Abstract
Although flies are important vectors of food-borne pathogens, there is little information to accurately assess the food-related health risk of the presence of individual flies, especially in urban areas. This study quantifies the prevalence and the relative risk of food-borne pathogens associated with the body surfaces and guts of individual wild flies. One hundred flies were collected from the dumpsters of 10 randomly selected urban restaurants. Flies were identified using taxonomic keys before being individually dissected. Cronobacter spp., Salmonella spp., and Listeria monocytogenes were detected using the PCR-based BAX system Q7. Positive samples were confirmed by culture on specific media and through PCR amplification and sequencing or ribotyping. Among collected flies were the housefly, Musca domestica (47%), the blowflies, Lucilia cuprina (33%) and Lucilia sericata (14%), and others (6%). Cronobacter species were detected in 14% of flies, including C. sakazakii, C. turicensis, and C. universalis, leading to the proposal of flies as a natural reservoir of this food-borne pathogen. Six percent of flies carried Salmonella enterica, including the serovars Poona, Hadar, Schwarzengrund, Senftenberg, and Brackenridge. L. monocytogenes was detected in 3% of flies. Overall, the prevalence of food-borne pathogens was three times greater in the guts than on the body surfaces of the flies. The relative risk of flies carrying any of the three pathogens was associated with the type of pathogen, the body part of the fly, and the ambient temperature. These data enhance the ability to predict the microbiological risk associated with the presence of individual flies in food and food facilities.
INTRODUCTION
Food-borne illnesses continue to be a serious public health problem. The Centers for Disease Control and Prevention (CDC) estimate that the consumption of contaminated food in the United States is the cause of illness in approximately one out of six people, leading to 128,000 hospitalizations and 3,000 deaths each year (13). These food-borne diseases are associated with 31 known pathogens (74) and with an assemblage of unspecified agents, including microbes, toxins, and other substances (73). The economic burden of these illnesses should not be underestimated. Recent assessments calculated the health care-related cost to be approximately $51 billion per year in the United States, including productivity losses and mortality (75).
There are several mechanisms whereby food products can become contaminated. Some examples include improper agricultural practices, such as the use of nonpotable water for irrigation of crops, the use of improperly sanitized manufacturing equipment and utensils, inappropriate storage and transportation temperatures, poor food handling practices, and lack of rodent and/or insect control, to name a few (26, 88). Insects have been recognized as playing an important role in the spread of food-related diseases as they can deliver viable pathogens either directly, through contact with food, or indirectly, through contact with food contact surfaces and utensils (91). Among insects, flies have long been implicated in the transmission of pathogens, contaminating food and water and causing some of the most devastating diseases affecting humans, such as typhoid fever (1) and cholera (52). Their association with decaying matter, garbage, feces, and other similar sources and their endophilic (entering buildings) and synanthropic (associated with humans) behaviors make them vectors of pathogens that cause enteric diseases, such as Salmonella spp. (67), Listeria monocytogenes (39), Campylobacter spp. (36), Escherichia coli O:157 (17), and the genus Cronobacter (formerly Enterobacter sakazakii) (60).
The indigenous bacterial community in the intestinal tracts of flies can be up to 10 times greater than the body surfaces (body, head, legs, and wings) (81). This indigenous microbiota has been shown to play an important role in the life cycle of flies and in their feeding, oviposition, and mating behaviors, possibly driving the evolution of new species (19, 79, 82). However, the role of food-borne pathogens on the body surfaces or in the intestinal tracts of flies is not totally understood. They can be opportunistic pathogens (89), but they can also establish symbiotic relationships with the host's commensal microbiota (56). Food-borne pathogens can remain in the intestines for a greater length of time than on the body surfaces (40, 71), where they are able to multiply (33) and colonize the fly's digestive tract (61), increasing the potential for dissemination.
Flies can be either mechanical or biological vectors of food-borne pathogens (22). As mechanical vectors, flies are vehicles of pathogens that do not develop or multiply in their body, contaminating food through contact with their body surfaces (57, 81). However, if the pathogen passes through the alimentary tract of the host and undergoes a stage of development or multiplication, the host is considered a biological vector (22, 64, 69). Since some food-borne pathogens have been shown to multiply in the intestinal tracts of the flies (33, 51, 61), flies can be considered biological vectors of these pathogens, contaminating food through regurgitation (vomit spots) and defecation (22, 64). Consequently, knowing if food-borne pathogens are located on the body surfaces or in the guts of an individual fly is important as this may have epidemiological implications. Unfortunately, most of the studies that have associated wild flies with food-borne pathogens have dealt with pooled groups of flies rather than individual specimens (27, 34, 39, 58, 67, 85), making food risk assessment on a per fly basis difficult. The few studies that have used single flies have macerated or homogenized the sample, making it impossible to differentiate if food-borne pathogens were on the body surfaces or in the guts of collected flies (43, 60).
From the 108 known families of flies, 29 are believed to be associated with the transmission of pathogens and even fewer with the transmission of food-borne pathogens (65). Four of these families, containing 12 fly species, were included in the list of pests that exhibit attributes contributing to the spread of food-borne pathogens. Such attributes include contacting potential sources of pathogens and endophilic, synanthropic, and communicative behaviors. Examples of pests exhibiting those attributes were published in the revised U.S. Food and Drug Administration (FDA) “filth strategy” (66) and have become known as the “Dirty 22.” Evidence of the presence of any of these pests in food, food processing areas, or food storage facilities is considered an indication of insanitation. Although flies are well documented for being a public health threat, and they comprise the majority of pests included in the “Dirty 22,” there is little information to accurately and quantitatively assess the food-related risk of the presence of an individual fly (65), especially in urban areas (42). Current food safety guidance lacks the scientific evidence required both to establish the prevalence of food-borne pathogens associated with these insects and to differentiate the species of flies that pose a factual threat for spreading food-borne pathogens (65).
The purpose of this study was to provide evidence for the prevalence of Cronobacter spp., Salmonella spp., and L. monocytogenes associated with the body surfaces and guts of individual flies collected from dumpsters outside restaurants in urban areas. Evidence for flies as a possible natural reservoir for Cronobacter spp. is also discussed. Additionally, the relative risk associated with a single fly transmitting these three food-borne pathogens was quantified. This information fills data gaps in the risk profile on the frequency of transmission of food-borne pathogens by different species of flies.
MATERIALS AND METHODS
Collection of flies.
A total of 100 flies were collected around the dumpsters of 10 randomly selected restaurants located in an 18-km radius from a centrally located point in the metropolitan area of Washington, DC. Each collection site was assigned a random number between 1 and 10. Flies were collected during the months of August and September 2011 between 10:00 a.m. and noon. The ambient temperature was recorded at the time of collection. Flies were collected individually using sterile entomological sweep nets. The nets were transferred to the lab in a cooler and placed at −20°C for 2 to 5 min until the flies were immobilized. All procedures, from collection to analysis, were performed on individual flies.
Identification and dissection of flies.
Using sterile forceps, each fly was transferred to 2-ml tubes containing 1 ml of buffered peptone water (BPW; Difco, Becton, Dickinson Company, Sparks, MD). Tubes were mixed gently by inversion for 2 min, allowing the whole body of the fly to be in contact with the medium. Flies were then removed from the BPW medium, placed in clean 2-ml tubes, and surface disinfected as follows: they were submerged in 70% ethanol for 1 min and rinsed with sterile water, followed by submersion in 0.05% bleach for 1 min and then two rinses in sterile water (90). To verify the effectiveness of the disinfection process, 100-μl aliquots of water from the last rinse were plated on Trypticase soy agar (TSA; Oxoid, Cambridge, United Kingdom) and incubated at 37°C for 24 h. Surface-disinfected flies were transferred to sterile petri dishes and identified to the species level using dichotomous keys for dipteran families (29, 32). The sex of the flies was determined through external morphology (21) before aseptic dissection of the guts. The guts of each fly were placed in 2-ml tubes containing 1 ml of BPW with 0.5-mm zirconia/silica beads (BioSpec Products, Inc., Bartlesville, OK) and vigorously shaken for 10 min in a Genie cell disruptor (Scientific Industries, Inc., Bohemia, NY). Vouchers of collected fly specimens were deposited at the Smithsonian National Museum of Natural History (USNM) (Diptera Unit, Alcohol Collection, lot no. 1205119) in Washington, DC.
Pathogen detection.
The PCR-based BAX system Q7 (DuPont Qualicon, Wilmington, DE) was used to screen for Salmonella (standard assay), the Listeria genus (24E assay), and E. sakazakii (standard assay; used to detect Cronobacter spp.) as per the manufacturer's protocols. Primary enrichment was performed by transferring ∼330 μl of BPW-S (surface) and BPW-G (guts) mixtures to the following media: (i) for Salmonella spp., 1 ml of prewarmed (42°C) BPW with incubation in a recirculating water bath at 42.5°C for 22 to 24 h; (ii) for Cronobacter spp., 1 ml of prewarmed (37°C) BPW with Novobiocin (10 mg/liter) with incubation at 37°C for 22 to 26 h; and (iii) for Listeria spp., 1 ml of freshly prepared room temperature 24 Listeria enrichment broth (24 LEB) with selective supplement (Oxoid, Cambridge, United Kingdom) with incubation at 37°C for 44 ± 5 h. Secondary enrichment was performed for the detection of Salmonella spp. and Cronobacter spp. For this, 100 μl of enriched samples was transferred to 400 μl of prewarmed (37°C) brain heart infusion (BHI) broth (Difco, Becton Dickinson Company, Sparks, MD) and the mixture was incubated at 37°C for 3 h. After enrichment was completed, all samples were prepared and processed according to the protocol described in the BAX system user's guide for each bacteria. To increase probability of detection, 20 μl of enriched samples (instead of 5 μl, as instructed by the manufacturer) were transferred to 200 μl of lysis buffer. Salmonella enterica subsp. arizonae, Cronobacter sakazakii, and L. monocytogenes from FDA bacterial collections were included as positive controls, and sterile medium was used as a negative control. Tubes from primary and secondary enrichments were kept either in the refrigerator (Salmonella spp. and Listeria spp.) or at room temperature (Cronobacter spp.) for further confirmation analysis of BAX-positive samples.
Confirmation of Cronobacter spp.
To confirm the presence of Cronobacter spp. in BAX-positive samples, a 3-mm loopful (10 μl) of BHI broth from the secondary enrichment was streaked on violet red bile glucose agar (VRBG; Difco, Becton, Dickinson Company, Sparks, MD) and on Brilliance Cronobacter agar, also known as the Druggan-Forsythe-Iversen formulation (DFI; Oxoid, Cambridge, United Kingdom), for isolation of single colonies. All plates were incubated at 36°C for 22 to 24 h. Five presumptive Cronobacter colonies from the plates described above were subcultured on DFI and incubated at 36°C for 22 to 24 h. One blue-pigmented colony on DFI was randomly selected and further streaked on TSA plates and onto two chromogenic media, DFI and ChromID (bioMérieux, S.A., Marcy l'Etoile, France). Plates were incubated at 25°C for 48 to 72 h (TSA) and at 36°C for 22 to 24 h (ChromID and DFI). The color of bacterial colonies on each medium was recorded. Additionally, presumptive identification of the selected colony was performed using the commercial Rapid API-20E biochemical identification system (bioMérieux, S.A., Marcy l'Etoile, France), according to the manufacturer's instructions. Because several other microorganisms exhibited blue-pigmented colonies on DFI agar and were shown to be non-Enterobacter spp. in the Rapid API-20E system, DNA from presumptive target bacteria was extracted for molecular identification.
Molecular identification of Cronobacter spp.
Bacterial DNA was extracted from TSA cultures using the PrepMan Ultra reagent (Applied Biosystems) as indicated by the manufacturer. Ribosomal 16S rRNA and 1,6-α-glucosidase genes were amplified from 1 μl of 2- to 10-fold-diluted DNA using the primer pairs P0-P6 (46) and EsAgf-EsAgr (55), respectively, at the recommended concentrations. The PCR mixtures were set up in a total volume of 50 μl using the EmeraldAmp GT-PCR master mix (TaKaRa Bio, Inc.). The PCR cycling conditions for each gene were the same as those described by the authors. PCR products were purified and sequenced by Retrogen, Inc. (San Diego, CA), using the respective primer pairs. Sequence files were imported into Sequencher 5.0 (GeneCodes, Ann Arbor, MI). The primers were trimmed off using default parameters, and bidirectional sequences were assembled into contigs with default settings. Sequences were compared against those in the NCBI nucleotide BLAST (nr/nt) database (http://blast.ncbi.nlm.nih.gov) using the megablast algorithm. Sequence similarities of ≥99% from presumptive Cronobacter isolates were considered reliable identifications for this organism.
E. sakazakii has been reclassified into the new genus Cronobacter (47). This genus contains seven species: C. sakazakii, C. malonaticus, C. turicensis, C. muytjensii, C. dublinensis, C. condimenti, and C. universalis (formerly Cronobacter genomospecies 1) (49). Full-length 16S rRNA sequences of 64 Cronobacter strains, representative of five species, along with two sequences from Pantoea species and one sequence from Enterobacter helveticus (included as an outgroup) were retrieved from GenBank (46, 47, 49). Retrieved sequences and sequences obtained in this study were aligned using the ClustalX software (Lasergene, Madison, WI). To clarify the taxonomic position of Cronobacter strains obtained in this study, phylogenetic analyses were performed using the neighbor-joining (NJ) method, combined with the Hasegawa, Kishino and Yano (HKY) nucleotide substitution model, found in Geneious Pro software (version 5.6). A consensus tree was created using the resample tree option and 1,000 bootstrap replicates.
Confirmation of Salmonella spp.
To confirm the presence of Salmonella, two 100-μl aliquots of BHI broth were removed from the secondary enrichment. The first aliquot was added to 10 ml of Rappaport-Vassiliadis (RV) medium (Oxoid, Cambridge, United Kingdom). The second one was added to 1 ml of tetrathionate (TT) broth (Difco, Becton, Dickinson Company, Sparks, MD). Both media were incubated at 42.5°C for 22 to 24 h. After incubation, samples were plated as recommended by the FDA Bacteriological Analytical Manual (BAM) protocol for Salmonella (2). Presumptive Salmonella colonies were identified using the commercial Rapid API-20E system according to the manufacturer's instructions. Ribotyping was performed on all presumptive Salmonella strains identified by the commercial Rapid API-20E system using the RiboPrinter microbial characterization system (DuPont Qualicon, Wilmington, DE) with the PvuII protocol, following the manufacturer's instructions. Identified Salmonella serovars were tested again using the BAX system with the Salmonella-2 standard assay.
Confirmation of the Listeria genus and L. monocytogenes strains.
BAX samples that were positive for the genus Listeria 24E assay were run using the L. monocytogenes BAX standard assay, according to the manufacturer's instructions. Ten microliters of primary enrichment medium from 24E Listeria genus BAX-positive samples was streaked on two Brilliance Listeria agar (BLA) plates (Oxoid, Cambridge, United Kingdom; formerly Oxoid chromogenic Listeria agar [OCLA]). The plates were incubated at 37°C for 22 to 26 h and then examined for both the presence of blue-green colonies and the presence of an opaque white halo indicating activity of lecithinase, an enzyme associated with virulence in Listeria spp. Two BAX-negative samples were randomly selected and cultured on BLA plates to corroborate the accuracy of the BAX assay. Presumptive Listeria species were confirmed by ribotyping as described before using the EcoRI protocol according to the manufacturer's instructions. Stocks of overnight cultures of all confirmed bacterial strains were placed in 1 ml of 30% glycerol–BPW at −20°C for long-term storage. Vouchers of bacterial strains isolated from flies in this study were also deposited at the Food and Drug Administration, Center for Food Safety and Applied Nutrition, Office of Regulatory Science, Division of Microbiology, College Park, MD (FDA/CFSAN/ORS/DM; collection no. SAL3538 to SAL3544, LIS0149 to LIS0156, and ENTB0350 to ENTB0368).
Data analysis.
To estimate the proportion of food-borne pathogens carried by a population of flies, a random sample size of 96 flies was the minimum sample size required to be 95% confident that the sample percentage was in error by no more than 10% (84). Hence, a sample of 100 flies was selected for this study. Statistical analyses of the presence of food-borne pathogens on the body surfaces and in the guts of collected flies were performed using the χ2 test (PROC FREQ; SAS v9.3, 2005; SAS Institute, Inc.). Overall associations between the presence of food-borne pathogens, the fly's body part, fly species, and collection sites were analyzed using the Fisher's exact probability test (PROC FREQ) (72). If significant overall associations were detected, pairwise comparisons among the levels of the significant variables were performed. The Fisher's exact test was used instead of the χ2 test, since χ2 analysis does not accurately calculate expected frequencies that are below 5. A two-tailed P value of <0.05 indicated statistical significance.
The predicted probability of a fly carrying food-borne pathogens on the body surfaces or in the guts was analyzed using the SAS logistic regression procedure (PROC LOGIT, 2005; SAS Institute, Inc.). Pathogen presence was the categorical dichotomous response variable (positive or negative; n = 600). The relationship between the response variable and the predictor variables, along with their interactions, was analyzed with the full logistic probability model described in equation 1
| (1) |
where logit (P) = ln [P/(1 − P)], ln is the natural log, P is the probability of the presence of bacterial food-borne pathogens, α is the P intercept, β1 to β5 are regression coefficients, and pathogen (Cronobacter spp., Salmonella spp., or L. monocytogenes), body part (surface and guts), fly species, fly sex (male or female), and the ambient temperature (°C) at the time of collection are the predictor variables. The stepwise variable selection method with analysis of maximum likelihood estimates based on a Wald χ2 P value of <0.05 was used to specify the best-fit, reduced model. The receiver operating characteristics (ROC) curve was used as a measurement of the goodness-of-fit of the model. The ROC curve quantifies the power of the predicted values using the area under the ROC curve (AUC). AUC values of >0.7 are considered to show acceptable discrimination, those of >0.8 are considered to show excellent discrimination, and those of >0.9 are considered to show outstanding discrimination (44).
Nucleotide sequence accession numbers.
Full-length sequences of 16S rRNA (>1,300 bp) and sequences from the 1,6-α-glucosidase gene of Cronobacter strains obtained from this study have been deposited in the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/index.html) under accession no. JQ963896 to JQ963914 and JX315535 to JX315553, respectively.
RESULTS
Four families were identified from collected flies: Calliphoridae (49%), Muscidae (48%), Sarcophagidae (2%), and Anthomyiidae (1%). All specimens but one were identified to the species level using taxonomic keys. Six different species of flies were found. The most abundant fly species was the housefly, Musca domestica (47%), followed by the blowflies, Lucilia cuprina (33%) and Lucilia sericata (14%). Other species included the secondary screwworm, Cochliomyia macellaria (2%), the red-tailed flesh fly, Sarcophaga haemorrhoidalis (2%), and the black garbage fly, Ophyra leucostoma (1%). One specimen (1%) was identified to the family level as Diptera: Anthomyiidae (Norman E. Woodley, Systematic Entomology Laboratory, Agricultural Research Service, U.S. Department of Agriculture). Sixty-nine flies were males, and 31 were females. The ambient temperature at the time of collection ranged between 16 and 36°C.
Detection and confirmation of Cronobacter spp.
Forty-five samples were positive for the E. sakazakii (Cronobacter) BAX standard assay. BAX-positive samples required several subculturing steps to obtain well isolated presumptive Cronobacter colonies on specific media. Nineteen out of the 45 BAX-positive samples exhibited typical pigmentation of the colonies on all three media: yellow, pale yellow, or cream-white pigmentation on TSA, blue-green pigmentation on DFI, and blue-black or blue-gray pigmentation on ChromID. However, the Rapid API-20E system identified 26/45 samples belonging to the genus Enterobacter: 24 samples were identified as E. cloacae, with percent identities ranging from 58.5 to 99.9%, and two samples were identified as E. aerogenes, with percent identities of 83.7 and 96% (see Table S1 in the supplemental material). The Rapid API-20E system also identified some typical Cronobacter pigmented colonies on chromogenic media as non-Enterobacter but belonging to some neighboring genera, such as Citrobacter freundii, Providencia stuartii, Proteus vulgaris, Pantoea spp., and Klebsiella pneumoniae. Therefore, the pigmentation of presumptive Cronobacter colonies on TSA or either of the two chromogenic media used, along with the identification of the Rapid API-20E system, was not a definitive criterion to confirm Cronobacter spp. on all BAX-positive samples from collected flies. DNA was subsequently obtained from individual colonies of all 45 E. sakazakii (Cronobacter) BAX-positive samples and PCR amplified using the 16S rRNA and 1,6-α-glucosidase (EsAg) primers. All 45 samples gave a PCR product of the expected size (>1,300 bp) when the 16S rRNA primers were used. However, just 19 samples amplified a specific fragment of approximately 1,500 bp when the EsAg primers were used (see Table S1).
Edited sequences from all amplified fragments were compared against those in the NCBI nucleotide database. Sixteen out of the 24 samples that were identified as E. cloacae using the Rapid API-20E system were identified as C. sakazakii after BLAST searches of the 16S rRNA sequences against those in the nucleotide database. One more sample was identified as C. turicensis and another as C. malonaticus (see Table S1 in the supplemental material). The remaining six samples were identified as Enterobacter spp. (4 samples), Enterobacter cancerogenus (1 sample), and E. cloacae (1 sample). Additionally, 16S rRNA sequences from the two bacterial strains identified as E. aerogenes with the Rapid API-20E system were identified as C. turicensis and Enterobacter spp. (see Table S1).
Overall, 16S rRNA sequences from 19 bacterial strains were identified as Cronobacter spp., with percent identities of ≥99%. Results were consistent as these 19 bacterial strains were the only ones that amplified the 1,6-α-glucosidase gene and were identified as C. sakazakii (17 strains) and C. turicensis (2 strains) using BLAST. The same 19 Cronobacter strains were the only ones showing typical pigmentation of the colonies on all three media used (see Table S1 in the supplemental material). Consequently, only these 19 samples were considered positive for Cronobacter spp. and included for further analysis.
A 16S rRNA NJ phylogenetic tree was constructed to clarify the taxonomic position of Cronobacter species obtained in this study (Fig. 1). Five well-defined clusters corresponding to five Cronobacter biogroups were delineated on the phylogenetic tree. Seventeen Cronobacter sequences from collected flies clustered within the C. sakazakii group. One sequence, isolated from the surface of S. haemorrhoidalis, clustered within the C. turicensis group, and another sequence, isolated from the surface of M. domestica, clustered within the C. universalis group (Fig. 1).
Fig 1.
Phylogenetic tree of 16S rRNA full-length sequences of Cronobacter strains. The neighbor-joining method combined with the model of Hasegawa, Kishino, and Yano (HKY) and 1,000 bootstrap replicates were used. Cronobacter strains obtained in this study and their GenBank accession numbers are in boldface. The bar indicates 1% estimated sequence divergence.
Detection and confirmation of Salmonella spp.
Seven samples were positive for the Salmonella BAX assay. Typical Salmonella colonies with black centers were observed on bismuth sulfite (BS) agar, xylose lysine desoxycholate (XLD) agar, and Hektoen enteric (HE) agar. From each sample, one to three typical and well-isolated Salmonella colonies were randomly selected for presumptive generic identification of Salmonella with the commercial Rapid API-20E system. All colonies were identified as Salmonella spp., with percent identities ranging from 67.5 to 99.5%. One colony from each sample was selected for ribotyping analysis using the PvuII restriction enzyme protocol.
Five ribotype patterns, corresponding to five Salmonella serovars, were obtained from colonies recovered from the seven Salmonella BAX-positive samples (Fig. 2a). Two Salmonella enterica serovar Senftenberg (DuPont ID pattern DUP-PUVII-1153) strains were isolated from the guts of two flies (M. domestica and L. cuprina), both collected from site 10, with identity similarities of 0.97 and 0.87, respectively. One S. enterica serovar Hadar strain (DUP-PUVII-3173), with identity similarity of 0.98, was isolated from the guts of one M. domestica fly collected from site 7. Three other distinctive Salmonella serovars were recovered from three flies collected from site 6: S. enterica serovar Poona (DUP-PUVII-3216) was recovered from both the body surfaces and guts of L. cuprina, with identity similarities of 0.94 and 0.96, respectively; S. enterica serovar Schwarzengrund/Bredeney (DUP-PUVII-1148) was recovered from the guts of L. cuprina, with 0.97 identity similarity; and S. enterica serovar Brackenridge (DUP-PUVII-3161) was recovered from the guts of L. sericata, with 0.96 identity similarity. All bacterial strains were also positive for the Salmonella-2 BAX assay.
Fig 2.
Ribotype patterns of Salmonella (a) and Listeria (b) strains isolated from flies collected from dumpsters outside restaurants in urban areas.
Detection and confirmation of the Listeria genus and L. monocytogenes.
Eight samples tested positive for the 24E Listeria genus BAX kit. Four of these were positive for the L. monocytogenes BAX standard assay. All Listeria BAX-positive samples were confirmed by cultures showing typical blue-green Listeria colonies on chromogenic BLA plates. However, none of the strains showed an opaque white halo. No bacterial growth was observed from Listeria genus BAX-negative samples.
The four samples that were BAX positive for both the Listeria genus and L. monocytogenes were confirmed as L. monocytogenes by the DuPont ID pattern DUP-1042 of the RiboPrinter database, with identity similarities ranging from 0.92 to 0.95 (Fig. 2b). The four samples that were BAX positive for the Listeria genus but BAX negative for the L. monocytogenes assay were confirmed as Listeria innocua and presented three different DuPont ID patterns—DUP-1007, DUP-1009, and DUP-1010—with identity similarities ranging from 0.89 to 0.98.
The four samples that were positive for L. innocua were from the guts of four different flies (two M. domestica flies, one L. cuprina fly, and one L. sericata fly) collected from four different sites (sites 1, 3, 5, and 7). The four samples that were positive for L. monocytogenes were isolated from three flies (two L. cuprina flies and one L. sericata fly) collected from site 6, with one of the L. cuprina flies carrying L. monocytogenes on both the body surfaces and the guts (Fig. 2b). Because L. innocua is not considered a human food-borne pathogen, only the four strains that were confirmed as L. monocytogenes were included for further statistical analysis.
Prevalence of food-borne pathogens.
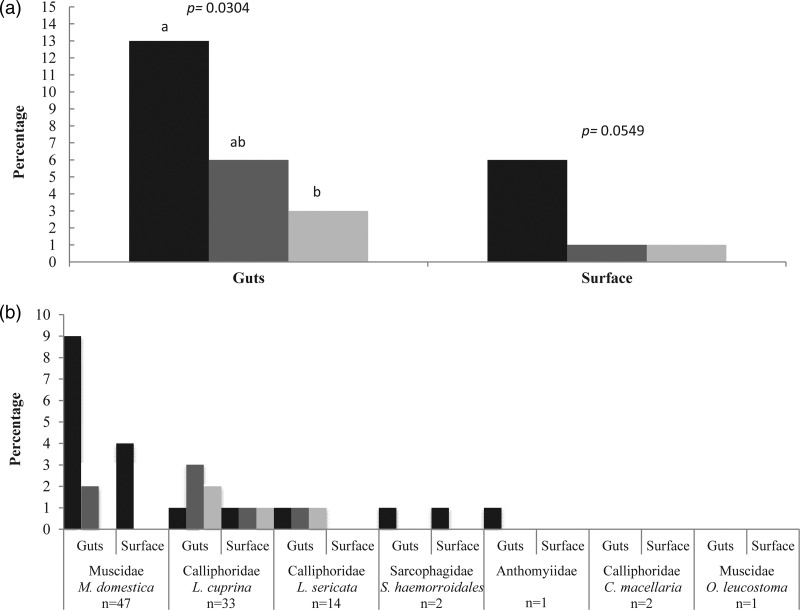
The percentages of flies that were found positive for the presence of Cronobacter spp., Salmonella spp., and L. monocytogenes, either in the guts or on the surface, were 14%, 6%, and 3%, respectively. The species of flies that were positive for Cronobacter spp. were M. domestica (10%), L. cuprina (1%), L. sericata (1%), and S. haemorrhoidalis (1%), along with the fly in the family Anthomyiidae (1%). The flies that tested positive for Salmonella spp. were M. domestica (2%), L. cuprina (3%), and L. sericata (1%), while the flies positive for L. monocytogenes were L. cuprina (2%) and L. sericata (1%).Thirty samples were positive for the presence of any of the three food-borne pathogens on either the body surfaces or the guts of collected flies. Aliquots of water from the last disinfection rinse of individual flies showed no bacterial growth on TSA plates, allowing us to conclude that there was no cross-contamination between the fly's body surfaces and the guts. The frequency of food-borne pathogens was statistically higher in the guts (22 positives), than on the body surfaces (8 positives; χ2 = 6.8772, df = 1, P = 0.0087) (Fig. 3a). A significant overall association was observed among bacteria detected in the flies' guts (Fisher's exact test, P = 0.0304) but not on the body surfaces (Fisher's exact test, P = 0.0549). Pairwise comparison among bacterial pathogens in the guts showed that the presence of Cronobacter spp. was statistically higher (13%) than that of L. monocytogenes (3%) (Fisher's exact test, P = 0.0165). However, no statistical differences were found between the presence of Cronobacter spp. and Salmonella spp. or between Salmonella spp. and L. monocytogenes (Fisher's exact test, P = 0.1464 and P = 0.4977, respectively) (Fig. 3a).
Fig 3.
Presence of Cronobacter spp. (black bars), Salmonella spp. (dark gray bars), and L. monocytogenes (light gray bars) on flies collected from dumpsters outside restaurants in urban areas. (a) Overall presence of pathogens on the body surfaces and in the guts of collected flies. (b) Presence of pathogens by fly species and body part. Means with the same letter are not statistically different from each other (Fisher's exact test, P < 0.05).
The overall association between the presence of pathogens and the species of flies was not statistically different (Fisher's exact test, P = 0.3275). Figure 3b shows the percentage of flies per species that were positive for pathogens found on the body surfaces and in the guts. The greatest prevalence of Cronobacter spp. was mostly found in the guts (9%) and on the body surfaces (4%) of M. domestica, whereas Salmonella spp. (3%) and L. monocytogenes (2%) were more abundant in the guts of L. cuprina. No bacterial pathogens were detected on C. macellaria and O. leucostoma.
A significant overall association was observed among the presence of pathogens and collection sites (Fisher's exact test, P = 0.0003). Multiple pairwise comparisons showed that sites 3 and 9, each containing one sample positive for Cronobacter spp. (Fisher's exact test, P = 0.0166 each), were significantly lower than site 10, where seven samples were positive for Cronobacter spp. and two samples were positive for Salmonella spp., and site 6, where four samples were positive for both L. monocytogenes and Salmonella spp. (Fisher's exact test, P = 0.0322 each).
Relative risk for the presence of food-borne pathogens.
Stepwise selection of the full logistic regression model gave rise to the final model shown in equation 2:
| (2) |
The estimates indicate that the probability of a single fly being positive for the presence of any of the three food-borne pathogens evaluated in this study was associated with the type of food-borne pathogen, the body part of the fly, and the ambient temperature. The reduced model showed good performance, as indicated by the AUC value of 0.73 (acceptable discrimination) with 95% Wald confidence limits of 0.6306 and 0.8296 (Table 1).
Table 1.
Analysis of maximum likelihood estimates of the logistic regression modela
| Parameter | β (SE) | Wald χ2 | P |
|---|---|---|---|
| Intercept | −5.3400 (1.0547) | 25.6335 | <0.0001 |
| Cronobacter spp. | 0.9208 (0.2689) | 11.7231 | 0.0006 |
| Salmonella spp. | −0.1694 (0.3196) | 0.2810 | 0.5960 |
| Guts | 0.5472 (0.2137) | 6.5598 | 0.0104 |
| Temp | 0.0743 (0.0357) | 4.3239 | 0.0376 |
For each parameter shown, df = 1. R2 = 0.04, maximum-rescaled R2 = 0.122, Kendall's τ-α = 0.044, Goodman-Kruskal γ = 0.47, Somers's Dxy = 0.46, and AUC = 0.73. Test values are as follows, with df = 4 for each: likelihood ratio, χ2 = 24.4707, P < 0.0001; score, χ2 = 24.3809, P < 0.0001; and Wald, χ2 = 21.4047, P = 0.0003.
From Fig. 4a to c, inferences can be made about the predicted probability of a fly carrying one of the three different pathogens at a given temperature. For example, at 27°C the probabilities of a fly carrying Cronobacter spp., Salmonella spp., or L. monocytogenes in the guts are 12.0%, 4.2%, or 3.0%, respectively. At the same ambient temperature, the probability of a fly carrying any of the three pathogens, on the body surfaces or in the guts is 2.8% or 4.4%, respectively (Fig. 4d). The ability to make these predictions is also confirmed by the positive coefficients associated with the predictor parameters, Cronobacter spp. (0.9208), Salmonella spp. (−0.1694), fly guts (0.5472), and temperature (0.0743) (Table 1).
Fig 4.
Relative risk of the presence of Cronobacter spp. (a), Salmonella spp. (b), L. monocytogenes (c), and any of the three pathogens (d) on body surfaces (gray triangles with dotted lines) and in guts (black squares with dotted lines) of wild flies according to ambient temperature (°C). Dotted lines represent the 95% upper and lower confidence intervals.
Our results also indicate that it is three times more likely to find any of the three bacterial pathogens in the guts than on the body surfaces of the flies (Table 2). Likewise, the presence of Cronobacter spp. in an individual fly was five times greater than that of L. monocytogenes and three times greater than that of Salmonella spp., while the presence of Salmonella spp. was two times greater than that of L. monocytogenes. These results are illustrated as changes in the probability of the presence of these pathogens on both the body surfaces and in the guts of a fly as a function of ambient temperature (Fig. 4a to c).
Table 2.
Odds ratio estimates of the presence of food-borne pathogens from collected flies
| Effect | Point estimate | 95% Wald confidence limits |
|---|---|---|
| Cronobacter spp. vs Salmonella spp. | 2.975 | 1.209, 7.322 |
| Cronobacter spp. vs L. monocytogenes | 5.325 | 1.762, 16.090 |
| Salmonella spp. vs L. monocytogenes | 1.790 | 0.512, 6.250 |
| Guts vs body surfaces | 2.988 | 1.293, 6.900 |
| Temp | 1.077 | 1.004, 1.155 |
DISCUSSION
Food-borne illness due to Cronobacter, a ubiquitous and opportunistic genus of bacteria, has historically been found among high-risk groups, especially neonates, making it a public health concern. It has been confirmed as the cause of severe systematic neonatal infection and mortality in premature newborns with underlying medical conditions (86) and also in apparently healthy full-term infants (7). Cronobacter spp. rarely affect adults, causing less severe infections (18, 38). Recorded cases and outbreaks in infants have been associated with the ingestion of milk-based powdered infant formula (20, 62, 80), but in a number of other cases involving infants and adults, the vehicle of transmission was not confirmed.
Species of Cronobacter have been isolated from a variety of foods, beverages (28, 48), and many other environmental sources, such as soil (50), air (63), household vacuum dust (48), and food preparation utensils, such as blenders (8). Cronobacter spp. have also been isolated from M. domestica, collected from dumpsters outside restaurants in Gainesville, FL (10), from urban areas in Maharashtra, India (35), and also from the guts of the stable fly, Stomoxys calcitrans (37, 60, 61), and the Mexican fruit fly, Anastrepha ludens (53). Despite the many sources from which Cronobacter spp. have been isolated, Cronobacter's natural habitat and primary reservoir still remain unknown.
Here we demonstrated that wild flies carry Cronobacter spp. both externally and internally, possibly serving as their natural reservoir. To be a natural reservoir of a particular microorganism, certain criteria need to be met (69). The host species must be capable of maintaining the pathogen in wild populations, without being negatively affected by the pathogen. Additionally, the pathogen must remain within the host species for a sufficient amount of time to be able to transmit it to the affected organisms (69). Our study showed that Cronobacter spp. were present either in the guts or on the body surfaces of 14% of the wild flies collected from dumpsters outside urban restaurants (see Table S1 in the supplemental material). This prevalence is greater than that reported by Mramba et al. (60) from wild stable flies collected in rural sites from Kansas and Florida, where only 2/928 (0.2%) flies were positive for Cronobacter spp. Occurrences of Cronobacter spp. greater than our findings have only been found in foods of plant origin, such as herbs and spices, seeds, organic breakfast cereals, and on animal feed or grain (41, 45, 48, 59), thus suggesting plants as another possible reservoir of this pathogen (76).
The flies collected in this study carried Cronobacter species from three distinctive groups (Fig. 1). Strains from two of these groups, C. sakazakii and C. turicensis, have been isolated from various food sources (41, 59). The third group, C. universalis, has been defined by five strains: its representative strain, NCTC 9529, was collected from a United Kingdom water sample in 1954, strain 96 was isolated from onion powder purchased in the United Kingdom, strain 1435 was isolated from rye flour purchased in Turkey, strain 731 was recovered from a leg infection of a 9-year-old boy (49), and strain E680 was recovered from an unknown source (46).
Seventeen of the strains isolated from collected flies belonged to the C. sakazakii group. These strains were isolated from M. domestica, S. haemorrhoidalis, L. cuprina, L. sericata, and Anthomyiidae. Strain Sh41s (JQ963910), isolated from the surface of S. haemorrhoidalis, belonged to the C. turicensis group, whereas strain Md1s (JQ963896), isolated from the surface of M. domestica, clustered with the five strains belonging to the C. universalis group. A BLAST search of the 16S rRNA nucleotide sequence of the Md1s strain, gave 99% identity to the C. universalis strain E680 (EF059861) (see Table S1 in the supplemental material). However, we did not perform further biochemical tests on the newly obtained Md1s strain, as indicated by Iversen et al. (47) and Joseph et al. (49).
There is evidence that flies are not negatively affected by harboring Cronobacter spp. in their guts. Despite the low prevalence of Cronobacter spp. in wild stable flies (60), it was demonstrated that this bacterial pathogen not only can persist in the guts of laboratory-reared stable flies for at least 20 days, but the bacteria can also support larval development in the absence of other microbes and can colonize the guts of newly emerged flies (61). Therefore, some food-borne pathogens such as Cronobacter spp. may be harbored in the flies' digestive tracts as normal gut flora that can be passed on to the next generation, enhancing their vector potential.
Cronobacter species are both thermo- and osmotolerant (9, 70, 78). These physiological characteristics suggest that they are probably able to survive on the body surfaces of the fly for longer periods of time than other food-borne pathogens, thus increasing the likelihood of mechanical transmission by flies. The environmental tolerance of Cronobacter spp. also makes them capable of withstanding food processing (3, 9, 68, 78), allowing flies to potentially contaminate food products, including those of plant origin, before or after processing.
Not only are the physiological characteristics of the bacteria conducive to survival on and inside the fly, but the flies themselves also exhibit the characteristics necessary for spreading food-borne pathogens, such as endophily, synanthropy, and communicative behaviors (65). Flies demonstrating the same characteristics that allow for the transmission of food-borne pathogens were shown to carry Salmonella spp. and L. monocytogenes. Both Salmonella (nontyphoidal) and L. monocytogenes are among the top five pathogens causing food-borne mortality in the United States (13), with nontyphoidal Salmonella being the leading cause of food-related hospitalizations (74).
Salmonella is commonly found in the environment and the gastrointestinal tracts of wild and farmed animals and humans. While it can be disseminated through a wide variety of routes, the consumption of food contaminated with animal feces is the most common route of dissemination. Outbreaks of Salmonella have implicated both animal- and produce-based products (25). Salmonella has also been associated with wild flies collected from animal farms (5, 32, 34, 39, 58, 67, 87) and, less frequently, with wild flies collected from urban areas (6, 16, 85). However, the reported frequencies of Salmonella spp. associated with flies vary greatly among studies. Some examples include 0% (27), 13.3% (16), 26.4% (87), 61.7% (85), and 100% (6). This great variability is explained mainly by the lack of systematic methods used and the use of either individual flies or pooled samples of flies, which varied from 10 to 50 flies per pool. In addition, only a handful of studies have identified Salmonella to the serovar level (16, 32, 67, 87).
In our study, Salmonella spp. were present in 6% of flies collected from urban areas, and a total of five serovars were identified: Seftenberg, Hadar, Poona, Brackenridge, and Schwarzengrund/Bredeney (Fig. 2a). S. enterica serovar Hadar was also reported from the internal contents and the surfaces of pooled samples of M. domestica collected from an animal facility in Malaysia (16), whereas S. enterica serovar Schwarzengrund and S. enterica serovar Senftenberg were recovered from M. domestica flies collected from swine farms in Taiwan (87). With the exception of S. enterica serovar Brackenridge, all Salmonella serovars isolated from the flies collected in this study have caused food-borne illness in the United States (14).
L. monocytogenes is the third leading cause of death from food-borne illness in the United States (74). A recent multistate outbreak of L. monocytogenes in the United States, linked to whole cantaloupes, led to the death of 30 people (15). L. monocytogenes is ubiquitous in agricultural settings (soil, decaying vegetation, plants, and water) and human and animal feces (24), and it can also persist in food manufacturing environments. Although L. monocytogenes has been found in many environments, 99% of human listeriosis cases appear to originate from food consumption (31), affecting mainly older adults, the immunocompromised, neonates, and pregnant women.
Even though flies have been implicated as vectors of L. monocytogenes, up to now, there is no scientific evidence associating this food-borne pathogen with wild flies collected from urban areas. L. monocytogenes was not found on M. domestica flies collected from an artisan cheese factory in Campinas, Brazil (11), but Listeria spp. were found on 5 out of 180 total M. domestica flies collected from 12 animal farms in Nuevo León, Mexico (39). However, the species of Listeria was not identified. Additionally, insects were shown to be the source for L. monocytogenes in an outbreak associated with Quargel cheese produced in Austria (D. Schoder and M. Wagner, presented at the IAFP European Symposium on Food Safety, Ede, The Netherlands, 19 May 2011). However, the specific type of insect that carried the pathogen was never identified.
To the best of our knowledge, this is the first evidence of flies acting as vectors of L. monocytogenes: 3% of collected flies were positive for this pathogen. Positive flies belong to the family Calliphoridae (L. cuprina and L. sericata) (Fig. 2b). L. innocua was also recovered from 4% of collected flies. It is possible to have false-negative results for the presence of L. monocytogenes when L. innocua is present. However, bacterial strains may behave differently, particularly when the concentrations of these two species in the sample differ (12). No further experiments were performed on strains isolated from collected flies to determine the possibility of inhibition among Listeria species and/or strains.
In our study, none of the flies was positive for all three pathogens evaluated. However, one L. cuprina fly (fly 81) carried Salmonella spp. and L. monocytogenes in the guts and on the body surfaces, and two more flies, L. cuprina (fly 82) and L. sericata (fly 90), had the same two pathogens in the guts. These flies were all collected from a single site (site 6), demonstrating that flies pick up bacterial pathogens from the surrounding environment (Fig. 2a and b). The same pattern was observed in site 10, where one M. domestica fly (fly 6) and one L. cuprina fly (fly 10) were found to carry both S. Seftenberg and C. sakazakii in their guts (Fig. 2a; see Table S1 in the supplemental material).
Studies on the interactions of the complex bacterial community that populates the fly's gut have mainly focused on nonpathogenic bacteria (19). We have demonstrated a statistically significant association between collection sites and the presence of food-borne pathogens, and it is assumed that microbiota in the fly's gut can be derived from their surrounding environment (18). The fact that some of the sites were “hot spots” for particular bacteria does not negatively affect the ability to assess the risk of pathogens associated with flies, because our sites were chosen at random. To more accurately assess how these “hot spots” may affect the ability of flies to transmit disease, more research on the persistence of ingested pathogenic bacterial strains by flies is needed. Preliminary studies have demonstrated that adult houseflies that ingested food with different concentrations of Salmonella and L. monocytogenes transmit these pathogens to their progeny (unpublished data).
Other bacteria identified from collected flies in this study, including Proteus spp., Proteus vulgaris, Proteus hauseri, Proteus penneri, Klebsiella pneumoniae, Providencia alcalifaciens, Providencia stuartii, Enterobacter spp., Enterobacter cloacae, Enterobacter cancerogenus, and Citrobacter freundii (see Table S1 in the supplemental material), have also been isolated from flies in other studies (10, 19, 35, 54, 83). These microorganisms are considered indigenous microbiota in flies as they can establish symbiotic relationships with the host, providing nutrients or other defensive compounds (56).
No false positives were detected when the BAX standard assays were used for Salmonella, the Listeria genus, and L. monocytogenes, and confirmation of viable pathogens on agar plates was in agreement with BAX-positive results. However, the E. sakazakii (Cronobacter) BAX standard assay showed 56% (25/45) false-positive presumptive Cronobacter spp., and cross-reactivity with other bacteria, such as C. freundii, was confirmed (data not shown). Hence, the BAX system alone is not sufficient to confirm the presence and the identity of Cronobacter spp. from individual flies. Additional steps, including determination of the phenotypic characteristics of the colonies in several specific media and 16S rRNA sequence analysis, should be included to confirm this microorganism.
This research also provides information on the relative risk of urban flies carrying Cronobacter spp., Salmonella spp., and L. monocytogenes. The predicted probability of the presence of these pathogens on either the body surfaces or in the guts of collected flies was positively correlated with ambient temperature (Fig. 4a to d). Higher ambient temperatures have been shown to increase the replication cycles of both food-borne pathogens (23, 77) and populations of flies (30), combined factors that could help to explain the reported increase of food-borne illnesses during the summer months (4).
Even though the relative risk of an individual fly carrying Cronobacter spp., Salmonella spp., and L. monocytogenes, externally or internally, has been correlated with ambient temperature, the question of whether a single fly could potentially deliver infectious or lethal doses of these food-borne pathogenic bacteria is still dependent on other factors. The behavioral pattern of the flies is one factor to consider, particularly because we have demonstrated that flies carry up to three times more bacterial food-borne pathogens in the guts than on the body surfaces. Thus, if a fly has sufficient time to feed, regurgitate, and defecate on the food, the chance of delivering those pathogens increases. Other factors include the physiological characteristics of the bacterial strain, the survival and/or growth of the pathogen on food, the conditions under which potentially contaminated food is maintained, and the susceptibility of the consumer population. Overall, this research provides quantitative data that contribute to assessment of the risk of the presence of flies in food or food facilities. Appropriate control of these insects may decrease the spread of food-borne pathogens and the risk to public health.
Supplementary Material
ACKNOWLEDGMENTS
We thank Norman E. Woodley (Systematic Entomology Laboratory, Agricultural Research Service, U.S. Department of Agriculture) for assistance in identifying unknown flies to the family level, Stuart J. Chirtel (FDA, CFSAN, Division of Public Health and Biostatistics) for assistance with the statistical analysis of the data, and Hannah Lee (Research Internship Program, Joint Institute for Food Safety and Applied Nutrition [JIFSAN], University of Maryland) for laboratory assistance.
This project was supported in part by an appointment to the Research Participation Program at the Center of Food Safety and Applied Nutrition administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration.
All authors reviewed and approved the manuscript. The use of specified instrumentation is not an endorsement by the FDA, and we certify that there is no conflict of interest with any financial organization regarding the material discussed in this article.
Footnotes
Published ahead of print 31 August 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Anderson JF. 1909. The differentiation of outbreaks of typhoid fever due to water, milk, flies and contact. Am. J. Public Hyg. 19: 251–259 [PMC free article] [PubMed] [Google Scholar]
- 2. Andrews WH, Jacobson A, Hammack TS. 2011. Salmonella. Bacteriological analytical manual. US Food and Drug Administration, Washington, DC [Google Scholar]
- 3. Arku B, Mullane N, Fox E, Fanning SA, Jordan K. 2008. Enterobacter sakazakii survives spray drying. Int. J. Dairy Technol. 61: 102–108 [Google Scholar]
- 4. Arthur A, Gournis E, McKeown D, Yaffe B. 2009. Toronto public health: foodborne illness in Toronto. Toronto Public Health, Toronto, Ontario, Canada: www.toronto.ca/health [Google Scholar]
- 5. Barber DA, Bahnson PB, Isaacson R, Jones CJ, Weigel RM. 2002. Distribution of Salmonella in swine production ecosystems. J. Food Prot. 65: 1861–1868 [DOI] [PubMed] [Google Scholar]
- 6. Barro N, Aly S, Tidiane OCA, Sababenedjo TA. 2006. Carriage of bacteria by proboscises, legs, and feces of two species of flies in street food vending sites in Ouagadougou, Burkina Faso. J. Food Prot. 69: 2007–2010 [DOI] [PubMed] [Google Scholar]
- 7. Biering G, et al. 1989. Three cases of neonatal meningitis caused by Enterobacter sakazakii in powdered milk. J. Clin. Microbiol. 27: 2054–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Block C, et al. 2002. Cluster of neonatal infections in Jerusalem due to unusual biochemical variant of Enterobacter sakazakii. Eur. J. Clin. Microbiol. Infect. Dis. 21: 613–616 [DOI] [PubMed] [Google Scholar]
- 9. Breeuwer P, Lardeau A, Peterz M, Joosten HM. 2003. Desiccation and heat tolerance of Enterobacter sakazakii. J. Appl. Microbiol. 95: 967–973 [DOI] [PubMed] [Google Scholar]
- 10. Butler JF, Garcia-Maruniak A, Meek F, Maruniak JE. 2010. Wild Florida house flies (Musca domestica) as carriers of pathogenic bacteria. Fla. Entomol. 93: 218–223 [Google Scholar]
- 11. Cardozo GMBQ, et al. 2009. Musca domestica L. as a vector of pathogenic microorganisms in ultra-filtered fresh Minas cheese. Braz. J. Food Technol. 12: 85–91 [Google Scholar]
- 12. Carvalheira A, Eusébio C, Silva J, Gibbs P, Teixeira P. 2010. Influence of Listeria innocua on the growth of Listeria monocytogenes. Food Control 21: 1492–1496 [Google Scholar]
- 13. CDC 7 February 2012, posting date. CDC estimates of foodborne illness in the United States. CDC 2011 estimates: findings. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/foodborneburden/2011-foodborne-estimates.html [Google Scholar]
- 14. CDC 16 January 2012, posting date. Foodborne Outbreak Online Database (FOOD). Centers for Disease Control and Prevention, Atlanta, GA: http://wwwn.cdc.gov/foodborneoutbreaks/Default.aspx [Google Scholar]
- 15. CDC 7 February 2012, posting date. Investigation update: multistate outbreak of listeriosis linked to whole cantaloupes from Jensen Farms, Colorado. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/listeria/outbreaks/cantaloupes-jensen-farms/101211/index.html [Google Scholar]
- 16. Choo LC, Saleha AA, Wai SS, Fauziah N. 2011. Isolation of Campylobacter and Salmonella from houseflies (Musca domestica) in a university campus and a poultry farm in Selangor, Malaysia. Trop. Biomed. 28: 16–20 [PubMed] [Google Scholar]
- 17. De Jesus AJ, Olsen AR, Bryce JR, Whiting RC. 2004. Quantitative contamination and transfer of Escherichia coli from foods by houseflies, Musca domestica L. (Diptera: Muscidae). Int. J. Food Microbiol. 93: 259–262 [DOI] [PubMed] [Google Scholar]
- 18. Dennison SK, Morris J. 2002. Multiresistant Enterobacter sakazakii wound infection in an adult. Infect. Med. 19: 533–535 [Google Scholar]
- 19. Dillon RJ, Dillon VM. 2004. The gut bacteria of insects: nonpathogenic interactions. Annu. Rev. Entomol. 49: 71–92 [DOI] [PubMed] [Google Scholar]
- 20. Drudy D, Mullane NR, Quinn T, Wall PG, Fanning S. 2006. Enterobacter sakazakii: an emerging pathogen in powdered infant formula. Clin. Infect. Dis. 42: 996–1002 [DOI] [PubMed] [Google Scholar]
- 21. Dübendorfer A, Hediger M, Burghardt G, Bopp D. 2002. Musca domestica, a window on the evolution of sex-determining mechanisms in insects. Int. J. Dev. Biol. 46: 75–79 [PubMed] [Google Scholar]
- 22. Ekdahl K, Normann B, Andersson Y. 2005. Could flies explain the elusive epidemiology of campylobacteriosis? BMC Infect. Dis. 5: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. FAO 2008. Climate change: implications for food safety, vol 1196 Food and Agriculture Organization of the United Nations, Rome, Italy [Google Scholar]
- 24. Farber JM, Losos JZ. 1988. Listeria monocytogenes, a foodborne pathogen. Can. Med. Assoc. J. 138: 413–418 [PMC free article] [PubMed] [Google Scholar]
- 25. Fatica MK, Schneider KR. 2011. Salmonella and produce survival in the plant environment and implications in food safety. Virulence 2: 573–579 [DOI] [PubMed] [Google Scholar]
- 26. FDA 2012. Bad bug book: foodborne pathogenic microorganisms and natural toxins handbook, 2nd ed US Food and Drug Administration, Washington, DC [Google Scholar]
- 27. Förster M, Sievert K, Messler S, Klimpel S, Feffer K. 2009. Comprehensive study on the occurrence and distribution of pathogenic microorganisms carried by synanthropic flies caught at different rural locations in Germany. J. Med. Entomol. 46: 1164–1166 [DOI] [PubMed] [Google Scholar]
- 28. Friedemann M. 2007. Enterobacter sakazakii in food and beverages (other than infant formula and milk powder). Int. J. Food Microbiol. 116: 1–10 [DOI] [PubMed] [Google Scholar]
- 29. Gagné R. 1991. Flies (Diptera), p 269–296 In Gorham JR. (ed), Insect and mite pests in food, an illustrated key, vol 1 US Department of Agriculture and US Department of Health and Human Services, Washington, DC [Google Scholar]
- 30. Goulson D, Derwent LC, Hanley ME, Dunn DW, Abolins SR. 2005. Predicting calyptrate fly populations from the weather, and probable consequences of climate change. J. Appl. Ecol. 42: 795–804 [Google Scholar]
- 31. Gray MJ, et al. 2004. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl. Environ. Microbiol. 70: 5833–5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Greenberg B. 1971. Flies and diseases, vol I Ecology and biotic associations Princeton University Press, Princeton, NJ [Google Scholar]
- 33. Greenberg B, Kowalski JA, Klowden MJ. 1970. Factors affecting the transmission of Salmonella by flies: natural resistance to colonization and bacterial interference. Infect. Immun. 2: 800–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Greenberg B, Varela G, Bornstein A, Hernandez H. 1963. Salmonellae from flies in a Mexican slaughterhouse. Am. J. Hyg. 77: 177–183 [DOI] [PubMed] [Google Scholar]
- 35. Gupta AK, et al. 2012. Phylogenetic characterization of bacteria in the gut of house flies (Musca domestica L.). FEMS Microbiol. Ecol. 79: 581–593 [DOI] [PubMed] [Google Scholar]
- 36. Hald B, Skovgård H, Pedersen K, Bunkenborg H. 2008. Influxed insects as vectors for Campylobacter jejuni and Campylobacter coli in Danish broiler houses. Poult. Sci. 87: 1428–1434 [DOI] [PubMed] [Google Scholar]
- 37. Hamilton JV, Lehane MJ, Braig HR. 2003. Isolation of Enterobacter sakazakii from midgut of Stomoxys calcitrans. Emerg. Infect. Dis. 9: 1355–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hawkins RE, Lissner CR, Sanford JP. 1991. Enterobacter sakazakii bacteremia in an adult. South. Med. J. 84: 793–795 [DOI] [PubMed] [Google Scholar]
- 39. Hernández-Escareño JJ, et al. 2012. Presence of Enterobacteriaceae, Listeria spp., Vibrio spp. and Staphylococcus spp. in house fly (Musca domestica L.), collected and macerated from different sites in contact with a few animal species. Rev. Cient. Fac. Cienc. Vet. 22: 128–134 [Google Scholar]
- 40. Hewitt CG. 1912. Houseflies and how they spread disease. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 41. Hochel I, Rùžičková H, Krásný L, Demnerová K. 2012. Occurrence of Cronobacter spp. in retail foods. J. Appl. Microbiol. 112: 1257–1265 [DOI] [PubMed] [Google Scholar]
- 42. Hogsette JR, Amendt J. 2008. Flies, p 209–237 In Bonnefoy X, Kampen H, Sweeney K. (ed), Public health significance of urban pests. World Health Organization, Geneva, Switzerland [Google Scholar]
- 43. Holt PS, Geden CJ, Moore RW, Gast RK. 2007. Isolation of Salmonella enterica serovar Enteritidis from houseflies (Musca domestica) found in rooms containing Salmonella serovar Enteritidis-challenged hens. Appl. Environ. Microbiol. 73: 6030–6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hosmer DW, Lemeshow S. 2000. Applied logistic regression, 2nd ed John Wiley and Sons, New York, NY [Google Scholar]
- 45. Iversen C, Forsythe S. 2004. Isolation of Enterobacter sakazakii and other Enterobacteriaceae from powdered infant formula milk and related products. Food Microbiol. 21: 771–777 [Google Scholar]
- 46. Iversen C, et al. 2007. The taxonomy of Enterobacter sakazakii: proposal of a new genus Cronobacter gen. nov. and descriptions of Cronobacter sakazakii comb. nov. Cronobacter sakazakii subsp. sakazakii comb. nov., Cronobacter sakazakii subsp. malonaticus subsp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov. and Cronobacter genomospecies I. BMC Evol. Biol. 7: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Iversen C, et al. 2008. Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactaridi subsp. nov. Int. J. Syst. Evol. Microbiol. 58: 1442–1447 [DOI] [PubMed] [Google Scholar]
- 48. Jaradat ZW, Ababneh QO, Saadoun IM, Samara NA, Rashdan AM. 2009. Isolation of Cronobacter spp. (formerly Enterobacter sakazakii) from infant food, herbs and environmental samples and the subsequent identification and confirmation of the isolates using biochemical, chromogenic assays, PCR and 16S rRNA sequencing. BMC Microbiol. 9: 225 doi:10.1186/1471-2180-9-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Joseph S, Cetinkaya E, Drahovska H, Levican A. 2012. Cronobacter condimenti sp. nov., isolated from spiced meat and Cronobacter universalis sp. nov., a novel species designation for Cronobacter sp. genomospecies 1, recovered from a leg infection, water, and food ingredients. Int. J. Syst. Evol. Microbiol. 62: 1277–1283 [DOI] [PubMed] [Google Scholar]
- 50. Khan AA, Jones RA, Cerniglia CE. 1998. Rapid method for the detection of genetically engineered microorganisms by polymerase chain reaction from soil and sediments. J. Ind. Microbiol. Biotechnol. 20: 90–94 [DOI] [PubMed] [Google Scholar]
- 51. Kobayashi M, et al. 1999. Houseflies: not simple mechanical vectors of enterohemorrhagic Escherichia coli O157: H7. Am. J. Trop. Med. Hyg. 61: 625–629 [DOI] [PubMed] [Google Scholar]
- 52. Kotenok YF, Chicherin YV. 1977. Houseflies (M. domestica L.) as transmitters of the agent of cholera. Zh. Mikrobiol. Epidemiol. December: 23–27 (In Russian.) [PubMed] [Google Scholar]
- 53. Kuzina LV, Peloquin JJ, Vacek DC, Miller TA. 2001. Isolation and identification of bacteria associated with adult laboratory Mexican fruit flies, Anastrepha ludens (Diptera: Tephritidae). Curr. Microbiol. 42: 290–294 [DOI] [PubMed] [Google Scholar]
- 54. Lam K, et al. 2007. Proliferating bacterial symbionts on house fly eggs affect oviposition behaviour of adult flies. Anim. Behav. 74: 81–92 [Google Scholar]
- 55. Lehner A, et al. 2006. Comparison of two chromogenic media and evaluation of two molecular based identification systems for Enterobacter sakazakii detection. BMC Microbiol. 6: 15 doi:10.1186/1471-2180-6-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Leroy PD, et al. 2011. The semiochemically mediated interactions between bacteria and insects. Chemoecology 21: 113–122 [Google Scholar]
- 57. Levine OS, Levine MM. 1991. Houseflies (Musca domestica) as mechanical vectors of shigellosis. Rev. Infect. Dis. 13: 688–696 [DOI] [PubMed] [Google Scholar]
- 58. Mian LS, Maag H, Tacal JV. 2002. Isolation of Salmonella from muscoid flies at commercial animal establishments in San Bernardino County, California. J. Vector Ecol. 27: 82–85 [PubMed] [Google Scholar]
- 59. Molloy C, et al. 2009. Surveillance and characterisation by pulsed field gel electrophoresis of Cronobacter spp. in farming and domestic environments, food production animals and retail foods. Int. J. Food Microbiol. 136: 198–203 [DOI] [PubMed] [Google Scholar]
- 60. Mramba F, Broce A, Zurek L. 2006. Isolation of Enterobacter sakazakii from stable flies, Stomoxys calcitrans L. (Diptera: Muscidae). J. Food Prot. 69: 671–673 [DOI] [PubMed] [Google Scholar]
- 61. Mramba F, Broce AB, Zurek L. 2007. Vector competence of stable flies, Stomoxys calcitrans L. (Diptera: Muscidae), for Enterobacter sakazakii. J. Vector Ecol. 32: 134–139 [DOI] [PubMed] [Google Scholar]
- 62. Mullane NR, et al. 2006. Enterobacter sakazakii: biological properties and significance in dried infant milk formula (IMF) powder. Int. J. Dairy Technol. 59: 102–111 [Google Scholar]
- 63. Mullane NR, et al. 2008. Dissemination of Cronobacter spp. (Enterobacter sakazakii) in a powdered milk protein manufacturing facility. Appl. Environ. Microbiol. 74: 5913–5917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nayduch D, Noblet GP, Stutzenberger FJ. 2002. Vector potential of houseflies for the bacterium Aeromonas caviae. Med. Vet. Entomol. 16: 193–198 [DOI] [PubMed] [Google Scholar]
- 65. Olsen AR. 1998. Regulatory action criteria for filth and other extraneous materials. III. Review of flies and foodborne enteric disease. Regul. Toxicol. Pharmacol. 28: 199–211 [DOI] [PubMed] [Google Scholar]
- 66. Olsen AR, Gecan JS, Ziobro GC, Bryce JR. 2001. Regulatory action criteria for filth and other extraneous materials. V. Strategy for evaluating hazardous and nonhazardous filth. Regul. Toxicol. Pharmacol. 33: 363–392 [DOI] [PubMed] [Google Scholar]
- 67. Olsen AR, Hammack TS. 2000. Isolation of Salmonella spp. from the housefly, Musca domestica L., and the dump fly, Hydrotaea aenescens (Wiedemann) (Diptera: Muscidae), at caged-layer houses. J. Food Prot. 63: 958–960 [DOI] [PubMed] [Google Scholar]
- 68. Osaili T, et al. 2008. Effect of environmental stresses on the sensitivity of Enterobacter sakazakii in powdered infant milk formula to gamma radiation. Lett. Appl. Microbiol. 47: 79–84 [DOI] [PubMed] [Google Scholar]
- 69. Putt SNH, Shaw APM, Woods AJ, Tyler L, James AD. 1988. Veterinary epidemiology and economics in Africa: a manual for use in the design and appraisal of livestock health policy. Epidemiology: some basic concepts and definitions. Veterinary Epidemiology and Economics Research Unit, Department of Agriculture, University of Reading, Reading, Berkshire, England [Google Scholar]
- 70. Riedel K, Lehner A. 2007. Identification of proteins involved in osmotic stress response in Enterobacter sakazakii by proteomics. Proteomics 7: 1217–1231 [DOI] [PubMed] [Google Scholar]
- 71. Rochon K, Lysyk TJ, Selinger LB. 2005. Retention of Escherichia coli by house fly and stable fly (Diptera: Muscidae) during pupal metamorphosis and eclosion. J. Med. Entomol. 42: 397–403 [DOI] [PubMed] [Google Scholar]
- 72. SAS Institute 2005. SAS 9.1.3 help and documentation. SAS Institute, Inc., Cary, NC [Google Scholar]
- 73. Scallan E, Griffin PM, Angulo FJ, Tauxe RV, Hoekstra RM. 2011. Foodborne illness acquired in the United States, unspecified agents. Emerg. Infect. Dis. 17: 16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Scallan E, et al. 2011. Foodborne illness acquired in the United States, major pathogens. Emerg. Infect. Dis. 17: 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Scharff RL. 2012. Economic burden from health losses due to foodborne illness in the United States. J. Food Prot. 75: 123–131 [DOI] [PubMed] [Google Scholar]
- 76. Schmid M, et al. 2009. Evidence for a plant-associated natural habitat for Cronobacter spp. Res. Microbiol. 160: 608–614 [DOI] [PubMed] [Google Scholar]
- 77. Semenza JC, Suk JE, Estevez V, Ebi KL, Lindgren E. 2012. Mapping climate change vulnerabilities to infectious diseases in Europe. Environ. Health Perspect. 120: 385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Shaker RR, Osaili TM, Abu Al-Hasan AS, Ayyash MM, Forsythe SJ. 2008. Effect of desiccation, starvation, heat, and cold stresses on the thermal resistance of Enterobacter sakazakii in rehydrated infant milk formula. J. Food Sci. 73: M354–M359 [DOI] [PubMed] [Google Scholar]
- 79. Sharon G, et al. 2010. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 107: 20051–20056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Simmons BP, Gelfand MS, Haas M, Metts L, Ferguson J. 1989. Enterobacter sakazakii infections in neonates associated with intrinsic contamination of a powdered infant formula. Infect. Control Hosp. Epidemiol. 10: 398–401 [DOI] [PubMed] [Google Scholar]
- 81. Steinhaus EA. 1967. Insect microbiology. Facsimile of 1947 edition. Hafner Publishing Co., Ltd., New York, NY [Google Scholar]
- 82. Su ZJ, et al. 2010. Comparison of bacterial diversity in wheat bran and in the gut of larvae and newly emerged adult of Musca domestica (Diptera: Muscidae) by use of ethidium monoazide reveals bacterial colonization. J. Econ. Entomol. 103: 1832–1841 [DOI] [PubMed] [Google Scholar]
- 83. Sulaiman S, Othman MZ, Aziz AH. 2000. Isolations of enteric pathogens from synanthropic flies trapped in downtown Kuala Lumpur. J. Vector Ecol. 25: 90–93 [PubMed] [Google Scholar]
- 84. Triola M. 2010. Elementary statistics using Excel, 4th ed, p 340–403 Pearson, Boston, MA [Google Scholar]
- 85. Ugbogu OC, Nwachukwu NC, Ogbuagu UN. 2006. Isolation of Salmonella and Shigella species from house flies (Musca domestica L.) in Uturu, Nigeria. Afr. J. Biotechnol. 5: 1090–1091 [Google Scholar]
- 86. van Acker J, et al. 2001. Outbreak of necrotizing enterocolitis associated with Enterobacter sakazakii in powdered milk formula. J. Clin. Microbiol. 39: 293–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wang YC, et al. 2011. Transmission of Salmonella between swine farms by the housefly (Musca domestica). J. Food Prot. 74: 1012–1016 [DOI] [PubMed] [Google Scholar]
- 88. WHO 2012. Food production to consumption. World Health Organization, Geneva, Switzerland: http://www.who.int/foodsafety/fs_management/en/ [Google Scholar]
- 89. Yano T, et al. 2008. Autophagic control of Listeria through intracellular innate immune recognition in Drosophila. Nat. Immunol. 9: 908–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zurek L, Denning SS, Schal C, Watson DW. 2001. Vector competence of Musca domestica (Diptera: Muscidae) for Yersinia pseudotuberculosis. J. Med. Entomol. 38: 333–335 [DOI] [PubMed] [Google Scholar]
- 91. Zurek L, Gorham JR. 2008. Insects as vectors of foodborne pathogens, p 1–16 In Voeller JG. (ed), Wiley handbook of science and technology for homeland security. Wiley Inc., Hoboken, NJ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.