Abstract
Bats belong to a wide variety of species and occupy diversified habitats, from cities to the countryside. Their different diets (i.e., nectarivore, frugivore, insectivore, hematophage) lead Chiroptera to colonize a range of ecological niches. These flying mammals exert an undisputable impact on both ecosystems and circulation of pathogens that they harbor. Pneumocystis species are recognized as major opportunistic fungal pathogens which cause life-threatening pneumonia in severely immunocompromised or weakened mammals. Pneumocystis consists of a heterogeneous group of highly adapted host-specific fungal parasites that colonize a wide range of mammalian hosts. In the present study, 216 lungs of 19 bat species, sampled from diverse biotopes in the New and Old Worlds, were examined. Each bat species may be harboring a specific Pneumocystis species. We report 32.9% of Pneumocystis carriage in wild bats (41.9% in Microchiroptera). Ecological and behavioral factors (elevation, crowding, migration) seemed to influence the Pneumocystis carriage. This study suggests that Pneumocystis-host association may yield much information on Pneumocystis transmission, phylogeny, and biology in mammals. Moreover, the link between genetic variability of Pneumocystis isolated from populations of the same bat species and their geographic area could be exploited in terms of phylogeography.
INTRODUCTION
Pneumocystis species are mostly airborne-transmitted, highly host-specific opportunistic microfungi responsible for severe pneumonia in a wide range of mammalian species (3). In humans, an unexpectedly high number of clustered cases of Pneumocystis pneumonia (PcP) occurring in 1981 revealed the AIDS pandemic (14). Nowadays, PcP is still quite frequently diagnosed in HIV-positive patients but also in patients with other causes of immunodeficiency, such as organ transplantation or anticancer therapy (5, 35, 62, 78). Still, Pneumocystis jirovecii remains a leading cause of high mortality in HIV patients, even after the introduction of highly active antiretroviral therapy (HAART) (11, 64). Moreover, new data have recently emerged implicating low burdens of Pneumocystis organisms as a cause of symptom worsening in patients with chronic obstructive pulmonary disease (42). Also Pneumocystis organisms may temporarily cause asymptomatic infection in immunocompetent hosts, hence constituting a potential infection source (16). Thus, immunocompetent Pneumocystis carriers could transmit the infection to immunocompromised individuals (16, 30, 32).
Pneumocystis organisms constitute a huge genus consisting of a large number of host-species-specific organisms found in different mammals (3, 17, 20, 21, 26, 27, 41, 49). Indeed, the narrow host specificity of Pneumocystis species emerged clearly from the failure of cross species infection among laboratory animals (1, 38) or between humans and SCID mice (33). Surveys in domestic, synanthropic, or wild species showed that mammals may harbor one or several host-specific Pneumocystis species or strains (3, 17, 27, 31, 41, 46).
Whether Pneumocystis organisms are able to survive or multiply in the environment remains an unanswered question. It is known that they can be transmitted by aerial route (16, 32, 45, 85) and likely by transplacental route, at least in some mammal species (15, 29, 63, 79). The unavailability of continuous in vitro culture systems has hindered research aiming at clarifying the role of each life cycle stage in Pneumocystis proliferation and transmission (44). However, ultrastructure (25, 99), short-term culture (2, 19), and cell sorting approaches (56, 57) led to quite heuristic life cycle hypotheses. Furthermore, using PcP animal models, molecular strategies and the exploring of Pneumocystis occurrence in wild mammals resulted in major advancements, like the notions of strong host species specificity (1, 27, 31, 33, 38, 41) and coevolution (3, 27, 31, 46).
Bats (Chiroptera) are widely distributed across various ecosystems and constitute one of the largest groups of mammals, second in number of species after the order Rodentia and first in number of individuals (69). We thus attempted to explore how Pneumocystis organisms can circulate and adapt within this singular clade of flying mammals. Indeed, about one-fourth of all the living mammalian species consists of bats, a group comprising almost 1,100 species throughout the world. Body weight of bats varies widely: from 2 g (Craseonycteris thonglongyai; hog-nosed or bumblebee bat) to 1,200 g (Pteropus vampyrus; large flying lemur) (10). The order of Chiroptera is divided into two suborders (66): megachiropters (megabats) and microchiropters (microbats). Megabats were reported to live in the Old World and microbats in the New and Old Worlds.
Chiropters occupy diversified habitats, from cities to the countryside, and exhibit the largest variety of diets among all mammalian orders (66). Most are insectivorous (about 700 species), but a significant number of bats feed almost exclusively on fruits (about 230 species), nectar or pollen (about 50 species), small vertebrates (7 species), or blood of mammals or birds (3 species) (66). Their different diets led Chiroptera to colonize diverse ecosystems, ranging from deserts to temperate forest or rainforests, of every continent of the world except Antarctica (9, 69). In some islands, for instance in New Zealand, the only native mammals are bats (22, 36, 43). Bats roost in caves, abandoned or occupied buildings, mines, tree canopies, and hollows and under leaves, bark, or rocks (9, 51). Some bat species are living solitary, whereas others form huge colonies comprising millions of individuals (9). Bats are long-lived mammals: the small 7-g Brandt's bat, Myotis brandtii, can live up to 38 years in the wild (98). Furthermore, some species of bats are able to migrate over long distances.
Interestingly, humans share many properties with bats: occupying diverse habitats, living in social organizations, being long-lived, able to perform long migrations, and harboring Pneumocystis (41). Bats may therefore represent interesting models to understand the circulation of the human-specific species P. jirovecii in human populations. Furthermore, bats have an unusual physiology. For instance, due to their large lungs and naked flight membranes, they are exposed to marked heat losses (66) that could influence the biology of bat-derived Pneumocystis organisms and reveal unknown adapting mechanisms of these singular parasitic fungi to their hosts.
The present study is based on the molecular detection of Pneumocystis in the lungs of bats sampled from diverse biotopes in New and Old Worlds. It explores the Pneumocystis infection frequency in chiropters and Pneumocystis genetic diversity in relation with environmental and behavioral conditions. The results suggest that wild bat populations constitute a good model for approaching the circulation of Pneumocystis organisms in mammal populations, host specificity, and coevolution events.
MATERIALS AND METHODS
Samples.
A total of 216 bat specimens from the New World (Mexico, Guyana, Argentina) and from the Old World (France) were examined for the presence of Pneumocystis in their lungs. A total of 155 wild microbats belonging to 17 species were collected in different areas of Mexico (88 specimens), French Guyana (13 specimens), Argentina (16 specimens), and France (38 specimens). A total of 61 megabat specimens belonging to 2 species (Rousettus aegyptiacus and Pteropus rodricensis) were sampled from two colonies held in the same enclosure in La Palmyre Zoological Park (France). The founders of these colonies came from Northern Africa and Rodrigues Island (southwest Indian Ocean, 19°43′00′′S and 63°25′00′′E), respectively. Nineteen and 20 Pteropus rodricensis individuals were, respectively, transferred in 1993 and 1994 out of Jersey Zoological Park (Jersey Island) to La Palmyre Zoological Park. The colony housed in Jersey was founded by 10 individuals captured in Rodrigues Island in 1967 and 1977. The population of Rousettus aegyptiacus in La Palmyre was founded in 1994, with 120 individuals from various locations.
After euthanasia, the lungs were removed and immediately stored at −20°C in sterile cryotubes until used. In all cases, national rules regulating bat species protection have been respected.
Size of bat colonies.
For the genus Tadarida, colony size was assessed by counting bat individuals. As no exact assessment of colony size was available for the other species, a semiquantitative estimation was performed on the basis of direct observational appreciation of the number of colony members. Such a method led to defined scores: big colony, >200 individuals; medium colony, 31 to 200 individuals; and small colony, 10 to 30 individuals.
DNA extraction.
DNA extraction from lung tissue samples was performed using a DNeasy tissue kit (Qiagen, Courtabœuf, France) by following the manufacturer's procedure with some modifications. Part of the lung tissue (25 mg) was lysed overnight in an incubator at 56°C with permanent rotation. In order to concentrate Pneumocystis DNA, the column was rehydrated with 100 μl of elution AE buffer. DNA was stored at −20°C. A negative control was systematically included in each series of DNA extraction.
DNA amplification and analyses.
The presence of Pneumocystis DNA in lungs was assessed by nested PCR at the mtLSU rRNA and mtSSU rRNA loci (91, 96), mitochondrial genes encoding rRNA. Primer sequences and PCR cycling conditions are shown in Table 1.
Table 1.
Primer sequences and cycling conditions of Pneumocystis nested PCR assays at either the mtSSU rDNA or mtLSU rDNA locus (modified from references 97 and 91)
| Loci or cycling step | Sequence or cycling conditions |
|
|---|---|---|
| mtSSU rRNA | mtLSU rRNA | |
| Loci | ||
| External primer pair | ||
| Forward | pAZ112-10F/R1: 5′-GGG AAT TCT AGA CGG TCA CAG AGA TCA G-3′ | pAZ102-H: 5′-GTG TAC GTT GCA AAG TAC TC-3′ |
| Reverse | pAZ112-10R/R1: 5′-GGG AAT TCG AAC GAT TAC TAG CAA TTC C-3′ | pAZ102-E: 5′-GAT GGC TGT TTC CAA GCC CA-3′ |
| Internal primer pair | ||
| Forward | pAZ112-13/R1: 5′-GGG AAT TCG AAG CAT GTT GTT TAA TTC G-3′ | pAZ102-X/R1: 5′-GGG AAT TCG TGA AAT ACA AAT CGG ACT AGG-3′ |
| Reverse | pAZ112-14/R1: 5′-GGG AAT TCT TCA AAG AAT CGA GTT TCA G-3′ | pAZ102-Y/R1: 5′-GGG AAT TCT CAC TTA ATA TTA ATT GGG GAG C-3′ |
| Cycling step | ||
| 1st round | ||
| Denaturation | 30 s, 94°C | 30 s, 94°C |
| Annealing | 1 min, 55°C | 1 min, 50°C |
| Elongation | 1 min, 65°C | 1 min, 65°C |
| No. of cycles | 40 | 30 |
| 2nd round (performed with 5% [vol/vol] of the first-round mix) | ||
| Denaturation 1 | 30 s, 94°C | 30 s, 94°C |
| Annealing 1 | 1 min, 52°C | 1 min, 55°C |
| Elongation 1 | 1 min, 65°C | 1 min, 72°C |
| No. of cycles | 10 | 30 |
| Denaturation 2 | 30 s, 94°C | |
| Annealing 2 | 1 min, 63°C | |
| Elongation 2 | 1 min, 65°C | |
| No. of cycles | 30 | |
Negative controls were included in each experiment of PCR amplification, to monitor for eventual contamination. When nonspecific bands were detected, amplification products of the expected size (about 250 to 300 bp) were extracted from a 2% agarose gel (run in Tris-borate-EDTA buffer) using a PCR purification kit (QIAEX II gel extraction kit; Qiagen, Courtabœuf, France). When a unique band of expected size was present, amplified products were directly sent for sequencing to GenoScreen (Pasteur campus, Genopole of Lille, France). Sequencing from both ends using sets of internal primers was performed on an automated DNA sequencer (3730XL DNA Analyser; Applied Biosystems). Amplification and sequencing of each sample were repeated at least twice. The mtLSU and mtSSU sequences were aligned with already known Pneumocystis sequences using the computer program CLUSTAL X (version 1.63b, December 1997) (89). Then, alignments were refined by visual optimization using the software Se-Al version 2.0a11 Carbon (75). The aligned sequences were converted to distance matrix (percentage of differences). The BLASTn (http://www.ncbi.nlm.nih.gov/BLAST/) program allowed the comparison of our query sequence with the sequences available from databases in order to confirm sequence novelty and absence of contamination. To infer phenetic relationships among Pneumocystis isolates of our data set, an improved version of the neighbor-joining algorithm based on a simple model of sequence data (BIONJ) (37) analyses using PAUP 4.0b9 software was run (87). Evaluation of statistical confidence in nodes was based on 1,000 bootstrap replicates in BIONJ (34).
Sensitivity of the PCR.
Amplification products of mtLSU and mtSSU rRNA from Pneumocystis carinii were cloned by using a TOPO TA cloning kit (Invitrogen). Fifteen separate colonies were selected from the transformant plates and examined for each positive sample. DNA extraction was performed with a QIAprep miniprep kit (Qiagen). To check that the insert contained in the plasmid was the expected sequence, we used the external primer set of each nested PCR mtLSU ribosomal DNA (rDNA) and mtSSU rDNA locus (Table 1). A range of dilution containing from 106 copies to 1 copy of the targeted gene was performed. To determine the PCR sensitivity, the presence of a Pneumocystis carinii gene fragment within plasmids was assessed by nested PCR at the mtLSU rRNA and mtSSU rRNA loci as described above. The amplification products were visualized on a 2% agarose gel containing ethidium bromide.
Statistical analysis.
Statistical analyses were performed using SAS (version 9; SAS Institute). Variables are described as counts (proportions). A generalized linear mixed-model approach (with PROC GLIMMIX) was used to investigate the association between Pneumocystis carriage and potential predictors. Models were built with a binomial error distribution and the logit link function, and capture site was included as a random factor. Covariates with a level of significance of <0.1 in the univariate analyses were entered in multivariate models, the variables being then considered significant with a P value of <0.05.
Nucleotide sequence accession numbers.
Pneumocystis mtLSU rDNA or mtSSU rDNA sequences were deposited in GenBank under accession numbers JQ039397, JQ061293 to JQ061303, and JQ061304 to JQ061318.
RESULTS
Detection of Pneumocystis DNA in the lungs of bats.
A sample was considered to be positive for the presence of P. carinii DNA when a specific band was amplified by PCR at the expected size either at the mtLSU rRNA or mtSSU rRNA locus or at both loci and then sequenced. Two hundred sixteen lung samples from 19 bat species were examined (Tables 2 and 3). None of the tested lung tissue samples gave a positive amplification after the first PCR round at either locus. At the second round, PCR amplification was positive in 71 animals (32.9%) belonging to 12 bat species: one megabat (Rousettus aegyptiacus) and 11 microbats (Table 3) species. Most positive lung samples were from neotropical bat specimens: 6/13 from French Guyana, 36/88 from Mexico, and 8/16 from Argentina. In metropolitan France, 14/38 lung samples from wild bats were found to be positive. In captive animals (North African bats housed in La Palmyre Zoological Park in metropolitan France), Pneumocystis DNA was detected in 6/61 lung samples. On the whole, a positive amplification at both mtSSU rDNA and mtLSU rDNA was obtained in 48/216 (22.2%) animals belonging to 9 bat species.
Table 2.
Rate of Pneumocystis carriage in bat species and ecological data
| Origin | Bat species | Rate of Pneumocystis carriagea | Size of bat coloniesb | Migratory | Day roost | Mating systemc | Close physical contact | Reference(s) or source |
|---|---|---|---|---|---|---|---|---|
| Wild microchiropters of the New World | Tadarida brasiliensis | 36/79 (45.6) | Big | Yes | Caves | MM/MF | Yes | 10, 59 |
| Artibeus hirsutus | 3/5 (60) | Small | No | Caves | Harem | ND | 72, 88 | |
| Mormoops megalophylla | 0/3 (0) | Big | No | Caves | Harem | No | Mormoops megalophylla, unpublished abstract compiled and edited by the Heritage Data Management System, Arizona Game and Fish Department, and reference 88 | |
| Myotis californicus | 1/1 (100) | Small | No | Caves | ND | ND | 88 | |
| Pteronotus parnellii | 1/3 (33.3) | NDd | No | Caves | ND | ND | 88 | |
| Natalus stramineus | 1/8 (12.5) | Big | No | Caves | ND | No | 88, 90 | |
| Pteronotus davyi | 0/1 (0) | ND | ND | Caves | ND | ND | ||
| Glossophaga soricina | 9/16 (56.5) | Small | No | Caves | Harem | Yes | 60, 66 | |
| Carollia perspicillata | 0/1 (0) | Small | Yes | Tree-dwelling, forest | Lek | Yes | 59 | |
| Wild microchiropters of the Old World | Nyctalus noctula | 6/20 (30) | Medium | Yes | Tree-dwelling, forest | Lek | Yes | 10, 60, 59 |
| Pipistrellus pipistrellus | 3/9 (33.3) | Medium | Yes | Anthropophilic | Harem | Yes | 59 | |
| Eptesicus serotinus | 2/2 (100) | Medium | No | Anthropophilic | ND | ND | 60 | |
| Nyctalus leisleri | 0/1 (0) | Small | Yes | Tree-dwelling, forest | Lek | Yes | 86 | |
| Myotis daubentoni | 0/1 (0) | Medium | Yes | Tree-dwelling, forest | MM/MF | Yes | 6, 60 | |
| Myotis myotis | 0/2 (0) | Big | Yes | Caves | MM/MF | Yes | 10, 60, 59 | |
| Plecotus austriacus | 1/1 (100) | Medium | No | Anthropophilic | ND | Yes | 80 | |
| Plecotus auritus | 2/2 (100) | Small | No | Tree-dwelling, forest | MM/MF | Yes | 10, 60, 59 | |
| Captive megachiropters of the Old World | Rousettus aegyptiacus | 6/17 (35.3) | Medium | No | Zoological park | |||
| Pteropus rodricensis | 0/44 (0) | Medium | No | Zoological park |
Number of Pneumocystis PCR-positive samples/total number of tested samples (%).
Bat colony size was scored as follows: small, 10 to 30 individuals; medium, 31 to 200 individuals; big, >200 individuals.
MM/MF, multi-male/multi-female.
ND, not done.
Table 3.
Pneumocystis carriage in batsa
| Origin | Bat species | Targeted loci | No. of Pneumocystis PCR-positive samples/total number of tested samples by sampling area |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| La Trinitaria cave, Chiapas, Mexico | Isla de Janitzio cave, Michoacán, Mexico | Juxtlahuacave, Colotlipa Guerrero, Mexico | El Salitre, cave, Hidalgo, Mexico | El Salitre, cave Santa Rosa, Morelos, Mexico | La Boca cave, Nuevo León, Mexico | Dique Escaba, Tucumán, Argentina | French Guyana | Natural History Museum Bourges, France | La Palmyre zoological park, France | All areas | |||
| Wild microchiropters of the New World | Tadarida brasiliensis | LSU | 0/10 | 2/6 | 5/20 | 16/27 | 7/16 | 30/79 | |||||
| SSU | 2/10 | 2/6 | 4/20 | 17/27 | 7/16 | 32/79 | |||||||
| Carrier | 2/10 | 2/6 | 5/20 | 19/27 | 8/16 | 36/79 | |||||||
| Artibeus hirsutus | LSU | 2/5 | 2/5 | ||||||||||
| SSU | 3/5 | 3/55 | |||||||||||
| Carrier | 3/5 | 3/5 | |||||||||||
| Mormoops megalophylla | LSU | 0/2 | 0/1 | 0/3 | |||||||||
| SSU | 0/2 | 0/1 | 0/3 | ||||||||||
| Carrier | 0/2 | 0/1 | 0/3 | ||||||||||
| Myotis californicus | LSU | 1/1 | 1/1 | ||||||||||
| SSU | 1/1 | 1/1 | |||||||||||
| Carrier | 1/1 | 1/1 | |||||||||||
| Pteronotus parnellii | LSU | 0/2 | 0/1 | 0/3 | |||||||||
| SSU | 0/2 | 1/1 | 1/3 | ||||||||||
| Carrier | 0/2 | 1/1 | 1/3 | ||||||||||
| Natalus stramineus | LSU | 1/8 | 1/8 | ||||||||||
| SSU | 0/8 | 0/8 | |||||||||||
| Carrier | 1/8 | 1/8 | |||||||||||
| Pteronotus davyi | LSU | 0/1 | 0/1 | ||||||||||
| SSU | 0/1 | 0/1 | |||||||||||
| Carrier | 0/1 | 0/1 | |||||||||||
| Glossophaga soricina | LSU | 3/4 | 6/12 | 9/16 | |||||||||
| SSU | 2/4 | 6/12 | 8/16 | ||||||||||
| Carrier | 3/4 | 6/12 | 9/16 | ||||||||||
| Carollia perspicillata | LSU | 0/1 | 0/1 | ||||||||||
| SSU | 0/1 | 0/1 | |||||||||||
| Carrier | 0/1 | 0/1 | |||||||||||
| Wild microchiropters of the Old World | Nyctalus noctula | LSU | 5/20 | 5/20 | |||||||||
| SSU | 6/20 | 6/20 | |||||||||||
| Carrier | 6/20 | 6/20 | |||||||||||
| Pipistrellus pipistrellus | LSU | 1/9 | 1/9 | ||||||||||
| SSU | 3/9 | 3/9 | |||||||||||
| Carrier | 3/9 | 3/9 | |||||||||||
| Eptesicus serotinus | LSU | 2/2 | 2/2 | ||||||||||
| SSU | 2/2 | 2/2 | |||||||||||
| Carrier | 2/2 | 2/2 | |||||||||||
| Nyctalus leisleri | LSU | 0/1 | 0/1 | ||||||||||
| SSU | 0/1 | 0/1 | |||||||||||
| Carrier | 0/1 | 0/1 | |||||||||||
| Myotis daubentoni | LSU | 0/1 | 0/1 | ||||||||||
| SSU | 0/1 | 0/1 | |||||||||||
| Carrier | 0/1 | 0/1 | |||||||||||
| Myotis myotis | LSU | 0/2 | 0/2 | ||||||||||
| SSU | 0/2 | 0/2 | |||||||||||
| Carrier | 0/2 | 0/2 | |||||||||||
| Plecotus austriacus | LSU | 0/1 | 0/1 | ||||||||||
| SSU | 1/1 | 1/1 | |||||||||||
| Carrier | 1/1 | 1/1 | |||||||||||
| Plecotus auritus | LSU | 2/2 | 2/2 | ||||||||||
| SSU | 1/2 | 1/2 | |||||||||||
| Carrier | 2/2 | 2/2 | |||||||||||
| Captive megachiropters of the Old World | Rousettus aegyptiacus | LSU | 4/17 | 4/17 | |||||||||
| SSU | 6/17 | 6/17 | |||||||||||
| Carrier | 6/17 | 6/17 | |||||||||||
| Pteropus rodricensis | LSU | 0/44 | 0/44 | ||||||||||
| SSU | 0/44 | 0/44 | |||||||||||
| Carrier | 0/44 | 0/44 | |||||||||||
| All areas | All species | Positive Pneumocystis samples/total samples | 2/10 | 2/6 | 4/17 | 6/21 | 3/5 | 20/29 | 8/16 | 6/13 | 14/38 | 6/61 | 71/216 |
| Pneumocystis carriage (%) | 20.0 | 33.3 | 23.5 | 28.6 | 60.0 | 69.0 | 50.0 | 46.2 | 36.8 | 9.8 | 32.9 | ||
DNA lung samples were screened by nested PCR at either the mtLSU rRNA (LSU) or mtSSU rRNA (SSU) locus or both loci for the presence of Pneumocystis. Results of Pneumocystis carriage in bats are given according to bat species, targeted locus, and sampling area. LSU, samples that are positive at the LSU locus, and some of these samples can also be positive at SSU; SSU, samples that are positive at the SSU locus, and some of these samples can also be positive at LSU; Carrier, a bat lung sample was considered positive for Pneumocystis DNA when nested PCR was positive at either mtLSU rRNA or mtSSU rRNA or at both loci.
The sensitivities of the nested PCR mtLSU and the nested PCR mtSSU indicate, respectively, a limit of detection of 10 copies and 1 copy. However, our experimental results indicate that the ratios of mtLSU-positive/mtSSU-negative samples and mtLSU-negative/mtSSU-positive samples are, respectively, 33.14% and 37.2% when P. rodricensis flying foxes were excluded. The sensitivity of Pneumocystis detection is not the same at both loci. Moreover, the Pneumocystis sequences are variable and primers hybridize differently according to the species of bats analyzed. Consequently, the rate of the Pneumocystis carriage is probably underestimated in this study.
Potential influence of host suborder and geography.
The frequency of Pneumocystis DNA detection was significantly higher in suborder Microchiroptera (41.9%) than in suborder Megachiroptera (9.8%) (P < 0.0001). However, only two species of megabats could be examined in the present work: Egyptian fruit bat (R. aegyptiacus) and Rodrigues Island flying fox (Pteropus rodricensis). The sampled specimens of these two species lived in the same enclosure since 1994 in La Palmyre Zoological Park. Pneumocystis DNA was detected in 35.3% of the 17 Egyptian fruit bats, but none of the 44 flying foxes was found to be positive.
With regard to geography, the frequencies of Pneumocystis carriage in bats from the New World and Old World were 43.6% and 20.2%, respectively. However, the presence of Pneumocystis DNA in bats from the Old World rose to 36.8% when P. rodricensis flying foxes were excluded (Table 4). No significant statistical association could be established between the presence of Pneumocystis DNA and Old or New World geographic location of the animals (P = 0.81) (Table 5). Consistently, no significant difference was found between microchiropters from the New (43.6%) and Old (35.6%) Worlds.
Table 4.
Ecological data on wild microchiropters and Pneumocystis carriage ratesa
| Characteristic | No. (%) |
||
|---|---|---|---|
| Whole sample (n = 155) | Noncarriers (n = 90) | Carriers (n = 65) | |
| Bat colony size | |||
| Big | 92 | 55 (59.8) | 37 (40.2) |
| Medium | 33 | 21 (63.6) | 12 (36.4) |
| Small | 26 | 11 (42.3) | 15 (57.7) |
| Migration | |||
| Yes | 113 | 68 (60.2) | 45 (39.8) |
| No | 41 | 21 (51.2) | 20 (48.8) |
| World | |||
| Old world | 38 | 24 (63.2) | 14 (36.8) |
| New world | 117 | 66 (56.4) | 51 (43.6) |
| Diet | |||
| Insectivorous | 133 | 80 (60.2) | 53 (39.8) |
| Nectarivorous | 16 | 7 (43.8) | 9 (56.2) |
| Frugivorous | 6 | 3 (50.0) | 3 (50.0) |
| Day roost | |||
| Tree-dwelling, forest | 25 | 17 (68.0) | 8 (32.0) |
| Cave | 118 | 67 (56.8) | 51 (43.2) |
| Anthropophilic | 12 | 6 (50.0) | 6 (50.0) |
| Mating system | |||
| Multi-male/multi-female | 84 | 46 (54.8) | 38 (45.2) |
| Harem | 33 | 18 (54.6) | 15 (45.4) |
| Lek | 22 | 16 (72.7) | 6 (27.3) |
| Contact in the colony | |||
| Crowding | 132 | 75 (56.8) | 57 (43.2) |
| Without contact | 11 | 10 (90.9) | 1 (9.1) |
| Climate | |||
| 1 = tropical | 10 | 8 (80.0) | 2 (20.0) |
| 2 = warm temperate | 49 | 34 (69.4) | 15 (30.6) |
| 3 = warm semi-arid | 29 | 9 (31.0) | 20 (69.0) |
| 4 = subtropical | 16 | 8 (50.0) | 8 (50.0) |
| 5 = equatorial | 13 | 7 (53.9) | 6 (46.2) |
| 6 = temperate | 38 | 24 (63.2) | 14 (36.8) |
| Humidity | |||
| No (climates 2, 3, 6) | 116 | 67 (57.8) | 49 (42.2) |
| Yes (climates 1, 4, 5) | 39 | 23 (59.0) | 16 (41.0) |
| Heat | |||
| No (climates 2, 6) | 87 | 58 (66.7) | 29 (33.3) |
| Yes (climates 1, 3, 4, 5) | 68 | 32 (47.1) | 36 (52.9) |
| Elevation | |||
| ≤800 m (origins d, f, g, h) | 96 | 48 (50.0) | 48 (50.0) |
| >800 m (origins a, b, c, e, i) | 59 | 42 (71.2) | 17 (28.8) |
Bat colony size was scored as follows: small, 10 to 30 individuals; medium, 31 to 200 individuals; big, >200 individuals. Data from P. rodricencis were excluded from analysis. Origin: a, La Trinitaria cave, Chiapas, Mexico; b, Isla de Janitzio cave, Michoacán, Mexico; c, El Salitre cave, Hidalgo, Mexico; d, La Boca cave, Nuevo León, Mexico; e, Juxtlahuaca cave, Colotlipa Guerrero, Mexico; f, Dique Escaba, Tucumán, Argentina; g, French Guyana; h, Natural History Museum of Bourges, France; i, El Salitre cave, Santa Rosa, Morelos, Mexico.
Table 5.
Pneumocystis carriage in bats: influence of ecological and behavioral factors (univariate analysis)a
| Characteristic | OR | 95% CI | P |
|---|---|---|---|
| Bat colony size | |||
| Medium or big | 1 | ||
| Small | 3.259 | 1.038–10.230 | 0.04 |
| Migration | |||
| Yes | 1 | ||
| No | 2.959 | 0.997–8.779 | 0.05 |
| Area | |||
| Old World | 1 | ||
| New World | 1.213 | 0.170–8.659 | 0.81 |
| Diet | |||
| Insectivorous or frugivorous | 1 | ||
| Nectarivorous | 3.948 | 0.864–18.045 | 0.07 |
| Day roost | |||
| Tree-dwelling, forest | 1 | ||
| Cavernicole | 1.299 | 0.286–5.902 | 0.71 |
| Anthropophilic | 2.148 | 0.518–8.903 | 0.29 |
| Mating system | |||
| Lek | 1 | ||
| Harem | 1.905 | 0.512–7.080 | 0.33 |
| MM-MF | 1.827 | 0.477–6.897 | 0.37 |
| Contact in the colony | |||
| No | 1 | ||
| Yes | 11.387 | 1.154–112.354 | 0.038 |
| Humidity | |||
| Yes | 1 | ||
| No | 1.064 | 0.285–3.972 | 0.92 |
| Heat | |||
| No | 1 | ||
| Yes | 2.030 | 0.685–6.020 | 0.16 |
| Elevation | |||
| >800 m | 1 | ||
| ≤800 m | 2.448 | 0.919–6.518 | 0.07 |
OR, odds ratio; CI, confidence interval. Significant results at P = 0.05.
Impact of ecological and behavioral factors.
Neither wild/captive state nor host phylogeny influenced Pneumocystis DNA detection significantly, with the obvious exception of P. rodricensis, in which Pneumocystis DNA was not found (Tables 2 and 3). In contrast, the size of the bat colony influenced the frequency of Pneumocystis DNA detection significantly (P = 0.04; Table 5). Thus, the probability of picking a Pneumocystis-infected bat was 3-fold higher in small colonies than in large ones, irrespective of species (Table 5). Likewise, migration and crowding influenced the frequency of Pneumocystis DNA detection significantly (Tables 5 and 6). The statistical multivariate analyses showed that migration and crowding are independent predictive factors of Pneumocystis carriage (Table 6). The probability of picking a Pneumocystis-infected bat was 5-fold higher in sedentary colonies than in migratory ones and 33-fold higher in crowding colonies (close physical contact between the colony members) than in noncrowding colonies (without close physical contact) (Table 6). When we limit analyses to bats in close physical contact (n = 132: 19 sedentary and 113 migratory bats), migration is the sole predictive factor, nonmigratory bats having an odds ratio (OR) of 5.091 (1.147 to 22.597) (P = 0.03) with regard to the migratory bats.
Table 6.
Pneumocystis carriage in bats: influence of ecological and behavioral factors (multivariate analysis)a
| Characteristic | OR | 95% CI | P |
|---|---|---|---|
| Migration | |||
| Yes | 1 | ||
| No | 4.759 | 1.15–20.310 | 0.036 |
| Contact in the colony | |||
| No | 1 | ||
| Yes | 33.313 | 2.687–412.963 | 0.007 |
OR, odds ratio; CI, confidence interval. Significant results at P = 0.05.
Cave temperature (°C) and relative humidity (%) were not recorded inside the main chamber. Temperature and relative humidity outside the cave were evaluated by general climate of the geographical area of the cave. The climate, roosting habits, and mating system did not seem to influence Pneumocystis carriage (Table 5). The elevation and food regimen did not seem to influence it either, although significance is borderline (P = 0.07).
The case of Tadarida brasiliensis, influence of elevation.
Focusing on a single species of bats allowed us to remove all the behavioral factors, to evaluate only the environmental factors. The largest number of sampled bats belonged to the species Tadarida brasiliensis. We have collected 79 specimens of T. brasiliensis in five locations from North and South America (Table 3, Fig. 1): four regions in Mexico (La Trinitaria cave Chiapas [1,460 m], Isla de Janitzio cave Michoacán [2,120 m], El Salitre cave Hidalgo [1,320 m], and La Boca cave Nuevo León [445 m to 600 m]) and one province in Argentina (Dique Escaba, Tucumán Province [650 m]). The Pneumocystis detection rate in T. brasiliensis was related to elevation (P = 0.04): in bats coming from locations situated lower than 800 m, the Pneumocystis DNA carriage rate was 5-fold higher than in bats coming from locations situated at an elevation superior to 800 m (Tables 7 and 8). On the other hand, the climate (humidity and heat) did not seem to have an impact on Pneumocystis DNA carriage.
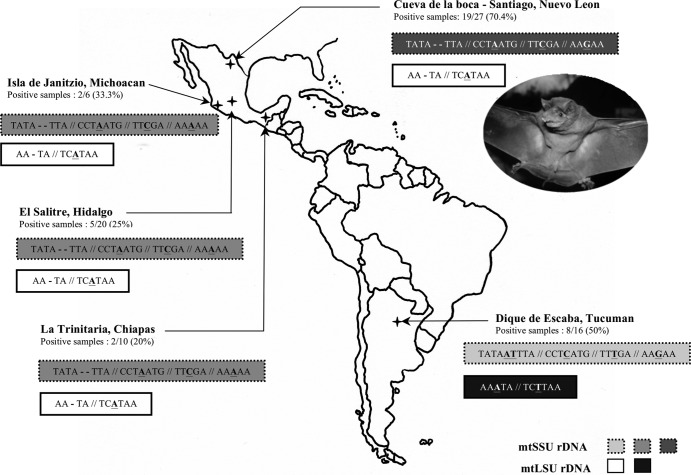
Fig 1.
Genetic polymorphism of Pneumocystis isolates detected in the lungs of Tadarida brasiliensis bats sampled from North and South America. For each geographic location, mtLSU rDNA and mtSSU rDNA Pneumocystis polymorphic sequences are indicated and framed: two mtLSU rDNA variants and three mtSSU rDNA variants. The number of positive samples over total number of analyzed samples is also indicated below each location. The photo represents one male bat belonging to the species Tadarida brasiliensis and originating from Dique Escaba in Argentina.
Table 7.
Pneumocystis carriage in Tadarida brasiliensis: influence of climate and elevation
| Characteristica | No. (%) |
||
|---|---|---|---|
| Whole sample (n = 155) | Noncarriers (n = 90) | Carriers (n = 65) | |
| Climate | |||
| 1 = tropical | 10 | 8 (80.0) | 2 (20.0) |
| 2 = warm temperate | 26 | 19 (73.1) | 7 (26.9) |
| 3 = warm semi-arid | 27 | 8 (29.6) | 19 (70.4) |
| 4 = subtropical | 16 | 8 (50.0) | 8 (50.0) |
| Humidity | |||
| No (climates 2, 3, 6) | 53 | 27 (50.9) | 26 (49.1) |
| Yes (climates 1, 4, 5) | 26 | 16 (61.5) | 10 (38.5) |
| Heat | |||
| No (climates 2, 6) | 26 | 19 (73.1) | 7 (26.9) |
| Yes (climates 1, 3, 4, 5) | 53 | 24 (45.3) | 29 (54.7) |
| Elevation | |||
| ≤800 m (origins d, f) | 43 | 16 (37.2) | 27 (62.8) |
| >800 m (origins a, b, c) | 36 | 27 (75.0) | 9 (25.0) |
Origin: a, La Trinitaria cave, Chiapas, Mexico; b, Isla de Janitzio cave, Michoacán, Mexico; c, El Salitre cave, Hidalgo, Mexico; d, La Boca cave, Nuevo León, Mexico; f, Dique Escaba, Tucumán, Argentina.
Table 8.
Pneumocystis carriage in Tadarida brasiliensis: influence of climate and elevation (univariate analysis)a
| Characteristic | OR | 95% CI | P |
|---|---|---|---|
| Humidity | |||
| Yes | 1 | ||
| No | 1.450 | 0.074–28.587 | 0.73 |
| Heat | |||
| No | 1 | ||
| Yes | 2.521 | 0.176–36.122 | 0.36 |
| Elevation | |||
| >800 m | 1 | ||
| ≤800 m | 5.049 | 1.096–23.252 | 0.04 |
OR, odds ratio; CI, confidence interval. Significant results at P = 0.05.
Pneumocystis genetic diversity in bats.
The genetic diversity of Pneumocystis from bats is examined by analyzing mtLSU rRNA and mtSSU rRNA sequences. PCR products ranged from 252 to 333 bp (nested PCR at the mtLSU rRNA gene) and from 334 to 480 bp (nested PCR at the mtSSU rRNA gene). The sequences under analysis are part of the ribosome and are coded by the mitochondrial genome, and no introns were detected in the mitochondrial genome (82). Direct sequencing of PCR products revealed that each Pneumocystis mtLSU rDNA or mtSSU rDNA sequence could be associated with a unique bat species (Tables 9 and 10; Fig. 2 and 3). Comparison of the mtLSU rRNA aligned sequences was carried out on 458 positions, including gaps, for a total of 14 taxa: 12 original sequences and two sequences already published, Pneumocystis murina (GenBank accession AF257179) and Pneumocystis oryctolagi (GenBank accession S42915), were chosen as outgroups. For the mtSSU rRNA locus, comparison of the aligned sequences was carried out on 530 positions, including gaps, for a total of 17 taxa: 15 original sequences and two sequences already published, Pneumocystis f. sp. Lepus europaeus (GenBank accession no. JF431106) and P. oryctolagi (47), were chosen as outgroups. Genetic divergence between Pneumocystis organisms isolated from different bat species, as assessed by Pneumocystis mtLSU rDNA or mtSSU rDNA sequence divergence, varied from 0.35 to 26.88% (Tables 9 and 10).
Table 9.
Divergence matrix of Pneumocystis mtLSU rDNA sequences amplified from 9 bat species (GenBank accession no. JQ039397 and JQ061293 to JQ061303), one rodent (P. murina from Mus musculus, GenBank accession no. AF257179), and one lagomorph species (P. oryctolagi from Oryctolagus cuniculus, GenBank accession no. S42915)a
| Host species | % of divergence from: |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
| 1. G. soricina FG | |||||||||||||
| 2. G. soricina JG | 00.35 | ||||||||||||
| 3. A. hirsutus ES | 09.64 | 08.87 | |||||||||||
| 4. T. brasiliensis M | 11.04 | 10.59 | 17.34 | ||||||||||
| 5. T. brasiliensis A | 10.35 | 09.92 | 17.57 | 00.78 | |||||||||
| 6. M. californicus LB | 11.88 | 12.33 | 14.82 | 08.15 | 09.21 | ||||||||
| 7. P. pipistrellus B(P6) | 14.11 | 13.12 | 14.37 | 12.15 | 12.72 | 07.42 | |||||||
| 8. N. noctula B(NO19) | 10.81 | 10.05 | 13.52 | 09.48 | 09.53 | 05.23 | 02.54 | ||||||
| 9. E. serotinus B(S1) | 11.22 | 10.57 | 13.84 | 06.91 | 07.91 | 02.00 | 06.83 | 04.76 | |||||
| 10. P. auritus B(OR3) | 12.75 | 12.03 | 15.09 | 08.93 | 09.30 | 03.53 | 05.44 | 05.52 | 01.94 | ||||
| 11. R. aegyptiacus P(34) | 15.60 | 15.19 | 20.58 | 18.30 | 18.40 | 20.19 | 23.19 | 17.68 | 18.22 | 20.14 | |||
| 12. R. aegyptiacus P(13) | 16.41 | 16.57 | 19.38 | 16.94 | 16.72 | 15.28 | 19.46 | 15.19 | 15.13 | 16.47 | 19.91 | ||
| 13. P. murina | 23.48 | 25.19 | 30.24 | 17.95 | 18.10 | 16.06 | 28.32 | 25.98 | 25.34 | 25.16 | 21.89 | 27.01 | |
| 14. P. oryctolagi | 19.91 | 21.96 | 28.98 | 18.24 | 17.75 | 16.04 | 27.29 | 24.37 | 26.72 | 26.19 | 18.59 | 28.66 | 17.48 |
Numbered column headings (1 to 13) correspond to numbered sources of host species DNA in the first column. Next to each bat species Latin name, the letter and number codes indicate geographic origin and reference number of samples, respectively. FG, French Guyana; B, Bourges, France; NL, Nuevo León, Mexico; M, Michoacán, Mexico; A, Argentina; LB, La Boca; JG, Juxtlahuaca Grotto, Mexico; ES, El Salitre, Mexico; P, Zoological Park of La Palmyre, France. Letters and numbers in parentheses indicate a particular sample.
Table 10.
Divergence matrix of Pneumocystis mtSSU rDNA sequences amplified from 15 bats (GenBank accession no. JQ061304 to JQ061318) and 2 lagomorph species (P. oryctolagi [47]; P. f. sp. Lepus europaeus, GenBank accession no. JF431106)a
| Host species | % of divergence from: |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | |
| 1. P. pipistrellus B(P6) | ||||||||||||||||
| 2. P. pipistrellus B(P3) | 00.46 | |||||||||||||||
| 3. N. noctula B(NO19) | 07.21 | 06.52 | ||||||||||||||
| 4. E. serotinus B | 13.34 | 13.26 | 14.65 | |||||||||||||
| 5. P. auritus B(OR3) | 12.64 | 12.01 | 14.15 | 02.71 | ||||||||||||
| 6. M. californicus NL | 12.05 | 11.71 | 13.45 | 04.29 | 02.73 | |||||||||||
| 7. P. austriacus B | 12.34 | 11.57 | 14.23 | 03.24 | 00.49 | 03.06 | ||||||||||
| 8. T. brasiliensis NL | 17.25 | 14.63 | 14.58 | 13.31 | 13.43 | 13.06 | 13.53 | |||||||||
| 9. T. brasiliensis M | 17.10 | 14.54 | 14.50 | 13.57 | 13.68 | 13.29 | 13.78 | 00.89 | ||||||||
| 10. T. brasiliensis A | 18.06 | 15.05 | 15.76 | 13.64 | 14.00 | 13.23 | 13.71 | 01.53 | 01.83 | |||||||
| 11. G. soricina JG | 14.76 | 13.67 | 15.02 | 12.68 | 12.44 | 13.25 | 12.72 | 16.28 | 15.81 | 16.24 | ||||||
| 12. A. hirsutus ES | 13.91 | 13.46 | 14.71 | 12.01 | 11.66 | 11.34 | 11.75 | 15.36 | 14.92 | 15.77 | 08.41 | |||||
| 13. P. parnellii ES | 13.83 | 14.04 | 14.64 | 13.71 | 14.33 | 13.36 | 13.74 | 16.93 | 16.45 | 16.62 | 08.81 | 08.45 | ||||
| 14. R. aegyptiacus P(12) | 21.50 | 20.68 | 21.83 | 17.84 | 19.05 | 18.23 | 18.32 | 20.99 | 20.38 | 19.96 | 14.38 | 13.93 | 12.87 | |||
| 15. R. aegyptiacus P(26) | 23.85 | 23.19 | 24.38 | 23.31 | 23.36 | 22.06 | 26.88 | 21.84 | 21.34 | 21.29 | 15.59 | 12.58 | 16.58 | 21.72 | ||
| 16. P. oryctolagi | 25.12 | 25.25 | 25.79 | 23.97 | 23.34 | 23.60 | 23.48 | 23.32 | 23.50 | 24.15 | 18.50 | 16.33 | 20.22 | 23.02 | 24.77 | |
| 17. P. f. sp. L. europaeus | 19.30 | 19.01 | 20.30 | 18.10 | 18.21 | 17.83 | 17.67 | 17.43 | 17.41 | 17.16 | 16.30 | 13.82 | 18.43 | 19.29 | 20.87 | 09.76 |
Numbered column headings (1 to 16) correspond to numbered sources of host species DNA in the first column. Next to each bat species Latin name, the letter and number codes indicate geographic origin and reference number of samples, respectively. B, Bourges, France; NL, Nuevo León, Mexico; M, Michoacán, Mexico; A, Argentina; JG, Juxtlahuaca Grotto, Mexico; ES, El Salitre, Mexico; P, Zoological Park of La Palmyre, France. Letters and numbers in parentheses indicate a particular sample.
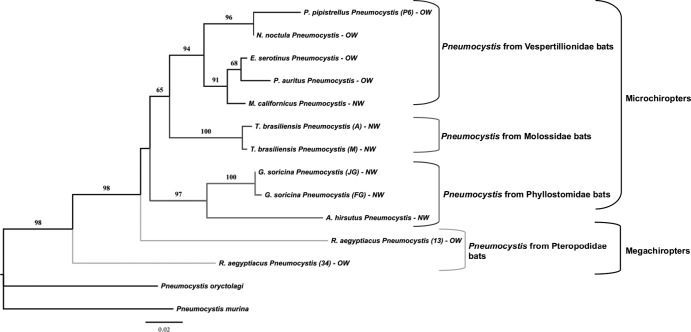
Fig 2.
Phenetic relationships of Pneumocystis organisms from bat species inferred from mtLSU rDNA sequences. The phylogram presented resulted from bootstrapped data sets obtained by using BIONJ analysis (heuristic search option in PAUP 4.0). The percentages above the branches are the frequencies with which a given branch appeared in 1,000 bootstrap replications. Bootstrap values below 50% are not displayed. Branch lengths correspond to the total nucleotide changes assigned to each branch by PAUP 4.0. Pneumocystis from rabbit (P. oryctolagi, GenBank accession S42915) and from mouse (P. murina, GenBank accession AF257179) were chosen as outgroups. Letter and number codes indicate geographic origin and reference number of samples, respectively. M, Michoacán, Mexico; A, Argentina; JG, Juxtlahuaca Grotto, Mexico; OW, Old World; NW, New World. Letters and numbers in parentheses indicate a particular sample (P6, 13, 34).
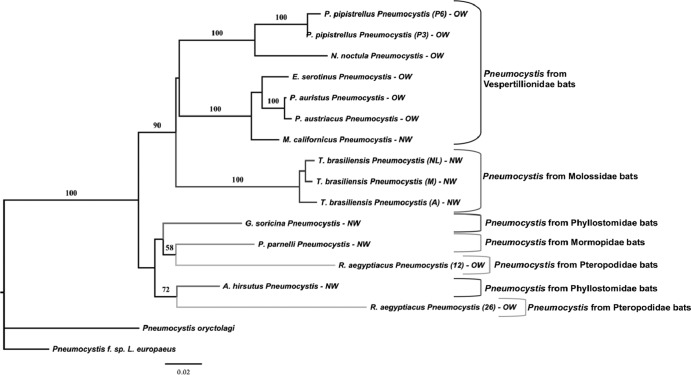
Fig 3.
Phenetic relationships of Pneumocystis organisms from bat species inferred from mtSSU rDNA sequences. The phylogram presented resulted from bootstrapped data sets obtained by using BIONJ analysis (heuristic search option in PAUP 4.0). The percentages above the branches are the frequencies with which a given branch appeared in 1,000 bootstrap replications. Bootstrap values below 50% are not displayed. Branch lengths correspond to the total nucleotide changes assigned to each branch by PAUP 4.0. Pneumocystis from rabbit (P. oryctolagi [47]) and from hare (P. f. sp. Lepus europaeus, GenBank accession JF431106) were chosen as outgroups. Letter and number codes indicate geographic origin and reference number of samples, respectively. NL, Nuevo León, Mexico; M, Michoacán, Mexico; A, Argentina; OW, Old World; NW, New World. Letters and numbers in parentheses indicate a particular sample (P3, P6, 12, 26).
With regard to the Pneumocystis mtLSU rRNA locus, we found 12 sequences in the lungs of 9 bat species (Table 9, Fig. 2). In 6 bat species (Myotis californicus, Nyctalus noctula, Artibeus hirsutus, Eptesicus serotinus, Pipistrellus pipistrellus, Plecotus auritus), one specific Pneumocystis mtLSU rDNA sequence per species was detected (Fig. 2). In contrast, Glossophaga soricina and Tadarida brasiliensis species each harbored 2 variants of Pneumocystis mtLSU rDNA depending on the collection area (31). The mtLSU rDNA sequences of Pneumocystis detected in G. soricina from French Guyana and Mexico diverged by 0.35%, corresponding to one nucleotide difference (Table 9), and grouped together on a phenetic branch that is supported by a 100% bootstrap value (Fig. 2). Likewise, a relatively close order of divergence (0.78%) was found between Pneumocystis mtLSU rDNA sequences amplified from T. brasiliensis samples collected from either Argentinean or Mexican colonies (Tables 9 and 11, Fig. 1). Consistently, these sequences are included in the same group supported by a 100% bootstrap value in phenetic analysis (Fig. 2). Interestingly, mtLSU rDNA polymorphism was also detected in R. aegyptiacus (from the La Palmyre Zoo), with two Pneumocystis variants that diverged by a much higher percentage (19.91%; Table 9). Both Megachiroptera-derived Pneumocystis organism sequences were placed in a basal position according to the phenetic analysis (Fig. 2). All mtLSU rDNA sequences of Pneumocystis organisms from Microchiroptera were included in the same clade. In addition, Pneumocystis sequences derived from the New World bats and those from the Old World bats are mixed (Fig. 2).
Table 11.
Pneumocystis polymorphism at the mtSSU and mtLSU rRNA loci in DNA lung samples from Tadarida brasiliensisa
| Sample origin | mtSSUrRNA sequence | mtLSUrRNA sequence |
|---|---|---|
| Mexico | ||
| Cueva de la Boca, Nuevo León | TATA–TTA//CCTAATG//TTCGA//AAGAA | AA-AT//TCATAA |
| Isla de Janitzio, Michoacan | TATA–TTA//CCTAATG//TTCGA//AAAAA | AA-AT//TCATAA |
| El Salitre, Hidalgo | TATA–TTA//CCTAATG//TTCGA//AAAAA | AA-AT//TCATAA |
| La Trinitaria, Chiapas | TATA–TTA//CCTAATG//TTCGA//AAAAA | AA-AT//TCATAA |
| Argentina | ||
| Dique de Escaba, Tucuman | TATAATTTA//CCTCATG//TTTGA//AAGAA | AAAAT//TCTTAA |
Results of Pneumocystis polymorphism in these bat specie are given according to sampling area. Bold nucleic acids indicate the mutations. Pneumocystis polymorphic sequences show two mtLSUrDNA variants (one from Mexico and the other from Argentina) and three mtSSUrDNA variants (two from Mexico and the third from Argentina).
Regarding the Pneumocystis mtSSU rRNA locus, we isolated 15 sequences from 11 bat species (Table 10, Fig. 3). In 8 bat species (M. californicus, G. soricina, N. noctula, A. hirsutus, E. serotinus, Plecotus austriacus, P. auritus, Pteronotus parnellii), we identified one specific Pneumocystis sequence per host species, while we detected two highly divergent mtSSU rRNA sequences in R. aegyptiacus (21.72%), two weakly divergent sequences in P. pipistrellus (0.46%), and three lowly divergent sequences in T. brasiliensis (0.89% to 1.83%) (Tables 10 and 11). In the phenetic tree displaying mtSSU rDNA sequences, both sequences of macrochiroptera-derived Pneumocystis organisms are mixed with Microchiroptera-derived Pneumocystis, but the bootstrap values are not significant (Fig. 3). The Pneumocystis sequences derived from the same host species (T. brasiliensis and P. pipistrellus) clustered together with 100% bootstrap values (Fig. 3).
Geography seemed to have a structuring effect on Pneumocystis mtSSU rRNA polymorphism. Thus, the two Pneumocystis sequences isolated from P. pipistrellus (number reference P3 and P6) identified two Metropolitan French Departments (Haute Marne [51] and Cher [17], respectively) (Table 10). Likewise, the 3 Pneumocystis sequences isolated from T. brasiliensis identified 3 geographical areas: (i) Michoacán-Hidalgo-Chiapas (Mexico), (ii) Nuevo León (Mexico), and (iii) Tucumán (Argentina) (Fig. 1, Tables 10 and 11).
DISCUSSION
Detection of Pneumocystis DNA in the lungs of bats.
The rate of the Pneumocystis carriage is probably underestimated in this study. Moreover, the abundance of Pneumocystis targets (mtLSU rRNA or mtSSU rRNA) in the bat samples studied here is generally low. Thus, chance can determine whether or not Pneumocystis gDNA is introduced in a given PCR. This random sampling could explain why some samples produced a positive amplification with one set of primers but not both.
Positive amplification requiring two PCR rounds is usually considered a case of Pneumocystis carriage (29, 30), i.e., healthy carriers harboring a low burden of Pneumocystis organisms. In the present work, the fact that single Pneumocystis PCR was negative in all cases suggests that no animal was heavily infected or developed pneumocystosis. In contrast, Pneumocystis carriage was a frequent event (about 33% of examined bats). Likewise, Laakkonen et al. reported high rates of Pneumocystis carriage (even if they used histological methods) in wild small mammals from California, but no animal showed histopathological changes typical of PcP (53). Indeed, PcP cases were only rarely reported in wild mammals (3).
Thus, mammals seem to usually develop mild though quite frequent subclinical Pneumocystis lung infections (17), suggesting that these microfungi develop efficient airborne circulation in natural ecosystems (3). Severe PcP would therefore be, as in humans, a rare event in the natural history of Pneumocystis infection (3).
This view is further consistent with the fine adaptation of Pneumocystis organisms to the alveolar microenvironment (8, 16, 23, 26), likely resulting from Pneumocystis-host species coevolution (46).
Consistently, it was experimentally shown that Pneumocystis organisms were able to replicate in the lungs of healthy hosts (16), which can subsequently transmit the microfungi by airborne route to susceptible or immunocompetent hosts (32, 39). These observations showed that healthy carriers could behave as a reservoir of Pneumocystis species, playing a critical role in the airborne circulation of Pneumocystis organisms in host populations (16, 17, 29, 30).
Prevalence of Pneumocystis colonization in bats.
The reported frequency of Pneumocystis DNA detection varied markedly between bat species (Table 2). This finding was in accordance with previous studies, where important differences in Pneumocystis prevalence between mammal species have been reported (52, 53). In micromammals collected in France (58), rodent infection rates varied according to the species (e.g., 67% in Apodemus sylvaticus, 78% in Eliomys quercinus). In the same study, the Pneumocystis global prevalence was as high as 68%. In Thailand, 58% of the collected Rattus norvegicus specimens harbored Pneumocystis DNA (17), but infection rates in other rodents from this country were quite divergent (M. Chabé and S. Morand, unpublished data). In primates, prevalence of Pneumocystis reached 33.6% in healthy macaques (Macaca fascicularis) maintained in partial release (30) and 26.5% in captive primates of 26 species taken as a whole (29).
All samples from megachiropters were collected from captive animals, and those from microchiropters were collected from wild animals. Thus, the significantly different Pneumocystis rates between megabats and microbats found in the present work may stem from bat taxonomy and/or from the captive/wild state of the animals as well. Within megachiropters, two species, namely, Egyptian roussette (R. aegyptiacus) and flying fox (P. rodricensis), were sampled, revealing strikingly different infection rates. Pneumocystis DNA was detected in 35.3% of Egyptian roussettes, while no P. rodricensis specimen was found to be carrying Pneumocystis. Egyptian roussette, a fruit bat species of the Old World, can be found throughout Africa (except in the desert regions of the Sahara) and Middle East, as far as Pakistan and Northern India. In this extensive distribution area, Egyptian roussettes exist as a relatively large wild population (12). In contrast, P. rodricensis is restricted to Rodrigues, the smallest of the Mascarene islands, and it is considered an endangered species mainly because of deforestation and cyclones that devastate the area from time to time. Only about 4,000 individuals are thought to exist (48). Rodrigues flying foxes seem to be the sole mammal species studied so far that revealed to be entirely negative for Pneumocystis DNA. A first hypothesis is that Pneumocystis DNA sequences at the studied loci would be so divergent that they could not be detected by our techniques. Another hypothesis is that the animals used to set up the captive population were totally free of Pneumocystis and thus suggests that Pneumocystis prevalence is very low or absent in wild Rodrigues flying fox populations or that the sampling of breeders that set up the colony was too small (40 individuals from Jersey Zoological Park). A third hypothesis is that Pneumocystis organisms had never colonized or had been lost by this insular host species. Furthermore, Rodrigues flying foxes and Egyptian roussettes are cohoused in the same enclosure in La Palmyre Zoological Park, and the cross-infection does not seem to occur between these 2 host species of megachiropters. This result confirms the strong host specificity in Pneumocystis strains (3, 27, 41).
If we exclude the absence of Pneumocystis DNA in Rodrigues flying foxes, the global infection rate is close between microchiropters (41.9%) and megachiropters (35.3%), suggesting a similar circulation pattern of Pneumocystis within both groups of either captive or wild chiropters. Likewise, Pneumocystis prevalence was found to be identical in bats from the New and Old Worlds. This may indicate similar intensity of circulation of Pneumocystis organisms in bat populations and a comparable pattern of Pneumocystis host-to-host transmission worldwide.
Crowding in the bat colony and the infection source issue.
Pneumocystis rates were found to increase when the individuals of the colony are in close contact independently of colony size (Tables 5 and 6). For example, in Natalus stramineus, a cave-dwelling bat living in big noncrowding colonies, the Pneumocystis DNA carriage rate is low (12.5%) (Table 2). This could be explained by the fact that bats hang individually without contact with each other, keeping a distance of 5 to 50 cm between themselves (90). While in Tadarida brasiliensis, a cave-dwelling bat living in big crowding colonies, the carriage rate is high (45.6%) (Table 2). A recent study in human beings showed that the level of Pneumocystis jirovecii DNA in exhaled air from infected patients decreased with increased distance from the patients (18). Furthermore, it has been shown that Pneumocystis organisms were able to multiply transiently in the lungs of immunocompetent hosts and to transmit the infection to either susceptible or immunocompetent hosts by the airborne route (16, 32, 39). Crowding could therefore favor Pneumocystis host-to-host airborne transmission in bats or in other mammals. This observation raises the question of the Pneumocystis infection source: do Pneumocystis hosts contract the infection from carrier (or infected) hosts or from hypothetical environmental forms of development?
The high rate of Pneumocystis carriage found in T. brasiliensis (Tables 2 and 3) suggests a highly active interhost airborne transmission. An intensive circulation of Pneumocystis organisms was also reported within the members of a social organization of healthy macaques (30). The occurrence of clustered cases of PcP in hospitals (68) and the reported evidence of human-to-human Pneumocystis transmission in the community (76) further suggest that interhost transmission could also be highly active in humans. In addition, the fact that Pneumocystis organisms are able to dwell and replicate in the lungs of immunocompetent hosts points out the healthy carrier hosts as the infection source and reservoir for Pneumocystis species (16).
Another underlying factor which may affect transmission efficiency of Pneumocystis is the proportion of newly born animals in the colony. In human communities, infants could constitute a major reservoir for Pneumocystis organisms (67, 94), and some data collected in other mammals, like domestic or wild rabbits (26, 40), pigs (50), and macaques maintained in partial release (30), seem to strengthen this hypothesis. A survey scheduled just after the breeding period in a bat colony could provide data on the levels of infection of young bats and on their potential role in the transmission of Pneumocystis organisms within the colony.
Alternatively, other observations suggested that Pneumocystis infection could be contracted from undefined environmental sources (45). Pneumocystis DNA was identified in air and water samples (7, 13, 73, 96). Wakefield was able to detect DNA from rat and human-derived Pneumocystis in air samples from rural locations in the United Kingdom (96). Furthermore, outdoor activities, such as gardening, camping, or hiking, have been reported to be associated with PcP in HIV-infected adults (65). However, on the whole, the active airborne host-to-host circulation of Pneumocystis organisms and the widening of the parasite reservoir to immunocompetent hosts (16, 24, 39) render the hypothesis of Pneumocystis environmental forms of infection less and less plausible. The detection of Pneumocystis DNA in air sampled either from the room of PcP patients (7, 71) or from facilities housing laboratory animals with PcP (54, 70) could attest to Pneumocystis dissemination with the exhaled air of infected hosts and, therefore, potential transmission to other hosts (7, 70, 83).
In the case of T. brasiliensis, the existence of migratory and nonmigratory populations could render the circulation of Pneumocystis organisms in the population more complex (77). Thus, migratory bats could carry significantly less Pneumocystis organisms than sedentary bats. Nevertheless, it is possible that the increased physiological stress and immunocompromise associated with migration might increase fungal growth within bats and increase transmission (4). But new research has also shown that migration allows hosts to escape from infected habitats, reduces disease levels in successful migrants when infected animals do not migrate successfully, and may lead to the evolution of less-virulent pathogens (4). So, between intervals of habitat use, unfavorable conditions (such as a lack of hosts) could eliminate most parasites, resulting in hosts returning to these habitats after a long absence to encounter largely disease-free conditions (4). Furthermore, parasites that decline in response to host migration may include specialist pathogens, as well as those with transmission stages that can build up in the environment (4). Actually, we can assume that when migrating, bats are less exposed to high fungal loads because (i) crowding is less important, (ii) the period of reproduction is over, and (iii) changing roosts implies renewal of ambient air and, likely, exposure to lower numbers of Pneumocystis infective airborne forms.
Climatic factors and altitude.
Temperature and humidity did not seem to impact the Pneumocystis DNA carriage in bats. However, the climatic factors used in our study represent global trends and did not necessarily reflect the actual conditions inside the caves. Samples from T. brasiliensis represent the biggest collection in the present study. In this species, Pneumocystis carriage rates ranged from 25% to 70% in five Latin American regions (Fig. 1). The factors accounting for such variability remain to be explored: location and sampling season but also direct exposure to weather changes versus living in a cave, where environmental conditions can be remarkably stable (66). Some studies explored the impact of climatic factors on Pneumocystis carriage in other host species. In Finland, an influence of seasonal changes on Pneumocystis carriage was reported in wild rodents and insectivores (52). In these host species, the highest prevalence of Pneumocystis organisms in lung samples was reported in late autumn (November), when the precipitation rate was high. The impact of environmental factors on Pneumocystis carriage was also evaluated in immunocompetent macaques (Macaca fascicularis) maintained in partial release (28). The number of macaques with detectable Pneumocystis DNA (assessed by nested PCR from deep nasal swab samples) was apparently correlated with mean precipitation rates (28). However, behavioral factors could also intervene. Actually, when it is raining, macaques group together, some against one another, or enter their shelters, which increases crowding and consequently the Pneumocystis host-to-host airborne transmission. Elsewhere, the detection rate of Pneumocystis DNA was higher in primates, which died (from any cause) during spring or summer (33.3%) rather than during the colder seasons (19.5%), though these differences did not reach statistical significance (29).
Regarding PcP, spontaneous pneumocystosis in domestic rabbits at weaning was usually found to be markedly less extensive in summer than in winter (74). A few studies reported seasonal variations in the occurrence of human cases of pneumocystosis (higher incidence of PcP with higher temperatures in London, Geneva, and Munich) (61, 84, 92). In contrast, surveys from Spain (93) or the United Kingdom (55) showed PcP incidence to peak in the winter months, as it occurs with other infectious respiratory diseases. In the Spanish survey, PcP incidence was negatively correlated with the mean temperature but not with rainfall activity or wind strength (93). These studies indicated that seasonal variation in PcP incidence may exist, albeit to different extents or tendencies. However, nonclimatic factors, such as human behavior or leisure activities, could also be associated with seasonal change and indirectly influence the PcP incidence (84). In bats, the present data suggest that host behavior (migration, crowding) as well as environmental factors such as climate or geographical area may influence Pneumocystis carriage. However, the roosting site did not seem to have a direct impact on the Pneumocystis DNA carriage, the elevation excepted. Altitude exposes the body to a set of constraints, the most important of which is hypoxia. The almost exponential decline of the atmospheric pressure is accompanied by a parallel decline of the oxygen pressure in the inspired air, which induces hyperventilation. Thus, altitude could directly impact on respiratory physiology and host defense mechanisms (95) and indirectly act on the development of Pneumocystis organisms in the alveolar microenvironment. These aspects were not explored in bats. In nonimmunocompromised laboratory rats, hypobaric hypoxia weakened host immune mechanisms and significantly impaired the surfactant composition. Such changes were not enough, however, to favor Pneumocystis growth or to inhibit Pneumocystis clearing from their lungs (95).
Genetic variability.
For each bat species carrying Pneumocystis DNA, at least one novel sequence was amplified at both mtLSU rRNA and/or mtSSU rRNA loci, suggesting that each species of bats could be harboring specific species of Pneumocystis (Fig. 2 and 3). The Pneumocystis sequences amplified from bat lung samples were markedly different from those of other host species registered in the GenBank database. Furthermore, no cross-infection occurred between Rodrigues flying foxes and Egyptian roussettes, although they were cohoused in the same enclosure. Present data strengthened therefore the host specificity concept of Pneumocystis (1, 3, 38) and were consistent with the strong host specificity demonstrated in previous studies dealing with other mammals (27, 41). A first investigation of primate-derived Pneumocystis demonstrated that Pneumocystis phylogeny mirrors its host phylogeny, suggesting a long-range physiological and genetic adaptation process leading to cospeciation (46). Pneumocystis species may have evolved together with their hosts. Likewise, present data showed that genetic divergence in bat-derived Pneumocystis organisms parallels phylogenetic divergence existing among the corresponding host, also suggesting coevolution (27, 41).
Unexpectedly, in the megabat R. aegyptiacus, we found two sequences highly divergent from each other at both mtLSU rRNA and mtSSU rRNA loci (19.9% and 21.7%, respectively). Interestingly, this divergence was comparable to divergence existing between Pneumocystis organisms harbored by M. californicus and rabbits (16.0% at the mtLSU rRNA locus and 23.6% at the mtSSU rRNA locus), as well as between G. soricina and rabbits (19.9% at the mtLSU rRNA locus and 18.5%, at the mtSSU rRNA locus). The presence of two Pneumocystis species in R. aegyptiacus could therefore be hypothesized. Two species of Pneumocystis, i.e., Pneumocystis carinii and Pneumocystis wakefieldiae (21), have been described in the same rat species (Rattus novergicus), though divergence was lower (9.6% and 8.9% at mtLSU rRNA and mtSSU rRNA loci, respectively) (26). In the case of T. brasiliensis, Pneumocystis genetic polymorphism was more limited and apparently related with host infraspecific variants. We found three mtSSU rDNA sequences: two were located in Mexico (Hidalgo/Michoacán/Chiapas region and Nuevo León), and the third one was located in Tucumán (Argentina) (Fig. 1; Table 11). In contrast, one mtLSU rDNA sequence type was amplified from T. brasiliensis lung samples from Mexico, and another one was amplified from lung samples of T. brasiliensis from Argentina. Interestingly, Pneumocystis polymorphism seemed to be related with T. brasiliensis subspecies, which have been described on the basis of geographical distribution and morphology (81). As we have previously suggested (31), Pneumocystis strain polymorphism could be used as a phylogeographic tool to be applied to host natural populations.
Conclusions.
Pneumocystis spp. form a group of parasitic microorganisms infecting a vast diversity of hosts in various ecosystems. A great number of mammalian species belonging to different orders of the clade Mammalia were found to be harboring Pneumocystis organisms. So far, all bat species (11 bat species belonging to 5 families of Chiroptera) examined in this study were found to harbor Pneumocystis DNA except for the flying fox species, Pteropus rodricensis. Interestingly, if confirmed, the absence of Pneumocystis organisms in 44 specimens of P. rodricensis is, as far as we know, the first report of a mammal population, in which no Pneumocystis organisms were detected. Globally, we found a high Pneumocystis carriage rate of 41.3% in bats. Social or behavioral factors (migration, breeding, crowding) may influence transmission of Pneumocystis within the colonies of bat species, while the environmental factors (such as the climate, the roosting habits, geographical place) do not seem to have of impact on carriage rate of Pneumocystis, with the exception of altitude.
In addition, genetic divergence existing among Pneumocystis DNA sequences isolated from different bat species illustrates the close host species specificity reported for Pneumocystis species (3, 38). A comprehensive phylogeny of Pneumocystis from bats is in progress, and comparison with bat phylogeny suggests coevolution (C. Demanche and C.-M. Aliouat-Denis, unpublished results) as it was reported in primates (27, 41, 46) and suggested in other mammalian groups (41). Finally, the link between genetic variability of Pneumocystis isolated from populations of the same bat species and their geographic localization could be exploited in terms of phylogeographical research (31).
ACKNOWLEDGMENTS
Thanks to Leonardo J. López and Alejandro Gómez Nisino, from the Instituto de Ecología, UNAM, for their help with accessing several Mexican caves, bat captures, and taxonomic determination, to Rubén Bárquez, from the Instituto Lillo, for his extraordinary help with accessing the Dique Escaba, San Miguel de Tucumán, Tucumán, Argentina, and to Laerte Ferreiro (Department of Pathology Veterinary Clinic, Veterinarian's University, Federal University of the Rio Grande of the South [UFRGS], Porto Alegre, RS, Brazil) as well as Michèle Lemaire and Laurent Arthur (Museum of Natural History, Bourges, France) for providing lung tissue samples from wild bats and for their sharp knowledge on the European bats. We acknowledge also Thierry Petit (Parc Zoologique de La Palmyre, France) and Dorothée Ordonneau (Parc Zoologique de Lille, France) for providing lung tissue samples from captive bats.
Haroon Akbar is on the “Overseas Scholarship Scheme for France” sponsored by HEC, Islamabad, Pakistan.
This study was developed in the framework of the ECOS-Nord Action “Pneumocystis spp. and Histoplasma capsulatum capsulatum coinfection in bats” (M05-A03) and was supported by the French Ministry of Research (EA4547 Lille-Nord-de-France University and Lille Pasteur Institute), by ANR in the framework of the “Biodiversité” program (CERoPath network), and by the ANR-ERANET “Pneumocystis” PathoGenoMics program (ANR-06-PATHO-009-01).
Footnotes
Published ahead of print 21 September 2012
REFERENCES
- 1. Aliouat EM, et al. 1994. Pneumocystis cross infection experiments using SCID mice and nude rats as recipient hosts showed strong host specificity. J. Eukaryot. Microbiol. 41:S71. [PubMed] [Google Scholar]
- 2. Aliouat EM, Dei-Cas E, Billaut P, Dujardin L, Camus D. 1995. Pneumocystis carinii organisms from in vitro culture are highly infectious to the nude rat. Parasitol. Res. 81:82–85 [DOI] [PubMed] [Google Scholar]
- 3. Aliouat-Denis CM, et al. 2008. Pneumocystis species, co-evolution and pathogenic power. Infect. Genet. Evol. 8:708–726 [DOI] [PubMed] [Google Scholar]
- 4. Altizer S, Bartel R, Han BA. 2011. Animal migration and infectious disease risk. Science 331:296–302 [DOI] [PubMed] [Google Scholar]
- 5. Amano K. 2006. Pulmonary infections in patients with rheumatoid arthritis who have received anti-TNF therapy. Intern. Med. 45:991–992 [DOI] [PubMed] [Google Scholar]
- 6. Atterby H, et al. 2010. Population genetic structure of the Daubenton's bat (Myotis daubentonii) in Western Europe and the associated occurrence of rabies. Eur. J. Wildl. Res. 56:67–81 [Google Scholar]
- 7. Bartlett MS, et al. 1994. Pneumocystis carinii detected in air. J. Eukaryot. Microbiol. 41:75S. [PubMed] [Google Scholar]
- 8. Beck JM, et al. 1998. Interaction of rat Pneumocystis carinii and rat alveolar epithelial cells in vitro. Am. J. Physiol. 275:L118–L125 [DOI] [PubMed] [Google Scholar]
- 9. Brunet-Rossinni AK, Austad SN. 2004. Aging studies on bats: a review. Biogerontology 5:211–222 [DOI] [PubMed] [Google Scholar]
- 10. Burland TM, Wilmer JW. 2001. Seeing in the dark: molecular approaches to the study of bat populations. Biol. Rev. Camb. Philos. Soc. 76:389–409 [DOI] [PubMed] [Google Scholar]
- 11. Calderón EJ, Gutiérrez-Rivero S, Durand-Joly I, Dei-Cas E. 2010. Pneumocystis infection in humans: diagnosis and treatment. Expert Rev. Anti Infect. Ther. 8:683–701 [DOI] [PubMed] [Google Scholar]
- 12. Carmi K, Ido I, Zeev A. 1999. Is the Egyptian fruit-bat Rousettus aegyptiacus a pest in Israel? An analysis of the bat's diet and implications for its conservation. Biol. Conserv. 88:301–306 [Google Scholar]
- 13. Casanova-Cardiel L, Leibowitz MJ. 1997. Presence of Pneumocystis carinii DNA in pond water. J. Eukaryot. Microbiol. 44:28S. [DOI] [PubMed] [Google Scholar]
- 14.CDC 1981. Pneumocystis pneumonia—Los Angeles. Morb. Mort. Wkly. Rep. 30:250–252 [PubMed] [Google Scholar]
- 15. Céré N, Drouet-Viard F, Dei-Cas E, Chanteloup N, Coudert P. 1997. In utero transmission of Pneumocystis carinii sp. f. oryctolagi. Parasite 4:325–330 [DOI] [PubMed] [Google Scholar]
- 16. Chabé M, et al. 2004. Immunocompetent hosts as a reservoir of Pneumocystis organisms: histological and RT-PCR data demonstrate active replication. Eur. J. Clin. Microbiol. Infect. Dis. 23:89–97 [DOI] [PubMed] [Google Scholar]
- 17. Chabé M, et al. 2010. Pneumocystis carinii and Pneumocystis wakefieldiae in wild Rattus norvegicus trapped in Thailand. J. Eukaryot. Microbiol. 57:213–217 [DOI] [PubMed] [Google Scholar]
- 18. Choukri F, et al. 2010. Quantification and spread of Pneumocystis jirovecii in the surrounding air of patients with Pneumocystis pneumonia. Clin. Infect. Dis. 51:259–265 [DOI] [PubMed] [Google Scholar]
- 19. Cushion MT, Ruffolo JJ, Walzer PD. 1988. Analysis of the developmental stages of Pneumocystis carinii, in vitro. Lab. Invest. 58:324–331 [PubMed] [Google Scholar]
- 20. Cushion M. 1998. Pneumocystis carinii, p 675–683. In Ajello L, Hay RJ. (ed), Topley and Wilson's microbiology and microbial infections, vol 4, 9th ed. Arnold, London, United Kingdom [Google Scholar]
- 21. Cushion MT, Keely SP, Stringer JR. 2004. Molecular and phenotypic description of Pneumocystis wakefieldiae sp. nov., a new species in rats. Mycologia 96:429–438 [PubMed] [Google Scholar]
- 22. Daniel MJ. 1990. Order Chiroptera, p 114–137. In King CM. (ed), The handbook of New Zealand mammals. Oxford University Press, Auckland, New Zealand [Google Scholar]
- 23. Dei-Cas E, et al. 1991. Ultrastructural observations on the attachment of Pneumocystis carinii in vitro. J. Protozool. 38:205S–207S [PubMed] [Google Scholar]
- 24. Dei-Cas E. 2000. Pneumocystis infections: the iceberg? Med. Mycol. 38:23–32 [PubMed] [Google Scholar]
- 25. Dei-Cas E, Aliouat EM, Cailliez JC. 2004. Pneumocystis cellular structure, p 61–95. In Walzer PD, Cushion M. (ed), Pneumocystis pneumonia, 3rd ed. Marcel Dekker, New York, NY [Google Scholar]
- 26. Dei-Cas E, et al. 2006. Pneumocystis oryctolagi sp. nov., an uncultured fungus causing pneumonia in rabbits at weaning: review of current knowledge and description if a new taxon on genotypic, phylogenetic and phenotypic bases. FEMS Microbiol. Rev. 30:853–871 [DOI] [PubMed] [Google Scholar]
- 27. Demanche C, et al. 2001. Phylogeny of Pneumocystis carinii from 18 primate species confirms host specificity and suggests co-evolution. J. Clin. Microbiol. 39:2126–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Demanche C, et al. 2003. Influence of climatic factors on Pneumocystis carriage within a socially organized group of immunocompetent macaques (Macaca fascicularis). J. Eukaryot. Microbiol. 50:611–613 [DOI] [PubMed] [Google Scholar]
- 29. Demanche C, et al. 2003. Assessment of Pneumocystis species carriage in captive primates. Vet. Rec. 152:811–813 [DOI] [PubMed] [Google Scholar]
- 30. Demanche C, et al. 2005. Molecular and serological evidence of Pneumocystis circulation in a social organization of healthy macaques (Macaca fascicularis). Microbiology 151:3117–3125 [DOI] [PubMed] [Google Scholar]
- 31. Derouiche S, et al. 2009. Pneumocystis as phylogeographic markers? Mem. Instit. Oswaldo Cruz 104:112–117 [DOI] [PubMed] [Google Scholar]
- 32. Dumoulin A, et al. 2000. Transmission of Pneumocystis carinii disease from immuno-competent contacts of infected hosts to susceptible hosts. Eur. J. Clin. Microbiol. Infect. Dis. 19:671–678 [DOI] [PubMed] [Google Scholar]
- 33. Durand-Joly I, et al. 2002. Pneumocystis carinii f. sp. hominis is not infectious to SCID mice. J. Clin. Microbiol. 40:1862–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791 [DOI] [PubMed] [Google Scholar]
- 35. Freeman AF, Davis J, Anderson VL, Barson W, Dirk N. 2006. Pneumocystis jirovecii infection in patients with hyper-immunoglobulin E syndrome. Pediatrics 118:1271–1275 [DOI] [PubMed] [Google Scholar]
- 36. Fujita MS, Tuttle MD. 1991. Flying foxes (Chiroptera: Pteropodidae): threatened animals of key ecological and economic importance. Conserv. Biol. 5:455–463 [Google Scholar]
- 37. Gascuel O. 1997. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 14:685–695 [DOI] [PubMed] [Google Scholar]
- 38. Gigliotti F, Harmsen AG, Haidaris PJ. 1993. Pneumocystis carinii is not universally transmissible between mammalian species. Infect. Immun. 61:2886–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gigliotti F, Harmsen AG, Wright TW. 2003. Characterization of transmission of Pneumocystis carinii f. sp. muris through immunocompetent BALB/c mice. Infect. Immun. 71:3852–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Guillot J, et al. 1999. Acquisition and biodiversity of Pneumocystis carinii in a colony of wild rabbits (Oryctolagus cuniculus). J. Eukaryot. Microbiol. 46:100S–101S [PubMed] [Google Scholar]
- 41. Guillot J, et al. 2001. Parallel phylogenies of Pneumocystis species and their mammalian hosts. J. Eukaryot. Microbiol. 48:113S–115S [DOI] [PubMed] [Google Scholar]
- 42. Gutiérrez S, et al. 2011. Pneumocystis jirovecii colonization in chronic pulmonary disease. Parasite 18:121–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hand S, Archer M, Godthelp H. 2005. Australian oligo-miocene mystacinids (Microchiroptera): upper dentition, new taxa and divergence of New Zealand species. Geobios 38:339–352 [Google Scholar]
- 44. Huang L, Morrison A, Limper AH, Beck JM. 2006. An official ATS workshop summary: recent advances and future directions in Pneumocystis pneumonia (PCP). Proc. Am. Thorac. Soc. 3:655–664 [DOI] [PubMed] [Google Scholar]
- 45. Hughes WT. 1982. Natural mode of acquisition for de novo infection with Pneumocystis carinii. J. Infect. Dis. 172:842–848 [DOI] [PubMed] [Google Scholar]
- 46. Hugot JP, Demanche C, Barriel V, Dei-Cas E, Guillot J. 2003. Phylogenetic systematics and evolution of primate-derived Pneumocystis based on mitochondrial or nuclear DNA sequence comparison. Syst. Biol. 52:735–744 [DOI] [PubMed] [Google Scholar]
- 47. Hunter JA, Wakefield AE. 1996. Genetic divergence at the mitochondrial small subunit ribosomal RNA gene among isolates of Pneumocystis carinii from five mammalian host species. Eukaryot. Microbiol. 43(5):24S–25S [DOI] [PubMed] [Google Scholar]
- 48.IUCN 2010 27 September 2010, accession date. IUCN red list of threatened species, version 3. IUCN, Gland, Switzerland: www.iucnredlist.org [Google Scholar]
- 49. Keely SP, Fischer JM, Cushion MT, Stringer JR. 2004. Phylogenetic identification of Pneumocystis murina sp. nov., a new species in laboratory mice. Microbiology 150:1153–1165 [DOI] [PubMed] [Google Scholar]
- 50. Kondo H, Hikita M, Ito M, Kadota K. 2000. Immunohistochemical study of Pneumocystis carinii infection in pigs: evaluation of Pneumocystis pneumonia and a retrospective investigation. Vet. Rec. 147:544–549 [DOI] [PubMed] [Google Scholar]
- 51. Kunz TH. 1982. Roosting ecology. In Kunz TH. (ed), Ecology of bats, p 1–55. Plenum Press, New York, NY [Google Scholar]
- 52. Laakkonen J, Henttonen H, Niemimaa J, Soveri T. 1999. Seasonal dynamics of Pneumocystis carinii in the field vole, Microtus agrestis, and in the common shrew, Sorex araneus, in Finland. Parasitology 118:1–5 [DOI] [PubMed] [Google Scholar]
- 53. Laakkonen J, Fisher RN, Case TJ. 2001. Pneumocystosis in wild small mammals from California. J. Wildl. Dis. 37:408–412 [DOI] [PubMed] [Google Scholar]
- 54. Latouche S, et al. 1997. Detection of Pneumocystis carinii f. sp. in air samples collected in animal rooms. J. Eukaryot. Microbiol. 44:46–47 [DOI] [PubMed] [Google Scholar]
- 55. Lubis N, et al. 2003. Prospective cohort study showing changes in the monthly incidence of Pneumocystis carinii pneumonia. Postgrad. Med. J. 79:164–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Martinez A, et al. 2009. High-speed cell sorting of infectious trophic and cystic forms of Pneumocystis carinii. J. Eukaryot. Microbiol. 56:446–453 [DOI] [PubMed] [Google Scholar]
- 57. Martinez A, et al. 2011. Ploidy of cell-sorted trophic and cystic forms of Pneumocystis carinii. PLoS One 6:e20935 doi:10.1371/journal.pone.0020935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mazars E, et al. 1997. Detection of Pneumocystis in European wild animals. J. Eukaryot. Microbiol. 44:39S. [DOI] [PubMed] [Google Scholar]
- 59. McCracken GF, Wilkinson GS. 2000. Bat mating systems, p 321–362. In Krutzch PH, Crichton EG. (ed), Reproductive biology of bats. Academic Press, New York, NY [Google Scholar]
- 60. McGuire LP, Ratcliffe JM. 2011. Light enough to travel: migratory bats have smaller brains, but not larger hippocampi, than sedentary species. Biol. Lett. 7:233–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Miller RF, Grant AD, Foley NM. 1992. Seasonal variations in presentation of Pneumocystis carinii pneumonia. Lancet 339:747–748 [DOI] [PubMed] [Google Scholar]
- 62. Monnet X, et al. 2008. Critical care management and outcome of severe Pneumocystis pneumonia in patients with and without HIV infection. Crit. Care 12:R28 (Erratum, 13:407.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Montes-Cano MA, et al. 2009. Vertical transmission of Pneumocystis jirovecii in humans. Emerg. Infect. Dis. 15:125–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Morris A. 2008. Is there anything new in Pneumocystis jirovecii pneumonia? Changes in P. jirovecii pneumonia over the course of the AIDS epidemic. Clin. Infect. Dis. 46:634–636 [DOI] [PubMed] [Google Scholar]
- 65. Navin TR, et al. 2000. Risk factors for community-acquired pneumonia among persons infected with human immunodeficiency virus. J. Infect. Dis. 181:158–164 [DOI] [PubMed] [Google Scholar]
- 66. Neuweiler G. 2000. The biology of bats. Oxford University Press, New York, NY [Google Scholar]
- 67. Nevez G, Totet A, Pautard JC, Raccurt C. 2001. Pneumocystis carinii detection using nested-PCR in nasopharyngeal aspirates of immunocompetent infants with bronchiolotis. J. Eukaryot. Microbiol. 48:122S–123S [DOI] [PubMed] [Google Scholar]
- 68. Nevez G, et al. 2008. Nosocomial Pneumocystis jirovecii infections. Parasite 15:359–365 [DOI] [PubMed] [Google Scholar]
- 69. Nowak RM. 1999. Walker's mammals of the world, 6th ed, p 253–489 The John Hopkins University Press, Baltimore, MD [Google Scholar]
- 70. Olsson M, Sukura A, Lindberg LA, Linder E. 1996. Detection of Pneumocystis carinii DNA by filtration of air. Scand. J. Infect. Dis. 28:279–282 [DOI] [PubMed] [Google Scholar]
- 71. Olsson M, et al. 1998. Identification of Pneumocystis carinii f. sp. hominis gene sequences in filtered air in hospital environments. J. Clin. Microbiol. 36:1737–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ortega J, Navarrete Vargas DA, Castellanos Morales G. 2009. Paternidad y relaciones de parentesco en una colonia de Artibeus hirsutus en la cueva del Salitre, Morelos, p 191–203. In Ortega J, Sedeño García JE, López López E. (ed), Setenta y cinco años de la Escuela National de Ciencias Biológica. Instituto Politécnico National, Mexico City, Mexico [Google Scholar]
- 73. Philippe L, et al. 1999. Impaction versus filtration for the detection of Pneumocystis carinii DNA in air. J. Eukaryot. Microbiol. 46:94S. [PubMed] [Google Scholar]
- 74. Rajagopalan-Levasseur P, et al. 1998. Response to Pneumocystis infection in an immunocompetent host. FEMS Immunol. Med. Microbiol. 22:107–121 [DOI] [PubMed] [Google Scholar]
- 75. Rambaut A. 2002. Se-Al: Sequence Alignment Editor, version 2.0 alpha 11 Carbon. University of Oxford, Oxford, United Kingdom: http://tree.bio.ed.ac.uk/software/seal/ [Google Scholar]
- 76. Rivero L, et al. 2008. Pneumocystis jirovecii transmission from immunocompetent carriers to infant. Emerg. Infect. Dis. 14:1116–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Russell AL, Medellin RA, Mccracken GF. 2005. Genetic variation and migration in the Mexican free-tailed bat (Tadarida brasiliensis mexicana). Mol. Ecol. 14:2207–2222 [DOI] [PubMed] [Google Scholar]
- 78. Russian DA, Levine SJ. 2001. Pneumocystis carinii pneumonia in patients without HIV infection. Am. J. Med. Sci. 321:56–65 [DOI] [PubMed] [Google Scholar]
- 79. Sanchez CA, et al. 2007. Exploring transplacental transmission of Pneumocystis oryctolagi in first-time pregnant and multiparous rabbit does. Med. Mycol. 45:701–707 [DOI] [PubMed] [Google Scholar]
- 80. Scheunert A, Zahn A, Andreas K. 2010. Phenology and roosting habits of the central European grey long-eared bat Plecotus austriacus (Fischer 1829). Eur. J. Wildl. Res. 56:435–442 [Google Scholar]
- 81. Schwartz A. 1955. Tadarida brasiliensis subspecies. J. Mamm. 36:108 [Google Scholar]
- 82. Sesterhenn TM, et al. 2010. Sequence and structure of the linear mitochondrial genome of Pneumocystis carinii. Mol. Genet. Genomics 283:63–72 [DOI] [PubMed] [Google Scholar]
- 83. Sing A, Wonhas C, Bader L, Luther M, Heeseman J. 1999. Detection of Pneumocystis carinii DNA in the air filter of a ventilated patient with AIDS. Clin. Infect. Dis. 29:952–953 [DOI] [PubMed] [Google Scholar]
- 84. Sing A, et al. 2009. Seasonal variation of Pneumocystis jirovecii infection: analysis of underlying climatic factors. Clin. Microbiol. Infect. 15:957–960 [DOI] [PubMed] [Google Scholar]
- 85. Soulez B, et al. 1991. Introduction of Pneumocystis carinii in a colony of SCID mice. J. Protozool. 38:123S–125S [PubMed] [Google Scholar]
- 86. Spada M, et al. 2008. Roost selection by non-breeding Leisler's bats (Nyctalus leisleri) in montane woodlands: implications for habitat management. Acta Chiropterol. 10:81–88 [Google Scholar]
- 87. Swofford DL. 2001. PAUP*: phylogenetica analyses using parsimony (* and other methods), version 4.0b10. Sinuaer, Sunderland, MA [Google Scholar]
- 88. Taylor ML, et al. 1999. Environmental conditions favoring bat infection with Histoplasma capsulatum in Mexican shelters. Am. J. Trop. Med. Hyg. 61:914–919 [DOI] [PubMed] [Google Scholar]
- 89. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Torres-Flores J, López-Wilchis R. 2010. Condiciones microclimáticas, hábitos de percha y especies asociadas a los refugios de Natalus stramineus en México. Acta Zool. Mex. 26:191–213 [Google Scholar]
- 91. Tsolaki AG, Beckers P, Wakefield AE. 1998. Pre-AIDS era isolates of Pneumocystis carinii f. sp. hominis: high genotypic similarity with contemporary isolates. J. Clin. Microbiol. 36:90–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Vanhems P, Hirschel B, Morabis A. 1992. Seasonal incidence of Pneumocystis carinii pneumonia. Lancet 339:1182. [DOI] [PubMed] [Google Scholar]
- 93. Varela JM, et al. 2004. Climatic factors and Pneumocystis jiroveci infection in southern Spain. Clin. Microbiol. Infect. 10:770–772 [DOI] [PubMed] [Google Scholar]
- 94. Vargas SL, et al. 2001. Search for primary infection by Pneumocystis carinii in a cohort of normal, healthy infants. Clin. Infect. Dis. 32:855–861 [DOI] [PubMed] [Google Scholar]
- 95. Vives MF, et al. 2008. Hypobaric hypoxia-related impairment of pulmonary surfactant proteins A and D did not favour Pneumocystis carinii Frenkel 1999 growth in non-immunocompromised rats. Parasite 15:53–64 [DOI] [PubMed] [Google Scholar]
- 96. Wakefield AE. 1996. DNA sequences identical to Pneumocystis carinii f. sp. carinii and Pneumocystis f. sp. hominis in samples of air spora. J. Clin. Microbiol. 34:1754–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wakefield AE. 1998. Genetic heterogeneity in Pneumocystis carinii: an introduction. FEMS Immunol. Med. Microbiol. 22:5–13 [DOI] [PubMed] [Google Scholar]
- 98. Wilkinson GS, South JM. 2002. Life history, ecology and longevity in bats. Aging Cell 1:124–131 [DOI] [PubMed] [Google Scholar]
- 99. Yoshida Y. 1989. Ultrastructural studies of Pneumocystis carinii. J. Protozool. 3:53–60 [DOI] [PubMed] [Google Scholar]