Abstract
Eubacterium cellulosolvens cleaved the flavone C-glucosides homoorientin and isovitexin to their aglycones luteolin and apigenin, respectively. The corresponding isomers, orientin and vitexin, or other polyphenolic C-glucosides were not deglycosylated. E. cellulosolvens also cleaved several O-coupled glucosides of flavones and isoflavones to their corresponding aglycones.
TEXT
Certain forage crops contain large amounts of polyphenols, including tannins, which may affect rumen fermentation by inhibiting the growth and activity of the ruminal microbiota (8, 10, 13). Currently, bioactive polyphenolic plant compounds are studied with respect to their suitability as enhancers of animal health, in particular, that of ruminants (11). Health effects could be achieved by favorably modulating the gut fermentation or by inhibiting pathogens. The latter effect has come into focus since the addition of antibiotics to animal feed was banned in 2006 in the European Union. Polyphenolics could also serve as substrates for ruminal bacteria, in particular, when sugars are attached. Several intestinal bacterial species from animals and humans are able to release sugar moieties from flavonoids, and a few may even degrade the flavonoid basic structure (1, 2, 12). Based on the observed biological activities, the resulting flavonoid aglycones and their ring cleavage products have been proposed to mediate some of the health effects reported for the parent flavonoids in mammals (5, 12, 16).
One of the fodder plants very rich in polyphenols is the legume tagasaste (Chamaecytisus proliferus), which has been investigated in more detail. The polyphenolic fraction of tagasaste, which has no negative effects on rumen fermentation, contains more than 70% glycosides of the flavones apigenin and luteolin (6). It has been assumed that these flavones are mainly present in the form of their C-glucosides (e.g., vitexin, orientin) and hypothesized that the glucose moiety could be released and fermented by rumen bacteria (6). In contrast to polyphenolic O-β-glucosides that may also be cleaved by mammalian glycosidases (5), the deglycosylation of C-glucosides appears to be exclusively catalyzed by bacterial enzymes.
Eubacterium cellulosolvens, an anaerobic cellulolytic bacterium, has been isolated from the rumen of sheep and cows as well as the gut of mice and rabbits (4, 7, 15). It appears to be a prominent ruminal bacterium in cattle (14). While the binding and degradation of cellulose by E. cellulosolvens have been studied in detail, nothing has been known of the cleavage of polyphenolic glycosides by this species. Here, we demonstrate that E. cellulosolvens is capable of deglycosylating certain flavone C-glucosides and of hydrolyzing O-glucosides of flavones and isoflavones.
Conversion of polyphenolic C-glucosides.
E. cellulosolvens ATCC 43171T was grown in 800 μl modified Reinforced Clostridial Medium (RCMmod) supplemented with 200 μM of the respective C-glucoside at 37°C for 144 h under strict anoxic conditions as described previously (3). C-Glucosides and bacteria incubated separately in RCMmod served as controls. Samples taken after 0, 24, 48, 72, 96, and 144 h were processed and analyzed by reversed-phase high-performance liquid chromatography–diode array detection (RP-HPLC/DAD) according to previously described methods (3). All incubations were carried out in triplicate, and each sample was extracted and analyzed once. E. cellulosolvens was tested for the conversion of homoorientin, orientin, isovitexin (all from PhytoLab, Vestenbergsreuth, Germany), vitexin (Roth, Karlsruhe, Germany), mangiferin, carminic acid (both from Sigma-Aldrich, Munich, Germany), and puerarin (LKT Laboratories, St. Paul, MN) (Fig. 1). Of these C-glucosides, only homoorientin (luteolin 6-C-glucoside) and isovitexin (apigenin 6-C-glucoside) were converted by E. cellulosolvens (Table 1). Most parts of both compounds were used up within 24 h of incubation. While homoorientin was deglycosylated within this time period to equimolar amounts of luteolin, the incubation of isovitexin with E. cellulosolvens resulted in the formation of only 26% of the corresponding aglycone apigenin based on the applied substrate. The amount of apigenin further increased to maximally 44% after 96 h of incubation. Luteolin and apigenin were not further degraded. The results show that only the glucose moiety attached to the C6 of flavones was removed by cleavage whereas that at C8 as present in orientin (luteolin 8-C-glucoside) and vitexin (apigenin 8-C-glucoside) remained unaffected. The C6 position is characterized by two adjacent hydroxyl groups, which appear to be essential for deglycosylation. However, neither the xanthone C-glucoside mangiferin nor the anthraquinone C-glucoside carminic acid was cleaved although adjacent hydroxyl groups were present. This indicates that additional structural features of the aglycone also play a role. However, so far the possibility cannot be excluded that the observed differences reflect differences in the transport of the individual compounds.
Fig 1.
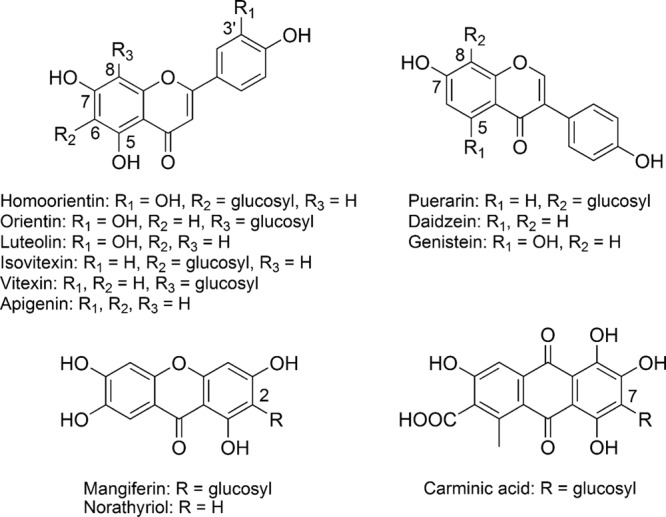
Structures of compounds used in the study.
Table 1.
Substrate depletion and metabolite formation within 24 h and 48 h of incubations with Eubacterium cellulosolvensa
| Substrate | % substrate depletion |
Metabolite | % metabolite formation |
||
|---|---|---|---|---|---|
| Within 24 h | Within 48 h | Within 24 h | Within 48 h | ||
| C-Glucosides | |||||
| Homoorientin | 79.9 ± 0.9 | 78.8 ± 1.2 | Luteolin | 82.9 ± 8.1 | 76.3 ± 11.9 |
| Orientin | −2.7 ± 0.9 | −8.1 ± 1.6 | ND | NA | NA |
| Isovitexin | 75.8 ± 2.3 | 90.7 ± 3.5 | Apigenin | 25.9 ± 5.2 | 27.8 ± 5.4 |
| Vitexin | 0.4 ± 6.7 | −0.4 ± 8.4 | ND | NA | NA |
| Mangiferin | 2.3 ± 1.6 | 4.2 ± 1.4 | ND | NA | NA |
| Puerarin | 2.8 ± 1.6 | 3.0 ± 3.3 | ND | NA | NA |
| Carminic acid | 8.3 ± 0.4 | 10.6 ± 1.4 | ND | NA | NA |
| O-Glucosides | |||||
| Luteolin 5-O-glucoside | 1.5 ± 1.4 | −0.9 ± 2.1 | ND | NA | NA |
| Luteolin 7-O-glucoside | 84.2 ± 1.7 | 98.2 ± 0.3 | Luteolin | 71.6 ± 2.4 | 85.7 ± 6.0 |
| Luteolin 3′-O-glucoside | 100.0 ± 0 | 100.0 ± 0 | Luteolin | 62.4 ± 1.9 | 63.4 ± 0.9 |
| Apigenin 7-O-glucoside | 90.1 ± 0.5 | 97.6 ± 0.2 | Apigenin | 38.3 ± 6.6 | 44.4 ± 4.3 |
| Daidzein 7-O-glucoside | 34.7 ± 5.6 | 67.7 ± 2.1 | Daidzein | 28.5 ± 1.9 | 53.3 ± 2.5 |
| Genistein 7-O-glucoside | 77.2 ± 2.1 | 100.0 ± 0 | Genistein | 73.4 ± 6.3 | 98.0 ± 1.3 |
Values are means of triplicate incubations ± SD. ND, not detected; NA, not applicable.
For comparison, the bacterial strain CG19-1 was incubated with selected C-glucosides using conditions identical to those described above for E. cellulosolvens. The strictly anaerobic strain CG19-1, which was recently isolated from human feces, deglycosylates puerarin and other aromatic C-glucosides (3). Moreover, strain CG19-1 cleaves the aglycones luteolin and apigenin, which result from deglycosylation of the corresponding flavone C-glucosides (3). In the present study, homoorientin, vitexin, and mangiferin were completely converted within 24 h to approximately equimolar amounts of 3-(3,4-dihydroxyphenyl)propionic acid, 3-(4-hydroxyphenyl)propionic acid, and norathyriol, respectively. These activities of strain CG19-1 confirmed previous work (3). The newly tested flavone C-glucosides orientin and isovitexin were completely transformed within 24 h of incubation to approximately equimolar amounts of 3-(3,4-dihydroxyphenyl)propionic acid and 3-(4-hydroxyphenyl)propionic acid, respectively. This indicates efficient deglycosylation of both C-glucosides as observed for their isomers, homoorientin and vitexin.
Thus, strain CG19-1 is capable of deglycosylating a more complex set of C-glucosides than E. cellulosolvens. However, one of the tested compounds, carminic acid, was not converted by either of the two organisms. This is probably due to its structural characteristics, which include a nonheterocyclic central ring and a second keto group on this ring. The difference observed between the two species may be explained by differences in substrate specificity of the C-deglycosylating enzymes or by differences in the uptake of the compounds. E. cellulosolvens and strain CG19-1 are only remote relatives within the Clostridia. While the former species is a member of the Eubacteriaceae, strain CG19-1 belongs to the Lachnospiraceae. The 16S rRNA gene sequence of E. cellulosolvens ATCC 43171 (accession number X71860) shows 90% identity to that of strain CG19-1 (accession number FJ711049).
The mechanism of C-deglycosylation by E. cellulosolvens and strain CG19-1 has yet to be clarified. Puerarin deglycosylation by another human intestinal bacterium was recently shown to occur by hydrolytic cleavage, yielding daidzein and glucose (9). E. cellulosolvens could take advantage of the released glucose moiety as a substrate for fermentation (15), whereas strain CG19-1 does not degrade glucose or other carbohydrates (3).
Conversion of flavonoid O-glucosides.
The ability of E. cellulosolvens to convert O-coupled flavonoid glycosides was tested as described above using the flavone derivatives luteolin 5-O-glucoside, luteolin 3′-O-glucoside (both available from previous work) (3), luteolin 7-O-glucoside, and apigenin 7-O-glucoside as well as the isoflavones daidzein 7-O-glucoside and genistein 7-O-glucoside (all from Roth, Karlsruhe, Germany). The aglycone structures of the compounds are given in Fig. 1. E. cellulosolvens deglycosylated all of the tested compounds to their corresponding aglycones except luteolin 5-O-glucoside. The conversion rates differed, as reflected by the percentage of the substrate converted after 24 h of incubation (Table 1). Luteolin 7-O-glucoside, luteolin 3′-O-glucoside, apigenin 7-O-glucoside, and genistein 7-O-glucoside were completely used up after 48 h, and that result was accompanied by increased product formation, while 32% of the initial daidzein 7-O-glucoside concentration was still present at this time point. The findings suggest that the glucose moiety attached to the hydroxyl group in the C5 position of the flavone structure prevents its cleavage or its uptake into the cell. The lack of the C5 hydroxyl group in daidzein 7-O-glucoside led to delayed deglycosylation and/or transport. It can be concluded that the free C5 hydroxyl group plays an important role in the cleavage and/or uptake of flavonoid O-glucosides, probably mediated by formation of a hydrogen bond with the carbonyl group at C4. All of the O-glucosides tested with E. cellulosolvens in the present study are converted by the human intestinal strain CG19-1 to their aglycones (in the case of the isoflavones) or even to the corresponding hydroxyphenylpropionic acids by cleavage of the resulting flavones (3).
In conclusion, E. cellulosolvens may benefit from the deglycosylation of flavonoid O- and C-glucosides by fermentation of the liberated glucose moiety. The resulting aglycones, which are not further degraded by E. cellulosolvens, may affect both the intestinal microbiota and the host organism. Thus, it is supposed that E. cellulosolvens contributes to the bioactivation of polyphenolic compounds of fodder plants in livestock.
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (grant BR 2269/4-1).
We thank Anke Gühler for technical assistance.
Footnotes
Published ahead of print 7 September 2012
REFERENCES
- 1. Aura AM. 2008. Microbial metabolism of dietary phenolic compounds in the colon. Phytochem. Rev. 7:407–429 [Google Scholar]
- 2. Blaut M, Schoefer L, Braune A. 2003. Transformation of flavonoids by intestinal microorganisms. Int. J. Vitam. Nutr. Res. 73:79–87 [DOI] [PubMed] [Google Scholar]
- 3. Braune A, Blaut M. 2011. Deglycosylation of puerarin and other aromatic C-glucosides by a newly isolated human intestinal bacterium. Environ. Microbiol. 13:482–494 [DOI] [PubMed] [Google Scholar]
- 4. Bryant MP, Small N, Bouma C, Robinson IM. 1958. Characteristics of ruminal anaerobic celluloytic cocci and Cillobacterium cellulosolvens n. sp. J. Bacteriol. 76:529–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Crozier A, Jaganath IB, Clifford MN. 2009. Dietary phenolics: chemistry, bioavailability and effects on health. Nat. Prod. Rep. 26:1001–1043 [DOI] [PubMed] [Google Scholar]
- 6. Edwards NJ. 2000. A review of tannins and other secondary metabolites in the fodder shrub tagasaste (Chamaecytisus proliferus), p 170–174. In Brooker JD. (ed), Tannins in livestock and human nutrition. Australian Centre for International Agricultural Research (ACIAR), Canberra, Australia [Google Scholar]
- 7. Fonty G, Gouet P. 1989. Fibre-degrading microorganisms in the monogastric digestive tract. Anim. Feed Sci. Technol. 23:91–107 [Google Scholar]
- 8. Jouany JP, Morgavi DP. 2007. Use of ‘natural’ products as alternatives to antibiotic feed additives in ruminant production. Animal 1:1443–1466 [DOI] [PubMed] [Google Scholar]
- 9. Nakamura K, et al. 2011. The C-glucosyl bond of puerarin was cleaved hydrolytically by a human intestinal bacterium strain PUE to yield its aglycone daidzein and an intact glucose. Chem. Pharm. Bull. (Tokyo) 59:23–27 [DOI] [PubMed] [Google Scholar]
- 10. Patra AK, Saxena J. 2011. Exploitation of dietary tannins to improve rumen metabolism and ruminant nutrition. J. Sci. Food Agric. 91:24–37 [DOI] [PubMed] [Google Scholar]
- 11. Rochfort S, Parker AJ, Dunshea FR. 2008. Plant bioactives for ruminant health and productivity. Phytochemistry 69:299–322 [DOI] [PubMed] [Google Scholar]
- 12. Selma MV, Espin JC, Tomas-Barberan FA. 2009. Interaction between phenolics and gut microbiota: role in human health. J. Agric. Food Chem. 57:6485–6501 [DOI] [PubMed] [Google Scholar]
- 13. Smith AH, Zoetendal E, Mackie RI. 2005. Bacterial mechanisms to overcome inhibitory effects of dietary tannins. Microb. Ecol. 50:197–205 [DOI] [PubMed] [Google Scholar]
- 14. van Gylswyk NO. 1990. Enumeration and presumptive identification of some functional groups of bacteria in the rumen of dairy cows fed grass silage-based diets. FEMS Microbiol. Lett. 73:243–261 [Google Scholar]
- 15. van Gylswyk NO, van der Toorn JJTK. 1986. Description and designation of a neotype strain of Eubacterium cellulosolvens (Cillobacterium cellulosolvens Bryant, Small, Bouma and Robinson) Holdeman and Moore. Int. J. Syst. Bacteriol. 36:275–277 [Google Scholar]
- 16. Williamson G, Clifford MN. 2010. Colonic metabolites of berry polyphenols: the missing link to biological activity? Br. J. Nutr. 104(Suppl 3):S48–S66 [DOI] [PubMed] [Google Scholar]


