Abstract
Although mangroves represent ecosystems of global importance, the genetic diversity and abundance of functional genes that are key to their functioning scarcely have been explored. Here, we present a survey based on the nifH gene across transects of sediments of two mangrove systems located along the coast line of São Paulo state (Brazil) which differed by degree of disturbance, i.e., an oil-spill-affected and an unaffected mangrove. The diazotrophic communities were assessed by denaturing gradient gel electrophoresis (DGGE), quantitative PCR (qPCR), and clone libraries. The nifH gene abundance was similar across the two mangrove sediment systems, as evidenced by qPCR. However, the nifH-based PCR-DGGE profiles revealed clear differences between the mangroves. Moreover, shifts in the nifH gene diversities were noted along the land-sea transect within the previously oiled mangrove. The nifH gene diversity depicted the presence of nitrogen-fixing bacteria affiliated with a wide range of taxa, encompassing members of the Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Firmicutes, and also a group of anaerobic sulfate-reducing bacteria. We also detected a unique mangrove-specific cluster of sequences denoted Mgv-nifH. Our results indicate that nitrogen-fixing bacterial guilds can be partially endemic to mangroves, and these communities are modulated by oil contamination, which has important implications for conservation strategies.
INTRODUCTION
Mangroves are widely distributed in the Earth's tropical and subtropical regions, covering approximately 60 to 75% of the global coastline (16, 33). Brazil, Indonesia, and Australia are the countries that possess the most mangrove areas (18). The importance of mangrove ecosystems lies in their great biological productivity, serving as a source of life for a high diversity of fish, crustaceans, mollusks, birds, reptiles, and mammals. In this context, microorganisms are important, as they are involved in the processing of essential compounds that sustain the higher organisms (1, 11, 18). Any environmental disturbance, depending on its frequency and intensity, caused either by natural or anthropogenic factors may put mangroves in an unsustainable situation, harming the key life support processes (12, 19, 20).
The availability of nitrogen limits primary production in many terrestrial and marine ecosystems. Biological nitrogen fixation is the dominant natural process by which atmospheric nitrogen enters ecosystems through its conversion into ammonia (30, 39, 40, 41). The provision of nitrogen to mangroves has been described as associated with organic matter decomposition. Marine environments often also have high rates of biological nitrogen fixation (BNF) (17, 24). For organisms to fix nitrogen, either in association with plants or not, the presence and activity of the nitrogenase enzyme complex is required (29). In mangrove sediments, the rate of nitrogen fixation is affected by the pool of dissolved inorganic nitrogen and by local conditions (21). In this context, N2 fixation is particularly significant in sediment settings, where prevailing anaerobic or microaerophilic conditions regulate nitrogen transformations as well as nitrogen losses to the atmosphere (4).
In order to monitor the microbial potential for nitrogen fixation, many studies have used the nifH gene as a proxy for abundance and diversity (8, 26, 28, 36). This gene has revealed sufficient variation to allow the detection of shifts in the community structure of nitrogen fixers in ecosystems under differing conditions (3), as well as shifts promoted by alterations over time and related to soil type and chemical properties (27).
In this study, we determined the abundance and diversity of diazotrophic bacterial communities present in the sediments of two distinct mangroves. Our primary purpose was to assess the effect of prior oil pollution on the diversity of these assemblages, identifying the major groups of organisms that are capable of contributing to N2 fixation. We used samples along transects in the two mangroves that differed in vegetation composition and some environmental parameters (see below), making this survey complementary to other assessments in the same area where shifts in archaeal (10) and fungal (13) communities were demonstrated. Indeed, the abundance of genes related to nitrogen transformations and the occurrence of nifH-related genes in these areas was also observed by a deep metagenomic survey carried out by our team (2), basing our approach on determining specific players of nutrient cycling in such areas.
MATERIALS AND METHODS
Mangroves and samples analyzed.
The two mangroves, near to the city of Bertioga, Sao Paulo, were selected based on their proximity to each other and their distinct history of contamination. They were denoted Bertioga Anthropogenic (AntMgv) (free of oil but near the city) and Bertioga Contaminated (OilMgv). The latter experienced an oil spill, having been contaminated with 35 million liters of oil in 1983. Both mangroves have as water sources mixtures of water from the sea and from the river Iriri, which crosses the area.
A transect of approximately 300 m was determined in each mangrove, oriented from the waterfront toward inland. Three easily distinguishable subregions were outlined, i.e., a first region 50 m from a local river (denoted site 1), a second in the center of the mangrove (denoted site 2), and a third far inland, called restinga, around 30 m from a transition between mangrove and forest. The two mangroves can also be divided into distinct areas based on their vegetation and characteristics (Table 1). The OilMgv is easily divided in two subregions separated by a small stream that crosses the mangrove. In the area landward of the river, oil effects were still present, while the area closer to the sea in the same mangrove did not show effects of the oil spill and vegetation was more similar to that of the other mangrove (AntMgv) (Table 1). A better visualization of these points and more details on vegetation composition and other parameters can be found in a previous study involving the same area (2).
Table 1.
Characteristics and history of contamination of the mangroves analyzed in this study
| Mangrove metagenome | Description | Coordinates | Contamination | Vegetation |
|---|---|---|---|---|
| OilMgv1 | Area free of oil contamination in the OilMgv | 23°53′49″ S, 46°12′28″ W | Small effect of oil spill | Presence of mangrove species,a predominance of R. mangle |
| OilMgv2-3 | Area highly affected by the oil contamination in the OilMgv | 23°53′49″ S, 46°12′28″ W | Highly affected by oil spill | Under recovery, low density of Rhizophora mangle |
| AntMgv | Mangrove near the city, under anthropogenic pressure | 23°54′06″ S, 45°15′03″ W | From human activity | Abundant existence of other species besides those typically found in mangroves |
In the state of Sao Paulo, the mangrove forest is composed mainly of three species: Avicennia shaueriana, Laguncularia racemosa, and Rhizophora mangle.
Triplicate samples were collected perpendicularly to the transect, separated from each other by at least 30 m. Sample grouping thus consisted of three replicates taken at each site within each mangrove. This yielded a total of 18 samples (2 mangroves at 3 points, with 3 replicates from each point). Each sediment sample was obtained with a 30-cm sampler, 7 cm in diameter, which was placed in the mangrove sediment and removed, resulting in sediment cores of approximately 1 kg.
All samples were subjected to physical and chemical analyses, determining the major soil characteristics (Table 2). These analyses were carried out in collaboration with the Laboratory of Soil Analysis at “Luiz de Queiroz” College of Agriculture (ESALQ/USP; Piracicaba, Brazil) according to the methodology described by Van Raij et al. (Table 2) (39a). Also, although oil components were not determined, the presence of oil is visible in the spilled area, mainly in the undersurface layers (30 to 50 cm depth).
Table 2.
Environmental data and results from global BEST testa
| Variable | Result by mangrove and test point |
|||||||
|---|---|---|---|---|---|---|---|---|
| OilMgv |
AntMgv |
Global R | P value | |||||
| P1 | P2 | P3 | P1 | P2 | P3 | |||
| pH | 6.9 | 6.8 | 6.4 | 7.1 | 6.8 | 6.9 | 0.253 | 0.001 |
| Density (g/cm3) | 0.96 | 0.96 | 0.95 | 0.96 | 0.95 | 0.96 | 0.148 | NS |
| Humidity (%) | 65.16 | 69.66 | 48.26 | 73.97 | 78.39 | 75.82 | 0.064 | NS |
| Organic matter (%) | 10.86 | 12.89 | 10.89 | 10.28 | 9.89 | 11.43 | 0.138 | NS |
| Total carbon (%) | 6.03 | 7.16 | 6.05 | 5.71 | 5.5 | 6.35 | 0.093 | NS |
| Total nitrogen (%) | 5.33 | 5.6 | 5.15 | 4.77 | 4.62 | 5.19 | 0.287 | 0.001 |
| Total sulfur (%) | 0.13 | 0.45 | 0.31 | 0.29 | 0.26 | 0.34 | 0.311 | 0.001 |
| Conductivity (mS) | 0.31 | 0.36 | 0.32 | 0.24 | 0.26 | 0.2 | 0.024 | NS |
| Salinity (%) | 7.77 | 8.01 | 8.84 | 5.56 | 8.22 | 8.36 | 0.197 | NS |
| Sandy (%) | 0.47 | 0.46 | 0.48 | 0.3 | 0.46 | 0.46 | 0.088 | NS |
Results from the global BEST test were determined in Primer-E software, which selects environmental variables best explaining community pattern as assessed by DGGE profiles. Values in bold indicate statistically significant variables. NS, not significant.
DNA extraction from sediment samples.
Total DNA was extracted from each sample using the PowerSoil DNA Isolation kit (Mobio, NY). After extraction, the quantity, integrity, and quality of the DNA obtained were checked by 1% (wt/vol) agarose gel electrophoresis, followed by staining in ethidium bromide and visualization in UV light. The extracted DNA was further quantified by NanoDrop spectrophotometer analysis (Thermo Scientific).
Quantification of nifH gene copies by qPCR.
The abundance of nitrogen fixers was quantified by quantitative PCR (qPCR) targeting the nifH gene. The reactions were performed in an ABI Prism 7300 Cycler (Applied Biosystems, Germany) in volumes of 25 μl and containing 1 μl of extracted DNA (approximately 50 ng), 12.5 μl of Power SYBR green master mix (Applied Biosystems), and 0.25 μM primers FGPH19 (35) and PolR (28). The parameters were those previously described in the literature (27, 36). Briefly, the amplifications were performed with one initial denaturation step at 94°C for 1 min, followed by 40 amplification cycles of 55°C for 27 s and an extension of 1 min at 72°C. The specificity of the amplification products was confirmed by melting curve analysis, and the expected sizes (450 bp) of the amplified fragments were checked in a 1.5% agarose gel stained with ethidium bromide. Standard curves were obtained using serial dilutions of the Escherichia coli-derived vector plasmid JM109 (Promega, Madison, WI) containing a cloned nifH gene from Bradyrhizobium liaoningense, using 102 to 107 gene copies μl−1. Threshold values obtained from sample amplification were interpolated in the standard curve determining the number of nifH genes found per gram of mangrove sediment.
Analysis of nitrogen-fixing communities in mangrove sediment using PCR-DGGE.
The total DNA extracted from all mangrove samples was subjected to amplification using specific primers described in the literature (28, 35) for denaturing gradient gel electrophoresis (DGGE) analysis of the nifH gene (27). The amplicons obtained were checked on agarose gels before DGGE analysis, which was performed in an Ingeny PhorU2 system (Ingeny, Goes, the Netherlands) with a 40 to 65% denaturing gradient (where 100% denaturant consisted of 7 M urea and 40% formamide) and 6.0% polyacrylamide. Electrophoresis was initiated at 60°C and maintained at 100 V for 16 h. After electrophoresis, gels were stained with SYBR Gold with a final concentration of 0.5 mg liter−1 (Invitrogen, Breda, the Netherlands) and photodocumented in UV light. Images of the gels were obtained with Image Master VDS (Amersham Biosciences, Buckinghamshire, United Kingdom) and stored as TIFF files.
DGGE patterns were compared by clustering the different lanes by Pearson's correlation implemented in GelCompar II software (Applied Maths, Sint-Martens Latem, Belgium) using the unweighted-pair group method with arithmetic means (UPGMA). Additionally, differences in multivariate aspects of community structure between treatments were determined with multidimensional scaling ordination (MDS) of square root-transformed species abundance data using the Bray-Curtis similarity measure, and their significance was assessed by two-way analysis of similarities (ANOSIM) in PRIMER (version 6; Primer-E Ltd., Plymouth, United Kingdom) (7). ANOSIM produces a test statistic, R, and a significance level, P. Values of R closer to 0 implied no differences between samples. The correlation between community structure and physicochemical parameters were tested using a global BEST test in Primer-E software (with a Spearman coefficient and 5,000 permutations), which selects environmental variables best explaining community patterns by maximizing a rank correlation between their respective resemblance matrices. The relationships between the physicochemical parameters were tested using Pearson's correlation in SPSS Statistics 20.
Construction and analysis of nifH gene clone libraries.
The DNA extracted from each sample was used to construct nifH-based clone libraries (one per subregion of sampling points due to the high similarity of replicates for DGGE analysis). The template DNA was subjected to amplification with specific primers for the nifH gene according to the methodology previously described (36). The amplification products were purified using the Wizard SV gel and PCR clean-up system (Promega, Madison, WI) and inserted into pGEM-T using the pGEM-T Easy Vector system kit (Promega, Madison, WI) according to the manufacturer's instructions. The colonies that exhibited vector inserts (white color) were selected for insert detection by amplification with the M13F and M13R primers. The resulting PCR products with the inserts were sent for sequencing at AGOWA (Berlin, Germany). Using this methodology, 458 clones containing nifH gene inserts were obtained. Sequences were trimmed to remove poor quality, and vector sequences were removed with the Lucy program, available from the Ribosomal Database Project.
Sequence analyses were performed at the level of nucleotides by phylogeny-based approaches and also by applying operational taxonomic unit (OTU)-based analyses using the MOTHUR program (34). The phylogeny-based analysis was made by comparing the obtained sequences to those from public databases (GenBank and Fungene) in neighbor-joining (31) trees constructed using MEGA 4.0 (38) with the Kimura 2-parameter method. The bootstrap consensus tree inferred from 1,000 replicates was taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in fewer than 50% bootstrap replicates were collapsed.
Nucleotide sequence accession numbers.
The sequences reported in this study were deposited in GenBank under the accession numbers JQ237691 to JQ237760.
RESULTS
Abundance of nifH in the mangrove sediments.
The nifH gene was detected and quantified across all sediment samples. The efficiency value for amplification of this target gene was 99.26%, and logarithmic regression curves (R2) reached a value of 0.99. The amplification specificity was found to be robust, with single peaks being detected during melting curve analysis.
Statistically similar nifH gene densities (P > 0.05) were observed between the two mangrove sample sets (oil polluted and free of oil) and among distinct sampling points (P > 0.05), with average values ranging from 6.81 to 8.26 log nifH copies per g sediment (Table 3). The lack of differences in nifH gene densities may have been due to the high variability across the replicate from point 1 in the oil-polluted mangrove (OilMgv). However, in spite of the lack of statistical support, a trend toward an increase of the abundance of nifH copies at point 1 was observed in the non-oil-affected mangrove, while numbers tend to be smaller at point 1 of the OilMgv (Table 3).
Table 3.
Quantification of nifH gene copies per sample site determined by qPCRa
| Mangrove | Copies of the target gene per gram of sediment (log x + 1) at point: |
||
|---|---|---|---|
| 1 | 2 | 3 | |
| OilMgv | 6.81 ± 0.54 | 7.26 ± 0.26 | 7.22 ± 0.21 |
| AntMgv | 7.40 ± 0.05 | 6.88 ± 0.07 | 6.87 ± 0.19 |
The R2 of the standard curve was greater than 0.99. The qPCR estimated efficiency was 99.26%. Values are means from three replicates (n = 3) ± standard errors.
Community structure analysis based on the nifH gene.
Analysis of the nifH-based PCR-DGGE profiles revealed that the numbers of bands per sample varied from 27 to 39. Furthermore, the profiles were qualitatively and quantitatively different, as different bands and/or band intensities were recorded in the different samples (Fig. 1a). The profile analysis by clustering (Pearson correlation) generated an external group encompassing all samples from the area near the sea in the oiled mangrove (OilMgv1). This cluster showed only 20% similarity to any of the other profiles. In this cluster, a separation between mangrove types was observed at 30% similarity, hence distinguishing other OilMgv samples from those obtained from AntMgv (Fig. 1a). In order to better visualize these clusters, an NMDS plot was generated in which three distinct groups emerged, encompassing profiles from oil site 1 (OilMgv1), oil sites 2 and 3 (OilMgv2-3), and the non-oiled mangrove (AntMgv) (Fig. 1b). ANOSIM analysis showed a significant effect of treatment (impacted versus not impacted; R = 0.576; P = 0.02), which is supported by the shifts observed in values of pH (R = 0.253; P = 0.001), total nitrogen (R = 0.287; P = 0.001), and sulfur content (R = 0.311; P = 0.001) (Table 2). Pearson's correlation revealed that these parameters did not interact significantly with each other or with the other measured environmental parameters.
Fig 1.
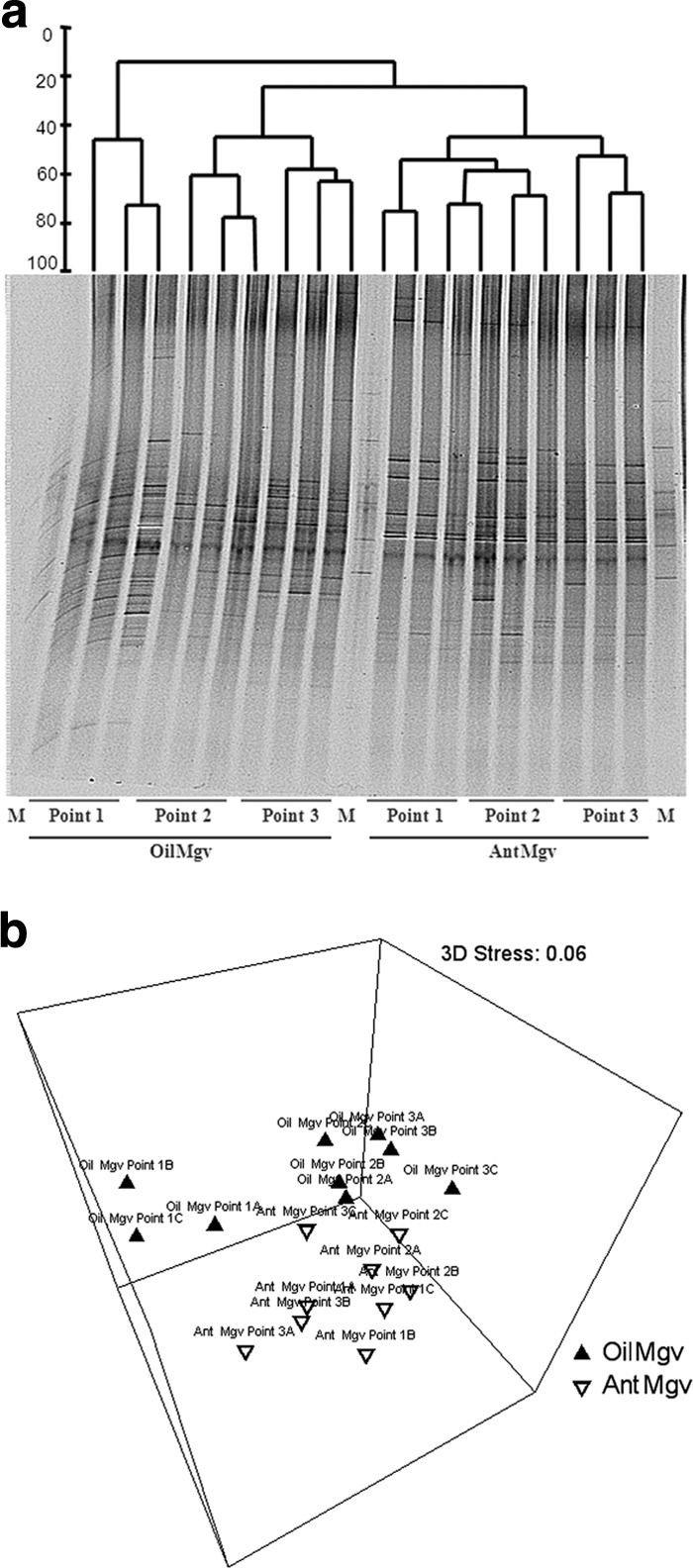
(a) PCR-DGGE profiles for nitrogen-fixing (nifH) bacteria. A standard nifH gene PCR-DGGE dendrogram was created using the UPGMA method based on the similarity calculated by densitometry. (b) Nonmetric multidimensional scaling (NMDS) analysis comparing the nifH community based on Bray-Curtis similarities.
Phylogenetic evaluation of nitrogen-fixer groups in the sample sites.
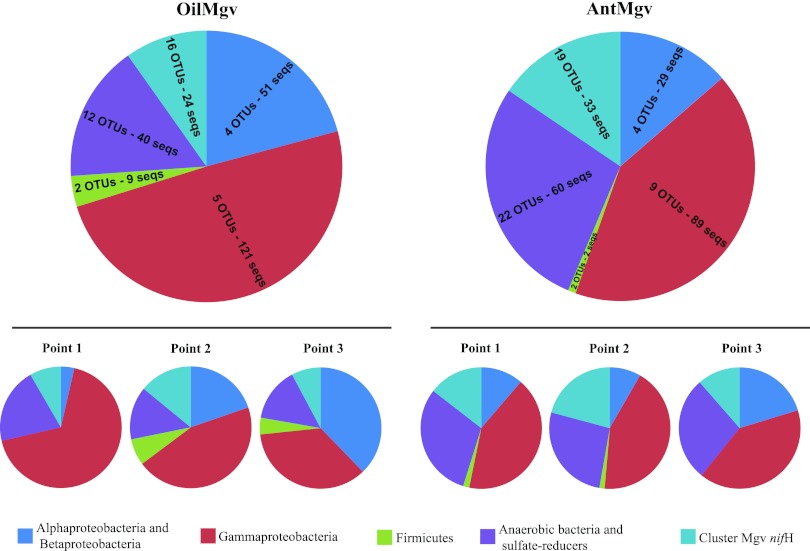
The six clone libraries based on the nifH gene encompassed a total of 458 trimmed sequences. Phylogenetic analyses showed a dominance of sequences affiliated with nifH of diverse Proteobacteria (290 sequences; 210 for Gammaproteobacteria and 80 for the cluster containing Alphaproteobacteria and Betaproteobacteria), followed by anaerobic bacteria related to sulfate reducers (100 sequences), Firmicutes (11 sequences) (Fig. 2), and a cluster of Mgv-nifH (57 sequences) encompassing novel nifH sequences which showed low similarity to any other nifH sequences.
Fig 2.
Taxonomic distribution of nifH-generated OTUs between each mangrove and among sites within mangroves. Sequences were clustered using a cutoff value of 80% similarity at the nucleotide level.
An OTU-based data treatment was then carried out in which all sequences were binned using a cutoff of 80% similarity at the nucleotide level. This data transformation yielded a total of 95 OTUs (encompassing all 458 sequences). While the two nifH groups within the Proteobacteria (Gammaproteobacteria and Alpha/Betaproteobacteria) appeared like low-diversity clusters (210 and 80 sequences fell into just 8 and 14 OTUs, respectively), anaerobic sulfate reducers and cluster Mgv-nifH represented high-diversity clusters (100 and 57 sequences, falling into 34 and 35 OTUs, respectively). Firmicutes showed an intermediate diversity (11 sequences falling into 4 OTUs) (Fig. 2).
The nifH clusters occurred differentially across the analyzed libraries. An analysis of the relative abundances of each cluster revealed that the Alpha/Betaproteobacteria, Firmicutes, and Mgv-nifH types showed similar average values, though Alpha/Betaproteobacteria abundances varied greatly across sites in the oiled mangrove. In contrast, differences were observed for the Gammaproteobacteria nifH type, which was more frequent in OilMgv (0.46) than in AntMgv (0.29). Also, the nifH group of anaerobic bacteria related to sulfate reduction was more frequent in AntMgv (0.42) than in OilMgv (0.16) (Fig. 2). Overall, in the more pristine mangrove, a relatively constant pattern was observed (AntMgv), whereas in the oil-polluted mangrove greater differences were found among samples. For instance, the frequency of Gammaproteobacteria-type nifH decreased and the frequency of nifH of Alpha/Betaproteobacteria increased along the transect from sea to inland (Fig. 2). Moreover, in the area most affected by the oil spill (points 2 and 3 in the OilMgv), increases of nifH sequences affiliated with Firmicutes were found.
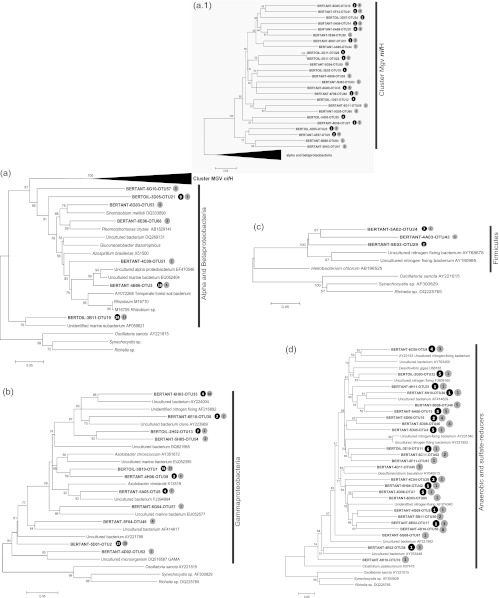
The phylogenetic affiliation of the sediment-derived sequences was then assessed in detail by including them in prestructured trees as described by Diallo et al. (8) and Mehta et al. (25). Many sediment nifH sequences turned out to be affiliated with those from uncultured organisms. In addition, others clustered with sequences without clear designation but that were obtained from genera described as nitrogen fixers, i.e., Sinorhizobium (Fig. 3a), Azotobacter (Fig. 3b), Desulfomicrobium (Fig. 3d), Azospirillum, Gluconacetobacter, Rhizobium, Desulfovibrio (Fig. 3), and nifH sequences in cluster Mgv-nifH (Fig. 3a.1). The nifH-like OTUs in the Alpha/Betaproteobacteria showed similarity to nifH genes of Sinorhizobium, Plemorphomonas, Gluconacetobacter, Rhizobium, and Azospirillum (Fig. 3a). Among the gammaproteobacterial and Firmicutes nifH types, the genera Azotobacter and Heliobacterium were predominantly found (Fig. 3b and c). Among the sulfate reducers and other anaerobes, nifH types of Desulfovibrio, Desulfumicrobium, and Clostridium (Fig. 3d) were dominant in the mangrove sediments.
Fig 3.
Phylogenetic analysis of nifH gene sequences retrieved from Bertioga mangroves. Bootstrap values (1,000 repetitions) above 50% are indicated next to the tree branches. Solid black and gray circles represent sequences retrieved from OilMgv and AntMgv, respectively. Trees display one representative sequence per OTU, and the total number of sequences within each OTU is numerically indicated in the circles. Shown are retrieved nifH sequences related to Alpha/Betaproteobacteria groups (a), Gammaproteobacteria (b), Firmicutes (c), and anaerobic and sulfate-reducing bacteria (d). Phylogenetic trees were constructed using the neighbor-joining method, and the evolutionary distances were computed using the Kimura 2-parameter method. There were a total of 416 nucleotide positions in the final data set, and the numbers of base substitutions per site are indicated in the scale bars.
Figure 3 also shows that the sequence types occurred in distinct abundance across the two mangroves. A clear example of differential incidence is provided by the OTUs of the Alpha/Betaproteobacteria. In the OilMgv samples, the alphaproteobacterial nifH gene type was prevalent, whereas the betaproteobacterial type dominated in the AntMgv.
Testing of the composition of libraries by UniFrac revealed that the diazotrophic communities at the six sampling sites differed significantly from each other (P < 0.05). Although branches are long, two clusters were observed (separated by high values in jack-knife clustering analysis), the first encompassing libraries derived from the oil-contaminated mangrove and the second encompassing those from the uncontaminated mangrove (Fig. 4).
Fig 4.

Dendrogram based on nifH gene clone libraries showing the differences in community structure at six sampling points. The distance matrices were generated with UNIFRAC and were used to group the points of the sediments analyzed using the unweighted-pair group method with arithmetic means, and jack-knife analysis was used to evaluate the robustness of each cluster related to the environment, size, and uniformity of the sample. The numbers indicate the frequency in the permutation test, which was supported by jack-knife analysis.
DISCUSSION
Biological nitrogen fixation is a key ecosystem process, and its activity is influenced by the resident functional microbial community and environmental conditions (13, 32). In mangroves, the sediment offers an outstanding habitat in which nitrogen fixation can occur, the main reason being the oxygen gradient, including anaerobic conditions and the presence of carbonaceous and other nutrients (2, 22). Here, we addressed questions about the abundance and diversity of diazotrophic communities across sediment samples and the possible effect of oil pollution. We used the nifH gene as a proxy for the nitrogen-fixing communities involved.
A few previous studies have addressed the microbial diversity in mangroves, most of them reporting taxonomical groups inhabiting this ecosystem and assessing shifts in the community structures in the face of environmental pressures (9, 15, 23, 42). However, the composition and dynamics of the mangrove microbiome are still scarcely explored and understood in terms of its functional groups (2, 13, 15, 32). In our study, the qPCR data revealed statistically similar diazotrophic community abundances across the samples of the two mangroves. This suggests that the carrying capacities for such microbial guilds were similar across the sediments sampled in the two transects. However, a trend toward an increased abundance in the area with higher oil impact (sites 2 and 3 within the oil-contaminated mangrove) was found. The oil contamination may have raised carbon input (see data for point 2 in Table 2) and limited O2 availability at sites 2 and 3, stimulating these populations compared to site 1 (unpolluted) in this mangrove. In addition, the vegetation at sites 2 and 3 was less dense and diverse (Table 1), leading to less intense cycling of organic matter, thus inducing the growth of diazotrophs to obtain inorganic forms of nitrogen. Such indications were also made by metagenomics analyses in the same area, where higher densities of anaerobes were found in the oiled mangrove (2). These observations are also underlined by Li et al. (22), who defined mangroves as environments with particular nutrient cycling and site-specific anaerobic and aerobic conditions, where distinct microbially mediated transformations can occur. It also helps explain why induced organisms in the oiled area were not affiliated with the anaerobic sulfate-reducer organisms but were affiliated with other (Alpha/Betaproteobacteria and Mgv-nifH) groups, evidencing a possible role of growth inhibition promoted by the oil under the sulfate-reducer organisms, an issue that should be better assessed later by a function-based approach (for example, metatranscriptomics).
Regarding the diversity of the nifH gene sequences, the differences in the PCR-DGGE profiles between mangroves and among sites within a mangrove showed the responsiveness of the diazotrophic communities to environmental contamination with oil. The differences between the mangrove sites corroborate previous data where other fractions of microbial communities were revealed to be responsive to oil pollution in mangrove sediments (10, 36). This allowed us to pinpoint the key nifH gene types as candidates for biological indicators of mangrove contamination. Other microbial groups have also been selected for this purpose, as recently described for the fungal (13) and archaeal (10) communities in the same area.
From the clone library analyses, we learned which nifH groups have the ability to cope with prevailing mangrove conditions, such as fluctuating salinity and oxia/anoxia (19, 20). Considering the phylogenetic placement of the nifH gene sequences, the mangroves exhibited a suite of nifH types affiliated with phyla across the bacterial tree. The sequences were, in part, similar to those described for diazotrophs in the marine environment, but they were also similar to those found in anaerobic and aerobic soils. Strikingly, several sequences grouped separately from those found in databases, forming the so-called Mgv-nifH cluster. This finding certainly merits further studies, as it puts us on the track of potentially novel diazotrophic types that abound in mangrove ecosystems and have putative ecological roles in this habitat. Mangroves offer very particular conditions to their inhabitants (mixed influences from marine and terrestrial environments) and hence can be seen as reservoirs for such genetic and ecological novelty. Similar findings are described in the literature, where specific clusters were composed of sequences retrieved from mangroves sediments for the gene dsrB (37) and archaeal amoA (10).
Many members of the Proteobacteria have been described as nitrogen fixers in marine ecosystems. Bagwell et al. (3) reported a set of nifH gene sequences affiliated with the Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria in oligotrophic tropical seagrass beds. Church et al. (6) detected nifH gene sequences identical to those previously described for an uncultivated gammaproteobacterial line in the north Pacific Ocean. Bird et al. (5) suggested that gammaproteobacterial nitrogen fixers are widespread and likely the most important component of the heterotrophic diazotrophic community of tropical and subtropical oceans. Together, these reports reinforce the role of Proteobacteria, in particular Gammaproteobacteria, as nitrogen fixers in marine ecosystems. This is consistent with the prominence of gammaproteobacterial nifH in our mangrove samples, particularly for the increase of such groups in the OilMgv.
Considering the contextual parameters that modulate the microbial assemblages in mangroves, physicochemical parameters and vegetation type stand out as the strongest agents. Here, parameters of nitrogen and sulfur contents, as well as pH values, were revealed to impose distinct conditions on the assemblage of nifH host communities. It can be suggested that these variables modulate specifically the activity of nitrogenase, possibly determining the essentiality of nifH presence in such communities. The contents of sulfur can play a role in supporting microbial metabolism (i.e., sulfate reduction) or exerting a toxic effect in specific microbial groups (in the form of H2S). However, the better combination of those factors is still to be elucidated, possibly by the use of some function-based approach, such as metatranscriptomics analysis. Flores-Meireles et al. (14) found the rhizosphere of Avicennia germinans to be a selective environment for diazotrophic microorganisms, being essential to determine the structure of diazotrophic communities associated with mangrove roots.
The results reported in this study provide fundamental insights into the structure and diversity of major nitrogen-fixing bacterial populations in mangrove sediments, revealing clusters of novel sequences that are uniquely found in the mangrove ecosystem. We propose the use of this Mgv-nifH mangrove nifH type as a protoprobe for a thorough future investigation of the involvement of the underlying microbial group in the sediment nitrogen fixation process, which in our view is presumably a free-living group.
ACKNOWLEDGMENTS
This work was financially supported by the São Paulo Research Foundation (FAPESP; number 2004/13910-6). We also thank the Dutch NWO-ERGO program for funding, and A. C. F. Dias received a graduate fellowship from FAPESP (no. 2008/54013-8).
We thank João L. Silva for his help with the mangrove expeditions and sampling.
Footnotes
Published ahead of print 31 August 2012
REFERENCES
- 1. Alongi DM. 1988. Bacterial productivity and microbial biomass in tropical mangrove sediments. Microb. Ecol. 15: 59– 79 [DOI] [PubMed] [Google Scholar]
- 2. Andreote FD, et al. 2012. The microbiome of Brazilian mangrove sediments as revealed by metagenomics. PLoS One 7: e38600 doi:10.1371/journal.pone.0038600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bagwell CE, et al. 2002. Molecular diversity of diazotrophs in oligotrophic tropical seagrass bed communities. FEMS Microb. Ecol. 39:113– 119 [DOI] [PubMed] [Google Scholar]
- 4. Beman JM, Francis CA. 2006. Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hypernutrified subtropical estuary: Bahia del Tobari, Mexico. Appl. Environ. Microbiol. 72: 7767– 7777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bird C, Martinez JM, O'Donnell AG, Wyman M. 2005. Spatial distribution and transcriptional activity of an uncultured clade of planktonic diazotrophic γ-proteobacteria in the Arabian Sea. Appl. Environ. Microbiol. 71: 2079– 2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Church MJ, Short CM, Jenkins BD, Karl DM, Zehr JP. 2005. Temporal patterns of nitrogenase gene (nifH) expression in the oligotrophic North Pacific Ocean. Appl. Environ. Microbiol. 71: 5362– 5370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clarke K, Gorley R. 2006. PRIMER v6: user manual/tutorial. Primer-E Ltd, Plymouth, United Kingdom: [Google Scholar]
- 8. Diallo MD, et al. 2004. Polymerase chain reaction denaturing gradient gel electrophoresis analysis of the N2-fixing bacterial diversity in soil under Acacia tortilis ssp. raddiana and Balanites aegyptiaca in the dryland part of Senegal. Environ. Microbiol. 6: 400– 415. [DOI] [PubMed] [Google Scholar]
- 9. Dias ACF, et al. 2010. The bacterial diversity in a Brazilian non-disturbed mangrove sediment. Antonie van Leeuwenhoek 98: 541– 551 [DOI] [PubMed] [Google Scholar]
- 10. Dias ACF, et al. 2011. Archaeal communities in sediments of three contrasting mangroves. J. Soils Sediments 11: 1466– 1476 [Google Scholar]
- 11. Dos Santos H, et al. 2011. Mangrove bacterial diversity and the impact of oil contamination revealed by pyrosequencing: bacterial proxies for oil pollution. PLoS One 6: e16943 doi:10.1371/journal.pone.0016943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Duke NC, Meynecke JO, Dittmann S. 2007. A world without mangroves? Science 317:41– 42 [DOI] [PubMed] [Google Scholar]
- 13. Fasanella CC, et al. 2012. The selection exerted by oil contamination on mangrove fungal communities. Water Air Soil Pollut 223: 4233– 4243 [Google Scholar]
- 14. Flores-Mireles AL, Winans SC, Holguin G. 2007. Molecular characterization of diazotrophic and denitrifying bacteria associated with mangrove roots. Appl. Environ. Microbiol. 73: 7308– 7321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gomes NCM, et al. 2008. Exploring the diversity of bacterial communities in sediments of urban mangrove forests. FEMS Microbiol. Ecol. 66: 96– 109 [DOI] [PubMed] [Google Scholar]
- 16. Gomes NC, et al. 2010. Mangrove microniches determine the structural and functional diversity of enriched petroleum hydrocarbon-degrading consortia. FEMS Microbiol. Ecol. 74: 276– 290 [DOI] [PubMed] [Google Scholar]
- 17. Holguin GB, et al. 1999. La microbiología de los manglares. Bosques en la frontera entre el mar y la tierra. Ciencia Desarrollo 25: 26– 35 [Google Scholar]
- 18. Holguin G, Vazquez P, Bashan Y. 2001. The role of sediment microorganisms in the productivity, conservation, and rehabilitation of mangrove ecosystems: an overview. Biol. Fertil. Soils 33: 265– 278 [Google Scholar]
- 19. Holguin G, et al. 2006. Mangrove health in an arid environment encroached by urban development—a case study. Sci. Total Environ. 363: 260– 274 [DOI] [PubMed] [Google Scholar]
- 20. Kathiresan K, Bingham BL. 2001. Biology of mangroves and mangrove ecosystems. Adv. Mar. Biol. 40: 81– 251 [Google Scholar]
- 21. Kristensen E, et al. 1998. Transformation and transport of inorganic nitrogen in sediments of a southeast Asian mangrove forest. Aquat. Microb. Ecol. 15: 165– 175 [Google Scholar]
- 22. Li M, Cao H, Hong Y, Gu JD. 2011. Spatial distribution and abundances of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) in mangrove sediments. Appl. Microbiol. Biotechnol. 89: 1243– 1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liang JB, et al. 2007. Recovery of novel bacterial diversity from mangrove sediment. Mar. Biol. 150: 739– 747 [Google Scholar]
- 24. Lovell CR, Friez MJ, Longshore JW, Bagwell CE. 2001. Recovery and phylogenetic analysis of nifH sequences from diazotrophic bacteria associated with dead aboveground biomass of Spartina alterniflora. Appl. Environ. Microbiol. 67: 5308– 5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mehta MP, Butterfield DA, Baross JA. 2003. Phylogenetic diversity of nitrogenase (nifH) genes in deep-sea and hydrothermal vent environments of the Juan de Fuca Ridge. Appl. Environ. Microbiol. 69: 960– 970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Patra AK, et al. 2006. Effects of management regime and plant species on the enzyme activity and genetic structure of N-fixing, denitrifying and nitrifying bacterial communities in grassland soils. Environ. Microbiol. 8: 1005– 1016 [DOI] [PubMed] [Google Scholar]
- 27. Pereira e Silva MC, Semenov AV, van Elsas JD, Salles JF. 2011. Seasonal variations in the diversity and abundance of diazotrophic communities across soils. FEMS Microbiol. Ecol. 77: 57– 68 [DOI] [PubMed] [Google Scholar]
- 28. Poly F, Monrozier LJ, Bally R. 2001. Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res. Microbiol. 152: 95– 103 [DOI] [PubMed] [Google Scholar]
- 29. Raymond J, Siefert JL, Staples CR, Blankenship RE. 2004. The natural history of nitrogen fixation. Mol. Biol. Evol. 21: 541– 554 [DOI] [PubMed] [Google Scholar]
- 30. Rosch C, Bothe H. 2009. Diversity of total, nitrogen-fixing and denitrifying bacteria in an acid forest soil. Eur. J. Soil Sci. 60: 883– 894 [Google Scholar]
- 31. Saitou N, Nei M. 1987. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406– 425 [DOI] [PubMed] [Google Scholar]
- 32. Santos HF, Carmo FL, Paes JE, Rosado AS, Peixoto RS. 2011. Bioremediation of mangroves impacted by petroleum. Water Air Soil Pollut. 216: 329– 350 [Google Scholar]
- 33. Schaeffer-Novelli Y, Cintron-Molero G, Adaime RR, Camargo TM. 1990. Variability of mangrove ecosystems along the Brazilian coast. Estuaries 13: 204– 218 [Google Scholar]
- 34. Schloss PD, et al. 2009. Introducing MOTHUR: open-source, platform-independent, community supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75: 537– 7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Simonet P, et al. 1991. Frankia genus-specific characterization by polymerase chain reaction. Appl. Environ. Microbiol. 57: 3278– 3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Taketani RG, dos Santos HF, van Elsas JD, Rosado AS. 2009. Characterization of the effect of a simulated hydrocarbon spill on diazotrophs in mangrove sediment mesocosm. Antonie van Leeuwenhoek 96: 343– 354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Taketani RG, Yoshiura CA, Dias ACF, Andreote FD, Tsai SM. 2010. Diversity and identification of methanogenic archaea and sulphate-reducing bacteria in sediments from a pristine tropical mangrove. Antonie van Leeuwenhoek 97: 401– 411 [DOI] [PubMed] [Google Scholar]
- 38. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596– 1599 [DOI] [PubMed] [Google Scholar]
- 39. Turk KA, et al. 2011. Nitrogen fixation and nitrogenase (nifH) expression in tropical waters of the eastern North Atlantic. ISME J. 5: 120– 1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a. Van Raij B, Cantarella H, Andrade JC, Quaggio JA. 2001. Análise química para avaliação da fertilidade de solos tropicais. Instituto Agronômico, Campinas, Brazil: (In Portuguese.) [Google Scholar]
- 40. Vlaeminck SE, Hay AG, Maignien L, Verstraete W. 2011. In quest of the nitrogen oxidizing prokaryotes of the early Earth. Environ. Microbiol. 13: 283– 295 [DOI] [PubMed] [Google Scholar]
- 41. Zehr JP, McReynolds LA. 1989. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl. Environ. Microbiol. 55: 2522– 2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou HW, Luanc TG, Zoub F, Tam NFY. 2008. Different bacterial groups for biodegradation of three- and four-ring PAHs isolated from a Hong Kong mangrove sediment. J. Hazard. Mater. 152: 1179– 1185 [DOI] [PubMed] [Google Scholar]