Abstract
The efficiency of nitrogen use is a key determinant of the completion of alcoholic fermentation. We analyzed the kinetics of consumption of 18 nitrogen compounds by 14 Saccharomyces cerevisiae strains of various origins in a synthetic medium that mimicked a grape must. The kinetic profiles of total nitrogen consumption were diverse, but the order of nitrogen source consumption was similar for all strains. The nitrogen compounds could be classified into three groups, according to their order of use: prematurely consumed (Lys), early consumed (Asp, Thr, Glu, Leu, His, Met, Ile, Ser, Gln, and Phe), and late consumed (ammonium, Val, Arg, Ala, Trp, and Tyr). The initial concentrations of these compounds did not alter the order in which they were consumed, except for arginine and ammonium. Early consumed amino acids are transported by specific permeases under Ssy1p-Ptr3p-Ssy5 (SPS)-mediated control that are expressed at the beginning of consumption. Most nitrogen compounds consumed late are transported by permeases under nitrogen catabolite repression (NCR), and others (Val, Trp, and Tyr) are transported by SPS-regulated low-affinity permeases. Therefore, the kinetic characteristics of transporters, as well as SPS and NCR, are likely key factors controlling the temporal sequence of consumption of nitrogen compounds and constitute a system highly conserved in S. cerevisiae species. This work sheds new light on the mechanistic basis of the sequential use of different nitrogen compounds in complex environments.
INTRODUCTION
Nitrogen availability is important for winemaking: it regulates the formation of yeast biomass and, in turn, the fermentation rate and the time to completion of fermentation (6, 7). Stuck or sluggish fermentation is often related to insufficient nitrogen in grape juice (8, 9). The nitrogen status of the must may also affect the production by Saccharomyces cerevisiae of many volatile compounds that contribute to wine flavor, as some amino acids are direct metabolic precursors for the synthesis of higher alcohols, short- to medium-chain fatty acids, and their ethyl ester or acetate ester derivatives (20, 48). The activities of metabolic routes involved in the production of glycerol and organic acids have also been shown to be dependent on the nitrogen source and concentration (1, 13, 42).
A wide range of compounds in grape juice contain nitrogen. Many factors have substantial effects on the quantitative and qualitative nitrogen content of musts, including grape variety and maturity, environmental features (such as soil fertility and climatic conditions), and viticultural practices (grape harvesting techniques) (3, 12). Nevertheless, ammonium ions and amino acids constitute on average 40% and 51 to 92%, respectively, of the yeast assimilable nitrogen (YAN) in juice during wine fermentation (3).
A wide variety of nitrogen-containing compounds, including ammonium, amino acids, and di- and tripeptides (11), can be assimilated by S. cerevisiae and provide the pools of polyamines, amino acids, nucleotide bases, and their derivatives that are required for the production of cell biomass. However, these nitrogen sources do not all support growth equally. Glutamine, glutamate, asparagine, and ammonium when the sole nitrogen source each allow a high specific growth rate and are considered to be good or preferred nitrogen sources. Conversely, S. cerevisiae grows slowly on proline, allantoin, or urea, typical poor or nonpreferred nitrogen sources. Yeast preferentially uses substrates that allow the best growth through a regulation mechanism called nitrogen catabolite repression (NCR); this mechanism operates at various levels to prevent the uptake of the poorest nitrogen sources if better sources are available (15, 33, 49). NCR involves the transcriptional repression of some genes encoding permeases, mediated by the four GATA-binding transcription factors encoded by GLN3, GAT1, DAL80, and GZF3 (16, 19, 33); it also involves selective inactivation by dephosphorylation of the corresponding products and their subsequent degradation (21, 47). The general amino acid carriers with broad substrate ranges, Gap1p (24) and Agp1p (23, 45), as well as the specific proline transporter Put4p (25) and the ammonium permeases (Mep1p, Mep2p, and Mep3p) (35, 36), are under NCR control.
Additional regulatory mechanisms involving the plasma membrane Ssy1-Ptr3-Ssy5 (SPS) sensor (18, 32) regulate the expression of genes encoding other specific permeases, according to the availability of the corresponding amino acids. The permeases involved include the branched-chain amino acid permeases encoded by BAP2 and BAP3, the high-affinity glutamine transporter Gnp1p, the tyrosine and tryptophan carriers Tat1p and Tat2p, the dicarboxylic amino acid permease Dip5p, and the high-affinity methionine permease Mup1p, and all are specifically induced in response to the presence of their substrate in the medium (22). These permeases are also able to catalyze with lower efficiency the import of several other amino acids (44).
The mechanisms favoring the uptake of one nitrogen source over another have been extensively investigated. However, few studies have explored the assimilation of nitrogen compounds when present as a complex mixture of ammonium and amino acids, as found in grape juice. These substrates are generally classified into four groups based on the time and the percentage of removal from the medium during industrial (brewing or wine) fermentations (7, 26, 27, 39, 40, 43). Nitrogen compounds of the first group are consumed almost totally during the first hours of the fermentation; those of the second group are removed gradually throughout the process; those of the third group are only used after depletion of the compounds of the first group. Proline, which is not assimilated under anaerobic conditions (10), constitutes the fourth group.
However, the sequence of assimilation of ammonium and amino acids has not been clearly established, and the assignment of some compounds, notably arginine, phenylalanine, tryptophan, tyrosine, and valine, differs between classifications. These discrepancies may be due to the use of different classification criteria (for example, time required to reach the half-concentration or the initial removal rate), of industrial media with diverse nitrogen source composition (wort/must), and of different yeast strains (brewing and wine yeast strains). Overall, very little information is available about the mechanistic basis, and particularly the underlying regulatory mechanisms and influence of amino acid concentrations, of the sequential assimilation of nitrogen compounds from complex media.
Nitrogen assimilation is important for yeast growth and completion of fermentation. A comprehensive exploration of assimilation of complex nitrogen resources during winemaking and the influence of the strain genetic background on nitrogen consumption would therefore be valuable.
We investigated the diversity of nitrogen assimilation from complex nitrogen compounds (ammonium and amino acids) by S. cerevisiae during fermentation and studied the mechanisms controlling the sequence of assimilation. We compared the consumption profiles of YAN and individual nitrogen substrates by a collection of S. cerevisiae strains from industrial and natural sources during wine fermentation. We analyzed these data according to both the availability of nitrogen compounds and the regulation of the permeases involved in their uptake.
MATERIALS AND METHODS
Yeast strains.
The Saccharomyces cerevisiae strains used in this study, listed in Table 1, were obtained from the Sanger Institute (eight strains) and from other laboratories (one strain) and companies (five strains). They were collected from ecologically diverse environments, including industrial processes, laboratories, and natural environments. All the strains were prototrophs and were selected (i) to cover a large genomic diversity of the Saccharomyces cerevisiae species (31) and (ii) on the basis of their different yields of biomass production from nitrogen consumed under enological conditions (14). For each strain, an aliquot of a reference stock, conserved at −80°C, was transferred to a yeast extract-peptone-dextrose (YPD) agar plate (1% Bacto yeast extract, 2% Bacto peptone, 2% glucose, 1.5% agar) 48 h before fermentation.
Table 1.
Origins and sources of S. cerevisiae strains studied
| Environment | Strain | Geographical origin | Collection |
|---|---|---|---|
| Industrial processes | |||
| Baker | YS2 | Australia | Sanger |
| Beer | NCYC361 | Ireland | Sanger |
| Palm wine | NCYC110 | Nigeria, West Africa | Sanger |
| Wine (commercial) | EC1118 | France | Lalvin |
| L2226 | France | Enoferm | |
| WE372 | South Africa | Anchor | |
| VL1 | France | Laffort | |
| ECA5 | France | Lallemand | |
| Laboratory | Y55 | France | Sanger |
| Natural environment | |||
| Bertam Palm | UWOPS05-227.2 | Malaysia | Sanger |
| Oak | YPS128 | Pennsylvania, USA | Sanger |
| YPS1009 | New Jersey, USA | Washington | |
| Vineyard | BC187 | Napa Valley, CA, USA | Sanger |
| L-1528 | Chile | Sanger |
Fermentation conditions.
Initial cultures in YPD medium were grown in 10-ml flasks at 28°C, with shaking (150 rpm), for 12 h and were then transferred to 10-ml flasks containing synthetic medium (SM) at an optical density at 600 nm (OD600) of 0.5, which mimics a grape must, and grown for 12 h at 28°C with shaking (150 rpm). These precultures were used to inoculate fermentations (OD600, 0.01) in SM containing 240 g liter−1 glucose, 6 g liter−1 malic acid, 6 g liter−1 citric acid, and 200 mg liter−1 nitrogen at pH 3.5, as well as vitamins and oligoelements found in grape juice (6). Ergosterol (1.875 mg liter−1), oleic acid (0.625 mg liter−1), and Tween 80 (0.05 g liter−1) were provided as anaerobic growth factors. The fermentations were performed in 0.33-liter fermentors equipped with fermentation locks to maintain anaerobiosis at 28°C, with continuous magnetic stirring (150 rpm).
Nitrogen (200 mg N liter−1) was supplied as one of various mixtures of amino acids along with NH4Cl (Table 2). The reference medium, SM200, simulated the composition of a typical grape juice; in SM200E medium, all the nitrogen compounds were included at equivalent amounts of nitrogen (10.53 mg N liter−1 for each compound). In SM200EM, the concentrations of arginine and ammonium were the same as those in SM200 and all the other amino acids were included at equivalent amounts of nitrogen (5.95 mg N liter−1). The relative distribution of nitrogen compounds in SM200M medium was the same as that of SM200, except that arginine and ammonium were included at low concentrations (equivalent to those in SM200E).
Table 2.
Initial concentrations of ammonium and amino acids in the media used in this study
| Nitrogen compound | Initial concn in medium (in various units) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SM200 |
SM200E |
SM200M |
SM200EM |
|||||||||
| mg/liter | mg N/liter | mmol/liter | mg/liter | mg N/liter | mmol/liter | mg/liter | mg N/liter | mmol/liter | mg/liter | mg N/liter | mmol/liter | |
| Lys | 0.6 | 0.1 | 0.00 | 54.9 | 10.5 | 0.38 | 1.1 | 0.2 | 0.01 | 31.5 | 6.0 | 0.22 |
| Tyr | 8.5 | 0.7 | 0.05 | 136.1 | 10.5 | 0.75 | 15.2 | 1.2 | 0.08 | 78.1 | 6.0 | 0.43 |
| Cys | 6.1 | 0.7 | 0.05 | 91.0 | 10.5 | 0.75 | 10.9 | 1.3 | 0.09 | 52.2 | 6.0 | 0.43 |
| Met | 14.6 | 1.4 | 0.10 | 112.0 | 10.5 | 0.75 | 26.1 | 2.5 | 0.18 | 64.4 | 6.1 | 0.43 |
| Phe | 17.7 | 1.5 | 0.11 | 124.1 | 10.5 | 0.75 | 31.5 | 2.7 | 0.19 | 71.1 | 6.0 | 0.43 |
| Gly | 8.5 | 1.6 | 0.11 | 56.4 | 10.5 | 0.75 | 15.2 | 2.8 | 0.20 | 32.2 | 6.0 | 0.43 |
| Ile | 15.3 | 1.6 | 0.12 | 98.5 | 10.5 | 0.75 | 27.2 | 2.9 | 0.21 | 56.4 | 6.0 | 0.43 |
| Asp | 20.7 | 2.2 | 0.16 | 100.0 | 10.5 | 0.75 | 36.9 | 3.9 | 0.28 | 57.4 | 6.0 | 0.43 |
| Leu | 22.6 | 2.4 | 0.17 | 98.5 | 10.5 | 0.75 | 40.2 | 4.3 | 0.31 | 56.4 | 6.0 | 0.43 |
| Val | 20.7 | 2.5 | 0.18 | 88.0 | 10.5 | 0.75 | 36.9 | 4.4 | 0.32 | 50.4 | 6.0 | 0.43 |
| His | 15.3 | 4.1 | 0.10 | 38.8 | 10.5 | 0.25 | 27.2 | 7.4 | 0.18 | 22.4 | 6.1 | 0.14 |
| Thr | 35.4 | 4.2 | 0.30 | 89.5 | 10.5 | 0.75 | 63.0 | 7.4 | 0.53 | 51.5 | 6.1 | 0.43 |
| Ser | 36.6 | 4.9 | 0.35 | 78.9 | 10.5 | 0.75 | 65.2 | 8.7 | 0.62 | 45.2 | 6.0 | 0.43 |
| Glu | 56.1 | 5.3 | 0.38 | 110.5 | 10.5 | 0.75 | 100.0 | 9.5 | 0.68 | 63.4 | 6.0 | 0.43 |
| Ala | 67.7 | 10.7 | 0.76 | 66.9 | 10.5 | 0.75 | 120.6 | 19.0 | 1.36 | 38.5 | 6.1 | 0.43 |
| Trp | 83.6 | 11.5 | 0.41 | 76.7 | 10.5 | 0.38 | 148.9 | 20.4 | 0.73 | 44.1 | 6.1 | 0.22 |
| Pro | 285.5 | 34.8 | 2.48 | 285.5 | 34.8 | 2.48 | 508.5 | 61.9 | 4.42 | 163.8 | 19.9 | 1.42 |
| Arg | 174.5 | 42.1 | 1.00 | 43.6 | 10.5 | 0.25 | 43.6 | 10.5 | 0.25 | 174.5 | 42.1 | 1.00 |
| Gln | 235.5 | 45.2 | 1.61 | 54.9 | 10.5 | 0.38 | 419.4 | 80.4 | 2.87 | 31.5 | 6.0 | 0.22 |
| NH4+ | 214.0 | 56.0 | 4.00 | 40.2 | 10.5 | 0.75 | 40.2 | 10.5 | 0.75 | 214.0 | 56.0 | 4.00 |
Analytic methods.
The population size was monitored by counting cells with an electronic particle counter (Multisizer 3 Coulter Counter; Beckman Coulter) fitted with a probe with a 100-mm aperture.
Residual ammonium ions in the supernatant (sample centrifugation, 3,000 × g, 4°C, 10 min) were assayed spectrophotometrically by using an Enzytec kit (catalog number 5380) according to the manufacturer's instructions.
Amino acids were quantified with a specific amino acid analyzer (Biochrom 30). Molecules with high molecular weights were precipitated by adding one volume of 25% (wt/vol) sulfosalicylic acid solution to 4 volumes of sample, and mixtures were incubated for 1 h at 4°C and centrifuged (4°C, 10 min, 3,000 × g). The sample was then filtered through a 0.22-μm-pore-size Millipore nitrocellulose membrane. Amino acids were separated by liquid chromatography on an ion-exchange column (Ultrapac-8 lithium form; Amersham Pharmacia Biotech) and revealed by the ninhydrin reaction, followed by absorbance measurements at 570 nm, except for proline, which was detected by measuring the absorbance at 440 nm. Norleucine (0.5 mmol ml−1) was added to the samples and used as an internal standard (29).
Estimation of population dynamics parameters.
The change over time of the cell number was analyzed using a standard logistic model within the R software (version 2.14.2), which describes exponential growth followed by a stationary phase (41) based on the following equation:
where Xt is the population size (in cells ml−1) at time t (in min), PopM is the maximum population size (in cells ml−1), X0 is the initial population size (in cells ml−1), and rXmax is the maximum rate of increase of the population (in min−1). This model was fit to experimental data points (growth kinetics, independently determined twice, with 15 measurements for each experiment), allowing the parameters rXmax and PopM to be estimated. The function calculated the parameters rXmax and PopM that minimized the sums of squares of the differences between experimental and fitted data points. For each replicate and strain, an average of 15 experimental data points was used to estimate PopM and rXmax (46).
Estimation of the parameters of the dynamics of nitrogen consumption.
The dynamics of nitrogen consumption during fermentation were fit by using a sigmoid or altered Gompertz decay function, as previously described by Tronchoni et al. in 2009 (50), using R, version 2.14.2 (41), with the following equation:
where Nt is the residual nitrogen concentration (in mg N liter−1) at time t (in min), Nmin is the lowest residual nitrogen concentration when t tends to infinity (t → ∞), ΔN is the difference between the upper and lower residual nitrogen concentrations, rN is the rate of nitrogen consumption (in min−1), and T50N is the half-time nitrogen consumption. For each replicate and strain, about 10 experimental data points were used to estimate the four parameters, minimizing the sums of squares differences between experimental and calculated data points. The equations obtained were then used to calculate the time necessary to consume 5% of the initial nitrogen source, at the concentration present in must, corresponding to the lag phase before YAN consumption: (TLAG), the time necessary to consume 50 and 95% of the initial nitrogen source at the concentration present in must (T50N and T95N) and the maximum rate of nitrogen cosumption (rNmax).
Statisical analyses.
Statistical analyses were performed using R software, version 2.14.2 (41).
To obtain a general overview of the growth and nitrogen consumption trait data, a principal component analysis (PCA) was performed using the FactoMineR package (30) with the following parameters: PopM, rXmax, TLAG, rNmax, T50N, and T95N. Each variable was then tested using a one-way analysis of variance (ANOVA) with the strain as a factor to describe the diversity between the 14 strains at a P value threshold of 0.05 without any multiplicity adjustment. For each parameter, normality of residual distributions and homogeneity of variance were studied using standard diagnostic graphics; no violation of the assumptions was detected.
To determine the order of consumption of the nitrogen compounds, we calculated the ratio between the time to consumption of 50% of each available nitrogen source and the time required for the consumption of 50% of the total available nitrogen (T50NR). T50NR was then tested using a one-way ANOVA with nitrogen source as a factor to detect a global effect. As the effect was significant at a P value threshold of 0.05, all pairwise comparisons for factor nitrogen compounds were tested using Tukey's honestly significant difference (HSD) test.
Pairwise correlations between initial concentrations and rNmax, TLAG, T50N, and T95N of nitrogen substrates were studied using Spearman's rank correlations, as distributions were not Gaussian.
The initial concentration of nitrogen compounds had a large effect on the rNmax, so we normalized this parameter using the ratio between rNmax and the initial concentration of nitrogen compounds (rNmaxR). For nitrogen-variable rNmaxR, a cox box transformation was necessary to obtain a normal distribution. To describe the diversity of the 14 strains and 18 nitrogen compounds, this variable was then tested using a two-way ANOVA with strain, nitrogen source, and their interactions as factors, using a P value threshold of 0.05. Normality of residual distributions and homogeneity of variance were studied using standard diagnostic graphics; no violation of the assumptions was detected.
RESULTS
Growth and nitrogen consumption of 14 S. cerevisiae strains from diverse origins.
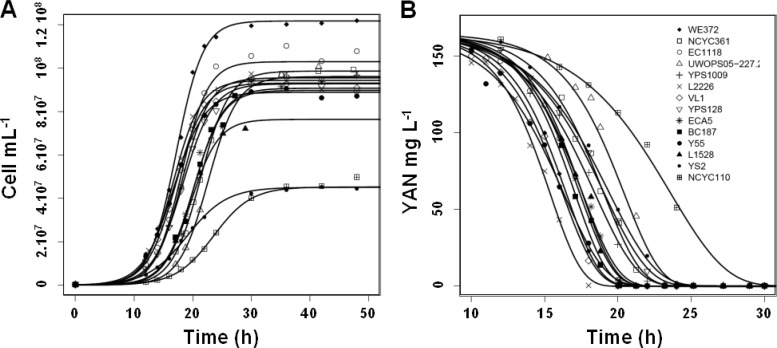
We first compared the growth and nitrogen assimilation profiles of 14 S. cerevisiae strains from a broad range of origins, including industrial processes (baking, beer, palm wine, and wine), natural environments, and laboratories (Table 1). All fermentations were carried out in duplicate, in a chemically defined medium (SM200) with a high glucose concentration (240 g liter−1) and 200 mg liter−1 of YAN (YAN is a mixture of 18 amino acids and ammonium ions at various concentrations [Table 2]). All the strains exhausted YAN during the growth phase (Fig. 1), consistent with nitrogen availability being the limiting factor for growth under these conditions. However, both the growth characteristics and the patterns of nitrogen consumption differed considerably between strains, with the maximum yeast population varying between 45 × 106 and 121 × 106 cells ml−1 and the time required for 95% nitrogen consumption varying from 17.5 to 27.9 h.
Fig 1.
Growth (A) and YAN consumption (B) during fermentation of 14 S. cerevisiae strains. Fermentations were carried out in duplicate on SM200 medium containing 240 g liter−1 glucose and 200 mg liter−1 YAN, consisting of a mixture of 18 amino acids and ammonium. For each strain, fitted data (dark lines) and experimental data are shown, as indicated in the symbol key in panel B. Mean values were calculated from 2 replicates.
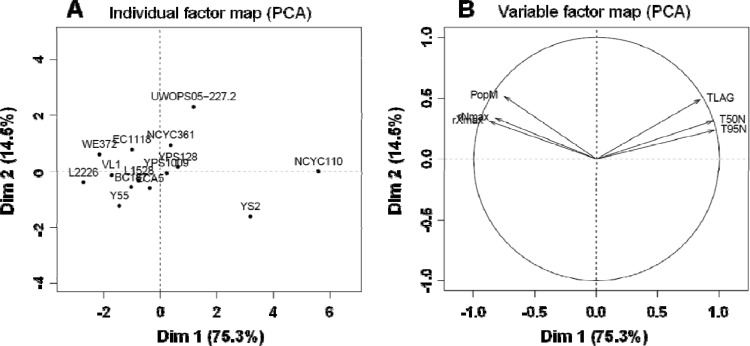
We compared six phenotypic traits related to growth dynamics (maximum population size, PopM, and maximal growth rate, rXmax) or distinction of nitrogen assimilation curves (duration of the lag phase before YAN consumption, TLAG; maximal rate of YAN consumption, rNmax; the time at which 50% of YAN was consumed, T50N; and the time at which 95% of YAN was consumed, T95N). All these measures differed substantially between yeast strains (Table 3); for example, the maximum rate of nitrogen consumption was between 16.8 and 27.8 mg N liter−1 h−1, depending on the strain. The contribution of each quantitative phenotypic trait to the differences between strains was investigated through PCA (Fig. 2). The first two axes accounted for 89.8% of the total variation and separated the strains mainly on the basis of rNmax, T50N, and T95N, which reflect the kinetic characteristics of the nitrogen consumption by yeast. The variables PopM and rXmax were notably correlated with variables describing nitrogen consumption (positively with rNmax and negatively with T50N and T95N). The strains exhibiting fast and early nitrogen assimilation, mostly those originating from industrial fermentative processes and including all the commercial wine strains, were also high biomass producers (Fig. 1 and 2). This suggests that the ability of a yeast to produce biomass depends on its nitrogen assimilation profile. Analysis of variance for each parameter confirmed that the strain effect was significant for growth variables (PopM, rXmax) as well as for variables relating to nitrogen consumption: TLAG, rNmax, T50N, and T95N (Table 3). For all phenotypic traits, more than 74% of the variance was explained by the strain variability (Table 3).
Table 3.
Growth and nitrogen consumption traits for the 14 S. cerevisiae strains during fermentation
| Strain | Mean ± SD value for traita |
|||||
|---|---|---|---|---|---|---|
| PopM (no. of cells/106 ml) | rXmax (no. of divisions/h) | rNmax (mg N/h) | TLAG (h−1) | T50N (h−1) | T95N (h−1) | |
| L2226 | 89 ± 3.1 | 0.44 ± 0.01 | 27.8 ± 0.438 | 8.7 ± 0.412 | 14.3 ± 0.377 | 17.5 ± 0.354 |
| YPS128 | 93 ± 5.7 | 0.39 ± 0.02 | 21.1 ± 1.124 | 10.5 ± 0.448 | 17.9 ± 0.165 | 22.0 ± 0.495 |
| VL1 | 93 ± 8.8 | 0.42 ± 0.02 | 25.8 ± 0.135 | 9.3 ± 0.012 | 15.4 ± 0.047 | 18.8 ± 0.059 |
| YS2 | 46 ± 2.3 | 0.27 ± 0.00 | 19.5 ± 0.426 | 11.7 ± 2.121 | 18.5 ± 0.354 | 22.4 ± 0.672 |
| NCYC110 | 45 ± 0.9 | 0.30 ± 0.03 | 16.8 ± 0.794 | 13.2 ± 1.037 | 22.6 ± 0.519 | 27.9 ± 0.200 |
| EC1118 | 105 ± 7.9 | 0.43 ± 0.02 | 24.3 ± 0.075 | 10.4 ± 0.024 | 16.7 ± 0.118 | 20.3 ± 0.200 |
| WE372 | 121 ± 9.5 | 0.44 ± 0.01 | 24.4 ± 0.079 | 9.2 ± 0.330 | 15.5 ± 0.082 | 19.1 ± 0.071 |
| YPS1009 | 95 ± 0.0 | 0.40 ± 0.00 | 21.1 ± 0.194 | 10.0 ± 0.000 | 17.4 ± 0.000 | 21.6 ± 0.000 |
| Y55 | 88 ± 6.2 | 0.42 ± 0.00 | 22.4 ± 0.887 | 8.2 ± 0.153 | 15.0 ± 0.354 | 18.9 ± 0.672 |
| L1528 | 74 | 0.46 | 23.1 | 9.6 | 16.5 | 20.4 |
| BC187 | 90 | 0.40 | 23.8 | 9.0 | 15.9 | 19.8 |
| NCYC361 | 102 | 0.40 | 22.4 | 11.2 | 18.1 | 22.0 |
| U227 | 96 | 0.40 | 25.6 | 13.3 | 19.7 | 23.3 |
| ECA5 | 91 | 0.40 | 21.4 | 9.3 | 16.5 | 20.5 |
| R2 (%)b | 93.0 | 91.6 | 96.1 | 74.3 | 98.4 | 98.0 |
| Pc | *** | *** | *** | ** | *** | *** |
Mean values and standard deviations were calculated from two replicates, except for strains L1528, BC187, NCYC 361, U227, and ECA5, for which only one experiment was conducted (and therefore data for these strains do not include standard deviations).
Adjusted R2 value.
**, significant at P < 0.01; ***, significant at P < 0.001.
Fig 2.
PCA of 14 strains for 6 phenotypic traits, based on mean values calculated from 2 replicates. (A) Individual factor map; (B) variable factor map. On the x axis is the percentage of variation explained by principal component 1 (Dim 1; 75.3%); on the y axis is the percentage of variation explained by principal component 2 (Dim 2; 14.5%).
Nitrogen source consumption. (i) Order for nitrogen source consumption.
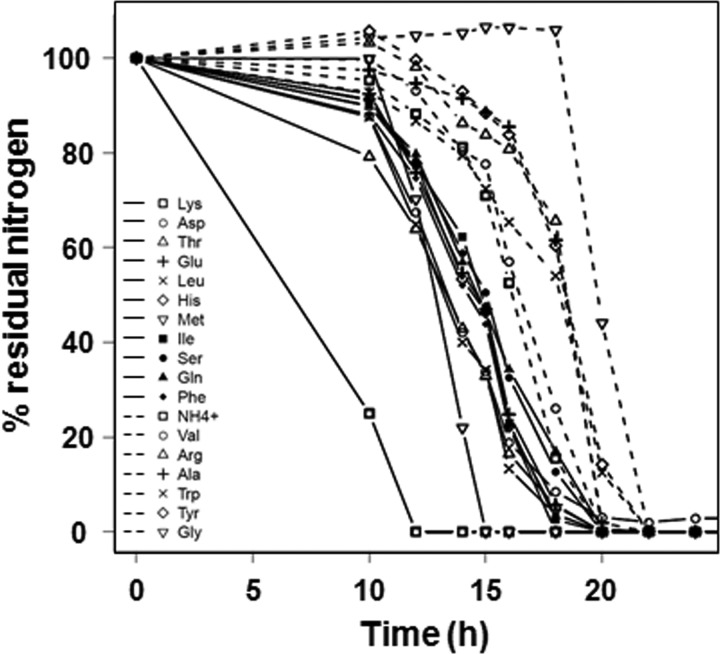
We then assessed the contribution of the 17 individual amino acids and of ammonium to the growth requirements during fermentation on synthetic medium containing 200 mg liter−1 YAN. Consumption profiles and variable characteristics of assimilation (rNmax, TLAG, T50N, and T95N) were determined for each nitrogen source and for each of the 14 strains (Fig. 3; see also Fig. S1 in the supplemental material). Nitrogen compounds were used sequentially throughout the growth phase. Substantial differences in their profiles of assimilation—rate, time, and duration of consumption—were observed. The times of consumption of the various sources of YAN were strain dependent. Nevertheless, all 14 strains displayed approximately the same order of assimilation of the nitrogen substrates.
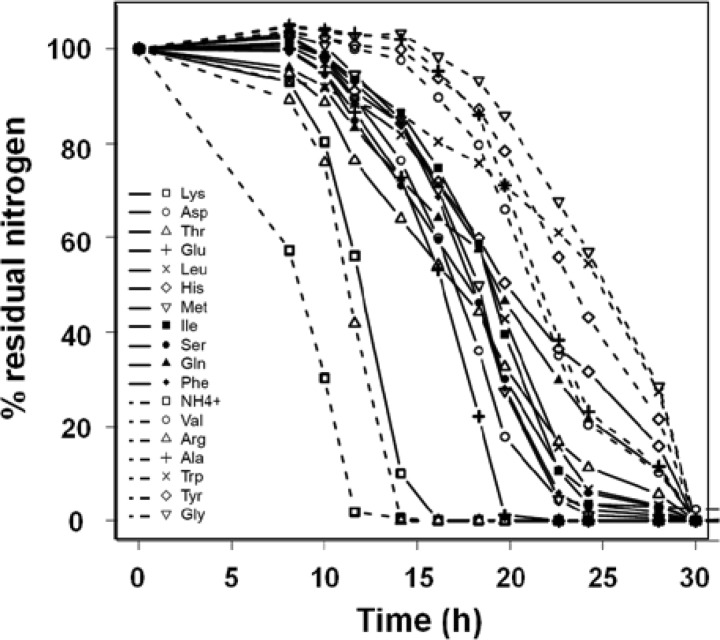
Fig 3.
Consumption of ammonium and amino acids by strain EC1118 during fermentation in SM200. Residual concentrations of Lys, Asp, Thr, Glu, Leu, His, Met, Ile, Ser, Gln, and Phe are shown by solid lines and the symbols indicated in the key. The residual concentrations of NH4+, Val, Arg, Ala, Trp, Tyr, and Gly are shown by dashed lines (and the symbols indicated in the key). Residual concentrations are expressed as percentages of the initial concentrations. Results of one representative experiment of two are shown.
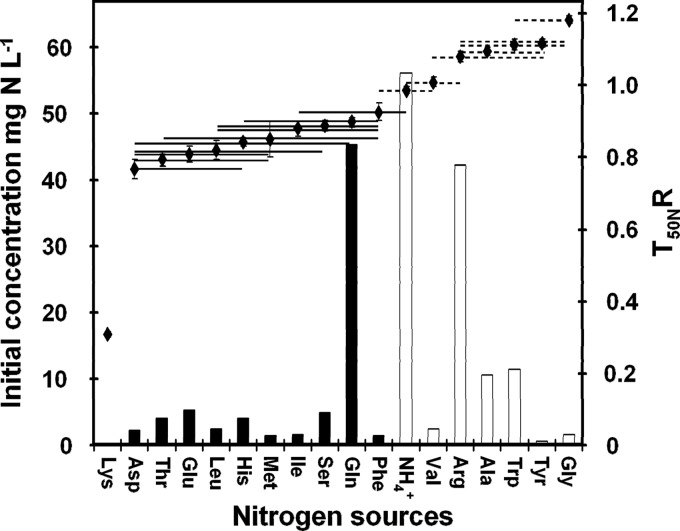
The mean for each of the 14 strains for the T50NR ratio between the time required to use 50% of each available nitrogen source and the time required for 50% consumption of total available nitrogen was calculated for each amino acid and for ammonium; one-way ANOVA revealed a global effect of amino acid (P < 0.001). Tukey's HSD test was used for post hoc comparisons of T50NR values at a P value threshold of 0.05, and this allowed ammonium and amino acids to be discriminated into three classes (Fig. 4). Lysine, which was provided at a very low concentration, was rapidly exhausted from the medium. Some nitrogen compounds, Asp, Thr, Glu, Leu, His, Met, Ile, Ser, Gln, and Phe, were used early, and others, NH4+, Val, Arg, Ala, Trp, Tyr, and Gly, were consumed at the end of the growth phase. One of the most abundant amino acids in the medium, Gln, and conversely, one of the least abundant, Lys, were both early consumed. Therefore, this sequential assimilation seemed to be independent of the relative concentrations of the individual amino acids and ammonium (0.12 mg N liter−1 to 56 mg N liter−1) (Fig. 4). Consistent with these findings, no correlation was found among the complete data set (14 strains and 18 nitrogen compounds) between the initial concentration and the variables describing the time of assimilation (TLAG, T50N, and T95N) of amino acids and ammonium.
Fig 4.
Initial concentrations and the order of assimilation of the nitrogen compounds in SM200 cultures. Initial concentrations of nitrogen compounds (in mg N liter−1) and the T50NR, which is the mean ratio for the 14 strains between the time for consumption of 50% of each available nitrogen source and the time required for the consumption of 50% of total available nitrogen. The standard errors of the means for the 14 strains are indicated by the vertical error bars. Horizontal bars depict the results (nonsignificant) of Tukey's HSD comparisons for nitrogen compounds at a P level of 0.05.
There were large differences between the rates of assimilation of the different amino acids, from 0.028 mg N liter−1 h−1 to 11.8 mg N liter−1 h−1. The nitrogen compounds present in the medium at the highest concentrations, in particular, ammonium, arginine, glutamine, alanine, and tryptophan, were removed at higher rates. Spearman's rank correlation indicated that the initial concentration accounted for a large part of the differences in consumption rate between amino acids (ρ, 0.87). Analysis of variance for rNmax did not detect an interaction between the nature of the nitrogen source and strain (P = 0.1426) and confirmed a significant effect for both of these two factors (P < 0.001 and P < 0.01, respectively).
(ii) Role of initial nitrogen source concentration for the order of amino acid consumption.
Although the profiles observed cannot be globally explained by a concentration effect, we cannot exclude the possibility that the consumption characteristics of some nitrogen compounds may specifically depend on their initial concentration in the medium. We therefore determined the profiles of assimilation of amino acids and ammonium by a representative wine yeast strain, EC1118, during fermentation on synthetic medium SM200E, containing 200 mg liter−1 YAN composed of equivalent amounts of each amino acid and ammonium (10.53 mg N liter−1 for each nitrogen compound). We showed that changes in the relative availabilities of nitrogen substrates did not alter the distribution into three classes, excepted in the case of arginine and ammonium, which were assimilated earlier in SM200E than in SM200 (Fig. 5).
Fig 5.
Consumption of ammonium and amino acids by strain EC1118 during fermentation in SM200E. Residual concentrations of Lys, Asp, Thr, Glu, Leu, His, Met, Ile, Ser, Gln, and Phe are shown by the solid lines and symbols indicated in the key, and concentrations of NH4+, Val, Arg, Ala, Trp, Tyr, and Gly are shown with dashed lines and the indicated symbols. Residual concentrations are expressed as percentages of the initial concentrations. Results of one representative experiment of five are shown.
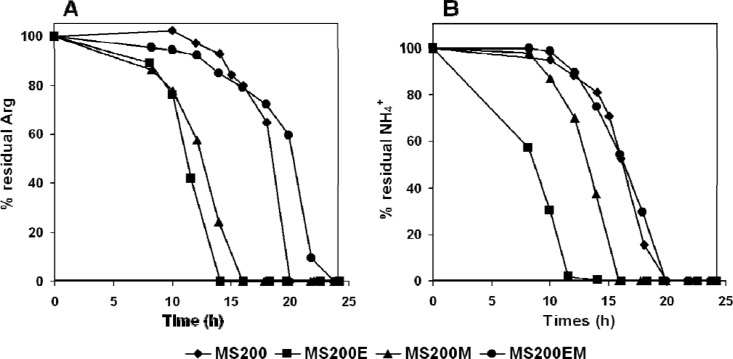
To study the relationships between the initial concentrations of arginine and ammonium and their times of consumption, we compared their assimilation profiles (Fig. 6) during growth on SM200, SM200E, SM200M (medium SM200 with arginine and ammonium provided at concentrations similar to those in SM200E), and SM200EM (medium SM200E with arginine and ammonium provided at concentrations similar to those of SM200). Arginine was consumed early when present in the medium at a low concentration (SM200E and SM200M) and later when provided at a higher concentration (SM200 and SM200EM). In contrast, no obvious relationship was observed between the initial amount of ammonium and the time of assimilation.
Fig 6.
Consumption of arginine (A) and ammonium (B) by strain EC1118 during fermentation in SM200, SM200E, SM200M, and SM200EM. Residual concentrations are expressed as percentages of the initial concentrations. Results of one representative experiment of two are shown.
Therefore, the order of consumption of nitrogen compounds by yeast during fermentation does not depend on their relative amounts in the medium, except for arginine.
(iii) Regulatory mechanisms of nitrogen source transporters controlling the order of consumption.
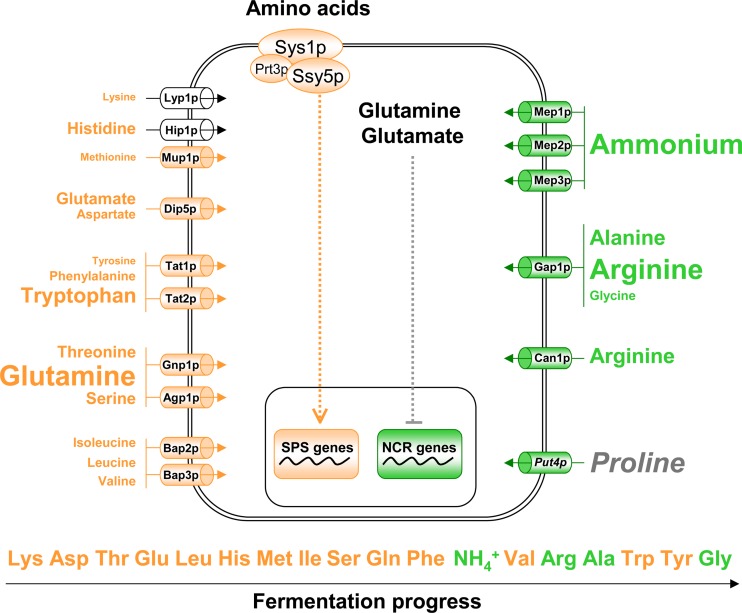
We then investigated a possible link between the sequence of assimilation of amino acids and ammonium during SM200 fermentation and the regulatory mechanisms controlling the activities of nitrogen transporters. In yeast, nitrogen permeases are controlled by two major regulatory mechanisms (summarized in Fig. 7): SPS-mediated control of specific permeases, and an NCR-mediated downregulation of general carriers, for example, the general permease Gap1p, in response to high intracellular concentrations of good nitrogen sources. Comparing the mode of regulation of nitrogen compound transporters (Fig. 7) with their order of assimilation (Fig. 3), we observed that the amino acids consumed early were those transported through SPS-regulated permeases. This was consistent with an induction of the expression of these permeases from the beginning of fermentation in response to the presence of extracellular amino acids. In contrast, amino acids imported by permeases under NCR control, including arginine, alanine, and glycine, were assimilated during the last part of the growth phase, after exhaustion of the other nitrogen compounds.
Fig 7.
List of the major nitrogen source transporters with their regulatory mechanisms. The transporters represented include carriers of one or a few specific amino acids (Lyp1p, Hip1p, Mup1, Dip5p, Tat1p, Tat2p, Gnp1, Agp1p, Bap2p, Bap3p, and Can1p) and the general amino acid carriers Gap1p, the arginine transporter Can1p, and the ammonium permeases (Mep1p, Mep2p, and Mep3) (35, 36). The sizes of the fonts for nitrogen compounds represent the relative abundance in the medium SM200. The regulatory mechanisms controlling the expression of nitrogen permease genes are indicated in orange (SPS) (32) or in green (NCR) (49). The order of nitrogen source consumption during wine fermentation is indicated at the bottom.
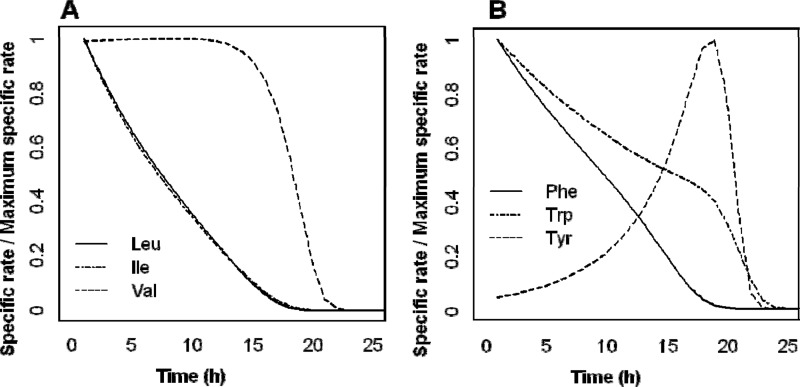
Therefore, the mode of regulation of nitrogen transporters can explain a large part of the pattern of sequential use of nitrogen compounds during fermentation, although there are some exceptions. Val, Trp, and Tyr were consumed late (Fig. 3 and 7), although the transporters mainly responsible for their uptake have been reported to be SPS regulated (32). Val, like Ile and Leu, is transported by the Bap2p and Bap3p permeases. The specific rates of Val, Leu, and Ile consumption during the fermentation (Fig. 8) revealed that Val was efficiently consumed only after Leu and Ile depletion. This suggested that Bap2p and Bap3p preferentially take up Leu and Ile to the detriment of Val. Similarly, Trp, Tyr, and Phe uptake is mediated by shared permeases, Tat1p and Tat2p. The specific rates of Trp and Phe consumption were high at the beginning of the fermentation and then decreased throughout the growth phase, whereas the rate of Tyr assimilation increased after 15 h of culture, by which time only low concentrations of Trp and Phe remained in the medium (Fig. 8). Therefore, the profiles of consumption of Trp, Tyr, and Phe are consistent with preferential uptake of Trp and Phe by Tat1p and Tat2p. Note that the concentration of Trp in the medium (11.5 mg N liter−1) was higher than that of Phe (1.5 mg N liter−1), and Trp was depleted later than Phe.
Fig 8.
Evolution of specific rates of amino acid consumption (in mg N liter−1 cell−1), normalized to the maximum specific rate (in mg N liter−1 cell−1) for Leu, Ile, and Val (A) and for Phe, Trp, and Tyr (B) as a function of time (h) for strain EC1118. Mean values were calculated from 2 replicates.
Overall, these results showed that the sequence of assimilation of the nitrogen compounds during fermentation is largely determined by both the kinetic characteristics and the regulation of the transporters of amino acids and ammonium.
DISCUSSION
S. cerevisiae strains display a large diversity in phenotypic traits related to growth, metabolite production, and fermentation capacity under industrial fermentation conditions (2, 14, 37, 38). Here, we report an analysis of the patterns of YAN assimilation during wine fermentation by yeast strains originating from various industrial and natural environments. These S. cerevisiae strains showed diverse dynamics of nitrogen consumption. This was consistent with the substantial variability in other characteristics of nitrogen assimilation, as previously described for commercial wine yeast. Indeed, important differences have been observed in the nitrogen requirements of strains with respect to their fermentative performances, either to complete fermentation in the presence of 20% sugar (3, 6) or to increase the fermentation rate during the stationary phase (28, 34). Likewise, industrial wine strains exhibit substantial differences in the amount of nitrogen consumed when nitrogen compounds are present in excess (26). We found that the profiles of N compound consumption, characterized by the rates and the times of assimilation, significantly affected the ability of strains to grow and produce biomass. It has been reported that the biomass formation governs the rate of fermentation under wine fermentation conditions, particularly in nitrogen-poor musts (51). Furthermore, the ability of strains to complete fermentation depends on the level of biomass production during the growth phase of fermentation (14). Therefore, the capacity of yeasts to assimilate the available nitrogen resource quickly may be a key factor contributing to the control of the course of fermentation. Despite the substantial diversity between the kinetics of nitrogen uptake by different strains, ammonium and amino acids were sequentially consumed by all the strains, with no marked differences in the order of assimilation of the nitrogen compounds between strains. Thus, the differences between yeast strains in the kinetics of nitrogen consumption were not due to differences in the ability of the strains to assimilate a particular nitrogen-containing compound.
The value of the normalized T50NR value, which depends both on the time and the rate of substrate consumption, reflects the whole profile of assimilation of a nitrogen compound. The nitrogen substrates could be clustered into three groups on the basis of this measure: compounds assimilated prematurely (Lys), early, or late during fermentation. We found that the association of an amino acid with early or late classes mainly depended on the regulation and the inherent kinetic properties of the transporters involved in its uptake; the affiliation did not appear to be related to its availability in the medium, excepted for lysine, which was present in trace amounts (0.61 mg liter−1) in the medium and was exhausted during the first hours of fermentation. The permeases involved in the uptake of amino acids consumed early (Asp, Thr, Glu, Leu, His, Met, Ile, Ser, Gln, and Phe) are encoded by genes that are under positive Ssy1p-mediated regulation (BAP2, BAP3, TAT1, TAT2, DIP5, MUP1, GNP1, and AGP1) (32). The assimilation of these nitrogen compounds is expected to result in the intracellular accumulation of glutamine and glutamate and, consequently, in the downregulation of NCR-sensitive genes. Consistent with this, the Gap1p NCR-controlled membrane transport protein has been reported to be repressed during the first stages of wine fermentation (4, 5). As a result, Ala, Arg, and Gly, which are mainly imported by Gap1p under these conditions, were only assimilated after exhaustion of the other nitrogen compounds, during the last part of the growth phase. On the basis of these data, we propose that the kinetic characteristics of these transporters contribute to the characteristics of sequential assimilation of nitrogen compounds. Indeed, the late consumption of Val appeared to be a consequence of differences in the affinity of the permeases Bap2p and Bap3p for branched amino acids (Val, Leu, and Ile). Similarly, a preferential uptake of Phe by Tat1p and Tat2p carriers probably led to the delayed assimilation of other aromatic amino acids (Tyr and Trp).
Only one amino acid, arginine, was assimilated at different times according to its availability in the medium. This may be a consequence of there being two distinct transporters for this amino acid. The general amino acid permease Gap1p, reported to be the major transporter of arginine (4, 5) and to be NCR controlled, may be responsible for the uptake of arginine under arginine-rich conditions, during the last stages of the growth phase. In the presence of low concentrations, arginine uptake may be mediated by the high-affinity arginine permease Can1p. As this carrier is regulated by NCR (17), this suggests that the level of catabolic repression on Can1p is not strong enough to totally abolish arginine transport. Consistent with this notion, CAN1 is only slightly repressed compared to GAP1 after an NH4+ pulse (T. Clement and C. Camarasa, unpublished data).
Finally, in S. cerevisiae, ammonium assimilation is mediated by the specific permeases Mep1p, Mep2p, and Mep3p, which exhibit different kinetic properties (Km values for NH4+ of 10 μM, 1 μM, and 2 mM, respectively) and are controlled differently by nitrogen (35). This may explain why ammonium was assimilated during the first hours of fermentation only when the concentration of glutamine, a key component of NCR regulation, was low.
This classification of amino acids according to their pattern of assimilation under wine-making conditions is in agreement with those reported in previous studies, which were carried out with various yeasts and culture medium and wine/wort fermentations (7, 26, 27, 40). These various observations indicate that the order of assimilation by S. cerevisiae of nitrogen substrates present as a complex mixture depends only slightly on the availability of these compounds in the medium and on the strain used. Most of the dissimilarities between studies in the assignment of amino acids concerned the allocation of tyrosine, tryptophan, and phenylalanine to the different groups. These discrepancies may be due to the empirical designation of the groups in previous studies, according to the time required to reach the half-concentration, the initial removal rate, and were generally determined using a very small number of strains (one or two). We compared the profiles of assimilation of amino acids and ammonium through a statistically based method for 14 S. cerevisiae strains and obtained consistent and reliable discrimination of nitrogen compounds into three classes. Discrepancies between studies concerning arginine and ammonium may be the result of the more complex patterns of assimilation of these nitrogen sources, which depend on conditions that differ between studies.
Our study revealed substantial differences between the rates of consumption of YAN, rNmax, by different strains that cannot be explained by preferential consumption of one or a few nitrogen compounds. Further investigations are now in progress to identify the causes of this variability.
Supplementary Material
ACKNOWLEDGMENTS
We thank Cécile Cadoux and Adeline Lopez for technical assistance.
This study was supported by the INRA Sys-Arômes program and Bioflavour Cost Action FA0907.
Footnotes
Published ahead of print 14 September 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Albers E, Larsson C, Lidén G, Niklasson C, Gustafsson L. 1996. Influence of the nitrogen source on Saccharomyces cerevisiae anaerobic growth and product formation. Appl. Environ. Microbiol. 62:3187–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Albertin W, et al. 2011. Population size drives industrial Saccharomyces cerevisiae alcoholic fermentation and is under genetic control. Appl. Environ. Microbiol. 77:2772–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bell SJ, Henschke PA. 2005. Implications of nitrogen nutrition for grapes, fermentation and wine. Aust. J. Grape Wine Res. 11:242–295 [Google Scholar]
- 4. Beltran G, Esteve-Zarzoso B, Rozes N, Mas A, Guillamón JM. 2005. Influence of the timing of nitrogen additions during synthetic grape must fermentations on fermentation kinetics and nitrogen consumption. J. Agric. Food Chem. 53:996–1002 [DOI] [PubMed] [Google Scholar]
- 5. Beltran G, Novo M, Rozès N, Mas A, Guillamón JM. 2004. Nitrogen catabolite repression in Saccharomyces cerevisiae during wine fermentations. FEMS Yeast Res. 4:625–632 [DOI] [PubMed] [Google Scholar]
- 6. Bely M, Sablayrolles JM, Barre P. 1990. Automatic detection of assimilable nitrogen deficiencies during alcoholic fermentation in oenological conditions. J. Ferment. Bioeng. 70:246–252 [Google Scholar]
- 7. Bisson LF. 1991. Influence of nitrogen on yeast and fermentation of grapes. Proceedings of the International Symposium on Nitrogen in Grapes and Wine. Am. J. Enol. Vitic. 42:78–89 [Google Scholar]
- 8. Bisson LF. 1999. Stuck and sluggish fermentations. Am. J. Enol. Vitic. 50:107–119 [Google Scholar]
- 9. Blateyron L, Sablayrolles JM. 2001. Stuck and slow fermentations in enology: statistical study of causes and effectiveness of combined additions of oxygen and diammonium phosphate. J. Biosci. Bioeng. 91:184–189 [DOI] [PubMed] [Google Scholar]
- 10. Brandriss MC, Magasanik B. 1979. Genetics and physiology of proline utilization in Saccharomyces cerevisiae: enzyme induction by proline. J. Bacteriol. 140:498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cai H, Kauffman S, Naider F, Becker JM. 2006. Genome-wide screen reveals a wide regulatory network for di/tripeptide utilization in Saccharomyces cerevisiae. Genetics 172:1459–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Callejón RM, Troncoso AM, Morales ML. 2010. Determination of amino acids in grape-derived products: a review. Talanta 81:1143–1152 [DOI] [PubMed] [Google Scholar]
- 13. Camarasa C, Grivet JP, Dequin S. 2003. Investigation by 13C-NMR and tricarboxylic acid (TCA) deletion mutant analysis of pathways for succinate formation in Saccharomyces cerevisiae during anaerobic fermentation. Microbiology 149:2669–2678 [DOI] [PubMed] [Google Scholar]
- 14. Camarasa C, Sanchez I, Brial P, Bigey F, Dequin S. 2011. Phenotypic landscape of Saccharomyces cerevisiae during wine fermentation: evidence for origin-dependent metabolic traits. PLoS One 6:e25147 doi:10.1371/journal.pone.0025147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cooper TG. 2002. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol. Rev. 26:223–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cunningham TS, Cooper TG. 1991. Expression of the DAL80 gene, whose product is homologous to the GATA factors and is a negative regulator of multiple nitrogen catabolic genes in Saccharomyces cerevisiae, is sensitive to nitrogen catabolite repression. Mol. Cell. Biol. 11:6205–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Daugherty JR, Rai R, el Berry HM, Cooper TG. 1993. Regulatory circuit for responses of nitrogen catabolic gene expression to the GLN3 and DAL80 proteins and nitrogen catabolite repression in Saccharomyces cerevisiae. J. Bacteriol. 175:64–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Forsberg H, Ljungdahl PO. 2001. Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids. Mol. Cell. Biol. 21:814–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Georis I, Feller A, Vierendeels F, Dubois E. 2009. The yeast GATA factor Gat1 occupies a central position in nitrogen catabolite repression-sensitive gene activation. Mol. Cell. Biol. 29:3803–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hazelwood LA, Daran JM, van Maris AJA, Pronk JT, Dickinson JR. 2008. The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 74:2259–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hein C, Springael JY, Volland C, Haguenauer-Tsapis R, André B. 1995. NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol. Microbiol. 18:77–87 [DOI] [PubMed] [Google Scholar]
- 22. Horák J. 1997. Yeast nutrient transporters. Biochim. Biophys. Acta 1331:41–79 [DOI] [PubMed] [Google Scholar]
- 23. Iraqui I, et al. 1999. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol. 19:989–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jauniaux JC, Grenson M. 1990. GAP1, the general amino acid permease gene of Saccharomyces cerevisiae. Nucleotide sequence, protein similarity with the other bakers yeast amino acid permeases, and nitrogen catabolite repression. Eur. J. Biochem. 190:39–44 [DOI] [PubMed] [Google Scholar]
- 25. Jauniaux JC, Vandenbol M, Vissers S, Broman K, Grenson M. 1987. Nitrogen catabolite regulation of proline permease in Saccharomyces cerevisiae. Cloning of the PUT4 gene and study of PUT4 RNA levels in wild-type and mutant strains. Eur. J. Biochem. 164:601–606 [DOI] [PubMed] [Google Scholar]
- 26. Jiranek V, Langridge P, Henschke PA. 1995. Amino acid and ammonium utilization by Saccharomyces cerevisiae wine yeasts from a chemically defined medium. Am. J. Enol. Vitic. 46:75–83 [Google Scholar]
- 27. Jones M, Pierce JS. 1964. Absorption of amino acids from wort by yeasts. J. Inst. Brew. 70:307–315 [Google Scholar]
- 28. Julien A, Roustan JL, Dulau L, Sablayrolles JM. 2000. Comparison of nitrogen and oxygen demands of enological yeasts: technological consequences. Am. J. Enol. Vitic. 51:215–222 [Google Scholar]
- 29. Kaspar H, et al. 2009. Urinary amino acid analysis: a comparison of iTRAQ®-LC-MS/MS, GC-MS, and amino acid analyzer. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 877:1838–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lê S, Josse J, Husson F. 2008. FactoMineR: an R package for multivariate analysis. J. Stat. Softw. 25:1–18 [Google Scholar]
- 31. Liti G, et al. 2009. Population genomics of domestic and wild yeasts. Nature 458:337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ljungdahl PO. 2009. Amino-acid-induced signalling via the SPS-sensing pathway in yeast. Biochem. Soc. Trans. 37:242–247 [DOI] [PubMed] [Google Scholar]
- 33. Magasanik B, Kaiser CA. 2002. Nitrogen regulation in Saccharomyces cerevisiae. Gene 290:1–18 [DOI] [PubMed] [Google Scholar]
- 34. Manginot C, Roustan JL, Sablayrolles JM. 1998. Nitrogen demand of different yeast strains during alcoholic fermentation. Importance of the stationary phase. Enzyme Microb. Technol. 23:511–517 [Google Scholar]
- 35. Marini AM, Soussi-Boudekou S, Vissers S, Andre B. 1997. A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:4282–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marini AM, Vissers S, Urrestarazu A, André B. 1994. Cloning and expression of the MEP1 gene encoding an ammonium transporter in Saccharomyces cerevisiae. EMBO J. 13:3456–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marullo P, et al. 2007. Single QTL mapping and nucleotide-level resolution of a physiologic trait in wine Saccharomyces cerevisiae strains. FEMS Yeast Res. 7:941–952 [DOI] [PubMed] [Google Scholar]
- 38. Masneuf-Pomarède I, et al. 2010. Reassessment of phenotypic traits for Saccharomyces bayanus var. uvarum wine yeast strains. Int. J. Food Microbiol. 139:79–86 [DOI] [PubMed] [Google Scholar]
- 39. Palmqvist U, Ayrapaa T. 1969. Uptake of amino acids in bottom fermentations. J. Inst. Brew. 75:181 [Google Scholar]
- 40. Perpete P, Santos G, Bodart E, Collin S. 2005. Uptake of amino acids during beer production: the concept of a critical time value. J. Am. Soc. Brew. Chem. 63:23–27 [Google Scholar]
- 41.R Development Core Team 2012. A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 42. Radler F. 1993. Yeasts: metabolism of organic acids, p 165–182. In Fleet GH, Wine microbiology and biotechnology. Chur, Philadelphia, PA [Google Scholar]
- 43. Ramos-Jeunehomme C, De Keyser L, Masschelein CA. 1979. Formation of aromatic and kinetic absorption substance from amino acid of wort. Proc. Congr. Eur. Brew. Conv. 12:505–510 [Google Scholar]
- 44. Regenberg B, Düring-Olsen L, Kielland-Brandt MS, Holmberg S. 1999. Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae. Curr. Genet. 36:317–328 [DOI] [PubMed] [Google Scholar]
- 45. Schreve JL, Sin JK, Garrett JM. 1998. The Saccharomyces cerevisiae YCC5 (YCL025c) gene encodes an amino acid permease, Agp1, which transports asparagine and glutamine. J. Bacteriol. 180:2556–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Spor A, et al. 2009. Niche-driven evolution of metabolic and life-history strategies in natural and domesticated populations of Saccharomyces cerevisiae. BMC Evol. Biol. 9:296 doi:10.1186/1471-2148-9-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stanbrough M, Magasanik B. 1995. Transcriptional and posttranslational regulation of the general amino acid permease of Saccharomyces cerevisiae. J. Bacteriol. 177:94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Swiegers JH, Bartowsky EJ, Henschke PA, Pretorius IS. 2005. Yeast and bacterial modulation of wine aroma and flavour. Aust. J. Grape Wine Res. 11:139–173 [Google Scholar]
- 49. ter Schure EG, van Riel NA, Verrips CT. 2000. The role of ammonia metabolism in nitrogen catabolite repression in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 24:67–83 [DOI] [PubMed] [Google Scholar]
- 50. Tronchoni J, Gamero A, Arroyo-López FN, Barrio E, Querol A. 2009. Differences in the glucose and fructose consumption profiles in diverse Saccharomyces wine species and their hybrids during grape juice fermentation. Int. J. Food Microbiol. 134:237–243 [DOI] [PubMed] [Google Scholar]
- 51. Varela C, Pizarro F, Agosin E. 2004. Biomass content governs fermentation rate in nitrogen-deficient wine musts. Appl. Environ. Microbiol. 70:3392–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.