Abstract
Intermediates of production of two batches of traditional mozzarella cheese were analyzed by culture-independent pyrosequencing. The quantitative distribution of taxa within the samples suggested that thermophilic lactic acid bacteria from the natural starter were mainly responsible for the fermentation, while microorganisms found in raw milk did not develop during fermentation.
TEXT
Mozzarella is perhaps the most popular nonripened cheese. Traditional mozzarella is mainly produced in southern Italy from water buffalo's milk, even though it is widely exported and also industrially produced in other countries. The technology of its manufacture has been described in detail in previous works (11, 14). The cheese is made from whole raw water buffalo's milk by adding a natural whey culture (NWC) (from the batch of the previous day) as starter in a 5-h curd fermentation. The specific characteristics of the final product arise mainly from the raw materials employed, the agri-ecosystem of the area of production, and the traditional technology of the manufacturing process. The use of raw buffalo's milk and the NWC in the process have so far been recognized as strong points of traditional mozzarella production, because premium-quality products arise as result of fermentation by the specific microbiota of raw milk and NWC. Due to the use of microbiologically complex raw materials, the traditional cheese-making processes are the most difficult to control, and it is of interest to develop reliable methods to monitor the fermentation in order to standardize the process for high-quality products while preserving their typical traits. The microbiota of mozzarella cheese has been studied in the past by culture-independent fingerprinting without identification of microbial taxa (9, 14). However, the microbiota involved in buffalo mozzarella production has never been thoroughly assessed by microbial species identification, although the complexity of the microbiota is recognized on the basis of culture-based microbiological determinations (10, 11, 21, 24). High-throughput-sequencing approaches were recently applied in a few cases to describe the microbiota of food products and have been shown to provide a thorough analysis of microbial diversity, producing much deeper output than more commonly used culture-independent approaches (1, 13, 16, 22, 23). In this study, intermediates of production of two batches of traditional mozzarella cheese were analyzed by culture-independent pyrosequencing in order to provide insights into the microbiota responsible for the production of this widely appreciated dairy product.
Samples were collected in May 2012 from two dairies producing top-quality traditional water buffalo mozzarella cheese, located in the Campania region (southern Italy) in the provinces of Salerno (Batch 1) and Caserta (Batch 2), respectively. Samples of raw milk (L), natural whey cultures (NWC), curd at the beginning (C0) and at end (CF) of the ripening, and final mozzarella cheese (M) were aseptically collected, cooled at 4°C, and analyzed within 6 h. Duplicate samples were collected from two batches within the same day of work, and equal amounts of samples were pooled prior to DNA extraction. Raw milk was taken from the vat before manufacturing started; curds at the beginning and end of ripening were taken after 20 min and 5 h after NWC addition, respectively; mozzarella cheese samples were collected from their governing liquid 20 min after molding.
Total DNA extraction from the dairy samples was carried out by using a Biostic bacteremia DNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA). The dairy samples were 2-fold diluted in one-quarter-strength Ringer's solution, and the protocol was applied to the pellet (12,000 × g) from 1 ml of suspension. The microbial diversity was studied by pyrosequencing of the amplified V1-V3 region of the 16S rRNA gene by using the primers Gray28F (5′-TTTGATCNTGGCTCAG) and Gray519r (5′-GTNTTACNGCGGCKGCTG), which amplify a fragment of 520 bp (2). 454 adaptors were included in the forward primer, followed by a 10-bp sample-specific multiplex identifier (MID). Each PCR mixture (final volume, 50 μl) contained 50 ng of template DNA, a 0.4 μM concentration of each primer, 0.50 mmol liter−1 of each deoxynucleoside triphosphate, 2.5 mmol liter−1 MgCl2, 5 μl of 10× PCR buffer, and 2.5 U of native Taq polymerase (Invitrogen, Milano, Italy). The following PCR conditions were used: 94°C for 2 min, 35 cycles of 95°C for 20 s, 56°C for 45 s, and 72°C for 5 min, and a final extension at 72°C for 7 min. After agarose gel electrophoresis, PCR products were first purified with a QIAquick gel extraction kit (Qiagen, Milano, Italy) and then with an Agencourt AMPure kit (Beckman Coulter, Milano, Italy) prior to further processing. Amplicons were used for pyrosequencing on a GS Junior platform (454 Life Sciences, Roche Diagnostics, Italy) according to the manufacturer's instructions by using Titanium chemistry. Sequences are available at the Sequence Read Archive (project SRP014821).
A first filtering of the results was performed by using the 454 Amplicon signal processing; then sequences were analyzed by using QIIME 1.5.0 software (5). In order to guarantee a higher level of accuracy in terms of OTU (operational taxonomic unit) detection, after the split library script performed by QIIME, the reads were excluded from the analysis if they had an average quality score lower than 25, if they were shorter than 200 bp, and if there were ambiguous base calls. Sequences that passed the quality filter were denoised (25), and singletons were excluded. OTUs defined by a 97% of similarity were picked using the Uclust method (12) and the representative sequences were submitted to the RDPII classifier (29) to obtain the taxonomy assignment and the relative abundance of each OTU using the Greengenes 16S rRNA gene database (19). Representative sequences for OTUs showing an incidence above 5% in at least one sample were double checked with the BLAST (BLASTN) search program (http://www.ncbi.nlm.nih.gov/blast/) to confirm the taxonomy assignment made by QIIME. Sequences that were aligned using PyNAST in QIIME (6) were used as input in Mothur 1.26.0 software (27) to generate Good's coverage, Chao1 richness (7), and Shannon diversity indexes (28). The OTU taxonomy table generated by QIIME was used to produce a heat map by using the clustering software TMeV v4.8 (26).
The run produced 90,500 reads after 454 amplicon signal quality control; after further filtering protocols, 77,277 reads were obtained, with an average length of 492 bp; the reads are distributed among the samples as reported in Table 1. The filtering of the sequences eliminated no more than 15% of the reads per sample on average. A total of 511 OTUs were obtained. The rarefaction analysis and the diversity indexes indicated that there was satisfactory coverage of the diversity for the all intermediates of production within the two batches (Table 1). The highest diversity was associated with raw milk samples that also had lower estimated sample coverage. Overall, despite the diversity of sequencing depth between samples, the rarefaction analysis indicated that a number of reads above 2,000 per sample would be advisable to obtain good coverage (Table 1).
Table 1.
Number of sequences analyzed, observed diversity, and estimated sample coverage for 16S rRNA amplicons from mozzarella cheese manufacturesa
| Sample | No. of reads | No. of OTUs | Chao1 richness | Shannon diversity index | ESC (%) |
|---|---|---|---|---|---|
| L1 | 7,860 | 192 | 1,963.52 (1,684.77, 2,326.65) | 3.45 (3.40, 3.51) | 93.7 |
| NWC1 | 14,066 | 11 | 790.98 (500.60, 1,336.95) | 1.46 (1.44, 1.48) | 99.1 |
| C01 | 10,459 | 26 | 332.40 (245.90, 490.22) | 1.07 (1.04, 1.10) | 99.2 |
| CF1 | 8,920 | 19 | 297.12 (229.89, 416.97) | 1.21 (1.18, 1.24) | 99.0 |
| M1 | 10,219 | 35 | 693.18 (535.76, 941.45) | 2.05 (2.02, 2.09) | 98.3 |
| L2 | 7,579 | 97 | 945.28 (782.52, 1,181.72) | 3.49 (3.45, 3.53) | 97.0 |
| NWC2 | 2,947 | 13 | 196.50 (152.98, 282.11) | 1.82 (1.76, 1.88) | 98.0 |
| C02 | 4,510 | 55 | 457.78 (371.12, 598.41) | 2.24 (2.17, 2.30) | 97.2 |
| CF2 | 5,661 | 33 | 402.12 (310.92, 558.83) | 1.72 (1.67, 1.77) | 98.1 |
| M2 | 5,056 | 30 | 304.29 (236.74, 425.68) | 1.73 (1.68, 1.78) | 98.3 |
Abbreviations: OTU, operational taxonomic unit; ESC, estimated sample coverage. Chao1 richness, Shannon diversity, and ESC were calculated with Mothur at the 3% distance level. Values in parentheses are 95% confidence intervals.
Read length represents a key issue for taxonomic assignment in studies using high-throughput sequencing to study microbial ecology. A taxonomic identification of food microbes to the putative species level is desirable in order to obtain useful information on the succession of the microbiota during food production and storage. To this purpose, long sequence reads, including more variable regions of the 16S rRNA gene, are required for an accurate assignment.
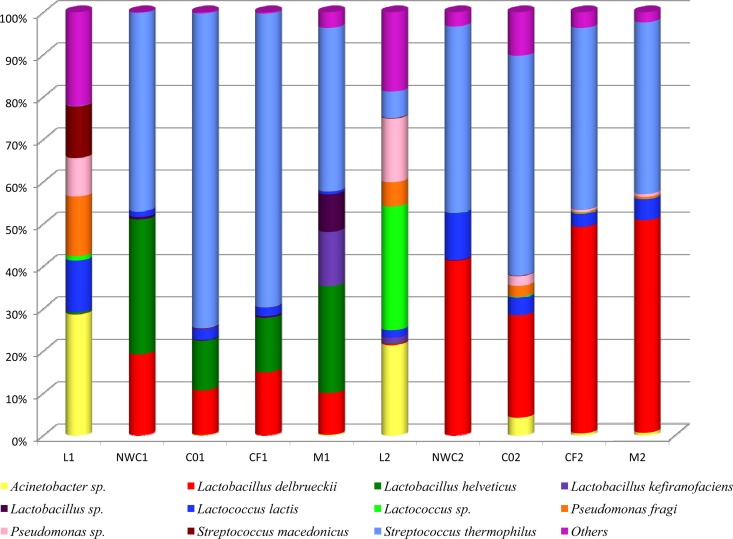
The application of 16S rRNA gene pyrosequencing allowed the determination of the microbial diversity in the intermediates of production of mozzarella cheese and also provided the relative abundance of the taxonomic levels of bacteria detected. The distribution of the OTUs with an incidence above 3% is reported in Fig. 1, where the evolution of the microbial diversity during the making of mozzarella cheese is shown. The raw water buffalo milk used for both batches displayed a complex microbiota composed of 192 (L1) and 97 (L2) total OTUs, respectively. The most abundant OTUs in both milks were the psychotropic organism Acinetobacter sp. and Pseudomonas sp., with incidences of about 21% and 20%, respectively (Fig. 1). Lactococcus lactis and Streptococcus macedonicus were also found, with an incidence above 10%, each in L1, whereas Lactococcus sp. represented almost 30% of the OTUs in L2 (Fig. 1). The most abundant OTUs in the natural starter NWC1 were Streptococcus thermophilus (47%), Lactobacillus delbrueckii (19%), and Lactobacillus helveticus (31%), while S. thermophilus (44%) and L. delbrueckii (41%) were the main OTUs in NWC2. The above OTUs were the most abundant even in the remaining samples collected during processing of each batch (Fig. 1). The trend of the lactic acid bacteria (LAB) populations were in overall agreement with viable counts of thermophilic and mesophilic LAB (data not shown).
Fig 1.
Incidence of OTUs based on 16S rRNA gene pyrosequencing analysis of all the DNA samples directly extracted from intermediates of production of two mozzarella cheese batches. Only OTUs with an incidence above 3% in at least one sample are shown.
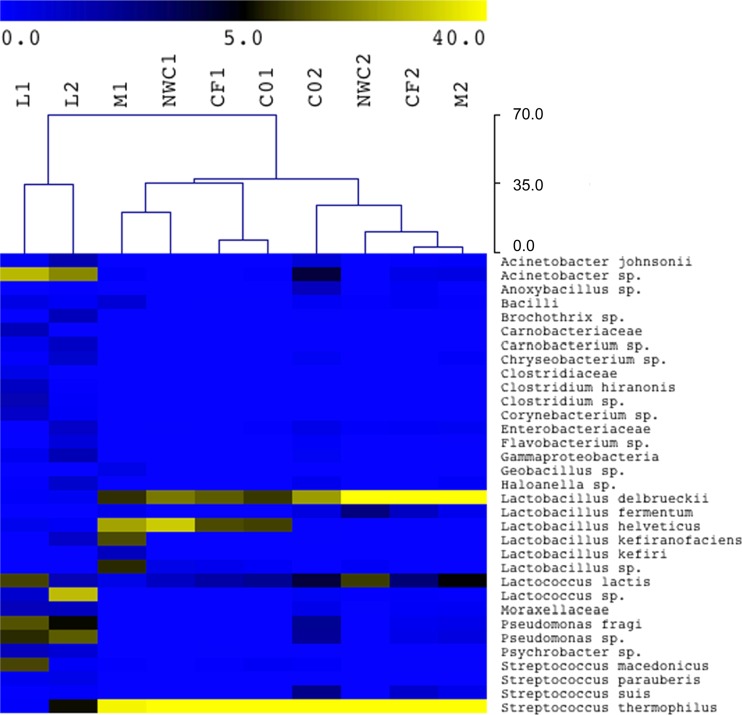
Given the minimum incidence of 0.5% in at least one sample, we considered 33 OTUs and used the percentage of abundance in each sample to generate the hierarchical clustering reported in Fig. 2. Raw milk samples with the most complex microbiota formed a cluster separated from the other samples, while the intermediates of production formed minor clusters, according to the specific dairy (Fig. 2). Most of the OTUs occurred in raw milk and not in the samples within each batch (Fig. 2). In addition, the OTUs associated with each fermentation, and determining the shown degree of similarity between samples are easily detected in the heat map (Fig. 2). The two batches studied here were from two different provinces in the main area of production. Accordingly, the microbiota associated with the production of buffalo mozzarella cheese and also the aroma profiles of mozzarella have been shown to be dependent on geographic origin (3, 17, 18). Abundant groups of Gram-negative OTUs, such as Pseudomonas spp. and Acinetobacter spp., as well as other contaminants, such as clostridia, carnobacteria, enterobacteria, and some other streptococci, characterized the microbiota of raw milk (Fig. 2). It was also noted that L. helveticus specifically occurred in batch 1, while Lactobacillus fermentum was found only in batch 2 (Fig. 2). Some accessory OTUs also occurred; L. lactis represented 12% of the OTUs in raw milk L1 but was never found beyond 2% during manufacture of batch 1. In contrast, in production 2, L. lactis originated from the NWC2 (11%) and occurred with an incidence of up to 5% in the other samples up to the final product. S. macedonicus and L. lactis occurring as dominant taxa in the raw milk L1 represented only minor populations during processing of the corresponding batch, similar to Lactococcus sp. in L2 (Fig. 2). Pseudomonas spp. coming from the raw milk also contaminated intermediates of production of batch 2, while some mesophilic lactobacilli occurred in the final mozzarella of dairy 1, including Lactobacillus kefiri and Lactobacillus kefiranofaciens (Fig. 2). The occurrence of such lactobacilli only in the final product can be due to their presence in the governing liquid of mozzarella cheese, which is composed of salted water and whey.
Fig 2.
Heat map depicting bacterial diversity and relative abundance in intermediates of production of two mozzarella cheese batches. The hierarchical dendrogram shows the distribution of bacteria based on average linkage clustering and Euclidean distance. The scale at the top defines the percentages of OTU in the samples as depicted by the colors in the heat map.
The deep sequencing approach was useful to identify the sources of bacteria in the cheese-making process and to ascertain whether they originated from the milk, the NWC starter, or other sources. The quantitative distribution of the OTUs within the samples suggests that the microbiota involved in the fermentation is carried by the NWC and that the microorganisms present in raw milk do not develop during fermentation. A limited amount of samples was analyzed in this study; in spite of the good sample coverage, the analysis of a greater number of samples from the same dairies and other dairies in the two provinces would further support the results obtained.
The NWCs used as natural starters were characterized in previous studies (10, 11, 15, 17), and they have been regarded as complex consortia of microorganisms with great importance for driving the fermentation and for determining the quality of the traditional product (14, 17). In this study, we assessed the microbiota of two premium-quality mozzarella batches representing the principal geographical area of production. Overall, we demonstrated that the microbiota associated with this dairy production is not as complex as previously thought and that a few thermophilic LAB drive the fermentation, while mesophilic LAB such as L. lactis are quantitatively less abundant during manufacture. Therefore, although diversity at the strain level can also play an important role, the complexity of the aroma profiles of mozzarella cheese likely does not arise exclusively from microbial fermentation. Other environmental factors, including farming, specific feeding, and raw milk quality, can impact the aroma of mozzarella cheese (4, 8, 20), which, together with fermentation, yields the typical traits of the end product.
As shown in this study, the sensitivity of high-throughput sequencing can reveal minor OTUs occurring during cheese making. This can represent an advantage for research purposes in some food-manufacturing processes, when low-quality products arise and reliable monitoring of the microbiological quality is needed in order to detect and identify microbial contaminants and their sources in production intermediates or final products.
ACKNOWLEDGMENTS
M.I. is an employee of Roche Diagnostics SpA, Italy. This does not affect the author's objectivity in the presentation of the results.
We thank Sabina Chiaretti, Department of Cellular Biotechnology and Haematology, University of Rome—La Sapienza, for laboratory availability and Francesca Dal Pero for technical collaboration.
Footnotes
Published ahead of print 31 August 2012
REFERENCES
- 1. Alegria A, Szczesny P, Mayo B, Bardowski J, Kowalczyk M. 2012. Biodiversity in Oscypek, a traditional Polish cheese, determined by culture-dependent and -independent approaches. Appl. Environ. Microbiol. 78:1890–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andreotti R, et al. 2011. Assessment of bacterial diversity in the cattle tick Rhipicephalus (Boophilus) microplus through tag-encoded pyrosequencing. BMC Microbiol. 11:6–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bonizzi I, Feligini M, Aleandri R, Enne G. 2007. Genetic traceability of the geographical origin of typical Italian water buffalo mozzarella cheese: a preliminary approach. J. Appl. Microbiol. 102:667–673 [DOI] [PubMed] [Google Scholar]
- 4. Brescia MA, Monfreda M, Buccolieri A, Carrino C. 2005. Characterization of the geographical origin of buffalo milk and mozzarella cheese by means of analytical and spectroscopic determinations. Food Chem. 89:139–147 [Google Scholar]
- 5. Caporaso JG, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caporaso JG, et al. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chao A, Bunge J. 2002. Estimating the number of species in a stochastic abundance model. Biometrics 58:531–539 [DOI] [PubMed] [Google Scholar]
- 8. Cifuni GF, et al. 2007. Effect of feeding systems on aromatic characteristics of buffalo mozzarella cheese. It. J. Animal Sci. 6:1147–1149 [Google Scholar]
- 9. Coppola S, Blaiotta G, Ercolini D, Moschetti G. 2001. Molecular evaluation of microbial diversity occurring in different types of mozzarella cheese. J. Appl. Microbiol. 90:414–420 [DOI] [PubMed] [Google Scholar]
- 10. Coppola S, Parente E, Dumontet S, La Peccerella A. 1988. The microflora of natural whey cultures utilized as starter in the manufacture of mozzarella cheese from water-buffalo milk. Lait 68:295–310 [Google Scholar]
- 11. Coppola S, Villani F, Coppola R, Parente E. 1990. Comparison of different starter systems for water-buffalo mozzarella cheese manufacture. Lait 70:411–423 [Google Scholar]
- 12. Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461 [DOI] [PubMed] [Google Scholar]
- 13. Ercolini D, et al. 2011. Monitoring of microbial metabolites and bacterial diversity in beef stored in different packaging conditions. Appl. Environ. Microbiol. 77:7372–7381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ercolini D, Mauriello G, Blaiotta G, Moschetti G, Coppola S. 2004. PCR-DGGE fingerprints of microbial succession during a manufacture of traditional water buffalo mozzarella cheese. J. Appl. Microbiol. 96:263–270 [DOI] [PubMed] [Google Scholar]
- 15. Ercolini D, Moschetti G, Blaiotta G, Coppola S. 2001. The potential of a polyphasic PCR-DGGE approach in evaluating microbial diversity of natural whey cultures for water-buffalo mozzarella cheese production: bias of “culture dependent” and “culture independent” approaches. Syst. Appl. Microbiol. 24:610–617 [DOI] [PubMed] [Google Scholar]
- 16. Leite AM, et al. 2012. Assessment of the microbial diversity of Brazilian kefir grains by PCR-DGGE and pyrosequencing analysis. Food Microbiol. 31:215–221 [DOI] [PubMed] [Google Scholar]
- 17. Mauriello G, Moio L, Genovese A, Ercolini D. 2003. Relationships between flavouring capabilities, bacterial composition and geographical origin of natural whey cultures (NWCs) used for traditional water-buffalo mozzarella cheese manufacture. J. Dairy Sci. 86:486–497 [DOI] [PubMed] [Google Scholar]
- 18. Mazzei P, Piccolo A. 2012. H-1 HRMAS-NMR metabolomic to assess quality and traceability of mozzarella cheese from Campania buffalo milk. Food Chem. 132:1620–1627 [DOI] [PubMed] [Google Scholar]
- 19. McDonald D, et al. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6:610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moio L, Dekimpe J, Etievant P, Addeo F. 1993. Volatile flavour compounds of water buffalo mozzarella cheese. It. J. Food Sci. 5:57–68 [Google Scholar]
- 21. Morea M, Baruzzi F, Cocconcelli PS. 1999. Molecular and physiological characterisation of dominant bacterial populations in traditional mozzarella cheese processing. J. Appl. Microbiol. 87:574–582 [DOI] [PubMed] [Google Scholar]
- 22. Nam Y-D, Yi S-H, Lim S-II. 2012. Bacterial diversity of cheonggukjang, a traditional Korean fermented food, analyzed by barcoded pyrosequencing. Food Control 28:135–142 [Google Scholar]
- 23. Nieminen TT, et al. 2012. Comparison of microbial communities in marinated and unmarinated broiler meat by metagenomics. Int. J. Food Microbiol. 157:142–149 [DOI] [PubMed] [Google Scholar]
- 24. Parente E, Rota MA, Ricciardi A, Clementi F. 1997. Characterization of natural starter cultures used in the manufacture of Pasta Filata cheese in Basilicata (southern Italy). Int. Dairy J. 7:775–783 [Google Scholar]
- 25. Reeder J, Knight R. 2010. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat. Methods 7:668–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saeed AI, et al. 2003. TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34:374–378 [DOI] [PubMed] [Google Scholar]
- 27. Schloss PD, et al. 2009. Introducing MOTHUR: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shannon CE, Weaver W. 1949. The mathematical theory of information. AT&T Tech. J. 27:359–423 [Google Scholar]
- 29. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]