Abstract
RoxA is an extracellular c-type diheme cytochrome secreted by Xanthomonas sp. strain 35Y during growth on rubber. RoxA cleaves poly(cis-1,4-isoprene) to 12-oxo-4,8-dimethyltrideca-4,8-diene-1-al (ODTD). Analysis of the RoxA structure revealed that Phe317 is located in close proximity (≈5 Å) to the N-terminal heme that presumably represents the active site. To find evidence of whether Phe317 is important for catalysis, we changed it to tyrosine, tryptophan, leucine, histidine, or alanine. All five RoxA muteins were expressed after integration of the respective gene into the chromosome of a Xanthomonas sp. ΔroxA strain. Residual clearing zone formation on opaque latex agar was found for Xanthomonas sp. strains expressing the Phe317Leu, Phe317Ala, or Phe317His variant (wild type > Leu > Ala > His). Strains in which Phe317 was changed to tyrosine or tryptophan were inactive. Phe317Ala and Phe312Leu RoxA muteins were purified, and polyisoprene cleavage activities were reduced to ≈3% and 10%, respectively. UV-visible spectroscopy of RoxA muteins confirmed that both heme groups were present in an oxidized form, but spectral responses to the addition of low-molecular-weight (inhibitory) ligand molecules such as imidazole and pyridine were different from those of wild-type RoxA. Our results show that residue 317 is involved in interaction with substrates. This is the first report on structure-function analysis of a polyisoprene-cleaving enzyme and on the identification of an amino acid that is essential for polyisoprene cleavage activity.
INTRODUCTION
Natural rubber [poly(cis-1,4-isoprene)] (NR) is an important naturally produced hydrocarbon polymer. Products made on the basis of NR have been in use on the scale of thousands of tons per year for more than a century. However, the fate of rubber materials in the environment is poorly understood. Only the fact that rubber is principally biodegradable is well-documented (2, 9, 10, 16, 26). The enzymes that are responsible for primary attack of polyisoprene in rubber-degrading microorganisms and the biochemical mechanisms by which these enzymes cleave rubber materials into low-molecular-weight products are almost unknown. Due to the high stability of carbon-carbon bonds, biodegradation of NR is a slow process compared to biodegradation of biopolymers, such as polypeptides, polysaccharides, and others, in which the polymer backbone contains heteroatoms such as nitrogen, oxygen, or other elements that facilitate enzymatic cleavage reactions. Heteroatoms are absent in polyisoprene; the C C double bond is the only functional group in polyisoprene that allows an attack by oxidizing enzymes. In fact, rubber-degrading organisms are widespread in nature (9, 10, 16). Some rubber-degrading bacteria, such as Xanthomonas sp. strain 35Y (23) and many actinomycetes, produce clearing zones on opaque latex agar, while others grow adhesively on rubber without clearing zone formation. Gordonia polyisoprenivorans and Gordonia westfalica belong to the latter group (see references 16 and 26 and references therein).
C double bond is the only functional group in polyisoprene that allows an attack by oxidizing enzymes. In fact, rubber-degrading organisms are widespread in nature (9, 10, 16). Some rubber-degrading bacteria, such as Xanthomonas sp. strain 35Y (23) and many actinomycetes, produce clearing zones on opaque latex agar, while others grow adhesively on rubber without clearing zone formation. Gordonia polyisoprenivorans and Gordonia westfalica belong to the latter group (see references 16 and 26 and references therein).
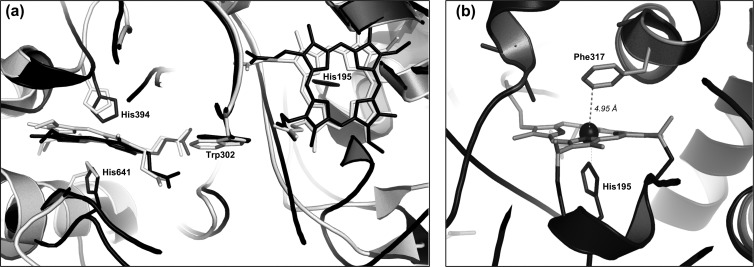
To date, two types of proteins that are essential for rubber degradation and that catalyze the primary attack of polyisoprene have been identified in rubber-degrading microorganisms. One is the latex clearing protein (Lcp) and was first described for Streptomyces sp. K30 but apparently is widely distributed in rubber-degrading bacteria (17). The other is the rubber oxygenase RoxA of Xanthomonas sp. 35Y (11), a potent rubber degrader isolated more than 20 years ago (23). Lcp and RoxA are completely different polypeptides without significant similarities in amino acid sequence. RoxA consists of 678 amino acids and is a c-type cytochrome that features two heme binding sites as well as a MauG motif (11). In contrast, Lcp (397 amino acids) does not have any metals or cofactors (17). Both Lcp and RoxA are responsible for cleavage of rubber to low-molecular-weight degradation products with aldehyde and keto groups at the molecule ends (4, 8, 22, 23), and therefore they apparently catalyze similar or even the same reaction. RoxA has been purified and studied in vitro (3, 4, 11). Purified RoxA is active in an aqueous environment if only the substrates, rubber and dioxygen, are present (4) and the physical conditions (pH and temperature) are appropriate. 12-Oxo-4,8-dimethyltrideca-4,8-diene-1-al (ODTD) was identified as the major degradation product. Isotope labeling experiments revealed that RoxA is a dioxygenase (3). Spectroscopic characterization of RoxA (18) showed that the two heme centers are present in an oxidized form and can be differentiated spectroscopically. Recently, our cooperation partners succeeded in solving the three-dimensional structure of RoxA (7; O. Einsle, University of Freiburg, personal communication) (Protein Data Bank [PDB] accession code 4B2N). The structure in the neighborhood of the two hemes is similar to that of bacterial cytochrome c peroxidases (CCPs), with two hemes buried deeply in the protein and arranged perpendicular to each other (Fig. 1a). However, unlike CCPs, RoxA does not need external reductants such as cytochrome c for activity (4), and consequently, all attempts to demonstrate peroxidase activity of RoxA were not successful (18). In conclusion, RoxA must have a reaction mechanism that is different from that of CCPs. Phe317 was identified in close proximity to the distal coordination site of the N-terminal heme, sloped opposite and at a short distance from the Fe ion (Fig. 1b). We assume that this heme represents the active site of RoxA and that Phe317 is involved in interaction with substrate molecules. Consequently, we investigated the importance of residue 317 by site-directed mutagenesis.
Fig 1.
(a) Similarity of RoxA structure to that of cytochrome c peroxidase of Nitrosomonas europaea (NEP). An overlay of the central regions of RoxA (black) and NEP (gray), including the two heme centers and the interheme region, is shown. The heme axial amino acid ligands and the tryptophan residues, located in the middle between the two hemes (Trp302 in RoxA), are also shown. (b) Structure around the N-terminal heme center of RoxA. Phe317 is located in close proximity (4.95 Å) to the sixth coordination site of the heme iron.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Table 1 shows strains, plasmids, and primers used in this study. Xanthomonas sp. 35Y (23) and related strains were grown in modified LB medium with a reduced concentration of yeast extract (10 g tryptone, 5 g NaCl, and 0.25 g yeast extract per liter) or in a mineral salts medium (MSM) with 0.1 to 0.2% purified rubber latex at 30°C for 10 to 12 days. For purification of recombinant RoxA, a Xanthomonas sp. ΔroxA-attB strain harboring the roxA variant of interest (Table 1) was grown in 0.5 liter of modified LB medium (20 individual cultures in 3-liter Erlenmeyer flasks) supplemented with 0.1% (wt/vol) l-rhamnose for ≈60 h at 30°C with continuous shaking. Cells were harvested (4°C) by centrifugation, and RoxA was purified from cell-free culture fluid as described below.
Table 1.
Strains, plasmids, and primers used in this study
| Strain or plasmid | Relevant characteristics or sequence (5′-3′) | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli strains | ||
| S17-1 | Conjugation strain | 19 |
| XL1-Blue | QuikChange transformation strain | |
| Xanthomonas sp. strains | ||
| 35Y | Growth on poly(cis-1,4-isoprene) latex, clearing zone formation | 23 |
| 35Y-CM | Chloramphenicol-resistant mutant of 35Y | 6 |
| 35Y-CM ΔroxA-attB (SN3727) | Chromosomal deletion of roxA, attB at former roxA site, no clearing zone formation on latex agar | This study |
| 35Y-CM ΔroxA-attB/pNH1-roxA-attP (in chromosome) (SN4230) | Expression of RoxA from rhamnose promoter; Kmr Cmr; clearing zone formation in the presence of rhamnose | This study |
| 35Y-CM ΔroxA-attB/pNH1-roxA-F317A-attP (in chromosome) (SN4784) | Expression of RoxA-F317A from rhamnose promoter; Kmr Cmr | This study |
| 35Y-CM ΔroxA-attB/pNH1-roxA-F317H-attP (in chromosome) (SN4785) | Expression of RoxA-F317H from rhamnose promoter; Kmr Cmr | This study |
| 35Y-CM ΔroxA-attB/pNH1-roxA-F317L-attP (in chromosome) (SN4786) | Expression of RoxA-F317L from rhamnose promoter; Kmr Cmr | This study |
| 35Y-CM ΔroxA-attB/pNH1-roxA-F317W-attP (in chromosome) (SN4787) | Expression of RoxA-F317W from rhamnose promoter; Kmr Cmr | This study |
| 35Y-CM ΔroxA-attB/pNH1-roxA-F317Y-attP (in chromosome) (SN4788) | Expression of RoxA-F317Y from rhamnose promoter; Kmr Cmr | This study |
| Plasmids | ||
| pIC20HE | Cloning vector | 1 |
| pSI17 | Source of PhiC31 integrase gene | 20 |
| pUC9::roxA-region | Source of roxA; Apr | 6 |
| pUC9::roxA-region-ΔroxA | Precise deletion of roxA coding sequence | This study |
| pUC9::roxA-region-ΔroxA-attB | PhiC31 attB site at position of former roxA gene | This study |
| pLO3 | sacB-containing suicide vector; Tcr | 13 |
| pLO3-Km | pLO3 with 1,108-bp aphII fragment of pBBR1MCS-2 in SmaI site; Tcr Kmr | This study |
| pLO3-Km-roxA-region | pLO3 with 3,603-bp SacI fragment including roxA gene | This study |
| pLO3-Km-roxA-region-ΔroxA | Precise deletion of roxA coding sequence | This study |
| pLO3-Km-roxA-region-ΔroxA-attB | Precise deletion of roxA coding sequence with attB at former roxA sequence | This study |
| pJOE6787.1 | pBR322 ori aphII (Kmr) mob int (PhiC31 integrase) attP | J. Altenbuchner |
| pNH1 | pJOE6787.1 with rhamnose regulation genes of p4782.1 | This study |
| pNH1-roxA | Coding sequence of roxA under rhamnose promoter control | This study |
| pNH1-roxA-attP (SN4230) | Coding sequence of roxA under rhamnose promoter control, with attP site | This study |
| pNH1-roxA-attP F317A (SN4784) | RoxA-F317A expression vector with attP site; Kmr | This study |
| pNH1-roxA-attP F317H (SN4785) | RoxA-F317H expression vector with attP site; Kmr | This study |
| pNH1-roxA-attP F317L (SN4786) | RoxA-F317L expression vector with attP site; Kmr | This study |
| pNH1-roxA-attP F317W (SN4787) | RoxA-F317W expression vector with attP site; Kmr | This study |
| pNH1-roxA-attP F317Y (SN4788) | RoxA-F317Y expression vector with attP site; Kmr | This study |
| Primers | ||
| s6549 | CACGTGATGAGTAAACTTGGTCTGACAGT | |
| s6550 | CCCCGTAGAAAAGATCAAAGGAT | |
| s6551 | GGGAATATTTCGAACCCCAGAGTCC | |
| s6552 | GGGCACGTGCAAGCGCAAAGAGAAAGC | |
| s6553 | ATCATTAATCAACCCTCAGCGGATGCCC | |
| s6554 | ATCATTAATCCCGTCTCAGCGCCTAACAGG | |
| s6555 | GGGACGTCCATCGTCCACATATCCAG | |
| s6556 | GGGACGTCCCTTGTCCAGATAGCCC | |
| 1fw | ACCGCTCGAGTGCCACCTGGGATGAATGTC | |
| 1rev | TCCGCTCGAGCGCTTGGTCGGTCATTTCG | |
| 2fw | GCTCTAGAGCTAGGGTTGGCTTCACGCCGCGTAATCGAT | |
| 2rev | GCTCTAGAGCGATTCCCCCTGTACGTCCCAACGAATGAGT | |
| 3fw | GTACAGGGGGAATCGCTCTAGAGTGCCAGGGCGTGCCCTTGGGCTCCCCG | |
| 3rev | CGCGGCGTGAAGCCAACCCTAGCCGCGCCCGGGGAGCCCAAGGGCACGCC | |
| 4fw | CGGGGTACCTTCAGGTTCATCATGCCGTTTGTG | |
| 4rev | TTCCGAGCTCCTCCACGGGGAGAGCCTGAGC | |
| 5fw | TCTCCTGCAAACTGCTTTTAC | |
| 5rev | GCGAATCTGAACTATCTCATCC |
Construction of a ΔroxA mutant of Xanthomonas sp.
In contrast to previous assumptions (6), expression of recombinant roxA in Xanthomonas sp. from plasmids provided in trans was not possible. Rather, expression of recombinant roxA required integration of the roxA gene into the chromosome. To avoid recombination of an introduced roxA copy with chromosomal roxA, it was necessary to remove the wild-type (WT) roxA copy from the chromosome. The plasmid used for deletion of roxA was constructed using the sacB-containing suicide plasmid pLO3 (13), into which the kanamycin resistance gene aphII of the plasmid pBBR1MCS-2 (12) was cloned by PCR (with primer pair 1), yielding pLO3-Km. A 3,603-bp SacI fragment of Xanthomonas sp. chromosomal DNA containing roxA and its up- and downstream regions was cloned into the SacI restriction sites of pLO3-Km and pBBR1MCS-2 as well as into the SmaI sites of pUC9 (via blunt ligation after restriction by the neoschizomer Ecl136II), yielding pLO3-Km::roxA-region, pBBR1MCS2::roxA-region, and pUC9::roxA-region, respectively. The coding sequences of roxA were removed from pLO3-Km::roxA-region and pUC9::roxA-region by PCR amplification using primer pair 2 and religation of the purified PCR products (via XbaI restriction in the case of pUC9::roxA-region and via blunt ligation in the case of pLO3-Km::roxA-region), yielding pUC9::roxA-region-ΔroxA and pLO3-Km::roxA-region-ΔroxA, respectively. The attB attachment site of PhiC31 was inserted into pUC9::roxA-region-ΔroxA by QuikChange PCR using primer pair 3. Finally, the attB site and the adjacent roxA up- and downstream DNA regions (1,145 bp) were cloned into pLO3-Km::roxA-region-ΔroxA via DraIII and SacII restriction, giving pLO3::roxA-region-ΔroxA-attB. The resulting construct was checked by DNA sequencing. The chromosomal roxA gene of the Xanthomonas sp. CM strain was removed by exchange with the attB site after conjugative transfer of the plasmid pLO3-Km-ΔroxA-attB (with selection of plasmid integrants on kanamycin agar and subsequent selection for double-crossover events by growth on 10% sucrose-containing solid medium). The success of roxA deletion in purified individual clones was confirmed by PCR amplification of the former roxA locus and by DNA sequencing.
Construction of a plasmid suitable for PhiC31-mediated integration into the chromosome of a Xanthomonas sp. ΔroxA strain and rhamnose-dependent expression of RoxA variants.
The PhiC31-dependent integration vector for roxA expression was constructed in a modular way. Starting from pIC20HE, a PmlI restriction site was first introduced downstream of the ampicillin resistance gene (bla) by PCR amplification of pIC20HE (with primers S6549 and S6550) and ligation of the linear DNA, giving plasmid pJOE6767.1. Subsequently, the bla gene was replaced by a kanamycin resistance gene (aphII) amplified from pBBR1MCS-2 (with primers S6551 and S6552). The aphII gene was inserted between the PmlI and SspI sites of pJOE6767.1, giving pJOE6772.2. In the next step, the PhiC31 integrase gene (int) together with the attP site was inserted into pJOE6772.2. The DNA fragment was obtained by a PCR using plasmid pSI17 as the template, with primers S6553 and S6554, and then was cut with AseI and inserted at the NdeI site of pJOE6772.2 to give pJOE6776.3. Finally, the mob gene and oriT region of pBBR1MCS-2 for conjugal transfer were PCR amplified (with primers S6565 and S6566) and the fragment inserted into the AatII site of pJOE6776.3. The new plasmid, pJOE6787.1, was the basis for anchoring of a cloned roxA copy into the chromosomal attB site of the Xanthomonas sp. ΔroxA-attB strain. For this purpose, the rhamnose regulation genes and the roxA coding sequence were PCR amplified using primer pair 4, with 4782.1::roxA as the template (6), to give pNH1-roxA-attP. The insertion was confirmed by DNA sequencing. In cases of roxA variants with mutations in the roxA sequence, pUC9::roxA was mutagenized via QuikChange PCR. The roxA sequence of pNH1-roxA-attP was replaced by the roxA variant sequence harboring the respective mutated codon by NdeI/ApaI ligation of pUC9::roxA (via replacement of respective NdeI-ApaI DNA fragments) into pNH1-roxA-attP. The plasmid harboring the roxA variant gene of interest was conjugatively transferred from Escherichia coli S17-1 to Xanthomonas sp. CM ΔroxA by spot mating on NB agar. Transconjugants were selected and purified on NB agar supplemented with 30 μg/ml kanamycin and 10 μg/ml chloramphenicol. Expression of PhiC31 integrase from pNH1-roxA-attP ensured instantaneous integration of the plasmid into the chromosome at the former position of the chromosomal roxA locus via the attB/attP sites (21, 25). Correct integration of the roxA variant was verified via PCR and DNA sequencing. The recombinant Xanthomonas sp. strains express the roxA variants only in the presence of rhamnose.
Purification of recombinant RoxA.
RoxA was purified at room temperature by use of an Aekta fast-performance liquid chromatography (FPLC) system (GE Healthcare, Uppsala, Sweden), using a modified protocol (18). In brief, cell-free concentrated (10-kDa cutoff) supernatant of a Xanthomonas sp. culture was passed through a Q-Sepharose Fast Flow column (Q-FF 50/11) preequilibrated with 20 mM Tris-HCl (pH 8.5) at a flow rate of 3 ml/min. RoxA was bound to the column and eluted in a step gradient at ≈50 mM NaCl. RoxA-containing fractions were combined and concentrated via ultrafiltration. An additional purification step was performed on a hydroxyapatite column. The column (CHT5-I) was equilibrated with potassium phosphate buffer (10 mM; pH 6.8) and was used after changing the buffer of the RoxA pool to 10 mM potassium phosphate, pH 6.8, by gel filtration on a HiPrep 26/10 column. RoxA was eluted with a linear gradient of 10 to 200 mM potassium phosphate, pH 6.8, at ≈40 mM. RoxA fractions were pooled and concentrated via ultrafiltration (30-kDa cutoff) to approximately 5 mg/ml with the addition of 300 mM NaCl on ice. High ionic strength prevented precipitation of RoxA at high protein concentrations. Purity was tested by SDS-PAGE and by determination of the quotient of absorption (optical densities [ODs]) at 406 nm and 280 nm, which was 1.35 for pure RoxA. Purified RoxA was frozen and stored in liquid nitrogen.
Assay of RoxA.
The following conditions were used for product analysis of RoxA-catalyzed polyisoprene cleavage by high-pressure liquid chromatography (HPLC) analysis. The reaction mixture contained 100 μl purified RoxA (2 to 20 μg/ml), rubber latex (0.2% [wt/vol] emulsion), and potassium phosphate buffer (100 mM; pH 7.0) in a total volume of 1 ml. The reaction was carried out at 37°C for 3 h in a 15-ml Falcon tube. The mixture was extracted with ethyl acetate, dried, dissolved in 100 μl methanol, and then subjected to HPLC analysis. An RP8 HPLC column (12 × 4 mm, 5-μm particle size) was operated at 0.7 ml/min with water (A) and methanol (B) as the mobile phase. The concentration of B was increased from 50% (vol/vol) to 100% (vol/vol) over 20 min. Products were detected at 210 nm, and ODTD eluted after 15.3 min. Purified ODTD (4) was used as a standard. Mixtures without RoxA or with heat-inactivated RoxA (10 min at 95°C) served as controls.
Semiquantitative clearing zone assay for RoxA activity determination.
Recombinant Xanthomonas sp. ΔroxA::attB strains harboring chromosomally anchored roxA variants under rhamnose control (Table 1) were grown in modified LB liquid culture. Five microliters of the respective culture was spotted onto a mineral salts agar plate with an opaque overlay agar (≈8 ml) of polyisoprene latex (0.25% [wt/vol] in agar) after purification (via 3 washes in 0.1% [wt/vol] Nonidet P-40) (kindly provided by Weber and Schaer, Hamburg, Germany). A Xanthomonas sp. ΔroxA::attB strain harboring chromosomally anchored wild-type roxA under rhamnose control served as a positive control. The plates were incubated at 30°C for 6 to 7 days. The intensity of clearing zone formation semiquantitatively indicated RoxA activity produced by the strain.
Heme staining and protein determination.
Heme staining was performed after separation of RoxA samples by nonreducing SDS-PAGE and subsequent assay for pseudoperoxidase activity of the RoxA heme groups as described recently (18). Protein concentration was determined by the Bradford method (5). Purified RoxA samples were determined by using the molar extinction coefficients of RoxA at 280 nm (ε280 = 1.53 × 105 M−1 cm−1) and 406 nm (ε406 = 1.8 × 105 M−1 cm−1).
RESULTS AND DISCUSSION
Construction of a Xanthomonas sp. strain for stable expression of recombinant RoxA.
Expression in E. coli of c-type cytochromes with MauG motifs, such as RoxA and MauG, has not been possible (6, 24). Apparently, RoxA (and MauG) holoenzymes are assembled by cytochrome c maturation systems that are different from that of E. coli. We therefore decided to express roxA in a homologous Xanthomonas sp. host. Recently, we showed that cloned roxA can be expressed in the homologous wild-type host Xanthomonas sp. (6). However, it turned out in this study that homologue expression did not occur from the introduced plasmid, as expected, but required integration of roxA into the chromosome. Chromosomal integration of the roxA-containing plasmid took long periods depending on clone length (weeks to months, with repeated transfers on solid or liquid medium) and could not be accelerated by increasing the size of cloned Xanthomonas DNA up- and downstream of roxA. Moreover, recombinant roxA with introduced site-specific mutations could recombine with the chromosomal wild-type copy of roxA in Xanthomonas sp., which sometimes resulted in elimination of the introduced mutation (data not shown). To avoid these disadvantages and to gain more rapid and reproducible integration of roxA into the chromosome, we deleted the chromosomal roxA locus and replaced it with the attB site of phage PhiC31 as described in Materials and Methods. The resulting Xanthomonas sp. ΔroxA-attB strain contained the attB site at the position of the former roxA gene, was sensitive to kanamycin and resistant to sucrose, and did not produce clearing zones on opaque latex agar. This result confirmed that functional homologue genes that can compensate for the absence of roxA are not present in Xanthomonas sp. A constructed plasmid (pNH1-roxA-attP) harboring the PhiC31 integrase gene (14, 15) plus the attP site, together with a roxA copy under the control of a rhamnose-dependent promoter, was introduced into the Xanthomonas sp. ΔroxA::attB mutant via conjugation. Both attP and attB sites represent short stretches of DNA, each having a different (imperfect) palindrome sequence and a central common TTG sequence at which recombination between both DNA strands by PhiC31 integrase occurs (21). The integration of the complete roxA-containing plasmid at the previous roxA site (now the attB site) was confirmed by PCR and by DNA sequencing of the roxA locus. Recombinant RoxA was reproducibly expressed in the presence of rhamnose by the transconjugant Xanthomonas strain but was not detectable on latex medium or any other medium in the absence of rhamnose. In conclusion, a reliable system for expression of recombinant RoxA variants in a homologous Xanthomonas sp. ΔroxA::attB host strain was now available and allowed the performance of structure-function analysis of RoxA. This expression system was superior to our previous system (6), which required repeated cultivation passages and waiting for rarely occurring recombination events.
Construction and expression of RoxA muteins.
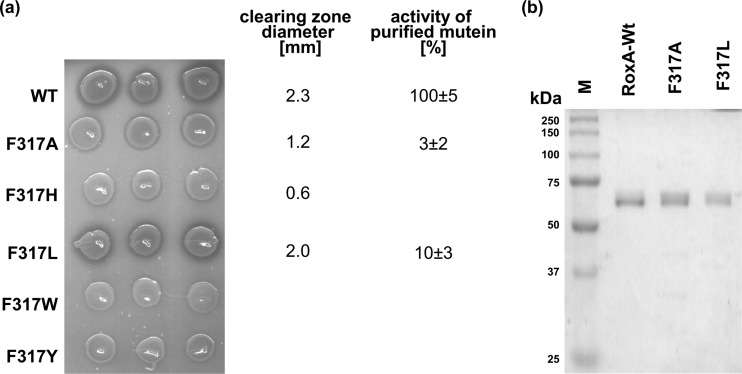
The three-dimensional structure of RoxA showed that the two hemes are arranged perpendicular to each other, similar to the case in bacterial CCPs (Fig. 1a). The C-terminal heme center of RoxA has two axial His ligands (His394 and His641), while the N-terminal heme has only one axial amino acid ligand (His195). The sixth coordination site is not occupied by an amino acid ligand. This finding suggested that this position could be accessible to external ligands or substrates. The amino acid residue next to the free coordination site of the N-terminal heme is Phe317 and is located ≈5 Å from and sloped opposite to the Fe ion (Fig. 1b). We hypothesized that the Fe center of this heme could represent the active site of the enzyme and that Phe317 is involved in interaction with substrate molecules. To find experimental support for this assumption, we replaced Phe317 by tyrosine, tryptophan, leucine, histidine, or alanine via site-directed mutagenesis of the roxA gene. All five roxA variants were separately integrated into the chromosome of the roxA deletion strain of Xanthomonas sp. via PhiC31 integrase-mediated attP/attB recombination. To test for the effects of the mutations, the Xanthomonas sp. ΔroxA strains harboring the wild-type roxA gene or one of the five mutant roxA variant genes were grown on solid latex overlay agar that had been supplemented with 0.1% rhamnose. The Phe317Leu mutant produced clearing zones that appeared later than those of the wild type and had a slightly smaller diameter (2.0 mm, compared to 2.3 mm for the wild type) (Fig. 2a), indicating that the replacement of the aromatic ring by an aliphatic residue of similar size had an effect on RoxA activity. However, the clearing zone of the Phe317Ala mutant was even more reduced (1.2 mm), and the Phe317His mutant formed only a hardly visible clearing zone near the detection limit (0.6 mm). No clearing zones were detected around colonies of the other two mutants (Phe317Trp and Phe317Tyr). In summary, changing the side chain of residue 317 resulted in stronger or weaker effects on RoxA activity. The replacement of phenylalanine by leucine had the weakest effect on activity. This was not surprising, since leucine has the same hydrophobic character as phenylalanine and occupies a similar space. Since RoxA with the Phe317Tyr mutation (or with Phe317His or Phe317Trp) was inactive, the presence of a hydrophilic functional group in the ring structure of tyrosine or of a bulky group apparently prevents activity.
Fig 2.
(a) Clearing zone formation of recombinant Xanthomonas sp. strains and activities of purified Phe317Leu and Phe317Ala muteins. Five-microliter aliquots of concentrated cell suspensions of Xanthomonas sp. ΔroxA strains harboring and expressing different RoxA muteins were spotted in triplicate on opaque latex overlay agar with l-rhamnose and then incubated for 6 days at 30°C. Clearing zone formation semiquantitatively indicates the activity of expressed RoxA muteins. The clearing zone diameters (averages for 3 biological replicates minus diameter of colony) are given in mm. The ODTD-forming activities of purified RoxA muteins are provided on the right for purified WT-RoxA (100%) and the Phe317Leu and Phe317Ala muteins. (b) Purification of recombinant muteins. WT-RoxA and the Phe317Ala and Phe317Leu muteins were purified as described in the text, separated by reducing SDS-PAGE, and stained with Coomassie blue.
These results confirmed our assumption that the side chain of residue 317 is important for RoxA activity. SDS-PAGE analysis of culture supernatants obtained from the respective strains after growth in the presence of rhamnose revealed RoxA bands for all strains (data not shown). Moreover, bands of comparable intensity were obtained after staining the gels for pseudoperoxidase activity (“heme staining”) and indicated that heme was stably incorporated in all RoxA variants (not shown). These results suggest that the observed differences in clearing zone formation resulted from different activities of the RoxA muteins, not from different degrees of expression.
RoxA activity and biochemical properties of Phe317Ala and Phe317Leu muteins.
To analyze the effect of the mutations more precisely, both RoxA muteins with reduced activity (Phe317Ala and Phe317Leu muteins) were purified as described in Materials and Methods. About 4 or 7 mg of purified RoxA mutein was obtained for each variant, with OD280/OD406 ratios of 1.13 (Phe317Ala) and 1.36 (Phe317Leu). This indicated a sufficient degree of purity of both RoxA muteins (>90% purity) that was confirmed by SDS-PAGE (Fig. 2b). No differences in the binding properties of RoxA muteins on Q-Sepharose or hydroxyapatite in comparison to wild-type RoxA were noticed during protein purification.
When purified Phe317Ala and Phe317Leu RoxA muteins were subjected to a quantitative polyisoprene cleavage assay, ODTD-forming activity was reduced to 3% ± 2% and 10% ± 3% residual activity, respectively, compared with that for purified wild-type RoxA. These values are in agreement with the reduced clearing zone diameters of the respective Xanthomonas sp. strains shown in Fig. 2a and confirmed that a residue with hydrophobic character at position 317 is important for RoxA activity. Significant amounts of products other than ODTD were not detected by HPLC analysis (not shown). Other biochemical properties, such as the temperature optimum (40°C) or pH optimum (pH 7), were also not changed detectably compared to wild-type properties. These results suggested that the mutations negatively influenced the reaction of the active center with substrate molecules but not the general properties of the enzyme.
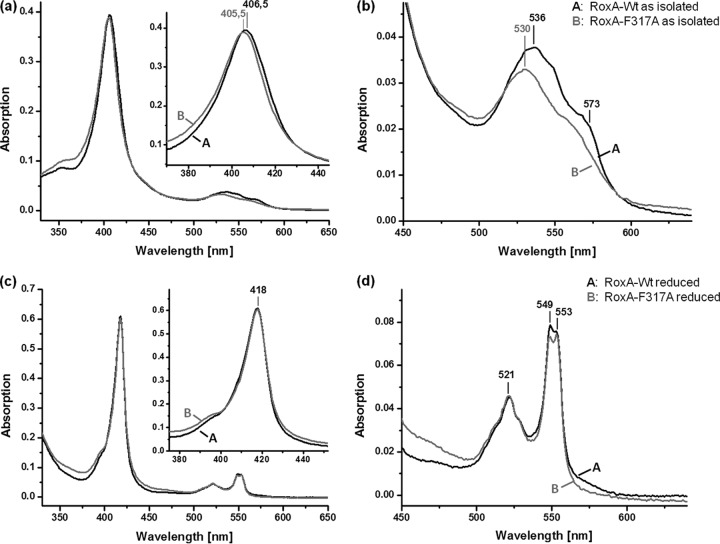
In our previous study, we showed that wild-type RoxA specifically responded to reducing agents and to the presence of low-molecular-weight inhibitory compounds such as imidazole and pyridine (18). Since residue 317 is the closest amino acid to the free coordination site of the N-terminal heme center, and since the chemical environment of heme is reflected by its spectroscopic properties, UV-visible (UV-vis) spectroscopy was performed with both muteins (Phe317Ala and Phe317Leu). The UV-vis spectra of both muteins, as isolated, showed typical c-type cytochrome spectra in an oxidized state (shown for Phe317Ala RoxA in Fig. 3). The Soret and Q bands were shifted by ≈1 nm, to a shorter wavelength (405 to 406 nm for the Phe317Ala RoxA mutein compared to 406 to 407 nm for WT-RoxA), and by 5 to 6 nm (530 nm for the Phe317Ala RoxA mutein compared to 535 to 536 nm for WT-RoxA), respectively (Fig. 3a). When the UV-vis spectrum of the chemically (dithionite) reduced Phe317Ala mutein was determined, the Soret band was shifted to 418 nm and two separated α-bands, with maxima at 549 nm and 553 nm, appeared that were very similar to those of wild-type RoxA, but with a slightly decreased intensity of the 549-nm α-band (Fig. 3b). Reoxidation of chemically reduced Phe317Ala RoxA by dioxygen resulted in a disappearance of the α-bands, as also found for wild-type RoxA (not shown). Thus, the spectral UV-vis properties of the Phe317Ala RoxA mutein, as isolated and in a reduced state, were similar but showed some differences from those of wild-type RoxA, as may be expected from an exchange of an amino acid that influences the electronic transitions of a porphyrin system. When the Phe317Leu mutein was analyzed by UV-vis spectroscopy, very similar results to those described for the Phe317Ala mutein were found (not shown).
Fig 3.
UV-vis spectra of WT-RoxA and RoxA-F317A. (a and b) Spectra of recombinant WT-RoxA and RoxA-Phe317Ala, as isolated (2 μM). The Soret-band region is enlarged in the inset of panel a; panel b shows the Q-band region from panel a. The Phe317Ala mutein had a 1-nm blue-shifted Soret maximum and less pronounced Q bands, with a blue-shifted maximum at 530 nm. (c and d) Spectra of recombinant WT-RoxA and RoxA-Phe317Ala, both reduced with Na+-dithionite under an N2 gas atmosphere. In panel c, the Soret-band region is enlarged in the inset; panel d shows the Q-band region from panel c. WT-RoxA and the Phe317Ala mutein both had a Soret band at 418 nm and exhibited only minor differences in intensity for the α-bands, which were of almost equal intensities. The 549-nm α-band appears to be slightly more intense than the 553-nm band in the WT spectrum and appears to be less intense in the Phe317Ala spectrum.
Low-molecular-weight heme ligands have different effects on WT-RoxA and on Phe317 muteins.
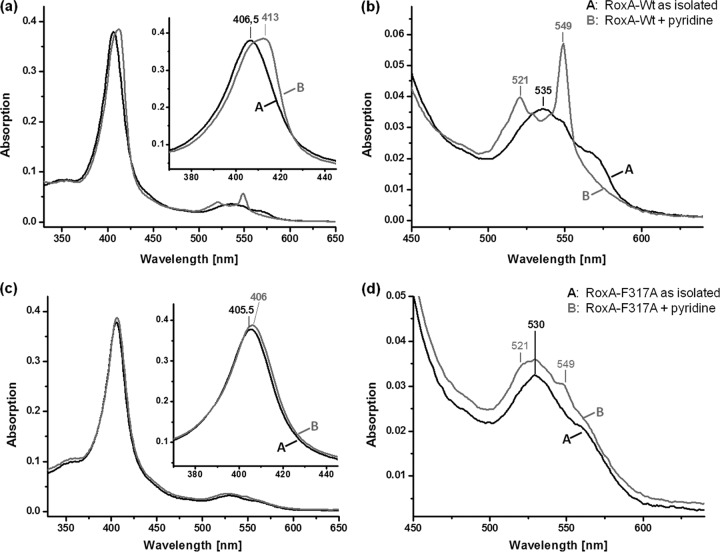
The results of UV-vis spectroscopy of RoxA muteins shown above suggest some differences in the biophysical properties and/or electronic environment of the N-terminal heme center of RoxA muteins compared to WT-RoxA. Reaction of WT-RoxA with imidazole, pyridine, or other related heterocyclic compounds with free electron pairs at the hetero atom resulted in a notable strong increase of the 549-nm α-band, even in the presence of dioxygen, and in almost complete inhibition of polyisoprene cleavage activity (Fig. 4a). Imidazole (in the form of histidine) is a typical axial heme ligand in many heme proteins. The effects of imidazole and related heterocyclic compounds mimicked the UV-vis effects of a partial reduction of the N-terminal heme center (increase of α-bands as in the presence of chemical reductants). In contrast to complete reduction (e.g., by dithionite), no increase of absorption of the other α-band at 553 nm appeared, confirming that the C-terminal bis-His-coordinated heme was not affected. We assume that low-molecular-weight heterocyclic compounds such as imidazole and pyridine have access to the free sixth coordination sphere of the N-terminal heme (active site), thereby leading to a competitive inhibition of polyisoprene cleavage activity of RoxA.
Fig 4.
UV-vis spectra of WT-RoxA and RoxA-F317A with heme ligands. (a and b) Spectra of recombinant WT-RoxA, as isolated (2 μM) and after addition of pyridine (5 mM; 1 h at room temperature), at the endpoint of the reaction. The Soret-band region is enlarged in the inset of panel a; panel b shows the Q-band region from panel a. WT-RoxA rapidly reacted with pyridine to give characteristic spectral changes as formerly reported (18), in accordance with a partial reduction of one heme center. (c and d) Spectra of recombinant RoxA-Phe317Ala, as isolated (2 μM) and after addition of pyridine (5 mM; 1 h at room temperature). In panel c, the Soret-band region is enlarged in the inset; panel d shows the Q-band region from panel c. Only minor changes appeared in the presence of pyridine and did not proceed through a prolonged incubation time. Similar weak effects on the UV-vis spectrum of RoxA-Phe317Ala were obtained by addition of other external heme ligands, such as imidazole (not shown). The characteristic changes in the WT-RoxA spectrum caused by imidazole have been reported previously (see Fig. 3a of reference 18).
Remarkably, and in contrast to the case for WT-RoxA, only minor effects on UV-vis spectra were observed when the Phe317Ala and Phe317Leu RoxA muteins were incubated with 1 mM imidazole or pyridine (Fig. 4b, results shown only for the Phe317Ala mutein). Only a very little broadening or shift of the Soret band and a marginal increase in the 549-nm α-band occurred upon incubation with imidazole or pyridine. The possibility that binding of imidazole or pyridine does not occur in the Phe317Ala and Phe317Leu muteins is unlikely, because the exchange of Phe against Ala (or leucine, to a minor extent) provides even more space around the sixth coordination sphere of heme. Binding of imidazole or pyridine to the active site of Phe317Ala and Phe317Leu muteins is additionally supported by the finding that both compounds minimized the rest activity of the Phe317Ala (≈3%) and Phe317Leu (10%) muteins to a hardly detectable level (<0.5%). Presumably, the distribution of π electron density along the Fe–N-ligand bond was affected in the RoxA muteins. These results indicate that the mutations either negatively influenced access to the active center of the two RoxA muteins for substrates and inhibitors or influenced an orbital overlapping the iron and the N-ligand that might be responsible for the intense 549-nm α-band in WT-RoxA in the presence of certain ligands. Both possibilities are in agreement with the conclusion that Phe317 is important for interaction of the enzyme with substrates, e.g., Phe317 is important for correct positioning of substrate molecules at the active site. Spectroscopic experiments with the true substrate (polyisoprene) are not possible because polyisoprene latex is opaque and absorbs light. The observation that a smaller residue at position 317 partially inhibits the enzyme might also be explained by the assumption that the substrate molecules have more degrees of freedom to bind and to orientate themselves in the active site in Phe317Ala RoxA and are not forced to bind in the correct position for latex cleavage or, in the case of external heme ligands, in their geometric position in WT-RoxA.
In summary, we have succeeded in providing all the tools necessary to study the enzymatic cleavage mechanism of polyisoprene systematically at the molecular level: (i) the 3-dimensional structure of RoxA can be investigated in silico and used for prediction of effects of structural changes by mutagenesis of RoxA, (ii) the roxA gene can be manipulated in E. coli, (iii) the roxA gene can be expressed in and purified from recombinant Xanthomonas sp. strains after integration into the chromosome, and (iv) the effects of manipulations on activity and structure can be studied by biochemical and biophysical methods. Here, for the first time, we identified an amino acid that is essential for polyisoprene cleavage activity and is part of the active site of RoxA.
ACKNOWLEDGMENTS
This work was supported by a grant from the Deutsche Forschungsgemeinschaft to D.J.
We thank the Weber and Schaer company for proving polyisoprene. The input of O. Einsle (University of Freiburg) and P. Kroneck (University of Konstanz) during discussions of RoxA function is gratefully acknowledged.
Footnotes
Published ahead of print 31 August 2012
REFERENCES
- 1. Altenbuchner J, Viell P, Pelletier I. 1992. Positive selection vectors based on palindromic DNA sequences. Methods Enzymol. 216:457–466 [DOI] [PubMed] [Google Scholar]
- 2. Bode HB, Kerkhoff K, Jendrossek D. 2001. Bacterial degradation of natural and synthetic rubber. Biomacromolecules 2:295–303 [DOI] [PubMed] [Google Scholar]
- 3. Braaz R, Armbruster W, Jendrossek D. 2005. Heme-dependent rubber oxygenase RoxA of Xanthomonas sp. cleaves the carbon backbone of poly(cis-1,4-isoprene) by a dioxygenase mechanism. Appl. Environ. Microbiol. 71:2473–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Braaz R, Fischer P, Jendrossek D. 2004. Novel type of heme-dependent oxygenase catalyzes oxidative cleavage of rubber (poly-cis-1,4-isoprene). Appl. Environ. Microbiol. 70:7388–7395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 6. Hambsch N, Schmitt G, Jendrossek D. 2010. Development of a homologous expression system for rubber oxygenase RoxA from Xanthomonas sp. J. Appl. Microbiol. 109:1067–1075 [DOI] [PubMed] [Google Scholar]
- 7. Hoffmann M, Braaz R, Jendrossek D, Einsle O. 2008. Crystallization of the extracellular rubber oxygenase RoxA from Xanthomonas sp. strain 35Y. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64:123–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ibrahim EMA, Arenskotter M, Luftmann H, Steinbüchel A. 2006. Identification of poly(cis-1,4-isoprene) degradation intermediates during growth of moderately thermophilic actinomycetes on rubber and cloning of a functional lcp homologue from Nocardia farcinica strain E1. Appl. Environ. Microbiol. 72:3375–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Imai S, et al. 2011. Isolation and characterization of Streptomyces, Actinoplanes, and Methylibium strains that are involved in degradation of natural rubber and synthetic poly(cis-1,4-isoprene). Enzyme Microb. Technol. 49:526–531 [DOI] [PubMed] [Google Scholar]
- 10. Jendrossek D, Tomasi G, Kroppenstedt RM. 1997. Bacterial degradation of natural rubber: a privilege of actinomycetes? FEMS Microbiol. Lett. 150:179–188 [DOI] [PubMed] [Google Scholar]
- 11. Jendrossek D, Reinhardt S. 2003. Sequence analysis of a gene product synthesized by Xanthomonas sp. during growth on natural rubber latex. FEMS Microbiol. Lett. 224:61–65 [DOI] [PubMed] [Google Scholar]
- 12. Kovach ME, et al. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 13. Lenz O, Friedrich B. 1998. A novel multicomponent regulatory system mediates H2 sensing in Alcaligenes eutrophus. Proc. Natl. Acad. Sci. U. S. A. 95:12474–12479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lomovskaia ND, Mkrtumian NM, Gostimskaya NL. 1970. Isolation and characterization of the actinophage of Streptomyces coelicolor. Genetica 6:135 [Google Scholar]
- 15. Lomovskaya ND, Mkrtumian NM, Gostimskaya NL, Danilenko VN. 1972. Characterization of temperate actinophage phi C31 isolated from Streptomyces coelicolor A3(2). J. Virol. 9:258–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rose K, Steinbüchel A. 2005. Biodegradation of natural rubber and related compounds: recent insights into a hardly understood catabolic capability of microorganisms. Appl. Environ. Microbiol. 71:2803–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rose K, Tenberge KB, Steinbüchel A. 2005. Identification and characterization of genes from Streptomyces sp. strain K30 responsible for clear zone formation on natural rubber latex and poly(cis-1,4-isoprene) rubber degradation. Biomacromolecules 6:180–188 [DOI] [PubMed] [Google Scholar]
- 18. Schmitt G, Seiffert G, Kroneck PMH, Braaz R, Jendrossek D. 2010. Spectroscopic properties of rubber oxygenase RoxA from Xanthomonas sp., a new type of dihaem dioxygenase. Microbiology 156:2537–2548 [DOI] [PubMed] [Google Scholar]
- 19. Simon R, Priefer U, Pühler A. 1983. A broad host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat. Biotechnol. 1:784–791 [Google Scholar]
- 20. Sladkova IA, Orekhov AV. 1994. Actinomycete plasmid and integrative vectors based on DNA of the temperate phi C31 actinophage, determining limitation of lytic development of phage phi C31, not dependent on repressor. Antibiot. Khimioter. 39:3–11 [PubMed] [Google Scholar]
- 21. Thorpe HM, Smith MC. 1998. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc. Natl. Acad. Sci. U. S. A. 95:5505–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tsuchii A, Suzuki T, Takeda K. 1985. Microbial degradation of natural rubber vulcanizates. Appl. Environ. Microbiol. 50:965–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tsuchii A, Takeda K. 1990. Rubber-degrading enzyme from a bacterial culture. Appl. Environ. Microbiol. 56:269–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Y, et al. 2003. MauG, a novel diheme protein required for tryptophan tryptophylquinone biogenesis. Biochemistry 42:7318–7325 [DOI] [PubMed] [Google Scholar]
- 25. Watanabe S, Nakamura S, Sakurai T, Akasaka K, Sato M. 2011. Improvement of a phiC31 integrase-based gene delivery system that confers high and continuous transgene expression. Nat. Biotechnol. 28:312–319 [DOI] [PubMed] [Google Scholar]
- 26. Yikmis M, Steinbüchel A. 2012. Historical and recent achievements in the field of microbial degradation of natural and synthetic rubber. Appl. Environ. Microbiol. 78:4543–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]