monocytogenes | 2 | 1.4 × 103 | 6.6 × 103 | 4.0 × 103 | 0 | <4.1 × 102 | 0 | <4.1 × 102 | ||||
| M. avium | 29 | <1.6 × 101 | 7.1 × 104 | 2.9 × 103 | 24 | <1.6 × 101 | 1.3 × 103 | 1.5 × 102 | 22 | <1.6 × 101 | 1.4 × 103 | 2.2 × 102 |
| S. enterica | 5 | 3.0 × 103 | 8.3 × 104 | 2.1 × 104 | 0 | <3.2 × 102 | 0 | <3.2 × 102 | ||||
No sample had detectable L. interrogans, Y. pseudotuberculosis, Cryptosporidium spp., Giardia spp., or stx2 gene of E. coli O157:H7 during any month. No. of ponds, number of wastewater ponds containing the indicated pathogen (n = 30 ponds). Min, minimum; Max, maximum.
To determine the effect of solids on the recovery and detection of pathogens, wastewaters with low and high total solids contents were spiked with G. lamblia cysts and C. parvum oocysts (Table 4). Regardless of the solids content, the mean levels of recovery of cysts from the low and high wastewater totals, at 15% and 19%, respectively, were very similar. In contrast, the recovery of oocysts from the high total content of wastewater solids was 210%, which is 3.4 times higher than in the low total content. Although this test was performed with only two pathogens, these results suggest that recovery of organisms might be slightly enhanced in some wastewaters with a higher solids content. Despite our efforts to quantify Giardia and Cryptosporidium spp. in the dairy wastewaters, no pond samples were found to contain them above their average detection limits of 156 cysts and 1,587 oocysts ml−1. Our results, however, do not exclude the possible presence of Giardia and Cryptosporidium spp. in the wastewaters, since they are generally found at relatively low concentrations in cattle feces (20, 21).
Table 4.
Recovery of and detection limits for Giardia and Cryptosporidium spp. in dairy wastewaters as determined by qPCR with low and high total solids
| Target organism | Total solids (g liter−1) | Wastewater titer range after spiking (log10 cysts/oocysts ml−1 wastewater) | Range of recovery values (%) | Mean recovery (%)a | qPCR detection limit (log10 cysts/oocysts ml−1) |
|---|---|---|---|---|---|
| Giardia spp. | 3.6 | 3–6 | 8–20 | 15 | 130 |
| 12.7 | 15–25 | 19 | 182 | ||
| Cryptosporidium spp. | 3.6 | 3–6 | 22–110 | 61 | 1,485 |
| 12.7 | 71–426 | 210 | 1,689 |
Data represent mean values as determined by spiking triplicate wastewater samples with pathogens at several titer levels.
The bacterial pathogen titers were generally consistent, with mean values of between 102 and 104 cells ml−1 (Table 3). The organisms detected at the highest titer levels were M. avium, S. enterica, and C. jejuni at 104 cells ml−1 in June, while C. jejuni was also detected at titer values of up to 104 cells ml−1 in August and October. The mean titer value for L. monocytogenes and stx1- and eaeA-positive E. coli was 103 cells ml−1, except in October, where the titer for eaeA-positive E. coli was lower by 1 order of magnitude. The fact that the stx1 and eaeA gene markers were detected suggests that some of the wastewaters were presumptive for enterohemorrhagic or enteropathogenic E. coli. While most studies do report the presence of virulence genes encoding Shiga toxin 2 in dairy manures (4, 11), the stx2 gene was not detected in the wastewater ponds. Although our qPCR detection limit for stx2 was 232 cells ml−1, the lack of detections suggests that our methodology may not have been sensitive enough, as the majority of Shiga toxin-producing E. coli isolates from dairy cattle are known to possess both stx1 and stx2 genes (24).
Concluding remarks.
Based on our results, the oligonucleotide primers and probes used in this study appeared to be suitable for quantitation of zoonotic bacterial pathogens in dairy wastewaters. While some of the key pathogens in the wastewaters were below their qPCR detection limits, this study successfully detected and quantified bacterial pathogens of public health concern at levels similar to those reported in the literature in related environments. In addition, the qPCR and culture-dependent results were in general agreement, further suggesting the suitability of using qPCR as an alternative to laborious culture-dependent techniques when analyzing dairy and (potentially) other livestock wastewaters. Once more, qPCR has turned out to be a reliable method to quantify pathogens in materials that are often difficult to analyze. Despite the fact that some of the values tended to be more conservative due to a possible codetection of irreversibly damaged or dead organisms, this approach allows the detection of putative pathogens in a dormant state which would not be detectable using culture techniques alone.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the efforts of Sheryl Verwey in conducting the laboratory analyses. We also thank the anonymous individuals who spent considerable amounts of time reviewing our manuscript and providing many helpful suggestions.
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA.
Footnotes
Published ahead of print 14 September 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Armon R, Payment P. 1988. A modified m-CP medium for enumerating Clostridium perfringens from water samples. Can. J. Microbiol. 34: 78–79 [DOI] [PubMed] [Google Scholar]
- 2. Budnick GE, Howard RT, Mayo DR. 1996. Evaluation of Enterolert for enumeration of enterococci in recreational waters. Appl. Environ. Microbiol. 62: 3881–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chapman PA, Siddons CA, Cerman Malo AT, Harkin MA. 1997. A 1-year study of Escherichia coli O157 in cattle, sheep, pigs and poultry. Epidemiol. Infect. 119: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cobbold RN, Rice DH, Szymanski M, Call DR, Hancock DD. 2004. Comparison of shiga-toxigenic Escherichia coli prevalences among dairy, feedlot, and cow-calf herds in Washington state. Appl. Environ. Microbiol. 70: 4375–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cook KL, Britt JS. 2007. Optimization of methods for detecting Mycobacterium avium subsp. paratuberculosis in environmental samples using quantitative, real-time PCR. J. Microbiol. Methods 69: 154–160 [DOI] [PubMed] [Google Scholar]
- 6. Cook KL, Bolster CH, Ayers KA, Reynolds DN. 2011. Escherichia coli diversity in livestock manures and agriculturally impacted stream waters. Curr. Microbiol. 63: 439–449 [DOI] [PubMed] [Google Scholar]
- 7. Dungan RS, Leytem AB. 2009. Qualitative and quantitative methodologies for determination of airborne microorganisms at concentrated animal-feeding operations. W. J. Microbiol. Biotechnol. 25: 1505–1518 [Google Scholar]
- 8. Dungan RS. 2010. Board-Invited Review: Fate and transport of bioaerosols associated with livestock operations and manures. J. Anim. Sci. 88: 3693–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dungan RS. 2012. Use of a culture-independent approach to characterize aerosolized bacteria near an open-freestall dairy operation. Environ. Int. 41: 8–14 [DOI] [PubMed] [Google Scholar]
- 10. Eckner KF. 1998. Comparison of membrane filtration and multiple-tube fermentation by the Colilert and Enterolert methods for detection of waterborne coliform bacteria, Escherichia coli, and enterococci used in drinking and bathing water quality monitoring in southern Sweden. Appl. Environ. Microbiol. 64: 3079–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Franz E, Klerks MM, De Vos OJ, Termorshuizen AJ, van Bruggen AHC. 2007. Prevalence of shiga toxin-producing Escherichia coli stx1, stx2, eaeA, and rfbE genes and survival of E. coli O157:H7 in manure from organic and low-input conventional dairy farms. Appl. Environ. Microbiol. 73: 2180–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gale P. 2005. Land application of treated sewage sludge: quantifying pathogen risks from consumption of crops. J. Appl. Microbiol. 98: 380–396 [DOI] [PubMed] [Google Scholar]
- 13. Gantzer C, et al. 2001. Monitoring of bacterial and parasitological contamination during various treatment of sludge. Water Res. 35: 3763–3770 [DOI] [PubMed] [Google Scholar]
- 14. Gilmore MS. 2002. The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC [Google Scholar]
- 15. Gurjar AA, Hegde NV, Love BC, Jayarao BM. 2008. Real-time multiplex PCR assay for rapid detection and toxintyping of Clostridium perfringens toxin producing strains in feces of dairy cattle. Mol. Cell. Probes 22: 90–95 [DOI] [PubMed] [Google Scholar]
- 16. Guy RA, Payment P, Krull UJ, Horgen PA. 2003. Real-time PCR for quantification of Giardia and Cryptosporidium in environmental water samples and sewage. Appl. Environ. Microbiol. 69: 5178–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hardy R, Schilling K, Fromm J, Dai X, Cook M. 2006. Technical background document: microbial risk assessment and fate and transport modeling of aerosolized microorganisms at wastewater land application facilities in Idaho. Idaho Department of Environmental Quality, Boise, ID: http://www.deq.idaho.gov/media/529643-microbial_risk_assessment.pdf Accessed 24 September 2012 [Google Scholar]
- 18. He JW, Jiang S. 2005. Quantification of enterococci and human adenovirus in environmental samples by real-time PCR. Appl. Environ. Microbiol. 71: 2250–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hein I, Flekna G, Krassnig M, Wagner M. 2006. Real-time PCR for the detection of Salmonella spp. in food: An alternative approach to a conventional PCR system suggested by the FOOD-PCR project. J. Microbiol. Methods 66: 538–547 [DOI] [PubMed] [Google Scholar]
- 20. Hutchison ML, Walters LD, Avery SM, Synge BA, Moore A. 2004. Levels of zoonotic agents in British livestock manures. Lett. Appl. Microbiol. 39: 207–214 [DOI] [PubMed] [Google Scholar]
- 21. Hutchison ML, Walters LD, Avery SM, Munro F, Moore A. 2005. Analyses of livestock production, waste storage, and pathogen levels and prevalences in farm manures. Appl. Environ. Microbiol. 71: 1231–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hutchison ML, Wlaters LD, Moore A, Avery SM. 2005. Declines of zoonotic agents in liquid livestock wastes stored in batches on-farm. J. Appl. Microbiol. 99: 58–65 [DOI] [PubMed] [Google Scholar]
- 23. Hutchison ML, Avery SM, Monaghan JM. 2008. The air-borne distribution of zoonotic agents from livestock waste spreading and microbiological risk to fresh produce from contaminated irrigation sources. J. Appl. Microbiol. 105: 848–857 [DOI] [PubMed] [Google Scholar]
- 24. Irino K, et al. 2005. Serotypes and virulence markers of shiga toxin-producing Escherichia coli (STEC) isolated from dairy cattle in São Paulo State, Brazil. Vet. Microbiol. 105: 29–36 [DOI] [PubMed] [Google Scholar]
- 25. Kim SG, et al. 2002. Development and application of quantitative polymerase chain reaction assays based on the ABI 7700 system (TaqMan) for detection and quantification of Mycobacterium avium subsp. paratuberculosis. J. Vet. Diagn. Invest. 14: 126–131 [DOI] [PubMed] [Google Scholar]
- 26. Klein M, et al. 2010. Diversity and abundance of zoonotic pathogens and indicators in manures of feedlot cattle in Australia. Appl. Environ. Microbiol. 76: 6947–6950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Klein M, et al. 2010. Monitoring bacterial indicators and pathogens in cattle feedlot waste by real-time PCR. Water Res. 44: 1381–1388 [DOI] [PubMed] [Google Scholar]
- 28. Klein M, Brown L, Ashbolt NJ, Stuetz RM, Roser DJ. 2011. Inactivation of indicators and pathogens in cattle feedlot manures and compost as determined by molecular and culture assays. FEMS Microbiol. Ecol. 77: 200–210 [DOI] [PubMed] [Google Scholar]
- 29. Kudva IT, Blanch K, Hovde CJ. 1998. Analysis of Escherichia coli O157:H7 survival in ovine or bovine and manure slurry. Appl. Environ. Microbiol. 64: 3166–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lebuhn M, Effenberger M, Gronauer A, Wilderer PA, Wuertz S. 2003. Using quantitative real-time PCR to determine the hygienic status of cattle manure. Water Sci. Technol. 48: 97–103 [PubMed] [Google Scholar]
- 31. Lleo MM, et al. 2005. Molecular vs culture methods for the detection of bacterial faecal indicators in groundwater for human use. Lett. Appl. Microbiol. 40: 289–294 [DOI] [PubMed] [Google Scholar]
- 32. Mete E, Kaleli I. 2006. Species distribution and antibiotic resistance of enterococci isolated from cattle farmers and cattles. Mikrobiyol. Bul. 40: 75–80. (In Turkish.) [PubMed] [Google Scholar]
- 33. National Agricultural Statistics Service 2009. 2007 census of agriculture. USDA-NASS, Washington, DC: http://www.agcensus.usda.gov/Publications/2007/Full_Report/index.asp Accessed 24 September 2012 [Google Scholar]
- 34. Nicholson FA, Groves SJ, Chambers BJ. 2005. Pathogen survival during livestock manure storage and following land application. Bioresour. Technol. 96: 135–143 [DOI] [PubMed] [Google Scholar]
- 35. Nogva HK, Rudi K, Naterstad K, Holck A, Lillihaug D. 2000. Application of 5′-nuclease PCR for quantitative detection of Listeria monocytogenes in pure cultures, water, skim milk, and unpasteurized whole milk. Appl. Environ. Microbiol. 66: 4266–4271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oliver DM, Clegg CD, Haygarth PM, Heathwaite AL. 2005. Assessing the potential for pathogen transfer from grassland soils to surface waters. Ad. Agron. 85: 125–180 [Google Scholar]
- 37. Oliver JD. 2010. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol. Rev. 34: 415–425 [DOI] [PubMed] [Google Scholar]
- 38. Pell AN. 1997. Manure and microbes: public and animal health problem? J. Dairy Sci. 80: 2673–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pelzer KD, Currin N. 2007. Zoonotic diseases of cattle. Virginia Cooperative Extension, publication 400-460. Virginia Tech, Blacksburg, VA: http://pubs.ext.vt.edu/400/400-460/400-460.html Accessed 24 September 2012 [Google Scholar]
- 40. Raizman EA, et al. 2004. The distribution of Mycobacterium avium ssp. paratuberculosis in the environment surrounding Minnesota dairy farms. J. Dairy Sci. 87: 2959–2966 [DOI] [PubMed] [Google Scholar]
- 41. Ravva SV, Sarreal CZ, Duffy B, Stanker LH. 2006. Survival of Escherichia coli O157:H7 in wastewater from dairy lagoons. J. Appl. Microbiol. 101: 891–902 [DOI] [PubMed] [Google Scholar]
- 42. Ravva SV, Korn A. 2007. Extractable organic components and nutrients in wastewater from dairy lagoons influence the growth and survival of Escherichia coli O157:H7. Appl. Environ. Microbiol. 73: 2191–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rico JL, Garcia H, Rico C, Tejero I. 2007. Characterisation of solid and liquid fractions of dairy manure with regard to their component distribution and methane production. Bioresour. Technol. 98: 971–979 [DOI] [PubMed] [Google Scholar]
- 44. Salgado M, et al. 2011. Fate of Mycobacterium avium subsp. paratuberculosis after application of contaminated dairy cattle manure in agricultural soils. Appl. Environ. Microbiol. 77: 2122–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. SAS Institute 2008. SAS/STAT 9.2 user's guide. SAS Institute, Cary, NC [Google Scholar]
- 46. Shannon KE, Lee DY, Trevors JT, Beaudette LA. 2007. Application of real-time quantitative PCR for the detection of selected bacterial pathogens during municipal wastewater treatment. Sci. Total Environ. 382: 121–129 [DOI] [PubMed] [Google Scholar]
- 47. Sharma VK, Dean-Nystrom EA. 2003. Detection of enterohemorrhagic Escherichia coli O157:H7 by using a multiplex real-time PCR assay for genes encoding intimin and Shiga toxins. Vet. Microbiol. 93: 247–260 [DOI] [PubMed] [Google Scholar]
- 48. Slana I, Pribylova R, Kralova A, Pavlik I. 2011. Persistence of Mycobacterium avium subsp. paratuberculosis at a farm-scale biogas plant supplied with manure from paratuberculosis-affected dairy cattle. Appl. Environ. Microbiol. 77: 3115–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Solomon EB, Yaron S, Matthews KR. 2002. Transmission of Escherichia coli O157:H7 from contaminated manure and irrigation water to lettuce plant tissue and its subsequent internalization. Appl. Environ. Microbiol. 68: 397–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stanley KN, Wallace JS, Currie JE, Diggle PJ, Jones K. 1998. The seasonal variation of thermophilic campylobacters in beef cattle, dairy cattle and calves. J. Appl. Microbiol. 85: 472–480 [DOI] [PubMed] [Google Scholar]
- 51. Stoddard RA, Gee JE, Wilkins PP, McCaustland K, Hoffmaster AR. 2009. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn. Microbiol. Infect. Dis. 64: 247–255 [DOI] [PubMed] [Google Scholar]
- 52. Thoerner P, et al. 2003. PCR detection of virulence genes in Yersinia enterocolitica and Yersinia pseudotuberculosis and investigation of virulence gene distribution. Appl. Environ. Microbiol. 69: 1810–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tyrrel SF, Quinton JN. 2003. Overland flow transport of pathogens from agricultural land receiving faecal wastes. J. Appl. Microbiol. 94: 87S–93S [DOI] [PubMed] [Google Scholar]
- 54. Unc A, Goss MJ. 2004. Transport of bacteria from manure and protection of water resources. Appl. Soil Ecol. 25: 1–18 [Google Scholar]
- 55. Venglovsky J, Saskova N, Placha I. 2009. Pathogens and antibiotic residues in animal manures and hygienic and ecological risks related to subsequent land application. Bioresour. Technol. 100: 5386–5391 [DOI] [PubMed] [Google Scholar]
- 56. Viau E, Bibby K, Paez-Rubio T, Peccia J. 2011. Toward a consensus view on the infectious risks associated with land application of sewage sludge. Environ. Sci. Technol. 45: 5459–5469 [DOI] [PubMed] [Google Scholar]
- 57. Wang L, Mankin KR, Marchin GL. 2004. Survival of fecal bacteria in dairy cow manure. Trans. ASAE 47: 1239–1246 [Google Scholar]
- 58. Yang C, Jiang Y, Huang K, Changqing Z, Yin Y. 2003. Application of real-time PCR for quantitative detection of Campylobacter jejuni in poultry, milk and environmental water. FEMS Immunol. Med. Microbiol. 38: 265–271 [DOI] [PubMed] [Google Scholar]
- 59. Zhang G, Brown EW, González-Escalona N. 2011. Comparison of real-time PCR, reverse transcriptase real-time PCR, loop-mediated isothermal amplification, and the FDA conventional microbiological method for the detection of Salmonella spp. in produce. Appl. Environ. Microbiol. 77: 6495–6501 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.