Abstract
N-Acylhomoserine lactones (AHLs) are used as quorum-sensing (QS) signal molecules by many Gram-negative bacteria. We have reported that Chryseobacterium sp. strain StRB126, which was isolated from the root surface of potato, has AHL-degrading activity. In this study, we cloned and characterized the aidC gene from the genomic library of StRB126. AidC has AHL-degrading activity and shows homology to several metallo-β-lactamase proteins from Bacteroidetes, although not to any known AHL-degrading enzymes. Purified AidC, as a maltose-binding fusion protein, showed high degrading activity against all tested AHLs, whether short- or long-chain forms, with or without substitution at carbon 3. High-performance liquid chromatography (HPLC) analysis revealed that AidC functions as an AHL lactonase catalyzing AHL ring opening by hydrolyzing lactones. An assay to determine the effects of covalent and ionic bonding showed that Zn2+ is important to AidC activity both in vitro and in vivo. In addition, the aidC gene could also be PCR amplified from several other Chryseobacterium strains. In conclusion, this study indicated that the aidC gene, encoding a novel AHL lactonase, may be widespread throughout the genus Chryseobacterium. Our results extend the diversity and known bacterial hosts of AHL-degrading enzymes.
INTRODUCTION
Quorum sensing (QS) is a cell-cell communication mechanism that allows bacterial populations to coordinate gene expression in response to cell density. It is believed that QS constitutes a central element in the social life of bacteria, conferring to the members of a bacterial community the ability to behave as an organized multicellular organism (2, 3). The majority of the QS systems thus far described in Gram-negative bacteria rely on N-acyl-l-homoserine lactones (AHLs) as signaling molecules; these QS systems have been found in more than 100 bacterial species, including many pathogens (10). AHL-mediated QS regulates the expression of many genes and is responsible for bioluminescence, production of pigments and antibiotics, biofilm formation, and many other factors that are important for pathogenicity (6, 24). In general, AHL-negative mutants show defects in pathogenicity; hence, it is expected that disrupting or manipulating the QS signals could inhibit the expression of virulence and infection of host cells (26).
Many AHL-degrading genes have been cloned and characterized from bacteria, fungi, mammalian cells, and metagenomic libraries constructed from environmental soil samples (20, 22). AHL-degrading enzymes have been divided into two functional groups: AHL lactonases, which catalyze AHL ring opening by hydrolyzing lactones, and AHL acylases, which hydrolyze the amide bond of AHL (22). The first demonstrated and best-studied AHL-degrading enzyme is AiiA lactonase, which belongs to the metallo-β-lactamase superfamily and was identified from Bacillus sp. strain 240B1 (8). Expression of aiiA in the plant pathogen Pectobacterium carotovorum subsp. carotovorum significantly attenuates pathogenicity on some crops (7). Transgenic plants expressing AHL lactonase exhibited significantly enhanced resistance to the infection of P. carotovorum subsp. carotovorum (7). The AHL lactonases, which also belong to the metallo-lactamase superfamily, were called “AiiA-type lactonases,” with respect to being first identified as AiiAs. AiiA-type lactonases have been identified from a vast range of bacterial original hosts, including Firmicutes, Acidobacteria, and Proteobacteria (22, 28). Several AiiA-type lactonases have been studied in depth and hold potential for application in the biocontrol of plant diseases (22, 28).
In previous studies, we have reported the putative AHL lactonase activities in the potato root-associated strains of Chryseobacterium sp., which is a member of the Cytophaga-Flavobacterium-Bacteroides (CFB) group (16). Recently, it was reported that the fish pathogen Tenacibaculum maritimum, also a member of the CFB group, produces and degrades AHLs (18). However, to our knowledge, there are no reports of the isolation and characterization of any AHL-degrading gene from the CFB group bacteria. In this study, we report the cloning of a gene that encodes a novel AiiA-type lactonase from the potato root-associated Chryseobacterium sp. strain StRB126 and characterization of the enzymatic kinetics of the novel AHL lactonase.
MATERIALS AND METHODS
Bacterial strains, plasmids, compounds, and growth conditions.
Selected bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli was grown at 37°C in Luria-Bertani (LB) medium (19). All other bacteria were grown at 30°C in tryptic soy broth (TSB; Nippon Becton, Dickinson, Tokyo, Japan). Solid bacterial media were made by adding agar at a final concentration of 1.5% to the liquid media. Antibiotics were added at final concentrations of 100 μg/ml ampicillin, 100 μg/ml chloramphenicol, 50 μg/ml kanamycin, and 10 μg/ml gentamicin as required. AHLs used in this study, N-hexanoyl-l-homoserine lactone (C6-HSL), N-octanoyl-l-homoserine lactone (C8-HSL), N-decanoyl-l-homoserine lactone (C10-HSL), N-dodecanoyl-l-homoserine lactone (C12-HSL), N-(3-oxohexanoyl)-l-homoserine lactone (3OC6-HSL), N-(3-oxooctanoyl)-l-homoserine lactone (3OC8-HSL), N-(3-oxodecanoyl)-l-homoserine lactone (3OC10-HSL), and N-(3-oxododecanoyl)-l-homoserine lactone (3OC12-HSL), were synthesized by a previously described method (4).
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F− supE44 ΔlacU169 (ϕ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Nippon Gene |
| C. violaceum | ||
| CV026 | ATCC 31532 derivative, cviI::Tn5xylE Kmr, Smr | 11 |
| VIR07 | ATCC 12472 derivative, cviI::Kmr, Apr | 13 |
| Chryseobacterium sp. | ||
| StRB126 | AHL-degrading strain isolated from potato root surface | 16 |
| StRB340 | AHL-degrading strain isolated from potato root surface | 16 |
| StRB341 | AHL-degrading strain isolated from potato root surface | 16 |
| StRB342 | AHL-degrading strain isolated from potato root surface | 16 |
| C. gleum | ||
| NBRC 15054 | Clinical isolate, type strain | NBRC |
| Plasmids | ||
| pUC118 | Cloning vector; Apr | TaKaRa Bio |
| pST126-E1 | 8.8-kb Sau3AI fragment from StRB126 genomic DNA in pUC118 | This study |
| pHSG398 | Cloning vector; Cmr | TaKaRa Bio |
| pHSG-aidC126 | pHSG398 containing aidC gene from StRB126 | This study |
| pLux28 | 8.8-kb SalI fragment from pHV200 in pSTV28 vector; Cmr | 26 |
| pUC118-aidC340 | pUC118 containing aidC gene from StRB340 | This study |
| pUC118-aidC341 | pUC118 containing aidC gene from StRB341 | This study |
| pUC118-aidC342 | pUC118 containing aidC gene from StRB342 | This study |
| pUC118-aidC15054 | pUC118 containing aidC gene from NBRC15054 | This study |
| pGEM-T easy | Cloning vector; Apr | Promega |
| pMAL-c2X | Cloning vector to make MBP fusions; Apr | New England Biolabs |
| pMAL-aidC | pMAL-c2X containing aidC gene from StRB126 | This study |
Cloning of an AHL-degrading gene from StRB126.
A standard protocol for genetic manipulation was used as described previously (19). The AHL-degrading gene of StRB126 was cloned by a previously described method (26). Briefly, both the genomic library of StRB126 and the AHL-responsive plasmid pLux28 were transformed into E. coli DH5α. The transformants were grown on LB agar plates containing ampicillin and chloramphenicol. The resulting colonies were inoculated into 200 μl of fresh LB medium containing ampicillin, chloramphenicol, and 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) in 96-well plates. After incubation at 30°C for 20 h with gentle shaking, the cell cultures in each well were evaluated for luminescence activities using a Luminescenser JNR-II (Atto, Tokyo, Japan). Positive clones were sequenced using a BigDye Terminator version 3.1 sequencing kit and an ABI Prism 3100 genetic analyzer (Applied Biosystems, Tokyo, Japan).
Cloning of the aidC gene and its homolog.
The aidC-coding region on the positive clone pST126-E1 was excised by EcoRV and PstI digestion and inserted into the SmaI-PstI sites of the pHSG398 cloning vector for construction of pHSG-aidC126. The aidC gene homologs from the root-associated Chryseobacterium sp. strains StRB340, StRB341, and StRB342 were amplified with GoTaq DNA polymerase (Promega, Tokyo, Japan) and the following primer set: 5′-CTC AGC TGG CAT TAG CAT GGG TCT TGA ATC-3′ and 5′-GCA GAC AGC TAT TCT GTT AGT TTT CAG CAG C-3′. The aidC gene homolog from Chryseobacterium gleum strain NBRC 15054 was also amplified with the same cycling parameters and the following primer set: 5′-AGC TTG CGC TAG CTT GGG TAT TGA ACC AGG-3′ and 5′-CTA CCT GTT ACC TAT CAT CTG CCT CTG TC-3′. PCR was performed using the following cycling parameters: 95°C for 30 s, 58°C for 30 s, and 72°C for 1 min for 30 cycles. The PCR products were cloned into the pGEM-T easy cloning vector (Promega), excised by SphI and PstI digestion, and inserted into the SphI-PstI sites of pUC118 for construction of pUC118-aidC340, pUC118-aidC341, pUC118-aidC342, and pUC118-aidC15054.
Purification of AidC as an MBP fusion.
The aidC-coding region in the genome of StRB126 was amplified with GoTaq DNA polymerase and the following primers, containing BamHI and PstI restriction sites (underlined), respectively: 5′-GGA TCC ATG AAT AGA AAG CGG GTT ATT GGC-3′ and 5′-CTG CAG AGC TAT TCT GTT AGT TTC AGC AGC-3′. PCR was performed using the following cycling parameters: 95°C for 30 s, 57°C for 30 s, and 72°C for 3 min for 30 cycles. The PCR products were cloned into the pGEM-T easy cloning vector, excised by BamHI and PstI digestion, and inserted into the BamHI-PstI sites of pMAL-c2X for construction of pMAL-aidC. For expression and purification of the maltose-binding protein (MBP)-AidC fusion protein, the complete preculture of E. coli DH5α harboring pMAL-aidC was inoculated into 300 ml of fresh LB medium and incubated for 2 h at 37°C, with shaking. Expression of the recombinant MBP-AidC fusion was induced upon the addition of 0.1 mM IPTG after 2 h, and expression was continued for an additional 8 h at 37°C. After incubation, cells were harvested by centrifugation and resuspended with 3 ml of BugBuster protein extraction reagent (Novagen, Inc., Madison, WI) and then incubated for 10 min at room temperature, with gentle shaking. Next, the suspension was sonicated, centrifuged at 10,000 × g for 5 min to remove the cell debris, and filtrated. Protein purification was performed using the same method described previously (26). As a negative control, we also purified an MBP-LacZα fusion from E. coli DH5α harboring pMAL-c2X. Expression and purification of recombinant MBP-AidC and MBP-LacZα were checked by SDS-PAGE analysis.
Detection of AHL-degrading activity of AidC.
The residual AHLs were detected using AHL biosensors Chromobacterium violaceum CV026 and VIR07, which respond to exogenous AHLs by producing the purple pigment violacein (11, 13). For detecting the AHL-degrading activity of AidC in vivo and in vitro, we used a previously described method (26). To determine the chemical structures of products from the reaction between AidC and AHLs, 50 μl of purified protein solution was mixed with 49 μl of column buffer [20 mM Tris-HCl buffer and 200 mM NaCl (pH 7.4)] and 1 μl of 200 mM C10-HSL stock solution (in methanol). After incubation at 30°C for 10 min, reactions were stopped with an equal volume of acetonitrile, and the mixture was vortexed and centrifuged to pellet the precipitated protein. The hydrolyzed C10-HSL, as a control, was made by incubating C10-HSL in 10 mM NaOH at room temperature for 30 min. Samples (25 μl) were analyzed by high-performance liquid chromatography (HPLC) according to a previously described method (26).
Characterization of enzymatic activity of AidC.
To determine the enzyme kinetics of AidC, purified MBP-AidC (final concentration, 1 μM) was added to a substrate solution in the column buffer, with a final volume of 40 μl γ-butyrolactone (γ-BL). l-Homoserine lactone (HSL), C6-HSL, C8-HSL, C10-HSL, C12-HSL, 3OC6-HSL, 3OC8-HSL, and 3OC12-HSL were selected as the substrates for an enzyme specificity analysis. The reaction mixtures were incubated at 30°C, stopped by adding 40 μl acetonitrile, and subjected to HPLC analysis. The residual AHL and its hydrolysis product were quantified by HPLC. All experiments were performed in triplicate, and all velocities were determined at time points at which no more than 20% of the substrate had been consumed. The kcat and Km values were calculated based on Michaelis-Menten equation. The effects of various metal ions and a metal-chelating reagent (EDTA) on AidC activity were examined both in vitro and in vivo. For the in vitro assay, 1 mM EDTA and 1 mM metal ions comprising Cu2+, Ca2+, Fe2+, Mn2+, Mg2+, Zn2+, and Co2+ were mixed with 1 μM purified AidC protein in column buffer (pH 7.4). After incubation at 30°C for 10 min, the remaining activity was measured under the standard conditions described above. For the in vivo assay, the full preculture of E. coli harboring the plasmid pMAL-aidC was diluted into fresh LB medium with 1 mM EDTA and 1 mM metal ions. IPTG was then added to induce AidC overexpression after 4 h cultivation. After an additional 8-h cultivation, crude cell extracts were prepared and adjusted to the same concentration. The AHL-degrading activities of crude cell extracts were examined by the same procedure as used in the in vitro assay.
Nucleotide sequence accession number.
The nucleotide sequences of aidC and its flanking open reading frames (ORFs) from Chryseobacterium sp. StRB126 have been deposited in the DDBJ/EMBL/GenBank databases under accession no. AB733663.
RESULTS AND DISCUSSION
Identification of AHL-degrading gene from the Chryseobacterium sp. StRB126.
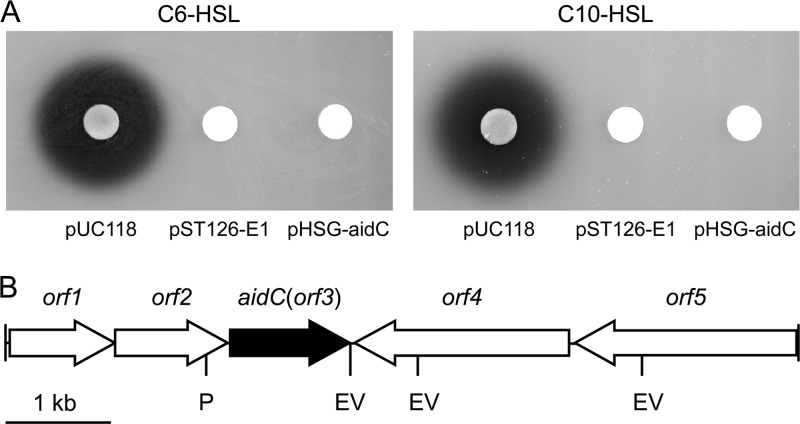
For cloning the AHL degradation gene, a pUC118-based StRB126 genomic library was constructed and used to transform E. coli DH5α harboring pLux28 (26). We then measured the luminescence activities of the resulting colonies on fresh LB medium. When approximately 2,300 transformants were screened, three clones expressed a low level of luminescence. To elucidate whether the reduction in luminescence resulted from degradation of AHL, E. coli DH5α cells harboring the plasmids from the positive clones were inoculated into LB medium containing 10 μM C6-HSL or C10-HSL. After incubation for 6 h, the residual AHL in the culture supernatant was detected by Chromobacterium violaceum reporters, which produced the purple pigment violacein in response to the presence of AHLs. Consequently, E. coli DH5α harboring a single plasmid, designated pST126-E1, showed clear AHL-degrading activity against both C6-HSL and C10-HSL (Fig. 1A).
Fig 1.
(A) AHL-degrading activity of E. coli DH5α harboring pUC118, pST126-E1, and pHSG-aidC. Subcultures of E. coli DH5α harboring these plasmids were mixed with 10 μM C6-HSL or C10-HSL and incubated at 37°C for 2 h. The residual AHL was detected using C. violaceum strain CV026 or VIR07. (B) Arrangement of the predicted ORFs on the original genomic clone pST126-E1. The scale represents a 1-kb length of nucleotides. The filled arrow indicates the aidC gene, and the open arrows indicate the other ORFs. EV, EcoRV; P, PstI.
The sequence of the genomic DNA fragment (8,784 bp), which was inserted into pST126-E1, contained five complete open reading frames (ORFs) (Fig. 1B). The third ORF (orf3) encoded a putative metallo-β-lactamase of 330 amino acids. The Orf3 amino acid sequence showed low-level similarity (13% identity) to the known AHL lactonases, including AiiA from Bacillus sp. 240B1 (7, 8). To test whether orf3 encodes an AHL-degrading enzyme, the complete orf3 was amplified by PCR and subcloned into the pHSG398 easy vector. E. coli DH5α harboring the orf3-expressing plasmid showed clear AHL-degrading activity at the same level as E. coli DH5α harboring pST126-E1 (Fig. 1A). These results suggested that orf3 encodes an AHL-degrading enzyme. Therefore, we named orf3 the autoinducer degrading gene from Chryseobacterium sp. (aidC) as being the first AHL-degrading gene identified from the genus Chryseobacterium.
AidC encodes an AHL lactonase.
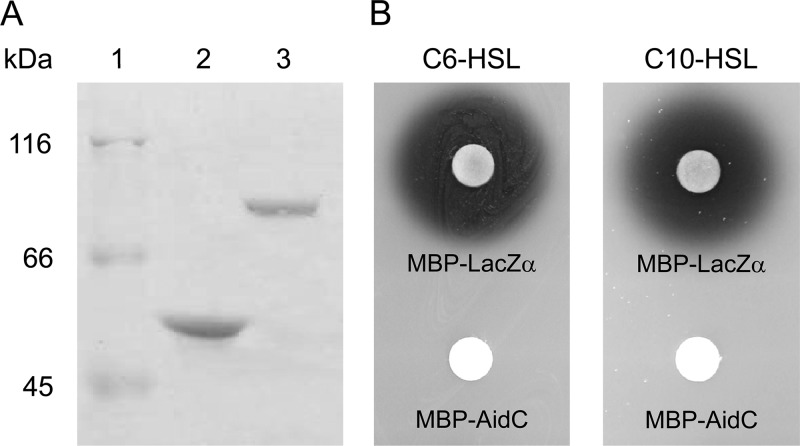
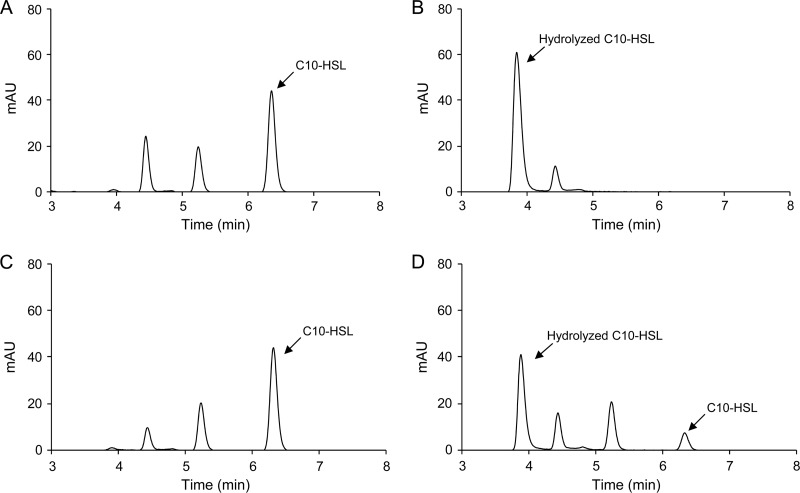
We purified AidC as an MBP fusion in an in vitro AHL-degrading assay. The MBP-AidC fusion protein was overproduced in E. coli DH5α and purified by maltose affinity chromatography, and the purified proteins were analyzed by 10% SDS-PAGE. The results revealed that the overexpressed protein was approximately 80 kDa in size (Fig. 2A), which was congruent with the predicted molecular weight of MBP-AidC, based on its amino acid sequence. When the AHL-degrading activity of the purified protein was examined, purified MBP-AidC completely degraded 100 μM C6-HSL and C10-HSL within 2 h, whereas MBP-LacZα did not (Fig. 2B). In our previous study, putative AHL lactonase activities were detected from StRB126 (16). Therefore, to determine whether AidC functions as an AHL lactonase, the structure of C10-HSL treated with MBP-AidC was analyzed by HPLC. Fractionation of the C10-HSL standard revealed a single major HPLC peak, with a retention time of approximately 6.3 min (Fig. 3A). To prepare the lactone ring-opened C10-HSL, C10-HSL was hydrolyzed by 10 mM NaOH. Fractionation of the hydrolyzed C10-HSL revealed one major HPLC peak with a retention time of approximately 3.9 min (Fig. 3B). To examine the enzymatic properties of AidC, solutions of MBP-LacZα and MBP-AidC were mixed with C10-HSL and incubated at 30°C for 10 min. Fractionation of MBP-LacZα-treated C10-HSL revealed a major HPLC peak, which corresponded to that of the C10-HSL standard (Fig. 3C). This result indicated that the MBP domain did not contain AHL-degrading activity. In contrast, fractionation of MBP-AidC-treated C10-HSL revealed two HPLC peaks, which corresponded to those of the C10-HSL standard and the lactone ring-opened C10-HSL (Fig. 3D). These results indicated that AidC functions as an AHL lactonase that catalyzes the opening of the AHL ring by hydrolyzing lactones.
Fig 2.
(A) Purification of MBP-LacZα and MBP-AidC fusion proteins. Lane 1, protein molecular mass marker (Fermentas, Hanover, MD); lane 2, purified MBP-LacZα; and lane 3, purified MBP-AidC. Samples were analyzed by SDS-PAGE using a 10% polyacrylamide gel. (B) AHL-degrading activity of MBP-LacZα and MBP-AidC. Solutions of purified MBP-LacZα and MBP-AidC were mixed with an equal volume of 1 mM C6-HSL or C10-HSL solutions and were then incubated at 30°C for 15 min. The residual AHL was detected using C. violaceum CV026 or VIR07.
Fig 3.
HPLC profiles of C10-HSL (A), C10-HSL hydrolyzed by 10 mM NaOH (B), C10-HSL treated with MBP-LacZα (C), and C10-HSL treated with MBP-AidC (D). The peaks corresponding to C10-HSL (a retention time of approximately 6.3 min) and hydrolyzed C10-HSL (3.9 min) are indicated by arrows.
AidC is a new member of the AiiA-type AHL lactonases.
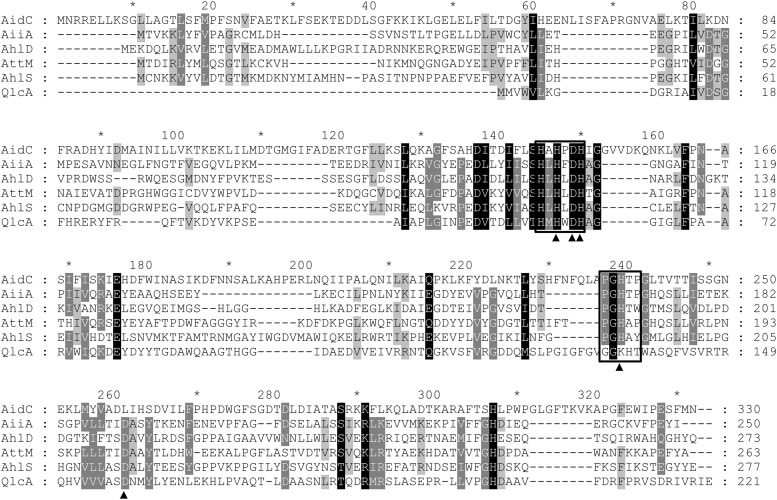
AiiA-type AHL lactonases, which belong to the metallo-β-lactamase superfamily, have been identified and characterized from various bacteria (8, 9, 14, 17, 22, 29). However, AidC has less than 13% identity to each of the known AiiA-type AHL lactonases, including 13% identity across the entire length of AiiA from Bacillus sp. 240B1, 12% identity across AttM from Agrobacterium tumefaciens strain C58 (29) and AhlD from Arthrobacter sp. strain IBN110 (15), 10% identity across AhlS from Solibacillus silvestris strain StLB046 (14), and 8% identity across QlcA from the soil metagenome (17). When the amino acid sequence of AidC was compared with those of the known AiiA-type lactonases (Fig. 4), the zinc-binding motifs HXHXDH∼H, which are commonly conserved sequences of the metallo-β-lactamase superfamily, were found in the known AiiA-type lactonases and in AidC (145HAHPDH150∼H239) (Fig. 4). Interestingly, D191 of AiiA, which has been proven to be essential for enzymatic activity by site-directed mutagenesis (8) and is shared by all the known AiiA-type lactonases (8, 14, 15, 17, 29), was replaced by L261 in AidC (Fig. 4).
Fig 4.
Comparison of amino acid sequences of AidC and five known AiiA-type lactonases. The amino acid sequence of AidC was compared with those of AiiA from Bacillus sp. 240B1, AhlD from Arthrobacter sp. strain IBN110, AttM from A. tumefaciens strain A6, AhlS from S. silvestris, and QlcA from the soil metagenome. Sequences were aligned using the ClustalW program (21) and shaded using the Genedoc program (http://www.nrbsc.org/gfx/genedoc/). The two zinc-binding motifs are boxed with rectangles. The amino acid residues that are essential for enzymatic activity of the known AHL lactonases are indicated by solid triangles.
The AHL lactonases can be grouped into three groups: the metallo-β-lactamase superfamily, the α/β-hydrolase-fold family (12, 26), and the phosphotriesterase (PTE) family (23). Among these, members from the metallo-β-lactamase superfamily have garnered more attention due to their wide distribution across the phyla of Firmicutes, Proteobacteria, and Acidobacteria (22, 28). To evaluate the novelty of AidC, we employed the amino acid sequence of AidC to determine the phylogenetic relationship between AidC and the known representative AHL lactonases from various bacteria using the neighbor-joining method (Fig. 5). Although AidC belongs to the same protein family as the known AiiA-type lactonases, we found that it is distant from the other lactonase clusters represented by the five other AiiA-type lactonases.
Fig 5.
Phylogenetic tree based on amino acid sequences of AidC, AiiA, AhlD, AttM, AhlS, QlcA, and AiiM from Microbacterium testaceum strain StLB037, AidH from Ochrobactrum sp. strain T63, and QsdA from Rhodococcus erythropolis strain W2. The dendrogram was constructed by the neighbor-joining method using the ClustalW program that is included with NJplot software. The scale bar represents 0.1 substitutions per amino acid position.
The substrate specificity and properties of AidC.
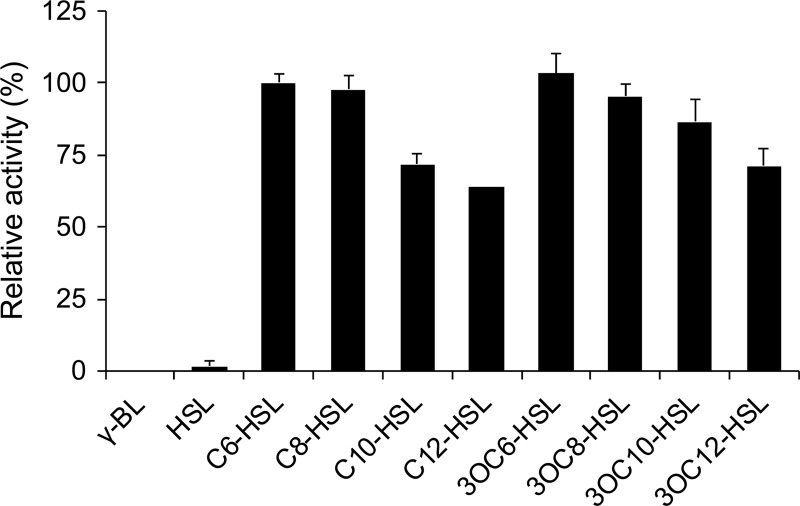
To assess the AHL substrate range of AidC, we assessed its ability to degrade various AHLs by HPLC. We found that MBP-AidC exhibited high relative activities toward all the tested AHLs, with or without 3-oxo substitution (Fig. 6). The 3-oxo substitution of AHLs is known to negatively affect the enzymatic activity of AiiA from Bacillus sp. 240B1 (25, 26). AidC, on the other hand, showed slightly higher degrading activity against 3-oxo-substituted AHLs than against unsubstituted AHLs (Fig. 6). In addition, although differences in acyl acid chain length did not significantly affect the enzymatic activity of AiiA (25), AidC was somewhat more effective at degrading C6-HSL and C8-HSL than C10-HSL and C12-HSL (Fig. 6). AidC did not show any degrading activity toward l-homoserine lactone or γ-butyrolactone, similar to AiiA (25).
Fig 6.
Substrate specificity of purified MBP-AidC. Purified MBP-AidC was mixed with substrate solutions in the column buffer (pH 7.4). After incubation at 30°C for 10 min, the residual AHL and its hydrolysis products were quantified by HPLC. We defined 100% relative activity as the activity toward C6-HSL. The data were reproduced at least three times, and error bars indicate standard deviations.
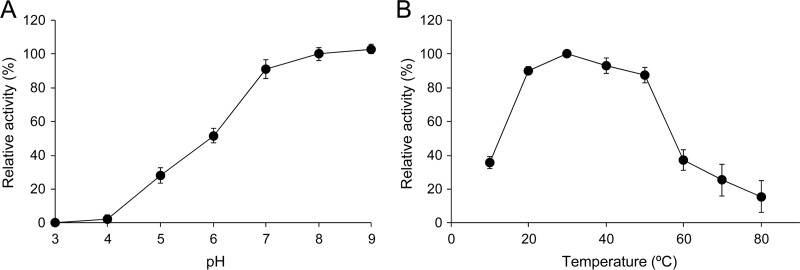
The optimal pH for AHL-degrading activity of MBP-AidC was examined using C10-HSL as the substrate at 30°C. AHL-degrading activity was enhanced as pH increased and reached a maximum at pH 8, but no or little activity was detected when pH was adjusted to pH 5 or below (Fig. 7A). The optimal temperature for the AHL-degrading activity of MBP-AidC was also examined using C10-HSL as the substrate. MBP-AidC displayed more than 80% of its maximum activity at 20°C to 50°C, but the relative activity was greatly reduced when temperature exceeded 50°C (Fig. 7B). Correspondingly, strain StRB126 could grow at temperatures ranging from 20°C to 50°C, reaching its optimum temperature at 30°C (data not shown). To determine the thermostability of AidC, purified MBP-AidC was preincubated at various temperatures for 2 h and the residual enzymatic activity was then determined. More than 90% of the activity remained after incubation at temperatures of 50°C or less, but the enzymatic activity was markedly reduced after incubation at temperatures over 60°C (data not shown).
Fig 7.
(A) Optimal pH of AHL-degrading activity of purified MBP-AidC. Purified MBP-AiiM was mixed with C10-HSL in reaction buffer with a pH ranging from pH 3 to pH 8 and was then incubated at 30°C. The data were reproduced at least three times, and error bars indicate standard deviations. (B) Optimal temperature of AHL-degrading activity of purified MBP-AidC. Purified MBP-AiiM was mixed with C10-HSL in the reaction buffer (pH 7.4) and incubated at temperatures ranging from 10 to 80°C. After incubation for 10 min, the residual substrate was quantified by HPLC. We defined 100% relative activity as the activity in the column buffer (pH 7.4) at 30°C. Control samples, which had been prepared under the same conditions but without AidC, were used to exclude the influence of autodegradation at high pH or high temperature. The data were reproduced at least three times, and error bars indicate standard deviations.
Kinetic analysis of AHL lactonase activity of AidC.
AHL hydrolysis kinetics of AidC was determined by a plot of the substrate concentration and velocity. The kcat and Km values were calculated by fitting the data to the Michaelis-Menten equation (Table 2). The AHL lactonase activity of AidC showed comparable catalytic activities against a range of structurally different AHLs, with kcat and Km values in the range of 1.97 to 2.35 s−1 and 0.046 to 0.072 mM, respectively. Within these narrow ranges, the enzyme showed higher affinities (Km), faster hydrolysis rates (kcat), and stronger catalytic efficiencies (kcat/Km) toward the AHLs with short acyl side chains than toward those with the longer derivatives. Additionally, AidC displayed somewhat lower kcat and kcat/Km values against the unsubstituted AHLs than against the 3-oxo-substituted AHLs (Table 2). Hydrolysis kinetics of the AiiA from Bacillus sp. 240B1 have been determined by plotting velocity versus substrate concentration (25). Compared with AiiA from Bacillus sp. 240B1, AidC showed lower Km and kcat values but higher catalytic efficiencies (kcat/Km) toward the AHLs (data not shown). In addition, Km values of AidC against AHLs are at levels of tens of micromolar and even higher for several other AHL lactonases (1, 5, 25), but AHLs can function as signals at multiple orders of magnitude below that level. Thus, in natural environments, the reaction rate should depend on the AidC concentration and the AHL concentration. The AidC/AHL reaction resembles a bimolecular reaction with a corresponding catalytic efficiency (kcat/Km). These results suggested that AidC may function much more effectively at quenching the bacterial QS systems which employ higher concentrations of AHLs than those employing lower concentrations of AHLs.
Table 2.
Kinetic parameters of AidC against AHLsa
| AHL | kcat (s−1) | Km (mM) | kcat/Km (mM−1 · s−1) |
|---|---|---|---|
| C6-HSL | 2.31 ± 0.16 | 0.055 ± 0.0043 | 42.0 |
| 3OC6-HSL | 2.35 ± 0.29 | 0.046 ± 0.0021 | 51.1 |
| C8-HSL | 2.01 ± 0.31* | 0.064 ± 0.0049* | 31.4 |
| 3OC8-HSL | 2.12 ± 0.24* | 0.064 ± 0.0037* | 33.1 |
| C10-HSL | ND | ND | ND |
| 3OC10-HSL | 1.97 ± 0.11* | 0.072 ± 0.0053* | 27.4 |
| C12-HSL | ND | ND | ND |
| 3OC12-HSL | ND | ND | ND |
The data are the means from triplicate experiments. The significance was determined by Student's t test (*, P < 0.05). ND, not determined due to poor solubility of the substrate in the column buffer.
Zinc is essential for AHL-degrading activity of AidC.
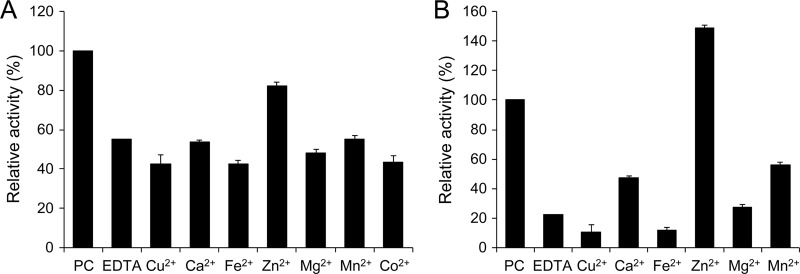
Sequence alignment of AidC and the known AiiA-type lactonases revealed that they share the zinc-binding motifs HXHXDH∼H in the central region (Fig. 4). To verify whether AidC requires any cofactor for its enzymatic activity, the effects of several divalent metal ions on the AHL-degrading activity of AidC were evaluated. For the in vitro assay, purified MBP-AidC was mixed with C10-HSL in column buffer containing EDTA and divalent metal ions. When 1 mM EDTA was added to this reaction mixture, the AHL-degrading activity of AidC was decreased by approximately 50% (Fig. 8A). However, AidC activity was successfully restored to 82% by the addition of Zn2+, but not by any other of the metal ions tested (Fig. 8A). For in vivo assays, E. coli harboring pMAL-aidC was cultivated in the LB medium containing EDTA and divalent metal ions, and the AHL-degrading activity of AidC in the crude cell extracts was then determined. The AHL-degrading activity of AidC was abolished by the addition of 1 mM EDTA but was restored by the addition of Zn2+ (Fig. 8B). These results suggested that Zn2+ is essential for the AHL-degrading activity of AiiA and the known AiiA-type lactonases. In addition, the AHL-degrading activity of AidC activity was markedly inhibited by the addition of Cu2+ (data not shown). This feature of AidC also corresponded to that of the known AiiA-type lactonases (25).
Fig 8.
The effects of EDTA and metal ions on AHL-degrading activity of AidC by in vitro (A) and in vivo (B) assays. For the in vitro assay, purified MBP-AidC was incubated in reaction buffer containing substrate and 1 mM each metal ion. After incubation at 30°C for 10 min, the residual activity of AidC was analyzed by HPLC. For the in vivo assay, E. coli harboring pMAL-aidC was cultivated with 1 mM EDTA and 1 mM each metal ion. Crude cell extracts were prepared and used for the AHL-degrading assay. We defined 100% relative activity as the activity in the absence of EDTA or metal ions; this was represented as PC (positive control). The data were reproduced at least three times, and error bars indicate standard deviations.
aidC is widely conserved in the genus Chryseobacterium.
In a previous study, we isolated 34 AHL-degrading Chryseobacterium strains from the roots of potato plants and divided them into eight related species groups (16). We now attempted to amplify by PCR procedures the aidC-homologous genes from the three AHL-degrading Chryseobacterium strains which belong to group VI, as well as StRB126 (16). The group VI strains, which were StLB126, StLB340, StLB341, and StLB342, had higher AHL-degrading activity than the other groups (16). Specific primers were designed based on the DNA sequences of aidC from StRB126. Following PCR amplification and DNA sequencing, aidC gene homologs were successfully cloned from all group VI strains. AidC from StLB126, StLB340, StLB341, and StLB342 had highly similar DNA sequences (more than 99% identity). In addition, E. coli DH5α harboring these aidC homologs showed significant AHL-degrading activity against various AHLs (data not shown). These results suggested that aidC sequences are highly conserved among group VI strains.
Based on BLAST search results, an aidC homologous gene was found in the whole-genome shotgun sequence of C. gleum NBRC 15054 (accession no. ACKQ00000000). However, we have previously reported that C. gleum NBRC 15054 did not show any AHL-degrading activity (16). To assess whether the AidC homolog (aidC15054) from NBRC 15054 shows AHL lactonase activity, aidC15054 was amplified from the chromosome of NBRC 15054 and cloned into the pUC118 cloning vector. E. coli DH5α harboring pUC118-aidC15054 showed a clear AHL-degrading activity level similar to AidC from StRB126. These results may suggest that the aidC gene homolog was not expressed in NBRC 15054 cells. NBRC 15054 appears distant from StRB126 in the phylogenetic analysis. Therefore, these results suggested that the aidC gene homolog may be widespread throughout Chryseobacterium strains, with or without AHL-degrading activity. In addition, in our previous study, aiiM homologous genes (encoding a group of AHL lactonases, AiiM) were only found from the potato leaf-associated Microbacterium isolates, but not found in strains from other resources (27). Correspondingly, although the aiiM homologous genes were highly similar to each other, their neighboring genes showed high diversity (27). When comparing the neighboring genes of aidC and aidC15054, we found it interesting that, although their downstream genes showed no identity, the nearest three upstream genes of aidC shared high identities with the corresponding genes located upstream of aidC15054 (data not shown). These results also suggested that the aidC gene may be widespread in the genus Chryseobacterium.
In summary, in this study, a novel AHL lactonase, AidC, was identified and characterized from a CFB group. Although AidC showed conserved zinc-binding motif sequences, as did the known AiiA-type lactonases, it showed little identity to known AHL lactonases; thus, we classified it into a novel group. In our previous study, AHL-degrading Chryseobacterium strains could be isolated from potato roots, which were collected from various areas (16). Our results suggested that CFB bacteria may also retain functional AHL lactonases, extending the diversity of AiiA-type lactonases and again highlighting the relationship between AHL lactonase and metallo-β-lactamases.
ACKNOWLEDGMENT
This work was supported by Grants-in-Aid from the Bio-oriented Technology Research Advancement Institution (BRAIN), Japan.
Footnotes
Published ahead of print 31 August 2012
REFERENCES
- 1. Afriat-Jurnou L, Jackson CJ, Tawfik DS. 2012. Reconstructing a missing link in the evolution of a recently diverged phosphotriesterase by active-site loop remodeling. Biochemistry 51: 6047–6055 [DOI] [PubMed] [Google Scholar]
- 2. Antunes LC, Ferreira RB. 2009. Intercellular communication in bacteria. Critic. Rev. Microbiol. 35: 69–80 [DOI] [PubMed] [Google Scholar]
- 3. Atkinson S, Williams P. 2009. Quorum sensing and social networking in the microbial world. J. R. Soc. Interface 6: 959–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chhabra SR, et al. 2003. Synthetic analogues of the bacterial signal (quorum sensing) molecule N-(3-oxododecanoyl)-l-homoserine lactone as immune modulators. J. Med. Chem. 46: 97–104 [DOI] [PubMed] [Google Scholar]
- 5. Chow JY, et al. 2010. Directed evolution of a thermostable quorum-quenching lactonase from the amidohydrolase superfamily. J. Biol. Chem. 285: 40911–40920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Kievit TR, Iglewski BH. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68: 4839–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dong YH, et al. 2001. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411: 813–817 [DOI] [PubMed] [Google Scholar]
- 8. Dong YH, Xu JL, Li XZ, Zhang LH. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. U. S. A. 97: 3526–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Flagan S, Ching WK, Leadbetter JR. 2003. Arthrobacter strain VAI-A utilizes acyl-homoserine lactone inactivation products and stimulates quorum signal biodegradation by Variovorax paradoxus. Appl. Environ. Microbiol. 69: 909–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Galloway WR, Hodgkinson JT, Bowden SD, Welch M, Spring DR. 2011. Quorum sensing in Gram-negative bacteria: small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem. Rev. 111: 28–67 [DOI] [PubMed] [Google Scholar]
- 11. McClean KH, et al. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143: 3703–3711 [DOI] [PubMed] [Google Scholar]
- 12. Mei GY, Yan XX, Turak A, Luo ZQ, Zhang LQ. 2010. AidH, an alpha/beta-hydrolase fold family member from an Ochrobactrum sp. strain, is a novel N-acylhomoserine lactonase. Appl. Environ. Microbiol. 76: 4933–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morohoshi T, Kato M, Fukamachi K, Kato N, Ikeda T. 2008. N-acylhomoserine lactone regulates violacein production in Chromobacterium violaceum type strain ATCC 12472. FEMS Microbiol. Lett. 279: 124–130 [DOI] [PubMed] [Google Scholar]
- 14. Morohoshi T, Tominaga Y, Someya N, Ikeda T. 2012. Complete genome sequence and characterization of the N-acylhomoserine lactone-degrading gene of the potato leaf-associated Solibacillus silvestris. J. Biosci. Bioeng. 113: 20–25 [DOI] [PubMed] [Google Scholar]
- 15. Park SY, et al. 2003. AhlD, an N-acylhomoserine lactonase in Arthrobacter sp., and predicted homologues in other bacteria. Microbiology 149: 1541–1550 [DOI] [PubMed] [Google Scholar]
- 16. Rashid R, Morohoshi T, Someya N, Ikeda T. 2011. Degradation of N-acylhomoserine lactone quorum sensing signaling molecules by potato root surface-associated Chryseobacterium strains. Microb. Environ. 26: 144–148 [DOI] [PubMed] [Google Scholar]
- 17. Riaz K, et al. 2008. A metagenomic analysis of soil bacteria extends the diversity of quorum-quenching lactonases. Environ. Microbiol. 10: 560–570 [DOI] [PubMed] [Google Scholar]
- 18. Romero M, Avendano-Herrera R, Magarinos B, Camara M, Otero A. 2010. Acylhomoserine lactone production and degradation by the fish pathogen Tenacibaculum maritimum, a member of the Cytophaga-Flavobacterium-Bacteroides (CFB) group. FEMS Microbiol. Lett. 304: 131–139 [DOI] [PubMed] [Google Scholar]
- 19. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 20. Schipper C, et al. 2009. Metagenome-derived clones encoding two novel lactonase family proteins involved in biofilm inhibition in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 75: 224–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Uroz S, Dessaux Y, Oger P. 2009. Quorum sensing and quorum quenching: the yin and yang of bacterial communication. Chembiochem 10: 205–216 [DOI] [PubMed] [Google Scholar]
- 23. Uroz S, et al. 2008. A Rhodococcus qsdA-encoded enzyme defines a novel class of large-spectrum quorum-quenching lactonases. Appl. Environ. Microbiol. 74: 1357–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Visick KL, Fuqua C. 2005. Decoding microbial chatter: cell-cell communication in bacteria. J. Bacteriol. 187: 5507–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang LH, Weng LX, Dong YH, Zhang LH. 2004. Specificity and enzyme kinetics of the quorum-quenching N-acyl homoserine lactone lactonase (AHL-lactonase). J. Biol. Chem. 279: 13645–13651 [DOI] [PubMed] [Google Scholar]
- 26. Wang WZ, Morohoshi T, Ikenoya M, Someya N, Ikeda T. 2010. AiiM, a novel class of N-acylhomoserine lactonase from the leaf-associated bacterium Microbacterium testaceum. Appl. Environ. Microbiol. 76: 2524–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang WZ, Morohoshi T, Someya N, Ikeda T. 2012. Diversity and distribution of N-acylhomoserine lactone (AHL)-degrading activity and AHL-lactonase (AiiM) in genus Microbacterium. Microbes Environ. 27: 330–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang HB, Wang LH, Zhang LH. 2007. Detection and analysis of quorum-quenching enzymes against acyl homoserine lactone quorum-sensing signals. Curr. Protoc. Microbiol. 1: 1C.3 doi:10.1002/9780471729259.mc01c03s05 [DOI] [PubMed] [Google Scholar]
- 29. Zhang HB, Wang LH, Zhang LH. 2002. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U. S. A. 99: 4638–4643 [DOI] [PMC free article] [PubMed] [Google Scholar]