Abstract
Despite the scientific and industrial importance of desiccation tolerance in Salmonella, knowledge regarding its genetic basis is still scarce. In the present study, we performed a transcriptomic analysis of dehydrated and water-suspended Salmonella enterica serovar Typhimurium using microarrays. Dehydration induced expression of 90 genes and downregulated that of 7 genes. Ribosomal structural genes represented the most abundant functional group with a relatively higher transcription during dehydration. Other main induced functional groups included genes involved in amino acid metabolism, energy production, ion transport, transcription, and stress response. The highest induction was observed in the kdpFABC operon, encoding a potassium transport channel. Knockout mutations were generated in nine upregulated genes. Five mutants displayed lower tolerance to desiccation, implying the involvement of the corresponding genes in the adaptation of Salmonella to desiccation. These included genes encoding the isocitrate-lyase AceA, the lipid A biosynthesis palmitoleoyl-acyltransferase Ddg, the modular iron-sulfur cluster scaffolding protein NifU, the global regulator Fnr, and the alternative sigma factor RpoE. Notably, these proteins were previously implicated in the response of Salmonella to oxidative stress, heat shock, and cold shock. A strain with a mutation in the structural gene kdpA had a tolerance to dehydration comparable to that of the parent strain, implying that potassium transport through this system is dispensable for early adaptation to the dry environment. Nevertheless, this mutant was significantly impaired in long-term persistence during cold storage. Our findings indicate the involvement of a relatively small fraction of the Salmonella genome in transcriptional adjustment from water to dehydration, with a high prevalence of genes belonging to the protein biosynthesis machinery.
INTRODUCTION
As part of its life cycle Salmonella can persist outside the host where it is exposed to various stresses (73), with desiccation being one of the most common and significant ones. Salmonella can survive from several weeks to several years on dry surfaces (21, 36, 48), eggshells (55), almond kernels (68), pecans (11), and dry confectionery raw materials (47). During the last decade, Salmonella has been involved in several national and international outbreaks related to consumption of low-water-activity foods such as snacks, almonds, peanut butter, chocolate, and paprika (4, 5, 6, 37, 45, 71).
In spite of the scientific importance and the practical implications of desiccation tolerance in Salmonella, the exact mechanisms of its cellular adaptation to desiccation remain relatively poorly understood. Several studies have investigated desiccation tolerance in Salmonella. Extracellular components such as thin aggregative fimbriae and cellulose as well as the lipopolysaccharide (LPS) core unit are apparently required for optimal desiccation tolerance (28, 29, 72). Exposure of Salmonella to desiccation stress increased cross-tolerance to a number of other stresses, including high temperature, osmotic stress, and biocides (30). We recently demonstrated that inhibition of protein synthesis by chloramphenicol significantly decreases the persistence of dried Salmonella cells (31), indicating a requirement for de novo protein synthesis for successful adaptation to desiccation stress.
In the present study, we have extended our studies and further analyzed global gene expression in hydrated and dehydrated Salmonella cells. Nine of the genes that were relatively upregulated in dehydration versus water were mutagenized, and their involvement in adaptation to desiccation stress was evaluated by determining the dehydration tolerance (DT) phenotype of each of the mutants.
MATERIALS AND METHODS
Bacterial strains, inoculum preparation, and growth conditions.
Salmonella enterica serovar Typhimurium SL1344 was used in this study. Bacteria were routinely grown on xylose-lysine-deoxycholate (XLD) agar (Difco Laboratories, Sparks, MD). For each experiment, a fresh colony was spread on a Luria-Bertani (LB) (Difco Laboratories) agar plate with a sterile swab and incubated for 20 h at 37°C. Cells were harvested by resuspension in sterile double-deionized water (SDDW) using a sterile rubber policeman and washed three times in SDDW (3,800 × g for 5 min) at room temperature, and the pellet was resuspended in SDDW to a final concentration of ∼2.0 × 109 CFU/ml (optical density at 600 nm [OD600] of 1.0).
Dehydration conditions.
Salmonella suspensions (13 ml) were placed in plastic 90-mm petri dishes (Miniplast, Ein-Shemer, Israel) at a concentration of ∼2.0 × 109 CFU/ml and air dried in a biosafety cabinet for 22 h at 25°C under ca. 40% relative humidity (RH). Nondesiccated (control) bacteria (at the same volume and concentration) were incubated for 22 h at 25°C in 50 ml polypropylene screw-cap tubes (Labcon, Petaluma, CA). Under these conditions essentially no evaporation occurred.
RNA extraction.
Desiccated cells were resuspended in the original volume (13 ml) of SDDW containing 20% stop solution (5% H2O-phenol and 95% ethanol). Cells were mixed gently, transferred to 50-ml polypropylene screw-cap tubes, incubated on ice for 30 min, and centrifuged for 10 min at 4°C at 3,800 × g. The pellet was resuspended in 5 ml of the supernatant and divided into four 1.5-ml vials. Aliquots were centrifuged at 4°C (3,800 × g, 5 min), and RNA was extracted from the cell pellet using the SV total RNA isolation system (Promega, Madison, WI) according to the manufacturer's instructions. Nondesiccated (control) cells were spun down by centrifugation (3,800 × g, 5 min), and the pellet was resuspended in 13 ml of SDDW supplemented with 20% stop solution. Cells were mixed gently by pipetting and treated as described above for the desiccated cells.
Transcriptomic analysis.
RNA labeling, hybridization to the microarrays, scanning, and data analysis were performed at the microarray core facility, Faculty of Medicine, Ein Kerem, The Hebrew University, Jerusalem. RNAs derived from dried and control Salmonella cells were labeled with Cy3 and Cy5 and hybridized to the Salmonella open reading frame (ORF) microarray platform STv7E (GPL11279 in the Gene Expression Omnibus [GEO] database at www.ncbi.nlm.nih.gov/geo), which represented 97.6% of all chromosomal ORFs (but only 37% of all plasmid ORFs) of Salmonella Typhimurium strain SL1344. A total of four separate experiments were performed with a Cy3 and Cy5 dye swap. Arrays were scanned using the Axon GenePix 4000B scanner (Molecular Devices LLC, Sunnyvale, CA). Image analysis and feature extraction were performed using the Axon GenePix Pro software (Molecular Devices). The data were analyzed using the LIMMA software package (67). Fold change was calculated by comparing averaged normalized signal intensities in dehydrated versus nondehydrated (control) cells. A moderated t test was performed in parallel, with the use of a false discovery rate correction for multiple testing (62). A P value of <0.05 was used to pinpoint significantly differentiated genes. A cutoff of a 1.8-fold change for up- or downregulated expression was chosen to define differential expression.
Bioinformatic analysis.
Functional analysis of the identified differentially expressed genes was performed using Blast2GO (18) at http://www.blast2go.com/b2ghome. Operon prediction and pathway analysis were performed using MicrobesOnline Operon Predictions (60) at http://www.microbesonline.org/operons/ and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database at http://www.genome.jp/kegg/pathway.html, respectively.
qRT-PCR.
Quantitative RT-PCR (qRT-PCR) was performed at GCA (Galil Genetics Ltd., Katzrin, Israel). Briefly, total RNAs (500 ng) of desiccated and nondesiccated bacteria were converted to cDNAs using the QuantiTect reverse transcription kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. RNA-DNA hybrids were degraded by RNase H activity. Quantitative PCR was performed in 48.48 Dynamic Array integrated fluidic circuits (IFCs) (Fluidigm, San Francisco, CA) using the EvaGreen DNA-binding dye (Biotium Inc., Hayward, CA). The primers for each of the target genes are listed in Table S1 in the supplemental material. Melting curves were used to allow assessment of the reaction quality. Expression values for both treatments (desiccated and control) were normalized to the expression levels of the rsuA gene, coding for 16S rRNA pseudouridylate synthase A, which did not exhibit differential expression in comparative microarray analyses. The reactions were performed with two independent RNA preparations, each with three repeats. Results were analyzed using the Fluidigm real-time PCR analysis software v.2.1.3 and the Fluidigm data collection software v.2.1.3. Fold change was calculated as the average relative quantification (RQ = 2−ΔΔCT) values for treated (dehydrated) and control (SDDW) samples from two independent experiments.
Generation of mutants and P22 phage transduction.
Target deletion mutations were generated in S. Typhimurium by the lambda red recombinase procedure (20), using primers listed in Table S1 in the supplemental material, with the pKD4 plasmid carrying a Kanr cassette as the template for PCRs. Mutations were verified using nearby locus-specific primers (see Table S1 in the supplemental material) with the respective primer k2 or kt (20) and by sequencing. To confirm that the phenotypes of the mutants were linked to the mutations, each mutation was reintroduced into a wild-type (WT) background via transduction, using phage P22 HT105/1 int-201 according to a previously published protocol (52). Transfer of each mutation was confirmed by PCR and sequence analysis, as described for the mutants.
DT and LTP.
Bacteria were dehydrated in 90-mm empty petri dishes for 22 h, as described above. Following dehydration, bacteria were resuspended in 13 ml of SDDW and incubated for 5 min at room temperature. Bacteria were collected, transferred into a 50-ml polypropylene screw-cap tube, and mixed by pipetting. Bacterial suspensions were serially diluted (1:10) and plated on LB agar. Viable counts were determined following incubation for 20 h at 37°C. Bacterial survival was determined at 22 h. Dehydration tolerance (DT) of the mutants is presented as percentage of the wild-type (WT) survival. For testing long-term persistence (LTP), cells were desiccated in a 96-well polystyrene plate (Greiner Bio-One, Frickenhausen, Germany) as described previously (30). The dried plates were incubated either at 4°C (40 to 45% RH) or in a climate-controlled 25°C incubator (Climacell; MMM Group, Munich, Germany) at 40% RH, and the viable counts were determined after 12 weeks of storage. LTP is presented as the total viable count (CFU) of surviving bacteria.
Survival in water.
Salmonella strains were stored in 13 ml SDDW for 22 h in 50-ml polypropylene screw-cap tubes, as described in “Dehydration conditions” above. The viable count was determined at time zero and after 22 h, as described above.
Microarray data accession number.
Data from the transcriptomic analysis experiments are accessible via GEO accession number GSE38475.
RESULTS
Identification of differentially expressed genes.
A total of 90 relatively upregulated and 7 downregulated genes were identified. The general functions of the differentially expressed genes are summarized in Table 1. Four out of seven downregulated genes are involved in maintenance of the Salmonella virulence plasmid; two others encode a putative catalase (STM1731) and a small RNA gene (tka1). The cutC copper homeostasis gene demonstrated the strongest decrease in gene expression (4.1-fold decrease).
Table 1.
Differentially regulated Salmonella Typhimurium SL1344 genes after 22 h of dehydrationa
| Category, gene, and/or locus tagb | Gene/protein annotation | Fold changec |
|---|---|---|
| Translation/ribosomal structure | ||
| deaD (STM3280.S) | Cysteine sulfinate desulfinase | 2.68 |
| ftsJ (STM3297) | 23S rRNA methyltransferase | 2.89 |
| rpsD (STM3416) | 30S ribosomal protein S4 | 2.40 |
| rpsK (STM3417) | 30S ribosomal protein S11 | 2.29 |
| rplO (STM3421) | 50S ribosomal protein L15 | 2.14 |
| rpsH (STM3426) | 30S ribosomal protein S8 | 2.39 |
| rplE (STM3428) | 50S ribosomal protein L5 | 3.16 |
| rplX (STM3429) | 50S ribosomal protein L24 | 2.69 |
| rpsQ (STM3431) | 30S ribosomal protein S17 | 2.25 |
| rplP (STM3433) | 50S ribosomal protein L16 | 2.92 |
| rpsC (STM3434) | 30S ribosomal protein S3 | 2.75 |
| rplB (STM3437) | 50S ribosomal protein L2 | 3.49 |
| rplW (STM3438) | 50S ribosomal protein L23 | 2.62 |
| rplD (STM3439) | 50S ribosomal protein L4 | 2.82 |
| rplC (STM3440) | 50S ribosomal protein L3 | 2.84 |
| rpsJ (STM3441) | 30S ribosomal protein S10 | 2.69 |
| fusA (STM3446) | Elongation factor EF-2 | 1.93 |
| rpsG (STM3447) | 30S ribosomal protein S7 | 2.16 |
| rplK (STM4149) | 50S ribosomal protein L11 | 2.33 |
| rplA (STM4150) | 50S ribosomal protein L1 | 2.02 |
| rplJ (STM4151) | 50S ribosomal protein L10 | 2.02 |
| miaA (STM4360) | tRNA delta(2)-isopentenylpyrophosphate transferase | 1.97 |
| Amino acid transport and metabolism | ||
| hisD (STM2072) | Histidinol dehydrogenase | 2.44 |
| hisC (STM2073) | Histidinol-phosphate aminotransferase | 2.67 |
| hisB (STM2074) | Imidazole glycerol-phosphate dehydratase/histidinol phosphatase | 2.23 |
| hisH (STM2075) | Imidazole glycerol phosphate synthase subunit HisH | 2.34 |
| hisA (STM2076) | 1-(5-Phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino]imidazole-4-carboxamide isomerase | 2.39 |
| hisF (STM2077) | Imidazole glycerol phosphate synthase subunit | 2.68 |
| glpB (STM2285) | Anaerobic glycerol-3-phosphate dehydrogenase subunit B | 2.18 |
| nifS (STM2543) | Cysteine desulfurase | 2.23 |
| argA (STM2992) | N-Acetylglutamate synthase | 2.30 |
| ygjU (STM3225) | Putative dicarboxylate permease | 2.04 |
| argG (STM3290.S) | Argininosuccinate synthase | 2.52 |
| gltB (STM3330) | Glutamate synthase large subunit | 2.42 |
| gltD (STM3331) | Glutamate synthase small subunit | 2.68 |
| prlC (STM3594) | Oligopeptidase A | 2.18 |
| argE (STM4120) | Acetylornithine deacetylase | 1.99 |
| argC (STM4121) | N-Acetyl-gamma-glutamyl-phosphate reductase | 2.26 |
| argB (STM4122) | Acetylglutamate kinase | 3.03 |
| argD (STM1303) | Bifunctional aconitate hydratase 2,2-methylisocitrate dehydratase | 1.83 |
| argI (STM4469) | Ornithine carbamoyltransferase | 1.83 |
| Stress response | ||
| dnaK (STM0012) | Molecular chaperone DnaK | 3.05 |
| hflB (STM3296) | ATP-dependent zinc-metallo protease | 2.84 |
| yhgI (STM3511) | Putative thioredoxin-like protein | 2.34 |
| ydaA (STM 1661) | Universal stress protein UspE | 1.90 |
| ibpA (STM3809.S) | Small heat shock protein | 2.75 |
| hslU (STM4091) | ATP-dependent protease heat shock protein | 2.19 |
| groEL (STM4330) | Chaperonin GroEL | 2.11 |
| sufD (STM1372) | Cysteine desulfurase modulator | 1.86 |
| Transcription | ||
| rpoE (STM2640) | RNA polymerase sigma factor | 2.53 |
| rpoA (STM3415) | DNA-directed RNA polymerase alpha subunit | 2.16 |
| rpoH (STM3568) | RNA polymerase sigma factor | 2.48 |
| rpoB (STM4153) | DNA-directed RNA polymerase beta subunit | 2.37 |
| rpoC (STM4154) | DNA-directed RNA polymerase beta subunit | 2.36 |
| rseA (STM2639) | Sigma factor RpoE negative regulatory protein RseA | 2.05 |
| fnr (STM1660.S) | Transcriptional regulator | 1.84 |
| greA (STM3299) | Transcription elongation factor | 1.82 |
| Energy production and conversion | ||
| icdA (STM1238) | Isocitrate dehydrogenase | 2.35 |
| fumA (STM1468) | Fumarase A | 2.17 |
| glpC (STM2286) | sn-Glycerol-3-phosphate dehydrogenase K small subunit | 2.43 |
| nifU (STM2542) | NifU-like protein | 2.70 |
| glpK (STM4086) | Glycerol kinase | 3.19 |
| aceB (STM4183) | Malate synthase | 2.92 |
| aceA (STM4184) | Isocitrate lyase | 3.34 |
| Inorganic ion transport | ||
| kdpC (STM0704) | Potassium-transporting ATPase C chain | 4.51 |
| kdpB (STM0705) | Potassium-transporting ATPase subunit B | 5.02 |
| kdpA (STM0706) | Potassium-transporting ATPase subunit A | 4.67 |
| cutC (STM1907) | Copper homeostasis protein | 0.24 |
| Carbohydrate transport and metabolism | ||
| glgA (STM3535) | Glycogen synthase | 1.89 |
| mglB (STM2190) | Galactose transport protein | 2.50 |
| pps (STM1349) | Phosphoenolpyruvate synthase | 2.00 |
| mtlA (STM3685) | Mannitol-specific enzyme IIABC component | 2.19 |
| Lipid transport and metabolism | ||
| ddg (STM2401) | Lipid A biosynthesis lauroyl acyltransferase | 3.69 |
| nlpD (STM2925) | Lipoprotein | 2.20 |
| nlpI (STM3281) | Lipoprotein | 1.97 |
| Other or unknown function | ||
| prpE (STM0371) | Putative acetyl coenzyme A synthetase | 1.93 |
| prpD (STM0370) | 2-Methylcitrate dehydratase | 1.83 |
| phoL (STM0669) | Putative phosphate starvation-inducible protein | 1.90 |
| yceD (STM1190) | Putative metal-binding protein | 2.08 |
| ycjX (STM1685) | Putative ATPase | 1.94 |
| sixA (STM2387) | Phosphohistidine phosphatase | 1.96 |
| iscA (STM2541) | Iron-sulfur cluster assembly protein | 3.13 |
| yggN (STM3107) | Putative periplasmic protein | 3.06 |
| exbB (STM3159) | Energy transduction protein | 2.05 |
| pckA (STM3500) | Phosphoenolpyruvate carboxykinase | 1.90 |
| slsA (STM3761) | Putative inner membrane protein | 3.35 |
| aceK (STM4185) | Bifunctional isocitrate dehydrogenase kinase/phosphatase protein | 2.69 |
| phoH (STM1126) | Phosphate starvation-inducible protein | 1.95 |
| minE (STM1816) | Cell division topological specificity factor MinE | 1.85 |
| yahO (STM0366) | Periplasmic protein of unknown function | 1.80 |
| ybeL (STM0653) | Hypothetical protein | 1.85 |
| tke1 (STM_sRNA) | Small RNA | 0.53 |
| STM1731 | Putative catalase | 0.54 |
| Virulence | ||
| parA (PSLT052) | Plasmid partition protein A | 0.43 |
| traN (PSLT095) | Mating pair stabilization protein | 0.44 |
| trbH (PSLT105) | Conjugative transfer protein | 0.41 |
| PSLT068 | Putative ParB-like nuclease | 0.51 |
All of the listed genes were significantly differentiated in expression compared to the control at a P value of <0.05.
Derived from S. Typhimurium strain LT2 (NCBI site).
The fold change is the ratio of the gene expression level of the dehydrated cells to that of the control maintained in water. Values lower than 1 represent decreased expression.
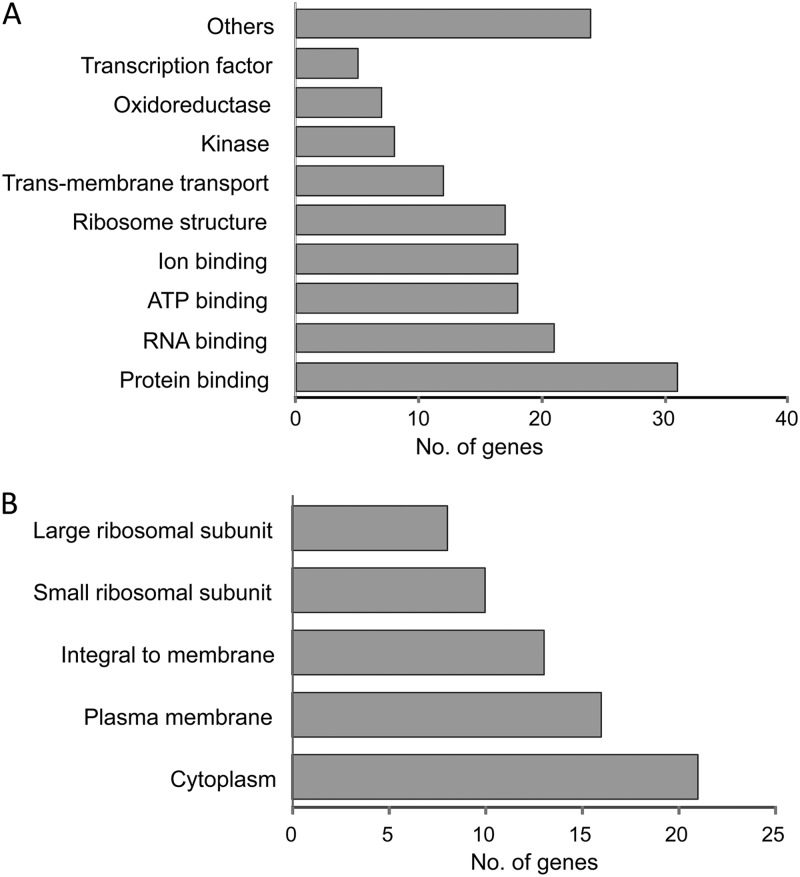
The largest group of upregulated genes (22 genes) belonged to the ribosome structure and biogenesis functional group, while the second-largest group (19 genes) included amino acid transport and metabolism genes. Five other functional groups involved genes associated with energy production and conversion (7 genes), stress response (8 genes), transcription (8 genes), and carbohydrate (4 genes) and lipid (3 genes) transport and metabolism. Sixteen genes had other or unknown functions. The highest induction (up to 5-fold) was observed in the kdpA, kdpB, and kdpC genes of the kdpFABC operon, encoding a potassium transport channel. The identified stress-induced genes included genes encoding three molecular chaperones (DnaK, GroEL, and IbpA), two proteases (HslU and HflB), a universal stress protein (YdaA), a putative thioredoxin-like protein (YhgI), and a cysteine desulfurase modulator (SufD). Other stress-related induced genes were rpoE and rpoH, encoding alternative sigma factors (σE and σ32).
Upregulated genes were also classified by either cellular process or cellular component (Fig. 1). Most of the identified genes had protein-binding (32 genes) or RNA-binding (21 genes) activities (Fig. 1A). Classification of the induced genes according to likely cellular compartment revealed that 29 gene products were membrane associated, 21 were located in the cytoplasm, and 18 represented ribosomal proteins.
Fig 1.
Clustering of dehydration-induced genes according to cellular function (A) or cellular component (B). Analysis was performed with Blast2GO using direct gene ontology (GO) count.
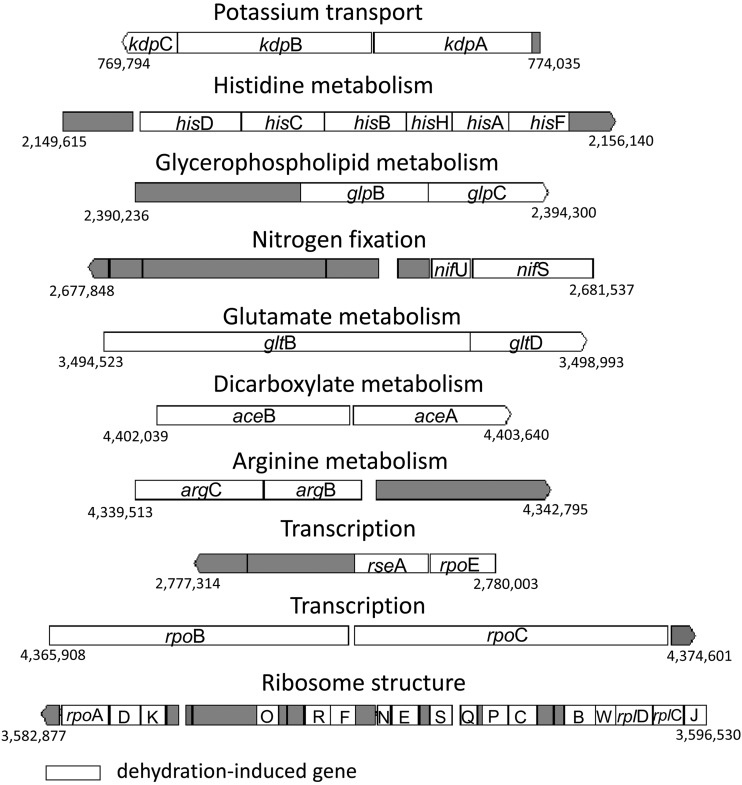
Forty-one out of the 90 upregulated genes belong to 10 distinct operons involved in metabolism of arginine, histidine, glutamate, dicarboxylate, and glycerophospholipid as well as potassium transport, nitrogen fixation, ribosome structure, and transcription (two operons) (Fig. 2). Because genes of distinct operons may be involved in the same metabolic pathways, the upregulated genes were also analyzed with the KEGG pathway tool. Seven genes belonging to six distinct operons were found to be associated with arginine metabolism (data not shown). The argBC, argA, argD, and argE genes are involved in biosynthesis of ornithine from glutamate, argI catalyzes the biosynthesis of citruline, and argG catalyzes synthesis of l-argino-succinate from aspartate.
Fig 2.
Operons containing more than one dehydration-induced gene. Analysis was performed using the MicrobesOnline Operon Prediction tool (60). Induced genes are marked with white background. The letters in the ribosomal operon denote the following: D, rpsD; K, rpsK; O, rplO; R, rplR; N, rpsN; E, rplE; S, rplN; Q, rpsQ; P, rplP; C, rpsS; B, rplB; W, rplW; and J, rpsJ. Numbers indicate the chromosomal location of the operon in S. Typhimurium strain LT2 (NCBI site).
Confirmation of microarray results by qRT-PCR.
Seven upregulated genes from different functional groups (ddg, glpK, dnaK, hisD, aceA, nifU, and kdpA), displaying the highest fold change in the microarray, were selected for further study. qRT-PCR analysis performed on RNA derived from two independent experiments confirmed the increased expression of these genes (Table 2). In general, the average expression levels of all tested genes were ∼5- to 23-fold higher in desiccated cells, compared to an unchanged control transcript.
Table 2.
Confirmation of selected dehydration-induced genes by qRT-PCR analysis
| Gene | Fold changea determined by: |
|
|---|---|---|
| Microarray analysis | qRT-PCR | |
| aceA | 3.3 | 4.7 (0.1) |
| glpK | 3.2 | 12.9 (1.6) |
| ddg | 3.7 | 13.5 (3.5) |
| nifU | 2.7 | 11.3 (1.6) |
| kdpA | 4.7 | 23.2 (4.2) |
| hisD | 2.4 | 17.1 (5.4) |
| dnaK | 3.0 | 23.4 (6.6) |
Calculation of fold change is described in Materials and Methods. Values in parentheses represent standard deviations from two independent qRT-PCR experiments, each performed in triplicate.
Dehydration tolerance in knockout mutants.
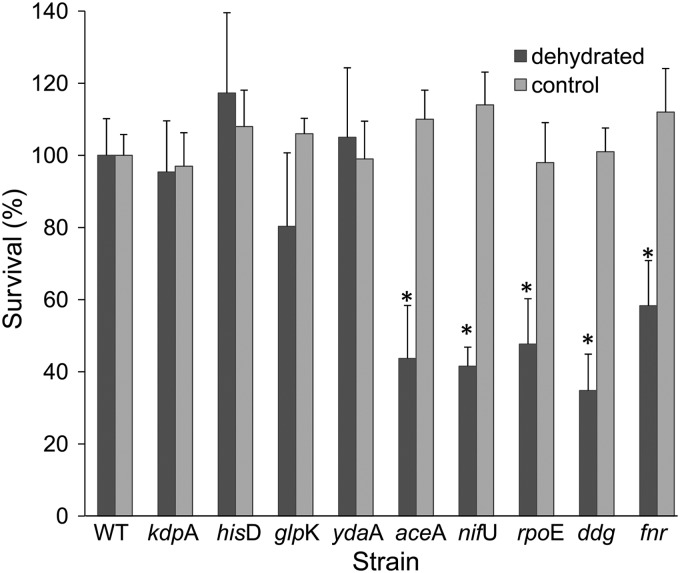
Knockout mutations were generated in 6 of the 7 genes. Mutagenesis of dnaK was unsuccessful, confirming previous studies reporting that this gene is under strong selection in rich media (14) and is essential in Salmonella Typhimurium strain 14028 (46). Knockout mutations were also generated in three other genes observed to be upregulated in arrays and that had previously been linked to environmental stresses. These included the universal stress UspE-like protein gene ydaA, the alternative sigma factor gene rpoE, and the global regulator gene fnr (2, 25, 49, 54, 63). Five of the nine mutants (the ΔaceA, ΔnifU, ΔrpoE, Δddg, and Δfnr mutants) were compromised in their dehydration tolerance, demonstrating ∼40% survival compared to the WT strain (Fig. 3). Notably, none of these mutants was impaired in survival in SDDW, suggesting that these mutations specifically affected dehydration tolerance. The survival of the ΔhisD, ΔglpK, ΔkdpA, and ΔydaA knockout mutants was comparable to that of the WT strain (Fig. 3). To confirm a genetic linkage between the mutation and the observed dehydration tolerance phenotype, each mutation was reintroduced back into the wild-type strain by P22 phage transduction. Survival of the transductants was comparable to that of the mutants (data not shown).
Fig 3.
Survival of Salmonella strains. Salmonella strains at a concentration of ∼2 × 109 CFU/ml were dehydrated at ambient temperature for 22 h or kept wet in SDDW under similar conditions (control). Survival is expressed as a percentage of viable cells relative to the WT strain (considered 100%) at 22 h. The viable count of the WT in SDDW (control) remained constant for 22 h, while the count of dehydrated WT cells was usually ∼1 × 109 CFU/ml. The data represent the mean percentage of survival in three independent experiments, each performed in triplicates on three separate days. Error bars denote the standard deviations of the means (n = 9). Asterisks indicate a significant difference (P < 0.05) compared to the WT strain.
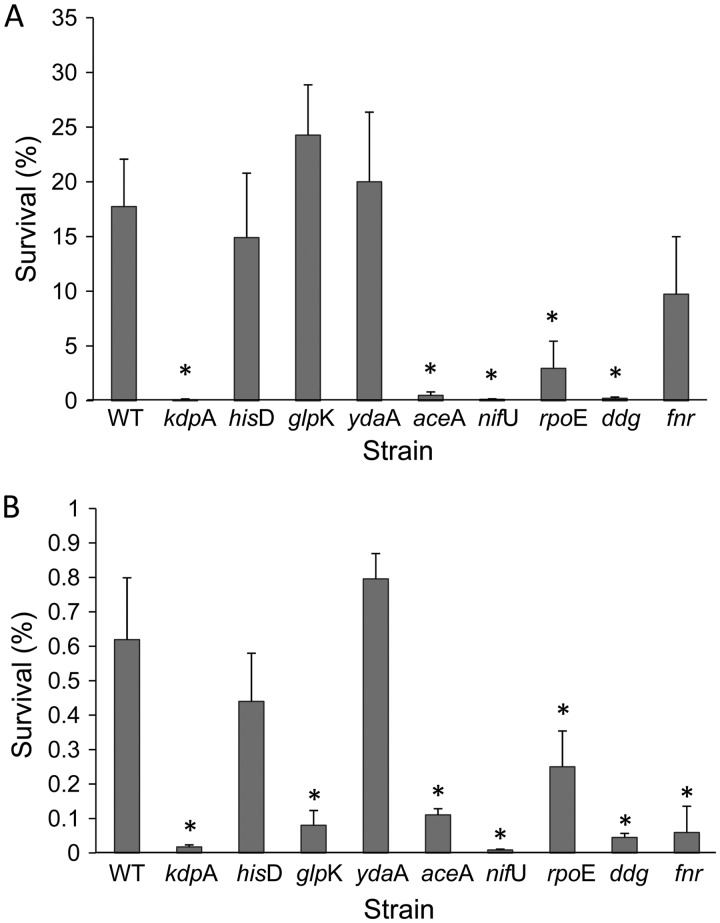
Long-term persistence during storage at 25°C and 4°C.
The long-term survival of the wild-type strain at 4°C was 17% (Fig. 4A), while storage at 25°C resulted in 0.6% survival (Fig. 4B). Most of the mutants, except for the ΔydaA and ΔhisD mutants, demonstrated significantly (P < 0.05) lower persistence at 25°C than the WT strain. Four of the five mutants (the ΔnifU, ΔrpoE, Δddg, and ΔaceA mutants), which were impaired in their dehydration tolerance, also had reduced long-term persistence at 4°C. Interestingly, the ΔkdpA mutant, which was not compromised in dehydration tolerance, had strongly impaired long-term persistence at 4°C.
Fig 4.
Long-term persistence of dehydrated Salmonella strains. Bacteria were dried for 22 h at 25°C and subsequently stored for 12 weeks at 4°C (A) or 25°C (B). For each strain, survival is expressed as the percentage of viable cells at 12 weeks relative to time zero (immediately following dehydration). The data represent the mean percentage of survival in three independent experiments, each performed in triplicate on three separate days. Error bars denote standard deviations of the means (n = 9). Asterisks indicate a significant difference (P < 0.05) in survival between the mutants and the WT strain.
DISCUSSION
In a previous study, we have reported that SDDW-suspended Salmonella develops cross-tolerance to multiple stresses following dehydration (30). In order to gain further insight into the mechanisms underlying Salmonella desiccation in SDDW, we have extended the study and identified genes which are differentially expressed in dehydrated Salmonella. The use of SDDW in treatment and control bacteria has several advantages over use of an isotonic medium in a dehydration model. First, we observed in preliminary experiments that Salmonella dehydrated in isotonic medium (saline; 0.85% NaCl) was less tolerant to desiccation than SDDW-suspended cells (data not shown). Second, using isotonic medium for desiccation experiments would have imparted a hyperosmotic stress at early stages of dehydration, due to the continuous increase in salt concentration that accompanies water loss, while the control bacteria will remain under isotonic conditions.
Four of the seven downregulated genes were found to be located on the Salmonella virulence plasmid (33). These genes are known to be induced during growth arrest, reduced nutrient supply, high temperature, iron limitation or under low pH (32, 51, 65), conditions representing the host environment. Because both the dehydrated and control cells were incubated in SDDW, a nutrient-deficient medium, the downregulation of these genes may indicate that the drying cells encountered physiological changes, likely affecting their expression. Previously, we have reported that under a similar dehydration regimen, Salmonella mortality reaches about 50%, while the viable number of bacteria incubated in SDDW remained constant (30). It is possible that the presence of nutrients released from the dead cells negatively affected the expression of the virulence plasmid genes. We note that only SL1344 plasmid 1 (SLP1) genes are represented on the microarray used for detection, while genes from plasmids SPL2 and SPL3 are not. Transcription of genes of those two plasmids, particularly those involved in plasmid maintenance, may also be downregulated in response to the desiccation conditions used here.
Under nutrient starvation and growth arrest, bacteria downregulate multiple genes involved in transcription and translation (16, 40, 58). Yet, in our study, the desiccation stress resulted in a considerable relative upregulation of numerous transcription- and translation-associated genes, including genes encoding DNA-directed RNA polymerase subunits (RpoA, RpoB, and RpoC) and ribosomal proteins. These findings are in agreement with the notion that the drying cells are exposed to nutrients released from dead cells, which might fuel the intense metabolism likely taking place in dehydrating cells. Consistent with this hypothesis, we observed an increase in the extracellular total organic carbon (TOC) in dehydrated compared to SDDW-suspended cells (data not shown). An additional source of nutrients might be residues released from the extracellular polymorphic substances (EPS) known to be produced in bacteria during desiccation (12, 59). Whether the drying cell can utilize the released organic matter for their metabolic rearrangement is yet to be determined.
The apparent relative increase in the machinery of transcription and translation in the drying cells indicates that de novo protein synthesis is necessary in order to adapt to the waterless environment. Whether the source of nutrients for these activities is exogenous or endogenous still needs to be determined. It has been previously reported that de novo protein synthesis takes place during carbon starvation via peptidase-dependent autophagy (57). The upregulation of the two proteases genes (hflB and hslU) likely to be involved in the degradation of misfolded proteins in the dehydrated bacteria may support this notion.
We found that the histidine and arginine biosynthetic pathways were induced during dehydration relative to bacteria maintained in water (Table 1), which perhaps indicates an additional role for histidine and arginine in adaptation of Salmonella to desiccation. Similarly, although most amino acid biosynthesis genes in E. coli are downregulated during transient growth arrest caused by glucose-lactose diauxie, the histidine and arginine biosynthesis genes were induced (16). Interestingly, both regulons are controlled by the argR repressor, while the biosynthesis genes of the other amino acids are controlled by the transcription factors Lrp and TrpR (17). Whether the histidine and arginine biosynthetic pathways were already upregulated during earlier dehydration stages or specifically induced under late-dehydration conditions remained to be studied.
We found that the glyoxylate shunt pathway genes encoding isocitrate lyase (aceA) and malate synthetase (aceB) were induced under dehydration. The glyoxylate shunt pathway is a latent pathway which is induced under glucose starvation (26). This pathway was also reported to be induced in Salmonella during anaerobic growth in the presence of hydrogen (50) and is required for persistence of the pathogen during chronic infection in mice (24). Increased activity of isocitrate lyase as well as the whole glyoxylate shunt pathway was observed in Escherichia coli exposed to superoxide (64). The authors hypothesized that increased metabolites flow through the glyoxylate shunt instead of the tricarboxylic acid (TCA) cycle, which reduces the amount of NADH produced from glucose, contributing to a decrease in reactive oxygen species production by respiration and thus helping to relieve oxidative stress. It is possible that induction of the glyoxylate shunt pathway plays a similar role in protecting Salmonella from oxidative radicals, which are known to be produced during desiccation (59). Indeed, mutation in the aceA gene lowered the persistence of Salmonella during dehydration, supporting the notion that the glyoxylate shunt pathway is involved in the adaptation of Salmonella to desiccation stress.
The induction of a number of oxidative stress response genes, such as the NifU-like protein gene iscA, the cysteine desulfurase modulator gene sufD, and the putative thioredoxin-like protein gene yhgI (8, 70), additionally supports the existence of oxidative stress during dehydration. IscA and SufD are members of two families of proteins involved in generalized iron-sulfur (Fe-S) protein maturation (8). Iron-sulfur clusters are ubiquitous and evolutionarily ancient prosthetic groups which participate in various cell processes, such as electron transfer, substrate binding/activation, iron/sulfur storage, regulation of gene expression, and enzyme activity (8). Two other genes involved in Fe-S cluster assembly, nifU and nifS, were also induced during dehydration. NifU and NifS are scaffolding proteins which act together to generate Fe-S cluster precursors destined for nitrogenase maturation (38). Mutation in the nifU gene resulted in decreased dehydration tolerance and compromised long-term persistence at both low and high storage temperatures. Due to the complexity of the cellular processes utilizing Fe-S clusters, it is difficult to point to a particular process affected by the nifU deletion in the dehydrated cells. Another gene which was induced during dehydration is the fnr gene. It encodes an oxygen-sensing transcriptional regulator. FNR (fumarate and nitrate reduction protein) is a DNA-binding global regulator which coordinates the transcription of diverse operons involved in motility, metabolism, and virulence under low-O2 growth in Salmonella and E. coli (25, 41). FNR oxygen sensing is mediated by oxygen-induced conversion of the DNA-binding dimeric [4Fe-4S]2+ cluster to a monomeric [2Fe-2S]2+ form (38). Knockout of fnr also diminished Salmonella survival following dehydration and after long-term storage at 25°C. However, unlike the ΔnifU mutant, the Δfnr mutant was not impaired in long-term persistence at 4°C, suggesting that the mechanisms by which nifU and fnr contribute to desiccation tolerance, at least at 4°C, do not necessarily overlap.
Recent findings demonstrating the ability of the NifU homolog IscU to bind and stimulate ATPase activity of Hsp70-type molecular chaperones in E. coli (66) may provide a clue regarding additional mechanisms involved in the adaptation of Salmonella to desiccation stress. We found that transcription of the dnaK gene, encoding Hsp70, was upregulated during dehydration. Other heat shock protein genes (groEL and ibpA) were also induced. Because these chaperones stabilize proteins during other stress (2, 68), it is likely that they also protect proteins during dehydration stress. Several other stress-related genes, including a universal stress protein gene (ydaA) as well as the alternative sigma factor genes rpoE and rpoH, were also induced during dehydration. The alternative sigma factor RpoE controls the expression of many genes involved in diverse stresses, including heat, oxidation, osmotic, and starvation stress (3, 35, 54, 61, 63). In agreement with the known role of RpoE in the general stress response, the ΔrpoE mutant exhibited a significant decrease in dehydration tolerance and long-term persistence. Salmonella possesses several universal stress proteins, designated Usp, which are induced in response to multiple stress, and their precise function is still not clear (49). Nonetheless, deletion of the ydaA gene, encoding a UspE-like protein, did not yield a desiccation tolerance phenotype.
Another gene which was induced during desiccation is ddg, encoding lipid A biosynthesis palmitoleoyl acyltransferase. Ddg was previously reported to be involved in cold shock in E. coli. Ddg modifies the lipid A composition in bacteria that are transferred from 30°C to 12°C, by replacing laurate with palmitoleate (15). This modification enables the cells to maintain optimal outer membrane fluidity under low-temperature conditions. Maintaining membrane fluidity is a key feature required for the cells to withstand desiccation stress. Dehydration of membrane bilayers increases van der Waal's interactions between adjacent phospholipids, contributing to membrane transition to the gel phase at ambient temperatures (59). This results in segregation of the membranes and aggregation of proteins, leading to leakage and loss of solutes (10). The finding that the Δddg mutant was impaired in dehydration tolerance and LTP supports the idea that modification of the lipid A structure is essential for Salmonella persistence in a low-water environment. It will be of interest to determine the lipid A compositions in hydrated and dehydrated Salmonella cells.
Among the dehydration-induced genes, the highest upregulation was observed in the kdpABC genes. This operon encodes a high-affinity K+ uptake system in many bacteria. The system consists of four subunits: the potassium transport channel protein (KdpA), a P-type ATPase (KdpB), and two additional subunits, KdpC and KdpF, which are involved in complex assembly and stabilization, respectively (9). This potassium transport complex is induced under K+-limiting growth conditions and in response to osmotic upshift (27, 39). Induction of the Kdp system was also observed in cyanobacteria in response to desiccation stress (43). Loss of water during dehydration increases intracellular ion strength and causes loss of turgor (59), two signals that trigger KdpFABC induction in enterobacteria (9, 23). It is likely that both signals are involved in kdpFABC induction during dehydration of Salmonella cells. Mutation in the kdpA gene, encoding a subunit that binds and moves K+ across the membrane, did not affect dehydration tolerance in Salmonella; however, a ΔkdpA mutant was significantly compromised in LTP during storage at 4°C and 25°C. The lack of a dehydration tolerance phenotype might be explained by the ability of the two constitutive K+ importer systems (Trk and Kup) (13) to compensate for the loss of the induced K+ channel. Nevertheless, the LTP-related phenotype of this mutant implies that the Kdp system is involved in Salmonella postdehydration persistence.
Previously, we reported that desiccated Salmonella acquires cross-tolerance to other stresses, including heat, high osmolarity, oxidation, and UV irradiation (30). Most of the dehydration-upregulated genes were previously reported to be induced under oxidative, osmotic, or heat stress (34, 35, 56, 70). These findings are concurrent with the idea that bacteria evoke a common stress response upon exposure to diverse environmental stresses (1, 2, 69). In contrast, only a few genes associated with acid stress in E. coli (7, 42, 44, 53) were also induced by desiccation. This finding may provide an explanation for the lack of cross-tolerance between dehydrated and acid-stressed Salmonella cells (30).
A transcriptomic analysis of Salmonella enterica serovar Enteritidis in peanut oil (a low-water-activity [aw] environment) compared to in LB broth was recently published (22). This study demonstrated a massive repression of all Salmonella genes in peanut oil compared to broth. The authors also followed gene expression at various time points following incubation in the oil. They found induction of 12 genes, mostly encoding heat shock and cold shock proteins, at 216 h compared to 72 h. Among these genes, only one (ibpA), encoding a heat shock protein, was upregulated in our study. This discrepancy might be explained by the different models tested in the two studies.
Our results demonstrated a greater similarity to those reported for dehydrated Bradyrhizobium cells (19). These included induction of transcriptional regulator genes, oxidative stress response genes, glyoxylate shunt genes, ribosomal structural genes, and heat shock protein genes. Induction of other genes in dehydrated Bradyrhizobium cells, such as those involved in synthesis of pili, flagella, exopolysaccharides, and trehalose, was not observed in our study. Similarly, our transcriptomic analysis did not identify several genes previously reported to be involved in desiccation tolerance of Salmonella, such as the O-antigen capsule synthesis genes yihO and yihQ (29), the O-chain assembly gene wzx (28), or genes involved in synthesis of curli and cellulose (72). One possible explanation is that transcription of these genes was also induced in our control cells incubated in SDDW. Another explanation might be related to the time point selected for mRNA extraction; it is possible that some of the desiccation stress response genes are induced only at an earlier or later phase of dehydration in Salmonella. Transcriptional analysis of Salmonella genes at different time points during dehydration and comparison to cells under different stress conditions will be useful in further understanding the desiccation response in this food-borne pathogen.
Supplementary Material
ACKNOWLEDGMENTS
This study was partially supported by the United States-Israel Binational Agricultural Research and Development (BARD) Fund (grant IS-4267-09 awarded to S.S.-S.). M.M. and S.P. were supported in part by NIH grants R21AI083964, R01AI083646, R56AI077645, and R01AI075093 and AFRI CSREES grant 2009-03579.
Footnotes
Published ahead of print 31 August 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Abee T, Wouters JA. 1999. Microbial stress response in minimal processing. Int. J. Food Microbiol. 50: 65–91 [DOI] [PubMed] [Google Scholar]
- 2. Aertsen A, Michiels CW. 2004. Stress and how bacteria cope with death and survival. Crit. Rev. Microbiol. 30: 263–273 [DOI] [PubMed] [Google Scholar]
- 3. Alba BM, Gross CA. 2004. Regulation of the Escherichia coli sigma dependent envelope stress response. Mol. Microbiol. 52: 613–619 [DOI] [PubMed] [Google Scholar]
- 4. Anonymous 2007. Multistate outbreak of Salmonella serotype Tennessee infections associated with peanut butter—United States, 2006–2007. MMWR Morb. Mortal. Wkly. Rep. 56: 521–524 [PubMed] [Google Scholar]
- 5. Anonymous 2010. Granola bars product recalls. http://www.fda.gov/Safety/Recalls/MajorProductRecalls/Granola/default.htm
- 6.Anonymous 2010. Salmonella Montevideo infections associated with salami products made with contaminated imported black and red pepper—United States, July 2009-April 2010. MMWR Morb. Mortal. Wkly. Rep. 59: 1647–1650 [PubMed] [Google Scholar]
- 7. Arnold CN, McElhanon J, Lee A, Leonhart R, Siegele DA. 2001. Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. J. Bacteriol. 183: 2178–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ayala-Castro C, Saini A, Outten FW. 2008. Fe-S cluster assembly pathways in bacteria. Microbiol. Mol. Biol. Rev. 72: 110–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ballal A, Basu B, Apte K. 2007. The Kdp-ATPase system and its regulation. J. Biosci. 32: 559–568 [DOI] [PubMed] [Google Scholar]
- 10. Beney L, Mille Y, Gervais P. 2004. Death of Escherichia coli during rapid and severe dehydration is related to lipid phase transition. Appl. Microbiol. Biotechnol. 65: 457–464 [DOI] [PubMed] [Google Scholar]
- 11. Beuchat LR, Heaton EK. 1975. Salmonella survival on pecans as influenced by processing and storage conditions. Appl. Microbiol. 29: 795–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Billi D, Potts M. 2001. Life and death of dried prokaryotes. Res. Microbiol. 153: 7–12 [DOI] [PubMed] [Google Scholar]
- 13. Buurman ET, McLaggan D, Naprstek J, Epstein W. 2004. Multiple paths for nonphysiological transport of K+ in Escherichia coli. J. Bacteriol. 186: 4238–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Canals R, et al. 2012. High-throughput comparison of gene fitness among related bacteria. BMC Genomics 13: 212 doi:10.1186/1471-2164-13-212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carty SM, Sreekumar KR, Raetz CRH. 1999. Effect of cold shock on lipid A biosynthesis in Escherichia coli. J. Biol. Chem. 274: 9677–9685 [DOI] [PubMed] [Google Scholar]
- 16. Chang DE, Smalley DJ, Conway T. 2002. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Mol. Microbiol. 45: 289–306 [DOI] [PubMed] [Google Scholar]
- 17. Cho BK, Federowicz S, Park YS, Zengler K, Palsson BÃ. 2012. Deciphering the transcriptional regulatory logic of amino acid metabolism. Nat. Chem. Biol. 8: 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Conesa A, et al. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676 [DOI] [PubMed] [Google Scholar]
- 19. Cytryn EJ, et al. 2007. Transcriptional and physiological responses of Bradyrhizobium japonicum to desiccation-induced stress. J. Bacteriol. 189: 6751–6762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de Cesare A, Sheldon BW, Smith KS, Jaykus LA. 2003. Survival and persistence. of Campylobacter and Salmonella species under various organic loads on food contact surfaces. J. Food Prot. 66: 1587–1594 [DOI] [PubMed] [Google Scholar]
- 22. Deng X, Li Z, Zhang W. 2012. Transcriptome sequencing of Salmonella enterica serovar Enteritidis under desiccation and starvation stress in peanut oil. Food Microbiol. 30: 311–315 [DOI] [PubMed] [Google Scholar]
- 23. Epstein W. 2003. The roles and regulation of potassium in bacteria. Prog. Nucleic Acids Res. Mol. Biol. 75: 293–320 [DOI] [PubMed] [Google Scholar]
- 24. Fang FC, Libby SJ, Castor MF, Fung AM. 2005. Isocitrate lyase (AceA) is required for Salmonella persistence but not for acute lethal infection in mice. Infect. Immun. 73: 2547–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fink RC, et al. 2007. FNR is a global regulator of virulence and anaerobic metabolism in Salmonella enterica serovar Typhimurium (ATCC 14028s). J. Bacteriol. 189: 2262–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fischer E, Sauer U. 2003. Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC-MS. Eur. J. Biochem. 270: 880–891 [DOI] [PubMed] [Google Scholar]
- 27. Frymier JS, Reed TD, Fletcher SA, Csonka LN. 1997. Characterization of transcriptional regulation of the kdp operon of Salmonella typhimurium. J. Bacteriol. 179: 3061–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Garmiri P, Coles KE, Humphrey TG, Kogan TA. 2008. Role of outer membrane lipopolysaccharides in the protection of Salmonella enterica serovar Typhimurium from desiccation damage. FEMS Microbiol. Lett. 281: 155–159 [DOI] [PubMed] [Google Scholar]
- 29. Gibson DL, et al. 2006. Salmonella produces an O-antigen capsule regulated by AgfD and important for environmental persistence. J. Bacteriol. 188: 7722–7730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gruzdev N, Pinto R, Sela S. 2011. Effect of desiccation on tolerance of Salmonella entericato multiple stresses. Appl. Environ. Microbiol. 77: 1667–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gruzdev N, Pinto R, Sela (Saldinger) S. 2012. Persistence of Salmonella enterica during dehydration and subsequent cold storage. Food Microbiol. 32: 415–422 [DOI] [PubMed] [Google Scholar]
- 32. Guiney DG, Libby S, Fang FC, Krause M, Fierer J. 1995. Growth-phase regulation of plasmid virulence genes in Salmonella. Trends Microbiol. 36: 275–279 [DOI] [PubMed] [Google Scholar]
- 33. Gulig PA. 1990. Virulence plasmids of Salmonella typhimurium and other salmonellae. Microb. Pathog. 8: 3–11 [DOI] [PubMed] [Google Scholar]
- 34. Gunasekera TS, Csonka LN, Paliy O. 2008. Genome-wide transcriptional responses of Escherichia coli K-12 to continuous osmotic and heat stresses. J. Bacteriol. 190: 3712–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harcum S, Haddadin F. 2006. Global transcriptome response of recombinant Escherichia coli to heat-shock and dual heat-shock recombinant protein induction. J. Ind. Microbiol. Biotechnol. 33: 801–814 [DOI] [PubMed] [Google Scholar]
- 36. Hiramatsu R, Matsumoto M, Sakae K, Miyazaki Y. 2005. Ability of Shiga toxin-producing Escherichia coli and Salmonella spp. to survive in a desiccation model system and in dry foods. Appl. Environ. Microbiol. 71: 6657–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Isaacs S, et al. 2005. An international outbreak of salmonellosis associated with raw almonds contaminated with a rare phage type of Salmonella Enteriditis. J. Food Prot. 68: 191–198 [DOI] [PubMed] [Google Scholar]
- 38. Johnson DC, Dean DR, Smith AD, Johnson MK. 2005. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74: 247–281 [DOI] [PubMed] [Google Scholar]
- 39. Jung K, Altendorf A. 2002. Towards an understanding of the molecular mechanisms of stimulus perception and signal transduction by the KdpD/KdpE system of Escherichia coli. J. Mol. Microbiol. Biotechnol. 4: 223–228 [PubMed] [Google Scholar]
- 40. Kaczanowska M, Rydén-Aulin M. 2007. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 7: 477–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kang Y, Weber KD, Qiu Y, Kiley PJ, Blattner FR. 2005. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J. Bacteriol. 187: 1135–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kannan G, et al. 2008. Rapid acid treatment of Escherichia coli: transcriptomic response and recovery. BMC Microbiol. 8: 37 doi:10.1186/1471-2180-8-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Katoh H, Asthana RK, Ohmori M. 2004. Gene expression in the cyanobacterium Anabaena sp. PCC 7120 under desiccation. Microb. Ecol. 47: 164–174 [DOI] [PubMed] [Google Scholar]
- 44. King T, Luccini S, Hinton JCD, Gobius K. 2010. Transciptomic analysis of Escherichia coli O157:H7 and K12 cultures exposed to inorganic and organic acids in stationary phase reveals acidulant- and strain-specific acid tolerance responses. Appl. Environ. Microbiol. 76: 6514–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kirk M, et al. 2004. An outbreak due to peanuts in their shell caused by Salmonella enterica serotypes Stanley and Newport—sharing molecular information to solve international outbreaks. Epidemiol. Infect. 132: 571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Knuth K, Niesalla H, Hueck CJ, Fuchs TM. 2004. Large-scale identification of essential Salmonella genes by trapping lethal insertions. Mol. Microbiol. 51: 1729–1744 [DOI] [PubMed] [Google Scholar]
- 47. Komitopoulou E, Peñaloza W. 2009. Fate of Salmonella in dry confectionery raw materials. J. Appl. Microbiol. 106: 1892–1900 [DOI] [PubMed] [Google Scholar]
- 48. Kusumaningrum HD, Riboldi G, Hazeleger WC, Beumer RR. 2003. Survival of foodborne pathogens on stainless steel surfaces and cross-contamination to foods. Int. J. Food Microbiol. 85: 227–236 [DOI] [PubMed] [Google Scholar]
- 49. Kvint K, Nachin L, Diez A, Nyström T. 2003. The bacterial universal stress protein: function and regulation. Curr. Opin. Microbiol. 6: 140–145 [DOI] [PubMed] [Google Scholar]
- 50. Lamichhane-Khadka R, Frye JG, Porwollik S, McClelland M, Maier RJ. 2011. Hydrogen-stimulated carbon acquisition and conservation in Salmonella enterica serovar Typhimurium. J. Bacteriol. 193: 5824–5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lucas RL, Lee CA. 2000. Unraveling the mysteries of virulence gene regulation in Salmonella Typhimurium. Mol. Microbiol. 36: 1024–1033 [DOI] [PubMed] [Google Scholar]
- 52. Maloy SR, Stewart VJ, Taylor RK. 1996. Genetic analysis of pathogenic bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 53. Maurer LM, Yohannes E, Bondurant SS, Radmacher M, Slonczewski JL. 2005. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J. Bacteriol. 187: 304–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. McMeechan A, et al. 2007. Role of the alternative sigma factors σE and σS in survival of Salmonella enterica serovar Typhimurium during starvation, refrigeration and osmotic shock. Microbiology 153: 263–269 [DOI] [PubMed] [Google Scholar]
- 55. Messens W, Grijspeerdt K, Herman L. 2005. Eggshell penetration by Salmonella. World Poult. Sci. J. 61: 71–86 [DOI] [PubMed] [Google Scholar]
- 56. Morgan RW, Christman MF, Jacobson FS, Storz G, Ames BN. 1986. Hydrogen peroxide-inducible proteins in Salmonella typhimurium overlap with heat shock and other stress proteins. Proc. Natl. Acad. Sci. U. S. A. 83: 8059–8063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Navarro Llorens JM, Tormo A, Martinez-Garcia E. 2010. Stationary phase in gram-negative bacteria. FEMS Microbiol. Rev. 34: 476–495 [DOI] [PubMed] [Google Scholar]
- 58. Paul BJ, Ross W, Gaal T, Gourse RL. 2004. rRNA transcription in Escherichia coli. Annu. Rev. Genet. 38: 749–770 [DOI] [PubMed] [Google Scholar]
- 59. Potts M. 1994. Desiccation tolerance of prokaryotes. Res. Microbiol. 58: 755–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Price MN, Huang KH, Alm EJ, Arkin AP. 2005. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 33: 880–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Raivio TL, Silhavy TJ. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55: 591–624 [DOI] [PubMed] [Google Scholar]
- 62. Reiner A, Yekutieli D, Benjamini Y. 2003. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19: 368–375 [DOI] [PubMed] [Google Scholar]
- 63. Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. 2006. Conserved and variable functions of the σE stress response in related genomes. PLoS Biol. 4: e2 doi:10.1371/journal.pbio.0040002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rui B, et al. 2010. A systematic investigation of Escherichia coli central carbon metabolism in response to superoxide stress. BMC Syst. Biol. 4: 122 doi:10.1186/1752-0509-4-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rychlik I, Gregorova D, Hradecka H. 2006. Distribution and function of plasmids in Salmonella enterica. Vet. Microbiol. 112: 1–10 [DOI] [PubMed] [Google Scholar]
- 66. Silberg JJ, Hoff KG, Tapley TL, Vickery LE. 2001. The Fe/S assembly protein IscU behaves as a substrate for the molecular chaperone Hsc66 from Escherichia coli. J. Biol. Chem. 276: 1696–1700 [DOI] [PubMed] [Google Scholar]
- 67. Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: 3. [DOI] [PubMed] [Google Scholar]
- 68. Uesugi AR, Danyluk MD, Harris LJ. 2006. Survival of Salmonella Enteritidis phage type 30 on inoculated almonds stored at 20, 4, 23, and 35°C. J. Food Prot. 69: 1851–1857 [DOI] [PubMed] [Google Scholar]
- 69. Vorob'eva LI. 2003. Stressors, stress reactions, and survival of bacteria: a review. Appl. Biochem. Microbiol. 40: 217–224 [PubMed] [Google Scholar]
- 70. Wang S, et al. 2009. Transcriptomic response of Escherichia coli O157:H7 to oxidative stress. Appl. Environ. Microbiol. 75: 6110–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Werber D, et al. 2005. International outbreak of Salmonella Oranienburg due to German chocolate. BMC Infect. Dis. 5: 7 doi:10.1186/1471-2334-5-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. White AP, Gibson DL, Kim W, Kay WW, Surette MG. 2006. Thin aggregative fimbriae and cellulose enhance long-term survival and persistence of Salmonella. J. Bacteriol. 188: 3219–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Winfield MD, Groisman EA. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 69: 3687–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.