Abstract
The γ-butyrolactone autoregulator signaling cascades have been shown to control secondary metabolism and/or morphological development among many Streptomyces species. However, the conservation and variation of the regulatory systems among actinomycetes remain to be clarified. The genome sequence of Kitasatospora setae, which also belongs to the family Streptomycetaceae containing the genus Streptomyces, has revealed the presence of three homologues of the autoregulator receptor: KsbA, which has previously been confirmed to be involved only in secondary metabolism; KsbB; and KsbC. We describe here the characterization of ksbC, whose regulatory cluster closely resembles the Streptomyces virginiae barA locus responsible for the autoregulator signaling cascade. Deletion of the gene ksbC resulted in lowered production of bafilomycin and a defect of aerial mycelium formation, together with the early and enhanced production of a novel β-carboline alkaloid named kitasetaline. A putative kitasetaline biosynthetic gene cluster was identified, and its expression in a heterologous host led to the production of kitasetaline together with JBIR-133, the production of which is also detected in the ksbC disruptant, and JBIR-134 as novel β-carboline alkaloids, indicating that these genes were biosynthetic genes for β-carboline alkaloid and thus are the first such genes to be discovered in bacteria.
INTRODUCTION
Members of the genus Streptomyces have been extensively studied due to their complex developmental life cycle and their ability to synthesize a vast array of important secondary metabolites used in human/veterinary medicine and agriculture. Recently, the rapid accumulation of genome information has enabled elucidation of the physiological mechanisms at the molecular level, in addition to pointing the hitherto-undiscovered ability to produce novel secondary metabolites, which are a promising source of new clinically useful compounds. Actinomycetes other than those in the genus Streptomyces are often called non-Streptomyces actinomycetes: these include actinomycetes of the genera Kitasatospora, Micromonospora, Actinoplanes, Amycolatopsis, and Nocardia, all of which produce useful natural compounds in the manner of the Streptomyces species. The genus Kitasatospora, members of which exhibit lifestyles and morphological development similar to those of Streptomyces species, is phylogenetically close to the genus Streptomyces, and belongs to the same family Streptomycetaceae. Kitasatospora setae NBRC 14216T produces bafilomycins A1 and B1 (compound 1 in Fig. 1A), specific inhibitors of vacuolar H+-ATPase commonly used as biochemical reagents to investigate molecular transport in eukaryotic cells (5, 30). The complete genome sequence revealed that K. setae has at least 24 genes or gene clusters for the biosynthesis of secondary metabolites, including bafilomycin (14). A vast majority of these genes and clusters play unknown roles in the biosynthetic processes and are presumably cryptic biosynthetic pathways. An improved understanding of the systems for regulating secondary metabolism in K. setae not only might reveal common features and differences of the genetic information between the genera Kitasatospora and Streptomyces but also could provide a great opportunity to discover novel natural compounds.
Fig 1.
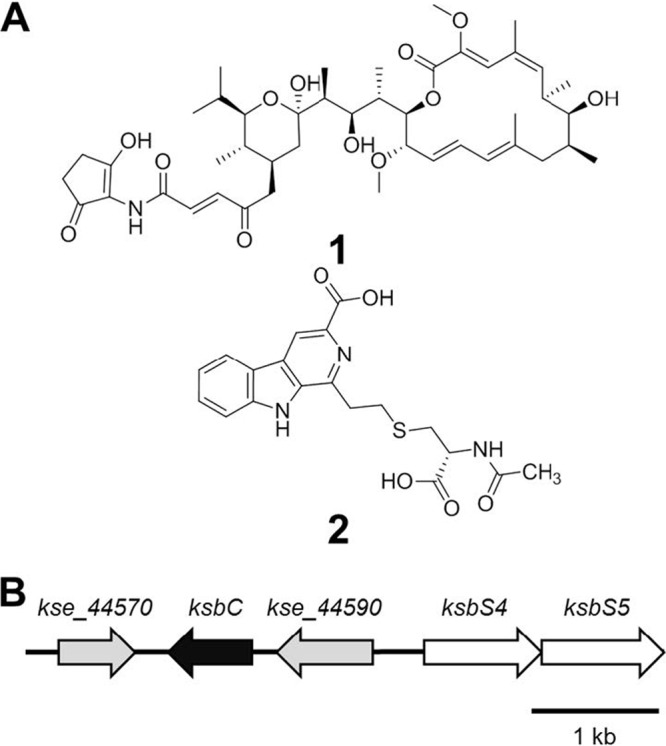
Chemical structures of metabolites of K. setae (A) and organization of the ksbC locus in K. setae (B). (A) Structures of bafilomycin B1 (compound 1) and kitasetaline (compound 2). (B) Gray arrows indicate putative regulatory genes, and white arrows indicate putative autoregulator biosynthetic genes.
γ-Butyrolactone autoregulator signaling cascades are known to be the major regulatory systems for Streptomyces secondary metabolism (3, 36). In this system, the autoregulator receptor binds to a specific DNA sequence called an autoregulatory element (ARE) in front of its target genes, repressing their transcription (10). Binding of the autoregulator prevents the receptor from interacting with DNA, allowing transcription of the target genes and in turn activating the coordinated expression of regulatory and enzymatic genes involved in secondary metabolism and sometimes in morphological development. KsbA is an autoregulator receptor of K. setae that is the only receptor identified in non-Streptomyces actinomycetes by the conventional method using degenerate PCR primers (9). KsbA functions as a negative regulator of bafilomycin production but has no influence on morphological differentiation. The DNA-binding activity of KsbA and the target genes remain to be elucidated. Searches of the genome sequence of K. setae demonstrated that, in addition to ksbA, two more putative autoregulator receptor genes, ksbB and ksbC, are encoded on the genome, together with afsA family genes encoding putative autoregulator synthase in the proximal region (14). The genomes of well-studied Streptomyces strains such as S. coelicolor A3(2), S. griseus, and S. avermitilis, have only one copy of afsA family genes. These findings prompted us to investigate the function of additional autoregulator receptors in K. setae, which might form a more complicated signaling network for secondary metabolism and/or morphological development compared to that of Streptomyces species.
In the present study, we report the role of KsbC in the regulation of secondary metabolism and morphological development and demonstrate that KsbC positively controls bafilomycin production and aerial mycelium formation. Moreover, the ksbC mutant showed precocious and abundant production of the metabolite, a novel β-carboline alkaloid named kitasetaline. We also identified the biosynthetic genes of kitasetaline and its derivatives JBIR-133 and JBIR-134 as new compounds by heterologous expression in a Streptomyces host strain and suggest a possible route for the supply of the β-carboline structure in bacteria.
(This research was conducted by A. Aroonsri in partial fulfillment of the requirements for a Ph.D.)
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
K. setae NBRC 14216T from the NITE Biological Resource Center (NBRC), Japan, was grown on ISP medium 2 (Becton Dickinson, Franklin Lakes, NJ). Escherichia coli DH5α was used for general DNA manipulation, the DNA methylation-deficient E. coli strain ET12567 containing pUZ8002 (31) was used for E. coli/Kitasatospora conjugation, and E. coli GM2929 hsdS::Tn10 was used to prepare unmethylated DNA for protoplast transformation in Streptomyces avermitilis SUKA22 (isogenic to S. avermitilis SUKA17 [21] but loxP sequences were replaced by mutant loxP sequences). E. coli BL21(DE3)/pLysS and the plasmid pET-15b, which were used for the expression of recombinant KsbC (rKsbC), were obtained from Novagen. The plasmids used were pUC19 for general cloning, and pKU451, pKU474, and pKU250 were used for gene disruption (21, 24). The genome-integrated vectors, pKU492Aaac(3)IV (see Fig. S1 in the supplemental material) and pKU503 (21), were used for subcloning a gene cluster for kitasetaline biosynthesis and the construction of BAC library for K. setae genome. For complementation, pENTR (Invitrogen) was used for DNA cloning, and pLT113 (24) was used to introduce DNA into Streptomyces. The medium conditions, antibiotic concentrations, and general E. coli and Kitasatospora/Streptomyces manipulations were as described previously (16). Spores (108 CFU) of K. setae strains were inoculated into 70 ml of YMM medium (2) in a 500-ml baffled flask, and mycelia were harvested after 36 h of cultivation at 28°C. The mycelia were washed, resuspended in fresh YMM medium, and stored at −80°C until use as a seed culture. All of the primers are listed in Table S1 in the supplemental material.
Disruption of the K. setae ksbC gene.
A 2.0-kb ksbC-upstream fragment was amplified using the primer pair ksbC-up-Fw and ksbC-up-Re, and a 2.2-kb ksbC-downstream fragment was amplified using the primer pair ksbC-down-Fw and ksbC-down-Re. The fidelity of the amplified regions was confirmed by sequencing. The two resultant fragments were digested with EcoRI and HindIII, and cloned together into the HindIII site of pKU451. The resultant plasmid was cleaved by EcoRI and ligated with a kanamycin-resistance gene amplified by the primer pair aphII-E-Fw/aphII-E-Re using pKU474 as a template to yield pLT430. A 5.6-kb HindIII fragment, recovered from pLT430, was inserted into the HindIII site of pKU250 to generate pLT433 for ksbC disruption. E. coli ET12567(pUZ8002) harboring pLT433 was conjugated with K. setae, and the wild-type (WT) gene was replaced with the disrupted allele (ΔksbC) by homologous recombination. The genotype of the ΔksbC candidates resistant to kanamycin and sensitive to thiostrepton was confirmed by PCR analysis.
Complementation of the ksbC deletion mutant.
The entire ksbC gene with its 504-bp upstream region was amplified by using the primer pair ksbC-comp-Fw/ksbC-comp-Re and then cloned into a pENTR vector to generate an entry clone. The entry clone was used with pLT113 in an LR reaction (LR Clonase Enzyme Mix; Invitrogen), resulting in pLT436. By intergenic conjugation and integration, the plasmid pLT436 was introduced into the ksbC-deletion mutant. The correct integration in the exconjugants was confirmed by PCR.
Analysis of K. setae secondary metabolites.
The seed culture was spread onto 2.5 ml of ISP medium 4 (Becton Dickinson), followed by incubation at 28°C for 5 days. The agar culture was diced and extracted with 2 volumes of methanol. The methanol-extract was collected by centrifugation and analyzed by using a high-pressure liquid chromatography (HPLC) system on an Inertsil ODS-3 column (4 μm; 4.6 by 250 mm; GL Sciences, Tokyo, Japan), using 80% CH3CN containing 0.1% trifluoroacetic acid (TFA) as a mobile phase and detection at 245 nm for bafilomycin production, and with a linear gradient system (eluents, H2O containing 0.075% TFA [medium A] and methanol containing 0.01% TFA [medium B]; gradient, 0 to 5 min 10% B and 5 to 55 min 10% B to 100% B; flow rate, 1 ml/min; UV detection at 276 nm) for kitasetaline production. Commercial bafilomycin A1 and bafilomycin B1 (Fluka, USA) and purified bafilomycin C1 were used as standards for HPLC analysis.
Molecular cloning of the gene cluster for kitasetaline biosynthesis and evaluation of the gene cluster by heterologous expression.
The mycelia of K. setae were embedded into 0.6% SeaPlaque GTG agarose (Lonza Group, Ltd., Switzerland) before digestion with 1 mg of lysozyme/ml for 16 h at 37°C. The resulting protoplasts were lysed by addition of 1% sodium N-lauroylsarcosinate and 1 mg of proteinase K/ml at 50°C for 24 h. After the inactivation of proteinase K by 0.1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (Nacalai Tesque, Inc., Japan) and removal of sodium N-lauroylsarcosinate by repeated washing with 50 mM EDTA (pH 8.0), DNA embedded in the agarose plug was partially digested with BamHI. Fragments of from 100 to 150 kb were purified by CHEF (contour-clamped homogeneous electric field) electrophoresis. The agarose gel containing DNA fragments were molten by NaI with a half weight of gel, and the molten agarose and NaI were removed by dialysis against TE (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]). The residual agarose were completely molten by using a GELase enzyme preparation (Illumina, Inc., USA). The DNA fragments were ligated with BamHI fragment of pKU503, and the ligated DNA was transformed into E. coli NEB10β. Each BAC clone was stored in six 384-well plates containing LB (100 μg of ampicillin/ml and 20% glycerol) at −80°C. A BAC clone, pKU503facP2-L12, containing putative gene cluster for kitasetaline biosynthesis, was introduced into S. avermitilis SUKA22 by protoplast transformation. For minimization of a gene cluster for kitasetaline biosynthesis, ∼500-bp homologous regions upstream and downstream of the gene cluster were prepared by PCR with pKU503facP2-L12 as a template using two primer pairs corresponding to upstream and downstream of the gene cluster: an upstream primer pair (forward, 5′-CTCGAGTCTAGAAGTTCTTCGACCTCGGCCTCACCTCC-3′ [bold characters indicate the XbaI site, and italic characters correspond to the region from nucleotides {nt} 8040461 to 8040486 of K. setae]; and reverse, 5′-CTCGAGGGATCCGGACGCTGGAGCAGATGAAGGACT-3′ [bold characters indicate the BamHI site, and italic characters correspond to the region from nt 8040873 to 8040897 of K. setae]) and a downstream primer pair (forward, 5′-CTCGAGGGATCCCGGGCTGCGCCGCATGGACCTC-3′ [bold characters indicate the BamHI site, and italic characters correspond to the region from nt 8051125 to 8051148 of K. setae]; and reverse, 5′-CTCGAGAAGCTTCAGGGACGGCGCCTGCCACAGCTCGAAGG-3′ [bold characters indicate the HindIII site, and italic characters correspond to the region from nt 8051601 to 8051573 of K. setae), respectively. The minimum cloning vector pRED (21) lacking XbaI site was also amplified by PCR using a primer pair: forward, 5′-CTCGAGTCTAGATGCCAGGAAGATACTTAACAG-3′ (bold characters indicate the XbaI site); and reverse, 5′-CTCGAGAAGCTTCCATTCATCCGCTTATTATC-3′ (bold characters indicate the HindIII site). After amplification of these segments, all segments treated with DpnI to remove a template DNA, the upstream segment digested with XbaI/BamHI, the downstream segment digested with BamHI/HindIII, and the vector segment digested with XbaI/HindIII were ligated together to yield pRED::upstream::downstream. The resulting plasmid was amplified by PCR with the forward primer of the downstream segment and the reverse primer of the upstream segment. The amplified fragment was treated with DpnI to remove a template DNA and digested with Bal31 for 30 s at 30°C. The biosynthetic gene cluster for kitasetaline in pKU503facP2-L12 was replaced by a linear molecule of pRED::upstream::downstream using the homologous regions using the λRED recombination system (E. coli DH10B and pKD119 were used [21]) to generate pRED::ksl (kitasetaline) cluster (11,141 bp, corresponding to nt 8040461 to 8051601 of K. setae). A XbaI/HindIII fragment, including an 11.1-kb ksl cluster, recovered from pRED::ksl, was inserted into the XbaI/HindIII sites of pKU492Aaac(3)IV, resulting in the pKU492Aaac(3)IV::ksl cluster, named pLT437. pLT437 was (i) digested with either HindIII/Bsu36I, HindIII/EcoRV, or XbaI/BstBI, (ii) treated with T4 DNA polymerase to yield blunt ends, and (iii) self-ligated, resulting in pLT438, pLT439, or pLT440, respectively. The constructed plasmids were transferred into E. coli GM2929 hsdS::Tn10, and the unmethylated plasmids were introduced into S. avermitilis SUKA22 by protoplast transformation. Integration of the plasmids was confirmed by apramycin resistance and PCR analysis. Spores (108 CFU) of SUKA22 derivative strains were inoculated on 2.5 ml of YMS-MC medium, followed by incubation at 28°C for 4 days. The agar culture was extracted with 2 volumes of methanol, and the analysis of the products was performed as described above in “Analysis of K. setae secondary metabolites.”
Isolation and structural elucidation of novel kitasetaline analogues.
The SUKA22 carrying pKU503facP2-L12 was cultivated in a 15-ml test tube containing 5 ml of seed medium (7). The test tube was incubated on a reciprocal shaker at 27°C for 2 days (320 rpm). An aliquot (2.5 ml) of the culture was then transferred to 500-ml baffled Erlenmeyer flasks, each containing 100 ml of a production medium (7). The sample was cultured on a rotary shaker (180 rpm) at 27°C for 5 days. After the fermentation broth (100 ml) was separated by centrifugation, the mycelial cake was extracted with acetone (40 ml) and concentrated in vacuo. The aqueous concentrate was successively washed with ethyl acetate and extracted three times with n-butanol (20 ml), after which the n-butanol layer was evaporated to dryness. The supernatant was washed with ethyl acetate and then extracted three times with n-butanol (50 ml), after which the organic phase was evaporated. The combined extracts (40.2 mg) were subjected to reversed-phase medium-pressure liquid chromatography (MPLC) eluted with a stepwise solvent system of water-methanol (100:0, 90:10, 80:20, 60:40, 40:60, 20:80, and 0:100). The fraction eluted with 10% methanol (12.3 mg) was further purified by RP-HPLC on a Capcell Pak C18 MGII column (5.0 μm, 20 mm [inner diameter] by 150 mm) developed with 35% aqueous methanol containing 0.1% formic acid (flow rate, 10 ml/min) to yield compounds 3 (2.4 mg, retention time = 22.6 min) and 2 (1.2 mg, retention time = 27.4 min) (see Fig. 4). Next, the 20% methanol eluate (2.0 mg) was purified by RP-HPLC on a Capcell Pak C18 MGII column (5.0 μm, 10 mm [inner diameter] by 150 mm) developed with a gradient solvent system of aqueous methanol (40 to 55% for 30 min) containing 0.1% formic acid (flow rate, 4 ml/min) to yield compound 4 (0.5 mg, retention time = 26.3 min). Ultraviolet (UV) and infrared (IR) spectra were measured on a Beckman Coulter DU730 UV/Vis spectrophotometer and a Horiba FT-720 spectrophotometer, respectively. Nuclear magnetic resonance (NMR) spectra were recorded on a Varian NMR System 600 NB CL. Chemical shifts were calibrated internally against the residual signal of the solvent in which the sample was dissolved (dimethyl sulfoxide [DMSO]-d6: δC 39.7, δH 2.49). HRESIMS data were recorded using a Waters LCT-Premier XE mass spectrometer. MPLC was carried out on a Purif-Pack ODS-100 column (Shoko Scientific). RP-HPLC was carried out using a CAPCELL PAK C18 MGII column (5.0 μm, 20 mm [inner diameter] by 150 mm, 10 mm [inner diameter] by 150 mm; Shiseido) with a Waters 2996 photodiode array detector and a Waters 3100 mass detector.
Fig 4.
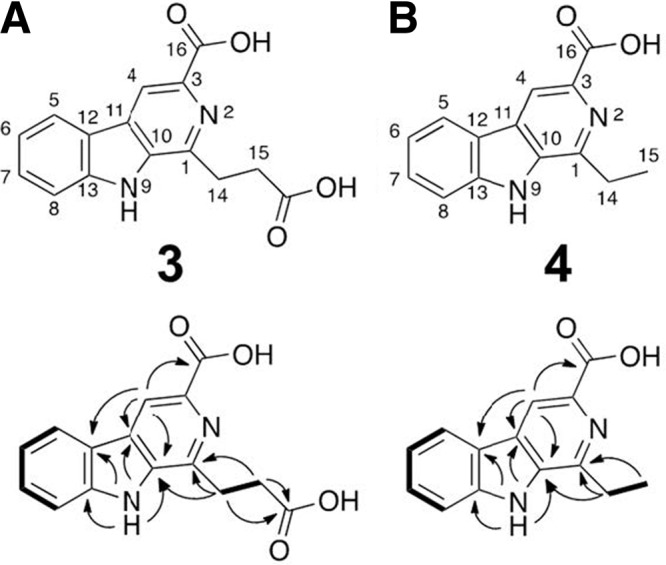
Chemical structures of JBIR-133 (A; compound 3) and JBIR-134 (B; compound 4). Lower column shows COSY and HMBC analysis. Bold lines represent 1H-1H correlations, and arrows indicate key HMBC correlations (1H ↔ 13C).
Physicochemical properties: JBIR-133 (compound 3).
Pale yellow amorphous solid; UV (methanol) λmax (log ε) 238 (4.41), 267 (4.57), 302 (3.97), 376 (3.41) nm; IR (KBr) vmax 3,143, 1,708, 1,241 cm−1; HRESIMS m/z 285.0869 [M+H]+ (calculated for C15H13N2O4, 285.0875); 13C NMR (150 MHz, DMSO-d6, δ ppm): 144.0 (C-1), 135.3 (C-3), 115.7 (C-4), 122.3 (C-5), 120.3 (C-6), 128.6 (C-7), 112.5 (C-8), 135.9 (C-10), 128.6 (C-11), 121.5 (C-12), 141.4 (C-13), 28.3 (C-14), 31.8 (C-15), 166.8 (C-16), 174.2 (15-CO2H). 1H NMR (600 MHz, DMSO-d6, δ ppm): 8.76 (1H, s, H-4), 8.34 (1H, d, J = 7.8 Hz, H-5), 7.29 (1H, t, J = 7.8 Hz, H-6), 7.58 (1H, t, J = 7.8 Hz, H-7), 7.64 (1H, d, J = 7.8 Hz, H-8), 12.12 (1H, s, H-9), 3.39 (2H, t, J = 7.2 Hz, H-14), 2.91 (2H, t, J = 7.2 Hz, H-15).
Physicochemical properties: JBIR-134 (compound 4).
Pale yellow amorphous solid; UV (MeOH) λmax (log ε) 237 (4.44), 269 (4.60), 302 (4.07), 377 (3.62) nm; IR (KBr) vmax 3,062, 1,701, 1,234 cm−1; HRESIMS m/z 241.0974 [M+H] + (calculated for C14H13N2O2, 241.0977); 13C NMR (150 MHz, DMSO-d6, δ ppm): 146.7 (C-1), 135.3 (C-3), 115.7 (C-4), 122.3 (C-5), 120.3 (C-6), 128.6 (C-7), 111.2 (C-8), 135.6 (C-10), 127.6 (C-11), 121.6 (C-12), 141.4 (C-13), 26.9 (C-14), 12.9 (C-15), 167.1 (C-16). 1H NMR (600 MHz, DMSO-d6, δ ppm): 8.76 (1H, s, H-4), 8.33 (1H, d, J = 7.8 Hz, H-5), 7.28 (1H, t, J = 7.8 Hz, H-6), 7.57 (1H, t, J = 7.8 Hz, H-7), 7.64 (1H, d, J = 7.8 Hz, H-8), 12.02 (1H, s, H-9), 3.17 (2H, q, J = 7.4 Hz, H-14), 1.38 (3H, t, J = 7.4 Hz, H-15).
Transcriptional analysis by semiquantitative RT-PCR and qRT-PCR.
Total RNA was prepared from mycelia on ISP medium 4 with cultivation for 5 days using an RNeasy minikit (Qiagen) and treated with DNase I (TaKaRa Bio). The cDNA was synthesized using Superscript III RNase H− reverse transcriptase (Invitrogen) and random primers (Invitrogen) according to the manufacturer's instructions. For semiquantitative reverse transcription-PCR (RT-PCR), the PCR amplification was performed with GoTaq green master mix (Promega KK) under the following conditions: 98°C for 3 min, followed by 35 cycles of 98°C for 30 s, 60°C for 30 s, and 72°C for 30 s. The absence of DNA contamination was confirmed by RT-PCR without reverse transcriptase. Quantitative RT-PCR (qRT-PCR) was performed using the Applied Biosystems 7300 Real-Time PCR System and SYBR green PCR master mix (Applied Biosystems) according to the supplier's recommendations. The reaction conditions were as follows: 95°C for 10 min, followed by 40 cycles of 15 s at 98°C for denaturation and 1 min at 60°C for annealing and extension. A final dissociation stage was run to generate a melting curve and consequently verify the specificity of the amplification products. Gene expression was measured in triplicate and normalized to the mRNA level of the hrdB gene (kse_54060) using the relative standard curve method.
Overexpression and purification of rKsbC.
The ksbC gene was amplified by PCR using genomic DNA of K. setae as a template and the primer pair rKsbC-Fw/rKsbC-Re. A 657-bp ksbC-containing DNA fragment was digested with NdeI and BamHI and then cloned in pET-15b digested with the same enzymes, resulting in pLT426, which was verified by sequencing. An LB culture of E. coli BL21(DE3)/pLysS harboring pLT426 containing ampicillin (50 μg/ml) and chloramphenicol (25 μg/ml) was grown at 37°C, until the optical density at 600 nm reached 0.6, at which time 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added for induction. After an additional 3 h of cultivation, the cells were harvested by centrifugation, resuspended in buffer A [50 mM Tris-HCl (pH 8.0) containing 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, and 1 mM p-(amidinophenyl)methanesulfonyl fluoride hydrochloride], and then disrupted by sonication. After centrifugation (6,230 × g, 15 min, 4°C), the supernatant was directly applied to a His Microspin purification module (GE Healthcare Bio-Sciences). Proteins containing rKsbC were eluted with 500 mM imidazole (in buffer A) according to the manufacturer's recommendations. The concentration of protein was measured with a Bio-Rad protein assay kit using bovine plasma gamma globulin as a standard. The purity of rKsbC was analyzed by SDS-PAGE.
Gel shift assay.
The fragments E-1, E-2, and E-3 were amplified by PCR with the primer pairs E1-Fw/E1-Re, E2-Fw/E2-Re, and E3-Fw/E3-Re, respectively, and cloned into the HincII site of pUC19. The DNA probes were labeled by PCR using these plasmids as templates with an fluorescein isothiocyanate (FITC)-labeled M13-47 primer and RV primer. After FITC-labeled probe (7.5 ng) and rKsbC (1 μg) were mixed and incubated at 25°C for 10 min, the reaction mixture was separated at 4°C by electrophoresis. The conditions used for the detection of DNA retardation were as previously described (19), and labeled DNA fragments were detected using an FMBIO II Multi-View (Hitachi Software Engineering) or Typhoon 9210 variable mode imager (GE Healthcare).
RESULTS
Bioinformatic analysis of autoregulator-receptor homologues of K. setae NBRC 14216.
In the genome sequence of K. setae NBRC 14216, we previously found two putative butyrolactone-autoregulator receptor genes, ksbB (kse_01050t and kse_75690t; identical genes encoded in the left and right terminal inverted repeats) and ksbC (kse_44580), in addition to ksbA, which acts as a negative regulator of bafilomycin production (14). A phylogenetic tree of KsbB and KsbC with other autoregulator receptors, including KsbA and pseudoreceptor regulators in Streptomyces (see Fig. S2 in the supplemental material), indicated that the KsbA/B/C proteins are grouped in the same branch but are positioned at the outmost clade of the Streptomyces autoregulator receptors and do not belong to the clade of pseudoreceptor regulators in Streptomyces species. The two groups (autoregulator receptors and pseudoreceptor regulators) can be distinguished easily by their pI values: autoregulator receptors have pI values of around 5 (pI 5.1 for ArpA [27] and pI 5.1 for BarA [26]), whereas pseudoreceptor regulators show very basic pI values (pI of 10.0 for CprB [28]). The observation that KsbA, KsbB, and KsbC have pI values of 4.7, 5.4, and 5.6, respectively, is consistent with the results of phylogenetic analysis, implying that these three proteins are most likely to be a set of autoregulator receptors in K. setae. More detailed analysis of the amino acid sequences indicated that, although all of the KsbA/B/C proteins have the N-terminal helix-turn-helix DNA-binding domain and KsbA contains the residues Pro-117 and Trp-121, which are regarded as important for DNA binding and autoregulator binding, respectively (29, 34), KsbB has an Ala at the position of Pro-117 with the conserved Trp-121, whereas KsbC contains a Ser at the position of Trp-121 with the conserved Pro-117 (see Fig. S2 in the supplemental material). These findings suggested that KsbB and/or KsbC may be involved in the regulation of secondary metabolism, but most likely in a different manner from typical Streptomyces autoregulator receptors.
The flanking regions of ksbC consist of two putative regulatory genes, kse_44570 and kse_44590 (Fig. 1B), with the former being homologous to various pseudo-receptor regulators, including ScbR2 of S. coelicolor A3(2) (37% identity), and the latter being homologous to various response regulator proteins of a bacterial two-component signal transduction system, including JadR1 of Streptomyces venezuelae (63% identity). The upstream region of the kse_44590 has two putative autoregulator biosynthetic genes, ksbS4 (kse_44600) and ksbS5 (kse_44610), that encode proteins belonging to the AfsA family proteins (including AfsA, which plays a role in A-factor biosynthesis [15]) and the NAD-dependent epimerase/dehydratase family (including BarS2, which plays a role in virginiae butanolide (VB) production [22]), respectively. The ksbC locus ranging from kse_44570 to ksbS5 closely resembles that of the VB-dependent regulatory island of S. virginiae.
KsbC acts as an activator of bafilomycin production and a repressor of kitasetaline production.
To assess the role of ksbC in the regulation of secondary metabolism, the ksbC gene was disrupted by insertion of a kanamycin resistance gene, resulting in a ksbC disruptant (see Fig. S3 in the supplemental material). The bafilomycin production in the ksbC disruptant decreased to 52% of the wild-type levels at 5 days of cultivation (Fig. 2A). To confirm that the lowered bafilomycin production resulted solely from the ksbC disruption, the genome-integrated plasmid pLT436 containing an intact copy of ksbC and its upstream region was reintroduced into the ksbC disruptant (Fig. 2B and see Fig. S3 in the supplemental material). Although site-specific integration of an empty vector pSET152 (4) alone (ΔksbC/pSET152 versus ΔksbC/−, or WT/pSET152 versus WT) increased the bafilomycin production (286 to 283% compared to that of the parental strain) for an unknown reason, it is clear that the ksbC disruption resulted in 52% reduction of the bafilomycin production and the ksbC complement recovered the production to the level of the corresponding WT strain. These results confirmed that ksbC plays a positive role in the regulation of bafilomycin biosynthesis. Next, to investigate whether KsbC controls the production of other secondary metabolites, we examined the HPLC profiles of the methanol extract from the WT strain and the ksbC disruptant (Fig. 3). After 48 h of cultivation on solid medium, when the aerial mycelium started forming in the wild-type strain, several peaks (elution times of 26.5, 27.7, 28.7, and 33.5 min) from the ksbC disruptant were larger than those from the wild-type strain. This pattern continued throughout the cultivation period and was observed until 15 days of cultivation. The differences became most apparent at 72 h of cultivation when the wild-type strain was in the early stage of sporulation but the ksbC disruptant aborted aerial mycelium formation (see below). After purification, 15 mg of pure compound eluted at 27.7 min (indicated by a black arrow in Fig. 3) was obtained from 1.25 liters of agar culture. Based on the physicochemical evidence, this compound (named kitasetaline [compound 2]; Fig. 1A) was a novel β-carboline alkaloid (2), which has not been isolated from any natural sources. The change of the kitasetaline production, attributable to the ksbC disruption, was partially restored by the introduction of the intact ksbC gene (see Fig. S4 in the supplemental material). Thus, KsbC can be concluded to be a negative regulator for the initiation and the amount of kitasetaline production.
Fig 2.
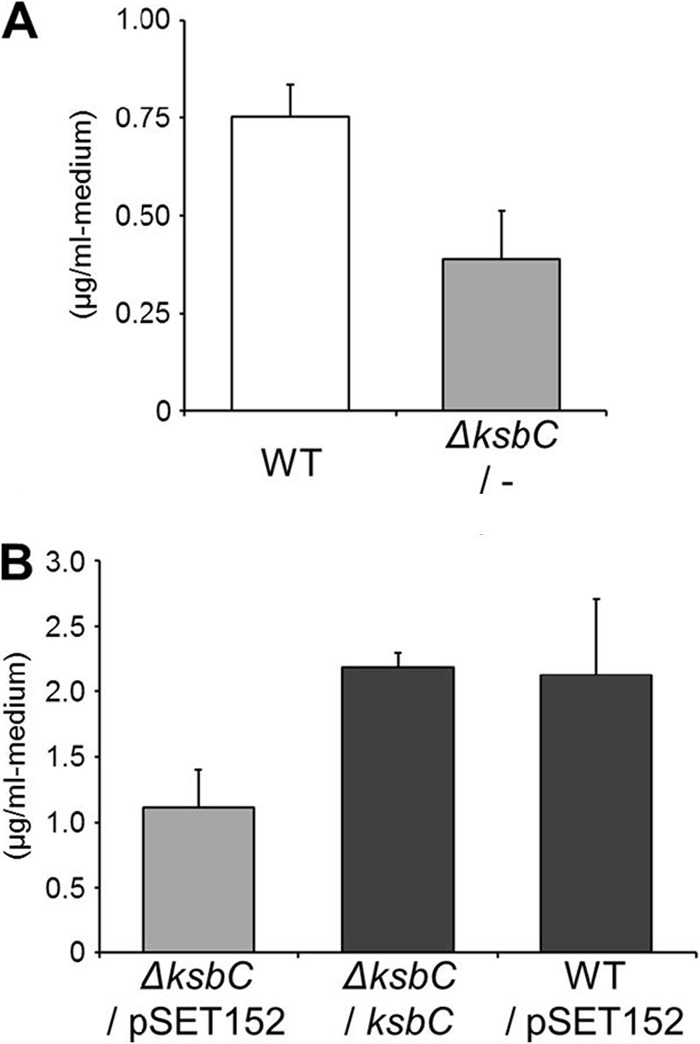
Bafilomycin production in the ksbC disruptant. The amount of bafilomycin was the total amount of bafilomycin derivatives (bafilomycin A1, B1, and C1). Error bars represent standard deviations from triplicate experiments. WT, wild-type strain; ΔksbC/−, ksbC disruptant; ΔksbC/pSET152, ΔksbC strain carrying pSET152; ΔksbC/ksbC, ksbC-complemented ΔksbC strain; WT/pSET152, the wild-type strain carrying pSET152. (A) Effect of ksbC disruption on bafilomycin production. (B) Complementation of the ksbC disruptant and effect of the pSET152 integration into the genome of K. setae on bafilomycin production.
Fig 3.

Kitasetaline production in the ksbC disruptant (A) and the wild-type strain (B). HPLC chromatograms of methanol extracts from each strain cultivated at the indicated time. mAU, milliabsorbance units at 276 nm. The peaks of kitasetaline and JBIR-133 are indicated as a black arrow and a gray arrow.
Heterologous expression and identification of the kitasetaline biosynthetic genes.
We recently initiated a screening program with a genomic BAC library of K. setae to discover cryptic natural compounds. Each BAC clone carrying a gene cluster for secondary metabolite biosynthesis estimated by bioinformatics analysis was introduced by protoplast transformation into a large-deletion mutant of S. avermitilis, SUKA22, a model host for heterologous expression of secondary metabolism, and their production profiles were explored by HPLC/DAD analysis. In the course of our extensive search for secondary metabolites, S. avermitilis SUKA22 carrying pKU503facP2-L12 exhibited a couple of peaks on ODS-HPLC analysis, which showed the characteristic absorptions of a β-carboline chromophore. As expected, no corresponding peak was observed in SUKA22 carrying pKU503 (empty vector) as a negative control. From 100 ml of culture of the SUKA22 carrying pKU503facP2-L12, two major components, kitasetaline (compound 2) and JBIR-133 (compound 3), and a minor component, JBIR-134 (compound 4) were detected and purified. The UV spectra of compounds 3 and 4 well agreed with the characteristic absorptions of a β-carboline chromophore at 238, 271, 305, and 379 nm, which were the same as those of kitasetaline (compound 2). The IR absorptions (1,708 and 1,701 cm−1, respectively) suggested the presence of carboxyl groups. The structure of compound 3 was mainly established by the analyses of the series of NMR spectra such as DQF-COSY, HSQC, and CT-HMBC. Eleven aromatic carbons (C-1 to C-13) were assignable to the β-carboline chromophore. The remaining substructure was elucidated as follows. The 1H-1H coupling between two methylene protons H2-14 (δH 3.39) and H2-15 (δH 2.91), which in turn 1H-13C long-range coupled to a nitrogen-bonded aromatic quaternary carbon C-1 (δC 144.0) and a carbonyl carbon 15-COOH (δC 174.2) established that a propionic acid moiety substituted at C-1. The 1H-13C long-range correlation from an aromatic proton H-4 (δH 8.76) to a carbonyl carbon C-16 (δC 166.8), together with the molecular formula indicated that the COOH group was attached to an aromatic quaternary carbon C-3 (δC 135.3). These results identified the structure of compound 3 as 1-(2-carboxyethyl)-β-carboline-3-carboxylic acid (Fig. 4A). The one-dimensional (1D) and 2D NMR data of a minor component (compound 4) led us to determine the substructure, β-carboline-3-carboxylic acid moiety, as the same as that of compound 3. The remaining partial structure was determined as follows. The 1H-1H coupling between methylene protons H2-14 (δH 3.17) and methyl protons H3-15 (δH 1.38), which in turn 1H-13C long-range coupled to a nitrogen-bonded aromatic quaternary carbon C-1 (δC 146.7) established an ethyl moiety substituted at C-1. Thus, the structure of compound 4 was determined as 1-ethyl-β-carboline-3-carboxylic acid (Fig. 4B). These two β-carboline alkaloids compounds 3 and 4 have not been isolated from any natural sources. Comparison of the UV/visible spectra and the retention time of JBIR-133 (compound 3) to those of the production profile of the K. setae ksbC mutant revealed that the ksbC disruption resulted in the abundant production of JBIR-133 (elution time at 26.5 min, indicated by a gray arrow in Fig. 3).
To identify genes responsible for the biosynthesis of kitasetaline and JBIR-133, a series of pKU492Aaac(3)IV containing shortened fragments from pKU503facP2-L12 was constructed (Fig. 5A), yielding pLT437-pLT440, which was then introduced into S. avermitilis SUKA22. After fermentation on solid medium, methanol extracts were subjected to HPLC analysis (Fig. 5B). SUKA22 carrying pLT437 (harboring eight genes, kse_70630 to kse_70700) produced kitasetaline as a minor metabolite and JBIR-133 as a major metabolite (Fig. 5B, trace I). However, deletion of kse_70700 (as in pLT438) led to a remarkable decrease of JBIR-133 production and no production of kitasetaline (Fig. 5B, trace II). Although SUKA22 carrying pLT439 still showed JBIR-133 production at a yield comparable to that of SUKA22 carrying pLT438 (Fig. 5B, trace III), deletion of three genes (kse_70630 to kse_70650), probably forming a single transcriptional unit (see below), failed to produce JBIR-133, which is similar to the production profile of SUKA22 carrying pKU492Aaac(3)IV as a negative control [Fig. 5B, traces IV and pKU492Aaac(3)IV]. These experimental evidences suggest that kse_70630, kse_70640, and kse_70650 would be mainly concerned with the formation of the β-carboline moiety in the biosynthesis of kitasetaline and its derivative, JBIR-133.
Fig 5.
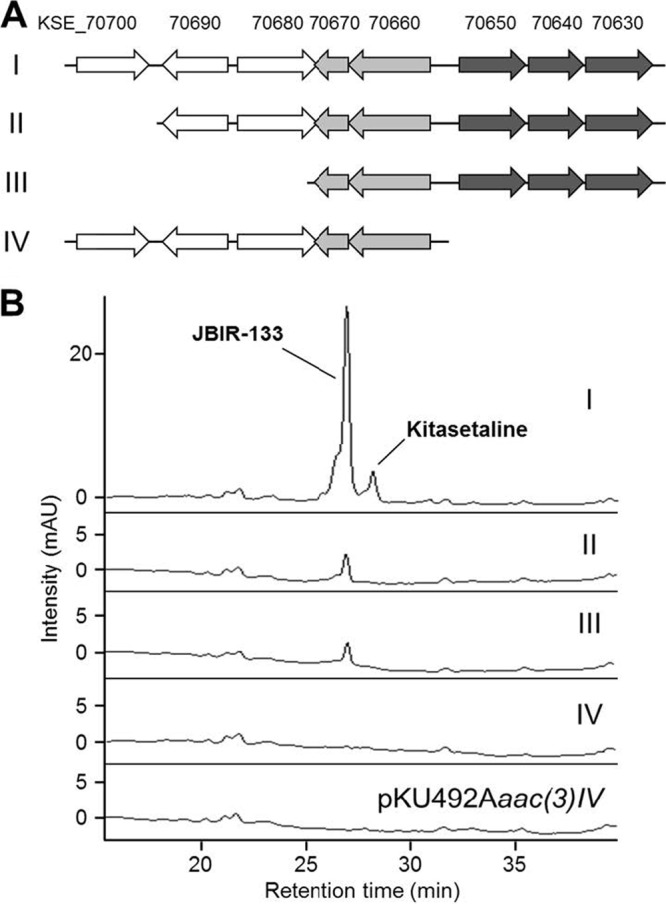
Heterologous expression of the β-carboline gene cluster in S. avermitilis SUKA22 and determination of the minimal gene cluster. (A) Schematic representation of the fragments used in this experiment. I, pLT437 containing genes from kse_70630 to kse_70700; II, pLT438 containing genes from kse_70630 to kse_70690; III, pLT439 containing genes from kse_70630 to kse_70670; IV, pLT440 containing genes from kse_70660 to kse_70700. The genes indicated by arrows are completely included in each fragment. (B) HPLC analyses (276 nm) of the metabolite profiles of S. avermitilis SUKA22 carrying pLT437 (I), pLT438 (II), pLT439 (III), pLT440 (IV), and pKU492Aaac(3)IV as an empty vector, respectively. JBIR-133 and kitasetaline are eluted at 26.7 and 28.0 min, respectively.
Organization of genes involving in β-carboline biosynthesis.
Three genes (kse_70630, kse_70640, and kse_70650 for the biosynthesis of β-carboline structure) are divergently located from the downstream kse_70620 encoding putative type I polyketide synthase and the upstream pabB (kse_70660) encoding putative p-aminobenzoate synthase. Interestingly, with narrow intergenic regions (48 bp between kse_70630 and kse_70640 and 74 bp between kse_70640 and kse_70650), the three genes seem to transcribe as a polycistron. The predicted 392-amino-acid (aa) gene product of kse_70630 shows 37% identity and 57% positive matches to CypX of Ktedonobacter racemifer DSM 44963 (ZP_06966380), 36% identity and 55% positive matches to CypX of Paenibacillus sp. strain Y412MC10 (YP_00324629), and 39% identity and 55% positive matches to Orf1, a probable cytochrome P450 monooxygenase, of Streptomyces eurythermus (ABV49609). The predicted 317-aa KSE_70640 shows moderate similarity to gene products of Gram-negative bacteria, Burkholderia and Agrobacterium species; Bxe_A1481 of Burkholderia xenovorans LB400 (YP_559525; 48% identity and 69% positive matches), BCh11DRAFT_2502 of Burkholderia sp. strain Ch1-1 (ZP_06841236; 48% identity and 69% positive matches), pRi2659_p014 of Agrobacterium rhizogenes (YP_001960988; 45% identity and 63% positive matches), cucumopine synthase of Agrobacterium rhizogenes (BAB13344; 45% identity and 63% positive matches), hypothetical protein of Agrobacterium rhizogenes (ABS11829; 45% identity and 64% positive matches), and cucumopine synthase of Agrobacterium rhizogenes (CAB65900; 42% identity and 59% positive matches). Among these proteins, two proteins (BAB13344 and CAB65900) have been characterized as cucumopine synthase that catalyzes a coupling between l-histidine and α-ketoglutaric acid by using NADH as a cofactor (35). However, these proteins have no apparent functional motifs. The predicted 377-aa gene product of kse_70650 belongs to an FAD-dependent oxidoreductase family with the highest similarity to Caci_2536 of Catenulispora acidiphila DSM 44928 (31% identity and 49% positive matches). There are only a few actinomycetes homologues of the KSE_70640 and KSE_70650 proteins with low similarity, and no locus resembling the region between kse_70650 and kse_70630 is found in the current database of the microbial genome.
Regulation of bafilomycin and β-carboline biosynthesis.
To elucidate the function of KsbC in transcriptional regulation of the gene clusters for bafilomycin and kitasetaline biosynthesis, the transcriptional levels were compared between the wild-type and the ksbC disruptant by semiquantitative RT-PCR analysis or quantitative analysis as described in Materials and Methods. The biosynthetic gene cluster for bafilomycin is predicted to be composed of 18 genes from kse_73410 to kse_73580, and the cluster includes bfmR, which is an essential pathway-specific positive regulator for the bafilomycin biosynthesis (H. Ikeda, unpublished data). Surprisingly, the transcriptional levels of bfmR, kse_70640, and kse_70630 in the ksbC disruptant were nearly identical to those of the wild-type strain (data not shown). These results indicated that KsbC has a pleiotropic effect on the production of bafilomycin and kitasetaline without direct transcriptional control of these biosynthetic genes.
Defect of aerial mycelium formation in the ksbC mutant.
Although KsbA did not participate in the control of morphological development under the conditions used here (9), a few autoregulator receptors in Streptomyces spp. control not only antibiotic production but also morphological development (10, 27, 33). To investigate whether KsbC is involved in the morphological control of K. setae, we carefully examined the morphological characteristics of the ksbC disruptant on two different solid media. On ISP medium 2, the wild-type strain and the ksbC disruptant were found to have identical phenotypes (data not shown). On ISP medium 4, however, a defect in the aerial mycelium was observed for the ksbC mutant and the ksbC mutant carrying pSET152 at 5 days of cultivation when the wild-type strain and the ΔksbC/ksbC strain showed aerial mycelium formation with an abundant amount of spores (see Fig. S5 in the supplemental material). After 15 days of cultivation, the ksbC disruptants were still defective in the formation of the aerial mycelium (data not shown), indicating that KsbC is involved in morphological development, although the exact cascade or mechanism is at present unknown and will require detailed analysis in the future.
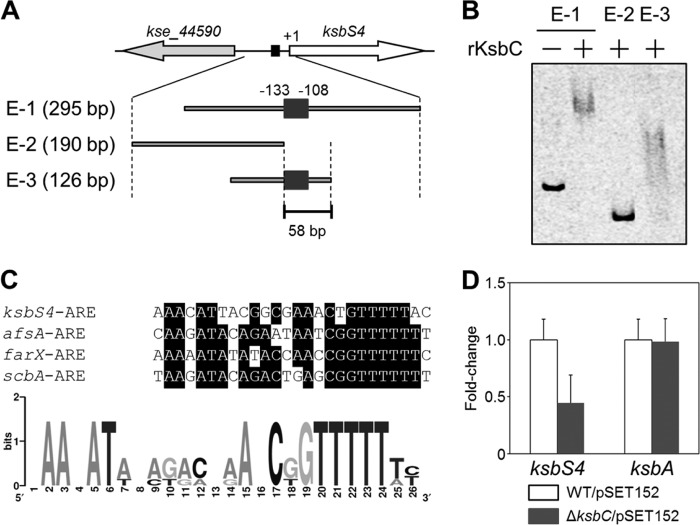
Binding of KsbC to the intergenic region of kse_44590 and ksbS4.
Many autoregulator receptors typically bind to ARE sequences that are found in the promoter region of the target genes (10). A candidate 26-bp ARE-like sequence is found in the 108-bp upstream region of the ksbS4 gene (Fig. 6A), suggesting that KsbC may bind to the ksbS4 upstream region. To examine whether KsbC has DNA-binding activity toward the ksbS4 upstream region, we performed a gel shift assay using a purified N-terminal His-tagged KsbC and DNA fragments encompassing the ARE-like sequence (Fig. 6B). Shift signals were only detected when this putative ARE sequence was included in the DNA probes, suggesting that KsbC probably recognizes the ARE-sequence upstream of ksbS4. As shown in Fig. 6C, this sequence, designated ksbS4-ARE, is highly similar to other ARE sequences that are present in the putative promoter region of the afsA family genes, and two conserved sequences [5′-AAXAT(A/T)-3′ and 5′-C(G/T)GTTTTT(T/A)-3′] emerged by the logo representation analysis. To investigate possible regulation of KsbC on the expression of ksbS4 and/or kse_44590, we compared the transcriptional levels of these two genes between the wild-type strain and the ksbC disruptant. The mRNA level of ksbS4 decreased significantly in the ksbC disruptant (45%) compared to that in the wild-type strain (Fig. 6D), whereas no remarkable differences in kse_44590 transcription were observed (data not shown). These results, together with the results of the gel shift assays, suggested that KsbC has a positive role on the transcriptional control of ksbS4 through the binding to ksbS4-ARE in the ksbS4 upstream region.
Fig 6.
Binding of KsbC to the intergenic region between kse_44590 and ksbS4 genes. (A) Location of probes used for the gel shift assay. The probes E-1 to E-3 used in the present study are shown, and their lengths are indicated on the left. The location of the putative 26-bp ARE sequence, situated 108 bp upstream of the ksbS4 gene, is indicated by a dark gray box. (B) Gel shift assay for the binding of purified His-tagged KsbC (rKsbC) to probes containing a plausible KsbC-binding site. (C) Comparison of the putative KsbC-binding sequence (ksbS4-ARE) with the ARE sequences located upstream of the afsA family genes (upper panel) and a logo-representation for conserved nucleotides using computational methods (lower panel). The genes in the list are afsA of S. griseus, farX of S. lavendulae FRI-5, and scbA of S. coelicolor A3(2). The consensus nucleotides in the ARE sequences are highlighted by black boxes. The logo representation was created using WebLogo analysis (http://weblogo.berkeley.edu/logo.cgi) based on the 26-bp ARE sequences. The relative sizes of the letters indicate their likelihood at the particular position. (D) Gene expression analysis of the ksbS4 and ksbA genes by qRT-PCR. WT/pSET152, the wild-type strain carrying pSET152; ΔksbC/pSET152, the ΔksbC strain carrying pSET152. The fold change is relative to the expression of each gene in the wild-type strain carrying pSET152. Error bars, SD from triplicate experiments.
DISCUSSION
The γ-butyrolactone autoregulator receptors and their homologues play a pivotal role in the production of secondary metabolites in many Streptomyces species, and sometimes on morphological development. In most cases, a single autoregulator receptor is found in one Streptomyces strain, and it negatively regulates the production of a specific secondary metabolite by controlling the proximally situated biosynthetic gene cluster by adjusting the transcriptional level of the in-cluster Streptomyces antibiotic regulatory protein (SARP) family gene (1, 32). We previously provided the first report of an autoregulator receptor in non-Streptomyces actinomycetes that K. setae has an autoregulator signaling cascade that uses KsbA as a negative regulator of bafilomycin production (9). In the present study, we verified that KsbC is the second autoregulator receptor of K. setae that has diverse regulatory functions on secondary metabolism (activation of bafilomycin and repression of kitasetaline and JBIR-133) in addition to a positive role on aerial mycelium formation that is not influenced by the first receptor KsbA. Two autoregulator receptors, KsbC and KsbA, have opposite properties for the regulation of bafilomycin production as the same target. The ksbC locus and the ksbA locus, neither of which have any biosynthetic gene cluster for secondary metabolites in the flanking regions, are 3.4 and 1.8 Mb, respectively, away from the biosynthetic gene cluster for bafilomycin, indicating that bafilomycin biosynthesis is controlled by two different autoregulator receptors encoded at a position distal to the gene cluster for bafilomycin biosynthesis. Transcriptional analysis demonstrated that KsbC does not control transcription of bfmR, which encodes a pathway-specific regulator for bafilomycin biosynthesis, and there appears to be no cross talk between the KsbC and KsbA regulatory pathways for bafilomycin production (no difference of ksbA transcription in the ksbC disruptant [Fig. 6D] or ksbC transcription in the ksbA disruptant [data not shown]). These findings indicated that, independent from the KsbA pathway, the KsbC pathway controls bafilomycin biosynthesis with no apparent control of BfmR.
With respect to the production of β-carboline alkaloids, we have identified plausible biosynthetic genes (kse_70630 to kse_70650) for the formation of the β-carboline structure, which are 3.1 and 1.5 Mb, respectively, away from the ksbC and ksbA loci. Unlike the case for the bafilomycin production, in the ksbA disruptant, the production level of kitasetaline and its derivatives, including JBIR-133, was at the same level as in the wild-type strain (data not shown), indicating that only KsbC is involved in the regulation of the β-carboline alkaloids biosynthesis. Because the substrates/precursors for the β-carboline alkaloids biosynthesis are distinct from those for the bafilomycins biosynthesis, precursor competition is unlikely to be the underlying mechanism for the increase in β-carboline alkaloid production and the decrease in bafilomycin production in the ksbC disruptant, suggesting that KsbC is a specific negative regulator for β-carboline production without direct transcriptional control on the β-carboline biosynthetic genes. Thus, KsbC has bidirectional characteristics for secondary metabolism, as an activator in one pathway and a repressor in another pathway, together with the regulation of morphological development. There have been few reports of autoregulator receptors exerting bidirectional characteristics like KsbC, except for FarA of the IM-2 signaling cascade in Streptomyces lavendulae FRI-5 (20).
Disruption of the ksbC gene led to the discovery of new compounds such as kitasetaline and JBIR-133, together with the finding of a novel biosynthetic route for microbial β-carboline alkaloid, with the aid of a heterologous expression system in the genome-minimized S. avermitilis strain. Genomes of microorganisms, especially actinomycetes, including Streptomyces species and K. setae, have numerous cryptic biosynthetic pathways in which genes appear to be expressed poorly, if at all, under the given cultivation condition. From the viewpoint of identifying new compounds, the K. setae wild-type strain produces a negligible amount of kitasetaline and JBIR-133, while the ksbC disruptant shows precocious and abundant production of kitasetaline and JBIR-133, enabling identification of the structure. There have been only a few reports in which the deregulation of a biosynthetic pathway through the alteration of actinomycetes regulatory genes allows sufficient materials, especially novel natural compounds, to be purified for structure elucidation. In S. coelicolor A3(2), Gottelt et al. disrupted a pseudoreceptor regulator gene scbR2 (see Fig. S2 in the supplemental material) located within a predicted biosynthetic gene cluster (cpk) and found a novel antibacterial activity of the unidentified product generated by the cpk gene cluster (12). In S. ambofaciens, Bunet et al. deregulated the pseudoreceptor regulator gene alpW (see Fig. S2 in the supplemental material) by gene disruption and identified that the alp gene cluster is responsible for the production of kinamycins (6), although in both cases the targeted regulatory genes are located in a proximal silent gene cluster and the linkage between the new product and the biosynthetic gene cluster seems readily available. As in the case of KsbC, genetic manipulation of an autoregulator receptor (or pseudoreceptor regulator) that has no proximal gene cluster for secondary metabolites will be the next stage in the discovery of novel natural products and new biosynthetic pathways.
β-Carboline alkaloids are a large group of natural and synthetic indole alkaloids and have diverse biological activities (including potent antitumor, antiviral, antimicrobial, and antiparasitic activities) (8). They are widely distributed in various plants, whereas only a few reports are available on the β-carboline compounds from bacterial origins (11). With respect to the biosynthetic pathway of β-carboline alkaloids, strictosidine synthase (STR1) of Rauvolfia serpentina (STR1; accession no. P68175), which catalyzes a coupling reaction between tryptamine and secologanin by way of a stereoselective Pictet-Spengler reaction, is the key enzyme in the biosynthesis of 2,000 indole alkaloids in plants. Our extensive search of the actinobacteria genome database revealed that similar homologues are frequently found to be embedded on the genome of Noca_2860 of Nocardioides sp. strain JS614 (YP_924049; 48% identity and 69% positive matches), GOARA_019_00250 of Gordonia araii NBRC 100433 (ZP_09210674; 30% identity and 46% positive matches), SSOG_07704 of Streptomyces hygroscopicus ATCC 53653 (ZP_07299621; 36% identity and 52% positive matches), SSMG_05546 of Streptomyces sp. strain AA4 (ZP_07281506; 28% identity and 42% positive matches), SSEG_06081 of Streptomyces sviceus ATCC 29083 (ZP_06915048; 28% identity and 43% positive matches), and SSQG_00525 of Streptomyces viridochromogenes DSM 40736 (ZP_07301638; 28% identity and 42% positive matches), but K. setae lacks such an orthologue, suggesting that simple computational analysis based on the sequence homology has not been sufficient to find a biosynthetic route of β-carboline compounds in K. setae. Here, we successfully identified three biosynthetic genes involved in the biosynthesis of kitasetaline and its derivatives by heterologous expression, although we cannot eliminate the possibility that PabA/B proteins (p-aminobenzoate synthases) might be involved in the biosynthesis in addition to these three proteins. The predicted gene product of kse_70640 with no identifiable motifs resembles cucumopine and mikimopine synthases from Agrobacterium rhizogenes. Cucumopine is a stereoisomer of mikimopine, which is synthesized from l-histidine and α-ketoglutaric acid via formation of Schiff base and its iminium cation by coupling α-amino residue of l-histidine and β-carbonyl residue of α-ketoglutarate (35). Therefore, the KSE_70640 protein presumably catalyzes a coupling between l-tryptophan and succinate semialdehyde, and the cyclization of the side chain by nucleophilic addition of aromatic ring to iminium cation generated through the Pictet-Spengler reaction to form a tryptoline derivative [1-(2-carboxyethyl)-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxylic acid]. Currently, an in vitro experiment is under way to further clarify the detailed reaction mechanism. Moreover, it is probable that the KSE_70650 protein (putative FAD-dependent oxidoreductase) is involved in the oxidation of piperidine ring in the tryptoline derivative to generate β-carboline derivatives. The KSE_70630 protein (putative P450 monooxygenase) would be concerned with the formation of kitasetaline. A significant decrease of the production of kitasetaline and JBIR-133 was observed by deletion of kse_70700 encoding putative major facilitator superfamily transporter, which often functions as an efflux pump of low molecular-weight compounds. Thus, the KSE_70700 protein is probably a transmembrane-type transporter for the export of kitasetaline and JBIR-133 from the cell.
AfsA family proteins are the principal enzymes for biosynthesis of γ-butyrolactone autoregulators in Streptomyces species (13, 15, 23). The single afsA family genes afsA, scbA, and avaA (17) are present on the genomes of S. griseus, S. coelicolor A3(2), and S. avermitilis, respectively. The flanking region of scbA has scbR (a receptor of an SCB1 autoregulator), and that of avaA has avaL1/L2 (receptors of an unidentified molecule), whereas the afsA locus is 3.9 Mb away from the locus of arpA (A-factor receptor gene). However, there have been many reports of afsA family genes and receptor genes being clustered at the same locus in various Streptomyces species (25). Interestingly, the genome of K. setae has four copies of afsA family genes, i.e., ksbS4 in the ksbC locus, ksbS2L/R (kse_01060t/kse_75680t) in the two ksbB loci, and ksbS3 (kse_22970) that is not accompanied by any autoregulator receptor gene. Together with the finding of two independent signaling pathways of KsbC and KsbA shown in the present study, these findings indicated that K. setae has multiple autoregulator signaling cascades for secondary metabolism. As shown in Fig. 6C, the ARE sequences located in the upstream region of afsA family genes are well aligned, suggesting that regulatory mechanisms for autoregulator biosynthesis might be conserved in the autoregulator signaling cascade. The observation that the ksbS4 transcription is downregulated in the ksbC disruptant is similar to the finding in the farA mutant of S. lavendulae FRI-5, in which farX, an afsA family gene for IM-2 biosynthesis, is not transcribed throughout the cultivation (18). These phenomena suggest that fine-tuning systems to control autoregulator production are also conserved beyond the genus in actinomycetes.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Harashima and M. Sugiyama for assistance with the qRT-PCR analysis.
This study was supported in part by a grant for the Joint Program in the Field of Biotechnology from the Japan Society for the Promotion of Science (JSPS), the National Research Council of Thailand, and the National Science and Technology Development Agency of Thailand to T.N.; by Grants-in-Aid for Scientific Research (B) (no. 21360404 and no. 24310157) and a Grant-in-Aid for Challenging Exploratory Research (no. 24651243) from the JSPS to T.N. and S.K.; by a Grant-in-Aid for Young Scientists (B) (no. 24780071) from JSPS to S.K.; by a Grant-in-Aid for Scientific Research on Innovative Areas (no. 22108006) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan to H.I.; by a Grant-in-Aid for Scientific Research from New Energy and Industrial Technology Development Organization to K.S.-Y. and H.I.; and by a scholarship from MEXT of Japan to A.A.
Footnotes
Published ahead of print 7 September 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Aigle B, Pang X, Decaris B, Leblond P. 2005. Involvement of AlpV, a new member of the Streptomyces antibiotic regulatory protein family, in regulation of the duplicated type II polyketide synthase alp gene cluster in Streptomyces ambofaciens. J. Bacteriol. 187:2491–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aroonsri A, Kitani S, Ikeda H, Nihira T. 2012. Kitasetaline, a novel β-carboline alkaloid from Kitasatospora setae NBRC 14216T. J. Biosci. Bioeng. 114:56–58 [DOI] [PubMed] [Google Scholar]
- 3. Bibb MJ. 2005. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8:208–215 [DOI] [PubMed] [Google Scholar]
- 4. Bierman M, et al. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49 [DOI] [PubMed] [Google Scholar]
- 5. Bowman EJ, Siebers A, Altendorf K. 1988. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc. Natl. Acad. Sci. U. S. A. 85:7972–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bunet R, et al. 2010. Characterization and manipulation of the pathway-specific late regulator AlpW reveals Streptomyces ambofaciens as a new producer of kinamycins. J. Bacteriol. 193:1142–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cane DE, He X, Kobayashi S, Omura S, Ikeda H. 2006. Geosmin biosynthesis in Streptomyces avermitilis: molecular cloning, expression, and mechanistic study of the germacradienol/geosmin synthase. J. Antibiot. 59:471–479 [DOI] [PubMed] [Google Scholar]
- 8. Cao R, Peng W, Wang Z, Xu A. 2007. β-Carboline alkaloids: biochemical and pharmacological functions. Curr. Med. Chem. 14:479–500 [DOI] [PubMed] [Google Scholar]
- 9. Choi S-U, Lee C-K, Hwang Y-I, Kinoshita H, Nihira T. 2004. Cloning and functional analysis by gene disruption of a gene encoding a γ-butyrolactone autoregulator receptor from Kitasatospora setae. J. Bacteriol. 186:3423–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Folcher M, et al. 2001. Pleiotropic functions of a Streptomyces pristinaespiralis autoregulator receptor in development, antibiotic biosynthesis, and expression of a superoxide dismutase. J. Biol. Chem. 276:44297–44306 [DOI] [PubMed] [Google Scholar]
- 11. Fotso S, et al. 2008. Furan oligomers and β-carbolines from terrestrial streptomycetes. J. Nat. Prod. 71:1630–1633 [DOI] [PubMed] [Google Scholar]
- 12. Gottelt M, Kol S, Gomez-Escribano JP, Bibb M, Takano E. 2010. Deletion of a regulatory gene within the cpk gene cluster reveals novel antibacterial activity in Streptomyces coelicolor A3(2). Microbiology 156:2343–2353 [DOI] [PubMed] [Google Scholar]
- 13. Hsiao NH, et al. 2007. ScbA from Streptomyces coelicolor A3(2) has homology to fatty acid synthases and is able to synthesize γ-butyrolactones. Microbiology 153:1394–1404 [DOI] [PubMed] [Google Scholar]
- 14. Ichikawa N, et al. 2010. Genome sequence of Kitasatospora setae NBRC 14216T: an evolutionary snapshot of the family Streptomycetaceae. DNA Res. 17:393–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kato JY, Funa N, Watanabe H, Ohnishi Y, Horinouchi S. 2007. Biosynthesis of γ-butyrolactone autoregulators that switch on secondary metabolism and morphological development in Streptomyces. Proc. Natl. Acad. Sci. U. S. A. 104:2378–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, England [Google Scholar]
- 17. Kitani S, et al. 2011. Avenolide, a Streptomyces hormone controlling antibiotic production in Streptomyces avermitilis. Proc. Natl. Acad. Sci. U. S. A. 108:16410–16415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kitani S, Doi M, Shimizu T, Maeda A, Nihira T. 2010. Control of secondary metabolism by farX, which is involved in the γ-butyrolactone biosynthesis of Streptomyces lavendulae FRI-5. Arch. Microbiol. 192:211–220 [DOI] [PubMed] [Google Scholar]
- 19. Kitani S, et al. 2008. Identification of genes involved in the butyrolactone autoregulator cascade that modulates secondary metabolism in Streptomyces lavendulae FRI-5. Gene 425:9–16 [DOI] [PubMed] [Google Scholar]
- 20. Kitani S, Yamada Y, Nihira T. 2001. Gene replacement analysis of the butyrolactone autoregulator receptor (FarA) reveals that FarA acts as a novel regulator in secondary metabolism of Streptomyces lavendulae FRI-5. J. Bacteriol. 183:4357–4363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H. 2010. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl. Acad. Sci. U. S. A. 107:2646–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee YJ, Kitani S, Kinoshita H, Nihira T. 2008. Identification by gene deletion analysis of barS2, a gene involved in the biosynthesis of γ-butyrolactone autoregulator in Streptomyces virginiae. Arch. Microbiol. 189:367–374 [DOI] [PubMed] [Google Scholar]
- 23. Lee YJ, Kitani S, Nihira T. 2010. Null mutation analysis of an afsA-family gene, barX, that is involved in biosynthesis of the γ-butyrolactone autoregulator in Streptomyces virginiae. Microbiology 156:206–210 [DOI] [PubMed] [Google Scholar]
- 24. Miyamoto KT, Kitani S, Komatsu M, Ikeda H, Nihira T. 2011. The autoregulator receptor homologue AvaR3 plays a regulatory role in antibiotic production, mycelial aggregation and colony development of Streptomyces avermitilis. Microbiology 157:2266–2275 [DOI] [PubMed] [Google Scholar]
- 25. Nishida H, Ohnishi Y, Beppu T, Horinouchi S. 2007. Evolution of γ-butyrolactone synthases and receptors in Streptomyces. Environ. Microbiol. 9:1986–1994 [DOI] [PubMed] [Google Scholar]
- 26. Okamoto S, Nakamura K, Nihira T, Yamada Y. 1995. Virginiae butanolide binding protein from Streptomyces virginiae: evidence that VbrA is not the virginiae butanolide binding protein and reidentification of the true binding protein. J. Biol. Chem. 270:12319–12326 [DOI] [PubMed] [Google Scholar]
- 27. Onaka H, et al. 1995. Cloning and characterization of the A-factor receptor gene from Streptomyces griseus. J. Bacteriol. 177:6083–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Onaka H, Nakagawa T, Horinouchi S. 1998. Involvement of two A-factor receptor homologues in Streptomyces coelicolor A3(2) in the regulation of secondary metabolism and morphogenesis. Mol. Microbiol. 28:743–753 [DOI] [PubMed] [Google Scholar]
- 29. Onaka H, Sugiyama M, Horinouchi S. 1997. A mutation at proline-115 in the A-factor receptor protein of Streptomyces griseus abolishes DNA-binding ability but not ligand-binding ability. J. Bacteriol. 179:2748–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Otoguro K, Nakagawa A, Omura S. 1988. Setamycin, a 16-membered macrolide antibiotic: identification and nematocidal activity. J. Antibiot. 41:250–252 [DOI] [PubMed] [Google Scholar]
- 31. Paget MS, Chamberlin L, Atrih A, Foster SJ, Buttner MJ. 1999. Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stratigopoulos G, Cundliffe E. 2002. Expression analysis of the tylosin-biosynthetic gene cluster: pivotal regulatory role of the tylQ product. Chem. Biol. 9:71–78 [DOI] [PubMed] [Google Scholar]
- 33. Stratigopoulos G, Gandecha AR, Cundliffe E. 2002. Regulation of tylosin production and morphological differentiation in Streptomyces fradiae by TylP, a deduced γ-butyrolactone receptor. Mol. Microbiol. 45:735–744 [DOI] [PubMed] [Google Scholar]
- 34. Sugiyama M, Onaka H, Nakagawa T, Horinouchi S. 1998. Site-directed mutagenesis of the A-factor receptor protein: Val-41 important for DNA-binding and Trp-119 important for ligand-binding. Gene 222:133–144 [DOI] [PubMed] [Google Scholar]
- 35. Suzuki K, Tanaka N, Kamada H, Yamashita I. 2001. Mikimopine synthase (mis) gene on pRi1724. Gene 263:49–58 [DOI] [PubMed] [Google Scholar]
- 36. Takano E. 2006. γ-Butyrolactone: Streptomyces signaling molecules regulating antibiotic production and differentiation. Curr. Opin. Microbiol. 9:287–294 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.