Abstract
With the aim of improving industrial-scale production of stable-isotope (SI)-labeled arginine, we have developed a system for the heterologous production of the arginine-containing polymer cyanophycin in recombinant strains of Ralstonia eutropha under lithoautotrophic growth conditions. We constructed an expression plasmid based on the cyanophycin synthetase gene (cphA) of Synechocystis sp. strain PCC6308 under the control of the strong PcbbL promoter of the R. eutropha H16 cbbc operon (coding for autotrophic CO2 fixation). In batch cultures growing on H2 and CO2 as sole sources of energy and carbon, respectively, the cyanophycin content of cells reached 5.5% of cell dry weight (CDW). However, in the absence of selection (i.e., in antibiotic-free medium), plasmid loss led to a substantial reduction in yield. We therefore designed a novel addiction system suitable for use under lithoautotrophic conditions. Based on the hydrogenase transcription factor HoxA, this system mediated stabilized expression of cphA during lithoautotrophic cultivation without the need for antibiotics. The maximum yield of cyanophycin was 7.1% of CDW. To test the labeling efficiency of our expression system under actual production conditions, cells were grown in 10-liter-scale fermentations fed with 13CO2 and 15NH4Cl, and the 13C/15N-labeled cyanophycin was subsequently extracted by treatment with 0.1 M HCl; 2.5 to 5 g of [13C/15N]arginine was obtained per fed-batch fermentation, corresponding to isotope enrichments of 98.8% to 99.4%.
INTRODUCTION
Stable-isotope (SI)-labeled biomolecules are increasingly in demand for various applications in quantitative mass spectrometry (19) and multidimensional nuclear magnetic resonance (NMR) (43). Of particular interest for quantitative proteomics are SI-labeled amino acids, especially SI-arginine, which are used for labeling of proteins in cultured cells, i.e., for the SILAC (stable-isotope labeling with amino acids in cell culture) method (30, 31). The beauty of this method is that cells growing on 13C/15N-labeled amino acids incorporate the label into their proteins without any perturbing effects on their metabolism. The production of 13C-labeled substances using algae and other autotrophic organisms that utilize 13CO2 as the sole carbon source is far more cost-effective than using heterotrophic organisms. Not only is 13CO2 per se cheaper than 13C-labeled sugars, but heterotrophs incorporate only a fraction of the label, the rest being lost due to respiration. SI-labeled compounds are high-value specialty chemicals that are produced in small batch sizes (e.g., 1 to 25 g).
The present study aims at improving the production of SI-labeled arginine. The novel approach taken by us revolves around the use of cyanophycin as an intermediate for arginine production. Cyanophycin (multi-l-arginyl-poly-l-aspartic acid) is an arginine-containing branched polypeptide found in cyanobacteria and in some other bacteria (16). Cyanophycin is synthesized nonribosomally by the action of a single enzyme, cyanophycin synthetase, in an ATP-consuming reaction (1, 40, 47). It serves as a nitrogen, carbon, and energy reserve and accumulates upon transition from the exponential to the stationary growth phase (25). Cyanophycin synthetases from cyanobacteria, including Anabaena variabilis (47) and Synechocystis sp. strain PCC6308 (2), have been purified and characterized. Related enzymes from some chemotrophic bacteria, such as Acinetobacter sp. strain ADP1, Bordetella sp., and Desulfitobacterium hafniense (16, 46), have also been studied. Cyanophycin has attracted considerable biotechnological interest as a potential source of polyaspartate, a biodegradable substitute for polyacrylate. Polyacrylate is a bulk chemical with various industrial and agricultural uses. With the bulk production of cyanophycin in mind, several studies have explored the heterologous expression of cyanobacterial cyanophycin synthetases in Escherichia coli (9), Pseudomonas putida (7), Ralstonia eutropha (7), Saccharomyces cerevisiae (41), Pichia pastoris (42), Rhizopus oryzae (27), and other organisms capable of growing on cheap feedstocks, such as agricultural waste (28). Cyanophycin has also been successfully produced in transgenic tobacco and potato plants (29). Cyanobacteria are themselves not suitable as cyanophycin production strains due to their fastidious growth requirements, low yields of biomass, and low cyanophycin content (3.5% of cell dry weight [CDW]) (13). Some of the microbial expression systems tested provided impressively high yields of cyanophycin (e.g., 46% of CDW for Acinetobacter calcoaceticus [8]). However, with the exception of R. eutropha, none of them are capable of autotrophic growth on CO2, a prerequisite for efficient labeling with 13CO2. For this reason, we chose R. eutropha H16 as the host strain for cyanophycin production, even though the yield in previous studies was much lower than in other strains.
R. eutropha H16 is able to grow with H2 and CO2 as sole sources of energy and carbon, respectively. Comprehensive information originating from previous genomic, transcriptomic, and proteomic studies is available for this strain (4, 6, 15, 20, 32, 33, 35, 36). Although the genome of R. eutropha harbors two homologous genes for cyanophycin synthetase (cphA and cphA′), no cyanophycin originating from the respective enzymes was detectable (37). R. eutropha H16 does not accumulate cyanophycin as a storage compound.
In this study, we report on the development of a heterologous expression system for R. eutropha H16 suitable for autotrophic production of cyanophycin as a basis for the production of the stable-isotope-labeled amino acids arginine and aspartate. Specifically, we describe efforts to advance strain optimization, including the integration of the cphA gene into a dispensable chromosomal gene and the development of a novel addiction system based on the essential transcription regulator for hydrogenase gene expression, HoxA. Other addiction plasmids have been used successfully in R. eutropha growing under heterotrophic conditions, but these vectors are not suitable for autotrophic culture on H2 and CO2 (45). Our new system mediates stable production of cyanophycin in lithoautotrophic fermentations without the use of antibiotics. SI-labeled cyanophycin can be easily extracted from the cells via a fast acid extraction method (7) and hydrolyzed to obtain the isotope-labeled amino acids [13C]arginine and [13C]aspartate.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The strains and plasmids used in this study are listed in Table 1. The strain HF884 is a derivative of R. eutropha H16 (DSM 428; ATCC 17699) carrying an in-frame deletion allele of the phaC1 gene (24). E. coli S17-1 (29) served as a donor for conjugative transfer. Strain HF950(pGE779) was used by our industrial partner, Silantes GmbH (Munich, Germany), for the technical production of SI-labeled cyanophycin.
Table 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Bacterial strains | ||
| R. eutropha | ||
| HF884 | ΔphaC1; mutant of H16 defective in PHB synthesis | 37 |
| HF950 | ΔphaC1 ΔhoxA; mutant of HF884 defective in lithoautotrophic growth | This study |
| HF952 | Mutant of HF884 with cphA6308 under the control of the PcbbL promoter integrated in the nor2 region of chromosome 2 | This study |
| E. coli | ||
| S17-1 | recA proA thi-1 hsdR; harbors the RP4 tra genes in the chromosome | 39 |
| Plasmids | ||
| pLO1 | Kmr sacB RP4 oriT ColE1 ori | 22 |
| pLO11 | Tcr RK2 ori Mob+; expression vector for Strep-tagged fusion proteins in R. eutropha | 38 |
| pCM62 | Tcr; bhr cloning vector | 26 |
| pSK::cphABsyn6308 | Derivative of pBluescript SK(−) carrying the genes cphA and cphB of Synechocystis sp. PCC6308 | 1 |
| pCH553 | pLO1 containing hoxA with 921-bp in-frame deletion | 23 |
| pCH700 | pLO1 containing a 2.6-kb EcoRV fragment with a 4.9-kb StuI deletion (ΔnorR2A2B2) | 34 |
| pCH1609 | 3.3-kb MspA1I fragment of pGE778 in pCH700 StuI | This study |
| pGE295 | Derivative of pVK101 carrying the hoxABCJ region of Alcaligenes sp. strain M50 | O. Lenz |
| pGE777 | 0.21-kb PCR fragment PcbbL sequence ligated to AflIII/NcoI-cut pLO11 | O. Lenz |
| pGE778 | pLO11 containing PcbbL-cphA6308 | This study |
| pGE779 | pLO11 containing PcbbL-cphA6308 and hoxABCJAlcaligenes sp. M50 | This study |
Media and growth conditions.
Strains of R. eutropha were grown in mineral salts medium as described previously (12). Heterotrophic cultures were grown in mineral salts medium containing 0.4% fructose (FN medium). Lithoautotrophic cultures were grown in mineral salts medium under an atmosphere of 80% H2, 10% CO2, 10% O2 in an explosion-proof 10-liter fermentor. Small-scale batch cultures were grown in 500-ml baffled conical flasks in glass desiccators gassed with a mixture of 3% H2, 10% CO2, 10% O2, 77% N2. Growth was monitored by measuring the optical density at 436 nm. Strains of E. coli were grown in Luria-Bertani broth containing 0.25% (wt/vol) sodium chloride (LSLB). Sucrose-resistant segregants of sacB-harboring strains were selected on LSLB plates containing 15% (wt/vol) sucrose (22). Solid media contained 1.5% (wt/vol) agar. Antibiotics were added as appropriate (for R. eutropha, 350 μg ml−1 kanamycin, 15 μg ml−1 tetracycline; for E. coli, 15 μg ml−1 tetracycline, 100 μg ml−1 ampicillin). In fed-batch cultivations, the nutrients NH4+, Fe3+, Ca2+, and Mg2+ were added at appropriate times to prevent growth limitation. To test stable isotopic labeling under production conditions, 12CO2 was replaced by 13CO2 (chemical purity, >99%; isotopic enrichment, >99%; Sigma-Aldrich) and 14NH4Cl was replaced by 15NH4Cl (chemical purity, >98%; isotopic enrichment, >99%; Shanghai Research Institute). Unless explicitly stated, all of the experiments reported in this paper were done with unlabeled CO2 and NH4Cl.
Plasmid and strain construction.
In order to construct expression vectors suitable for use under lithoautotrophic growth conditions, a 0.21-kb PCR fragment carrying the PcbbL promoter sequence (18) and appropriate restriction sites was amplified via PCR using primers cbbL fwd (AflIII) (5′-CGAACATGTGCAACTGGCGAAGGGTAAGG-3′) and cbbL rev (NcoI) (5′-CGTCCATGGTTGTCTCCTTGCGTGGTTG-3′) from R. eutropha strain HF210. The resulting PCR fragment was cut with AflIII and NcoI and ligated to pLO11 (38), a mobilizable expression vector for Strep-tagged fusion proteins in R. eutropha, resulting in pGE777. This plasmid was a kind gift of O. Lenz, Humboldt-Universität, Berlin, Germany. The Synechocystis PCC6308 cyanophycin synthetase gene was amplified using primers cphA PciI (5′-GTCACATGTTAATCCTCAAAACACAAACCC-3′) and cphA BglII (5′-TAAGATCTTTCACTACTGAGATGATATTTCTCAA-3′) and plasmid pSK::cphAco (1) as the template. The latter plasmid was a kind gift of A. Steinbüchel (Westfälische Wilhelms-Universität, Münster, Germany). The PCR fragment was inserted into pGem-T (Promega) and verified by sequencing. A 2.6-kb PciI-BglII fragment of the appropriate pGem-T clone was inserted into the NcoI-BglII-cut vector pGE777, resulting in plasmid pGE778 carrying cphA6308 with a Strep tag-encoding sequence at the 3′ terminus. A 9.1-kb HindIII-BglII fragment of pGE295 carrying the hoxABCJ region of Alcaligenes sp. strain M50 (DSM 2625) was ligated to BglII-HindIII-cut pGE778, resulting in pGE779.
For the integration of the PcbbL-cphA expression cassette into chromosome 2, a 3.3-kb MspA1I fragment of pGE778 was introduced into the StuI site of pCH700 (34), resulting in pCH1609.
The suicide plasmid pCH553 (18) containing a 921-bp deletion in hoxA was used to delete the hoxA gene in HF884. The mutant strain was designated HF950.
Mobilizable plasmids were transferred from E. coli S17-1 to R. eutropha by a spot mating technique, as described previously (39). Transconjugants were selected on mineral salts agar plates containing 0.4% fructose and the appropriate antibiotic. Mutants harboring an allelic exchange within the chromosome were isolated as described previously (22).
Plasmid stability.
For the determination of plasmid stability, strains were grown in medium without antibiotic, and samples were withdrawn from the cultures at different times over the course of cultivation. The samples were diluted and spread on mineral salts medium with and without antibiotic. The CFU on plates without antibiotic were counted and set to 100%, and the numbers of CFU obtained from plates with antibiotic were referenced to them.
Cyanophycin extraction and arginine purification.
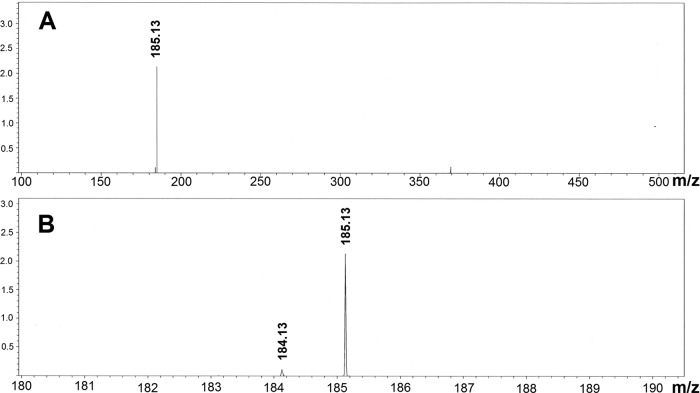
Laboratory scale extraction of cyanophycin from cells was done by the acid extraction procedure described by Frey et al. (9), including two acid solubilization and neutralization cycles. Finally the purified cyanophycin was vacuum dried. Cyanophycin was determined gravimetrically, i.e., the cyanophycin extracted from a given amount of biomass was weighed after drying. Yields are given as percentages of CDW. For a fast qualitative test of cyanophycin accumulation during fermentation, the extraction protocol was shortened as follows. One milliliter of cell culture was harvested, and cell pellets were extracted with 1 ml 0.1 M HCl at 37°C for 2 h. Ten microliters of the supernatant was mixed with 10 μl of a 0.2% SDS and 0.2% mercaptoethanol mixture, heated at 56°C for 5 min, and analyzed via SDS-PAGE. For purification of arginine from cyanophycin, the polymer was hydrolyzed with 6 N HCl at 110°C for 24 h. Excess HCl was removed under vacuum. The hydrolysate was decolorized with active charcoal and applied to a Dowex 50 column. Impurities and aspartate were eluted by flushing with 0.25 M ammonium acetate, pH 7. Arginine was eluted with 0.5 M NH4OH. Ammonia was removed by evaporation and freeze-drying. The resulting product is a white powder >95% chemically pure. The chemical purity was assessed by amino acid analysis with postcolumn ninhydrin staining on a Sykam amino acid analyzer. The major impurity was ammonium salt. To assay the isotopic enrichment of the purified arginine, samples were examined via mass spectrometry on a Brucker Daltonics microTOF mass spectrometer in positive mode. The isotopic enrichment of [13C/15N]arginine (MW, 185.13 for the protonated form) was calculated using the areas under the peaks at MW 185.13, 184.13, and 183.13.
RESULTS AND DISCUSSION
Plasmid-based expression of cyanophycin synthetase under autotrophic growth conditions.
We constructed an expression vector on the basis of the broad-host-range vector pCM62 (26), in which we cloned a Strep-tagged version of the cphA gene from Synechocystis sp. PCC6308 under the control of the strong PcbbL promoter of the R. eutropha H16 cbbc operon (17). The cbbc operon contains the genes encoding most of the enzymes of the Calvin-Benson-Bassham cycle, by which R. eutropha H16 fixes CO2 during lithoautotrophic growth. PcbbL-controlled transcription is repressed during cultivation on pyruvate and succinate, partially derepressed on fructose and gluconate, and fully derepressed during autotrophic growth on CO2 and formate (3, 11, 17).
The resulting construct, designated pGE778, was transferred via conjugation from E. coli S17-1 to R. eutropha HF884 (37), a mutant deficient in the synthesis of the storage polymer poly(3-hydroxybutyrate) (PHB). Wild-type strains of R. eutropha produce copious amounts of PHB and therefore are not suitable as production strains. When the transconjugants were cultivated heterotrophically on 0.4% fructose, the cells appeared as optically uniform rods under the light microscope. In contrast, cells cultivated lithoautotrophically on a mixture of H2, CO2, and O2, i.e., under conditions that lead to induction of the PcbbL promoter, contained prominent light-scattering inclusions (Fig. 1). The number and size of the light-scattering granules increased in the course of cultivation. These observations suggested to us that the granules consisted of cyanophycin, also known as cyanophycin granule polypeptide (CGP), the product of cyanophycin synthetase (CphA). Immunodetection specific for the Strep tag fused to the C terminus of CphA6308 clearly showed the heterologous overproduction of cphA6308 under lithoautotrophic growth conditions (data not shown). Cyanophycin extracted from the cells formed a polydisperse band on SDS-PAGE with sizes ranging from 22 to 30 kDa (data not shown).
Fig 1.
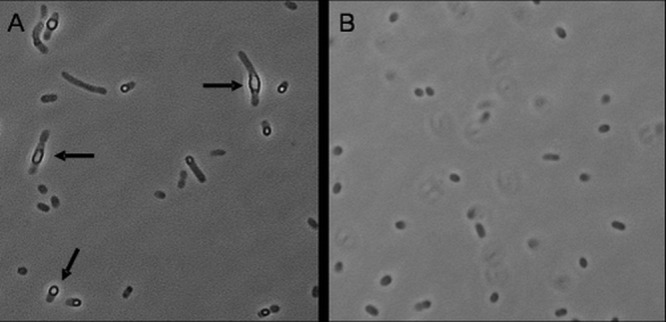
Micrographs of R. eutropha HF950(pGE779) under cyanophycin-inducing and noninducing conditions. Samples were withdrawn at an optical density at 436 nm (OD436) ranging from 8 to 10. (A) Strain HF950(pGE779) cultivated lithoautotrophically on H2 and CO2 (in a mixture of 3% H2, 10% CO2, 10% O2, and 77% N2). Light-scattering inclusions (arrows) indicate the presence of CGP. (B) Strain HF950(pGE779) cultivated heterotrophically in mineral salts medium with 0.4% fructose as a carbon source.
Cyanophycin production in the absence of antibiotic selection.
The yield of cyanophycin as a percentage of CDW was determined in cultures of strain HF884(pGE778) with and without antibiotic. Cells grown in the presence of 15 μg/ml tetracycline accumulated cyanophycin up to 5.5% of CDW. In cultures without antibiotic, the cells accumulated less than 3% cyanophycin based on the CDW. Microscopic examination revealed that the number of cells with a light-scattering cyanophycin inclusion decreased in the course of the cultivation. Plasmid loss would be an obvious explanation for this observation. In order to monitor plasmid loss, we scored tetracycline-resistant cells in parallel cultures grown in minimal medium containing fructose with and without the addition of tetracycline. Not surprisingly, the cultures grown in the absence of tetracycline contained significantly more tetracycline-sensitive cells. After a 24-hour incubation, only about 70% (71.4% ± 8.5%; n = 3) of the cells were tetracycline resistant compared to control cultures in the same medium with tetracycline. This is in line with the conjecture that plasmid-free cells were increasing in number, since there was no selective pressure for plasmid maintenance.
Chromosomal integration of PcbbL-cphA.
Our first strategy to overcome the problem of plasmid loss was to integrate the PcbbL-cphA6308 expression cassette into the chromosome of HF884. We chose to integrate the construct into the dispensable nor2 region located on chromosome 2, resulting in strain HF952. The integration was confirmed by PCR amplification using primers specific for flanking sequences. HF952 showed a stable but low-level accumulation of cyanophycin during lithoautotrophic growth (<2% based on CDW). The lower levels of cyanophycin than in strain HF884(pGE778) are most likely due to low gene dosage, i.e., a single copy of cphA (Fig. 2). As a consequence of the above finding, we did not pursue this approach further.
Fig 2.
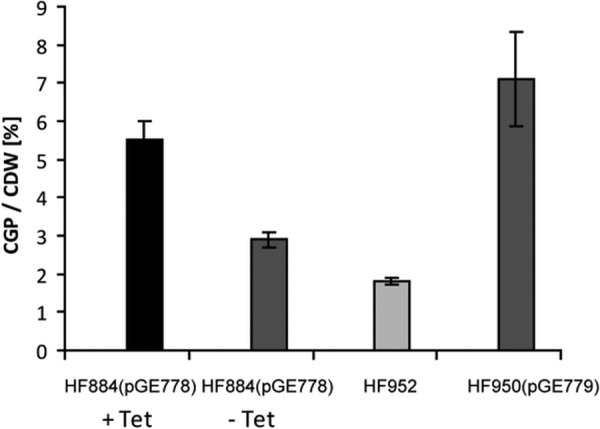
Comparison of cyanophycin yields for different overproducing strains. Cells of HF884(pGE778) were cultivated with and without antibiotic, and the yields of cyanophycin were compared with those for HF952 (chromosomally integrated copy of the cphA gene) and HF950(pGE779) (hoxA-based plasmid addiction system). The cells were grown in batch mode under an H2-CO2-O2-N2 (3:10:10:77) atmosphere to an OD436 of 10 to 12, i.e., 1.5 to 1.8 g/liter CDW. The error bars represent standard deviations of two biological replicates.
Construction of a hoxA-based plasmid addiction system.
Previous studies demonstrated the efficacy of a plasmid addiction system for maintaining an expression vector in cells of R. eutropha grown heterotrophically in the absence of antibiotic selection (45). We decided to adopt a similar strategy to ensure stable maintenance of a cphA expression plasmid under lithoautotrophic growth conditions. Our addiction system is based on the regulatory mechanism governing hydrogenase gene expression in R. eutropha. Hydrogenase expression is controlled by four components encoded by the genes hoxA, -B, -C, and -J (5, 10). HoxB and HoxC constitute subunits of a regulatory hydrogenase responsible for hydrogen sensing. HoxA and HoxJ form a two-component system, where HoxJ is a histidine kinase and HoxA is a response regulator (21). The dephosphorylated form of HoxA is active as a transcription regulator, promoting the expression of the dual operons encoding the soluble and membrane-bound hydrogenases. Mutants lacking hoxA are not able to grow on H2 due to a lack of hydrogenase.
First, we engineered an appropriate R. eutropha strain as a host for the addiction plasmid. We generated an in-frame deletion in the hoxA gene of HF884 via an allelic-exchange protocol (22). The resulting strain, designated HF950, grew normally on organic substrates but was unable to grow lithoautotrophically on H2 and CO2. Next, we proceeded to construct the corresponding vector plasmid. We inserted a fragment of DNA carrying the hoxA gene of Alcaligenes sp. M50 into the cphA expression vector, resulting in plasmid pGE779 (Fig. 3). The latter strain is a very close relative of R. eutropha H16. In fact, it should probably be designated R. eutropha (14). Subtle differences in the nucleotide sequences of the homologous regulatory genes of strains M50 and H16 are correlated with deregulated and excessive hydrogenase expression in strain H16 (23). We deliberately used the regulatory components of Alcaligenes sp. M50 instead of the autochthonous components of R. eutropha H16. The addiction plasmid was transferred via conjugation into HF950. The resulting strain had its ability to grow lithoautotrophically restored, with a doubling time of approximately 6.1 h under an atmosphere of H2, CO2, and O2 (80%, 10%, and 10%, respectively), which is somewhat slower than the wild-type R. eutropha H16 (4.2 h).
Fig 3.
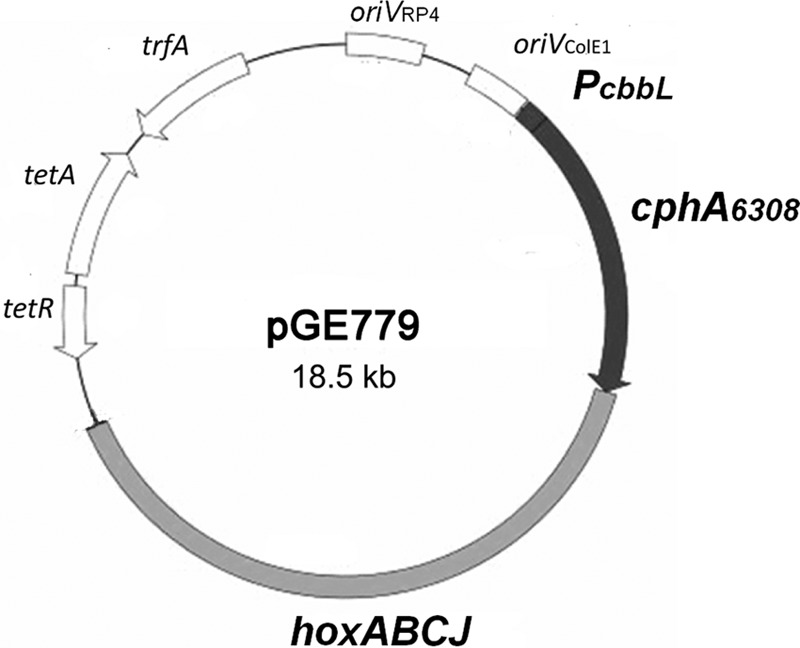
Expression plasmid pGE779 harboring the cyanophycin synthetase gene (cphA6308) from Synechocystis sp. 6308 under the control of the cbbL promoter (PcbbL) and the hydrogenase regulator region from Alcaligenes sp. strain M50 (hoxABCJ). oriVcolE1, colE1 origin of replication; oriVRP4, RK2/RP4 origin of replication; trfA, RK2/RP4 replication initiator; tetA and tetR, tetracycline resistance determinants.
Yields of cyanophycin from different heterologous production strains.
In a lithoautotrophic batch cultivation with cultures grown in a glass desiccator containing a defined gas mixture (H2/O2/CO2/N2; 3%:10%:10%:77%), HF950(pGE779) produces reproducible amounts of cyanophycin, with a maximum yield of 7.1% ± 1.2%. This is slightly more than HF884(pGE778) in the presence of tetracycline (5.5% ± 0.5%) and significantly more than the amounts yielded by antibiotic-free cultivation of HF884(pGE778) (2.9% ± 0.2%) and HF952 (1.8% ± 0.2%) (Fig. 2). Plasmid stability tests showed practically no plasmid loss even after prolonged cultivation, confirming that pGE779 is highly stable (Fig. 4). In contrast, the plasmid pGE778 is rapidly lost from HF884 when no antibiotic is present in the growth medium (see above).
Fig 4.
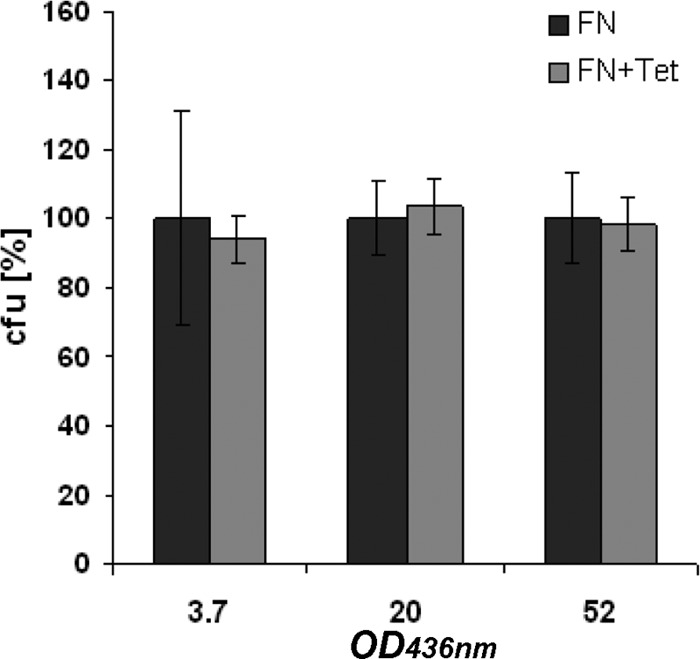
Stability of plasmid pGE779 in strain HF950. Cultures were grown lithoautotrophically in a fed-batch fermentation. Samples withdrawn at different times (OD436 = 3.7, 20, and 52) were spread on mineral salts medium agar plates containing 0.4% fructose (FN) either with (+Tet) or without antibiotic. CFU on agar plates without antibiotic were set to 100%, and CFU obtained from the plates with antibiotic were referenced to them. The error bars represent standard deviations of triplicates.
As mentioned above, several studies on the heterologous expression of cyanophycin synthetase have been published in recent years. Different cyanophycin synthetase genes and various vector-host systems have been tested. Expression of the cphA gene from Synechocystis sp. PCC6803 in E. coli DH1 resulted in cyanophycin yields of 12% (based on CDW) for cells grown in minimal medium and 24% for cells grown in complex medium (9). Cells of P. putida KT2440 and R. eutropha H16 expressing the cphA1 gene from A. variabilis produced as much as 23.0% and 20.0% cyanophycin (relative to CDW), respectively (44). Mutants defective for the synthesis of polyhydroxyalkanoates made even more cyanophycin: 24.0% and 22.0%, respectively. Other studies have focused on cyanophycin production in fungi. Steinle and coworkers investigated the expression of the cphA gene from Synechocystis sp. PCC6308 in S. cerevisiae (41). The authors showed that cyanophycin accumulated up to 6.9% of CDW. Another study from the same laboratory demonstrated the production of cyanophycin up to 23.3% in P. pastoris carrying a mutant cphA gene (42). Recently, cphA genes from Synechocystis and Anabaena were expressed in R. oryzae, resulting in the synthesis of cyanophycin up to 0.5% of CDW (27). All of the studies cited above are based on heterotrophic culture conditions and, hence, are not relevant to the special case of producing SI-labeled amino acids. Nevertheless, the high levels of cyanophycin obtained in some of these systems are enticing and suggest that cyanophycin yields in autotrophic production based on the system reported here can be improved. We have begun a program of experiments aimed at the systematic optimization of our expression plasmid. Preliminary results indicate that transferring the PcbbL-cphA expression cassette to a replicon with a higher copy number leads to a doubling of the cyanophycin yield (S. Lütte and B. Friedrich, unpublished data). In a recent systematic study on heterotrophically grown R. eutropha, Lin and coworkers showed that by adjusting various physicochemical parameters of the fermentation, the yield of cyanophycin could be increased to 47.5% (24). It is not unlikely that a similar optimization of the conditions of autotrophic fermentation will also lead to improved yields.
Yield and purity of SI-labeled arginine produced on an industrial scale.
In order to test our new expression system under real-life production conditions, strain HF950(pGE779) was grown lithoautotrophically on 13CO2 and 15NH4Cl in a 10-liter closed fed-batch fermentor. Samples were withdrawn at various points during the course of fermentation and analyzed for cyanophycin accumulation by SDS-PAGE. Cyanophycin was present in the cells and appeared to increase during the exponential growth phase (Fig. 5). Yields of SI-labeled arginine varied between 2.5 and 5 g per 10-liter fermentation. After harvesting the cells, cyanophycin was isolated by acid extraction and subsequently hydrolized, yielding [13C/15N]arginine and [13C/15N]aspartate. The [13C/15N]arginine was purified via ion-exchange chromatography. The isotopic enrichment of 13C plus 15N in the [13C/15N]arginine, as determined by mass spectrometry, varied between 98.8% and 99.4% (Fig. 6). These results are proof of principle demonstrating that enrichment of cyanophycin in lithoautotrophically grown cells is an attractive approach to the production of SI-labeled arginine.
Fig 5.
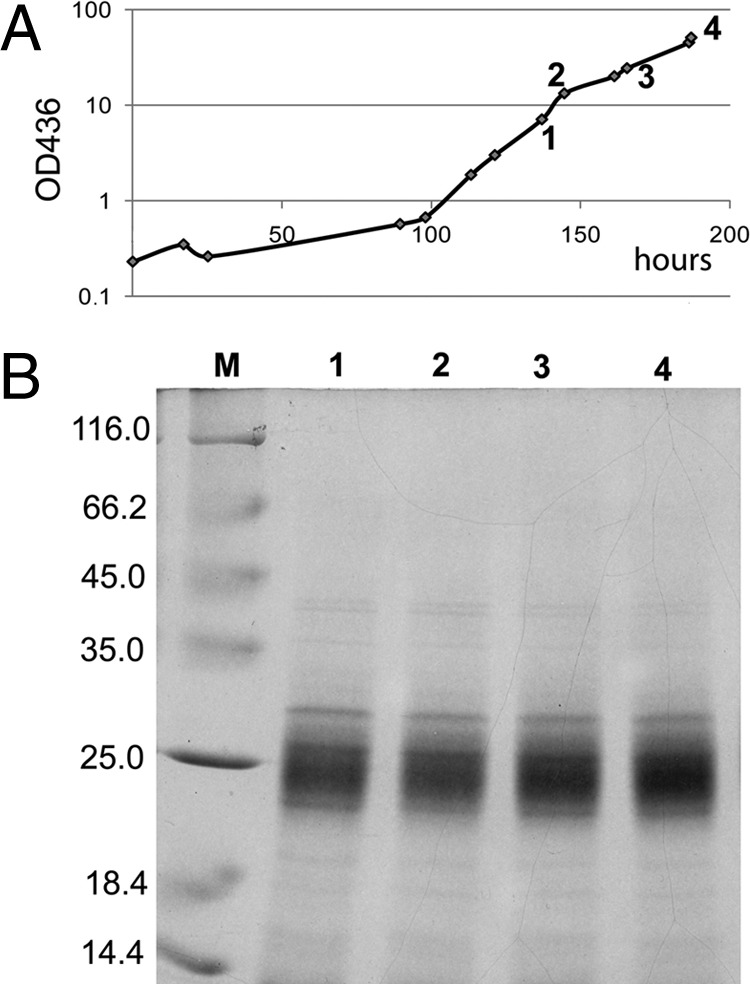
Cyanophycin overproduction during lithoautotrophic fed-batch cultivation of R. eutropha HF950(pGE779) grown on 13CO2 and 15NH4Cl. (A) Growth curve of HF950(pGE779). Samples were taken at the time points indicated in panel A (1 to 4) and subjected to SDS-PAGE. (B) SDS-PAGE. Equal amounts of cells (based on the optical density at 436 nm) were harvested and extracted with 0.1 M HCl. The acid-extracted cyanophycin was dissolved in buffer containing SDS and mercaptoethanol and separated on a 15% SDS-PAGE gel. M, protein size standards. Molecular weights of the standard proteins are given at the left.
Fig 6.
Mass spectrum of [13C/15N]arginine obtained from overproduced cyanophycin. Cyanophycin from R. eutropha HF950(pGE779) grown on 13CO2 and 15NH4Cl was hydrolyzed, yielding a mixture of SI-labeled arginine (mass, 184.2 Da) and SI-labeled aspartate (not shown), which were chromatographically separated. Mass spectra were measured in positive mode. (This means that the molecular mass is 1 Da higher due to the protonation.) (A) Full spectrum. (B) Zoom into the mass range of 185 Da.
ACKNOWLEDGMENT
This work was supported by the Bundesministerium für Bildung und Forschung (BMBF) in the framework of the Competence Network GenoMik-Transfer (application-oriented research on nonpathogenic microorganisms for health, nutrition, and resource-efficient industrial production).
Footnotes
Published ahead of print 31 August 2012
REFERENCES
- 1. Aboulmagd E, Oppermann-Sanio FB, Steinbüchel A. 2000. Molecular characterization of the cyanophycin synthetase from Synechocystis sp. strain PCC6308. Arch. Microbiol. 174:297–306 [DOI] [PubMed] [Google Scholar]
- 2. Aboulmagd E, Oppermann-Sanio FB, Steinbüchel A. 2001. Purification of Synechocystis sp. strain PCC6308 cyanophycin synthetase and its characterization with respect to substrate and primer specificity. Appl. Environ. Microbiol. 67:2176–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bowien B, Kusian B. 2002. Genetics and control of CO2 assimilation in the chemoautotroph Ralstonia eutropha. Arch. Microbiol. 178:85–93 [DOI] [PubMed] [Google Scholar]
- 4. Brigham CJ, et al. 2010. Elucidation of β-oxidation pathways in Ralstonia eutropha H16 by examination of global gene expression. J. Bacteriol. 192:5454–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buhrke T, Lenz O, Porthun A, Friedrich B. 2004. The H2-sensing complex of Ralstonia eutropha: interaction between a regulatory [NiFe] hydrogenase and a histidine protein kinase. Mol. Microbiol. 51:1677–1689 [DOI] [PubMed] [Google Scholar]
- 6. Cramm R. 2009. Genomic view of energy metabolism in Ralstonia eutropha H16. J. Mol. Microbiol. Biotechnol. 16:38–52 [DOI] [PubMed] [Google Scholar]
- 7. Diniz SC, Voss I, Steinbüchel A. 2006. Optimization of cyanophycin production in recombinant strains of Pseudomonas putida and Ralstonia eutropha employing elementary mode analysis and statistical experimental design. Biotechnol. Bioeng. 93:698–717 [DOI] [PubMed] [Google Scholar]
- 8. Elbahloul Y, Krehenbrink M, Reichelt R, Steinbüchel A. 2005. Physiological conditions conducive to high cyanophycin content in biomass of Acinetobacter calcoaceticus strain ADP1. Appl. Environ. Microbiol. 71:858–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Frey KM, Oppermann-Sanio FB, Schmidt H, Steinbüchel A. 2002. Technical-scale production of cyanophycin with recombinant strains of Escherichia coli. Appl. Environ. Microbiol. 68:3377–3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Friedrich B, Buhrke T, Burgdorf T, Lenz O. 2005. A hydrogen-sensing multiprotein complex controls aerobic hydrogen metabolism in Ralstonia eutropha. Biochem. Soc. Trans. 33:97–101 [DOI] [PubMed] [Google Scholar]
- 11. Friedrich CG, Friedrich B, Bowien B. 1981. Formation of enzymes of autotrophic metabolism during heterotrophic growth of Alcaligenes eutrophus. J. Gen. Microbiol. 122:69–78 [Google Scholar]
- 12. Gottschalk G, Eberhardt U, Schlegel HG. 1964. Verwertung von Fructose durch Hydrogenomonas H 16 (I). Arch. Mikrobiol. 48:95–108 [PubMed] [Google Scholar]
- 13. Hai T, Frey KM, Steinbüchel A. 2006. Engineered cyanophycin synthetase (CphA) from Nostoc ellipsosporum confers enhanced CphA activity and cyanophycin accumulation to Escherichia coli. Appl. Environ. Microbiol. 72:7652–7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jenni B, Realini M, Arani M, Tamer Ü. 1988. Taxonomy of non H2-lithotrophic, oxalate-oxidizing bacteria related to Alcaligenes eutrophus. Syst. Appl. Microbiol. 10:126–133 [Google Scholar]
- 15. Kohlmann Y, et al. 2011. Analyses of soluble and membrane proteomes of Ralstonia eutropha H16 reveal major changes in the protein complement in adaptation to lithoautotrophy. J. Proteome Res. 10:2767–2776 [DOI] [PubMed] [Google Scholar]
- 16. Krehenbrink M, Oppermann-Sanio FB, Steinbüchel A. 2002. Evaluation of non-cyanobacterial genome sequences for occurrence of genes encoding proteins homologous to cyanophycin synthetase and cloning of an active cyanophycin synthetase from Acinetobacter sp. strain DSM 587. Arch. Microbiol. 177:371–380 [DOI] [PubMed] [Google Scholar]
- 17. Kusian B, Bednarski R, Husemann M, Bowien B. 1995. Characterization of the duplicate ribulose-1,5-bisphosphate carboxylase genes and cbb promoters of Alcaligenes eutrophus. J. Bacteriol. 177:4442–4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kusian B, Bowien B. 1995. Operator binding of the CbbR protein, which activates the duplicate cbb CO2 assimilation operons of Alcaligenes eutrophus. J. Bacteriol. 177:6568–6574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lahm HW, Langen H. 2000. Mass spectrometry: a tool for the identification of proteins separated by gels. Electrophoresis 21:2105–2114 [DOI] [PubMed] [Google Scholar]
- 20. Lawrence AG, et al. 2005. Transcriptional analysis of Ralstonia eutropha genes related to poly-(R)-3-hydroxybutyrate homeostasis during batch fermentation. Appl. Microbiol. Biotechnol. 68:663–672 [DOI] [PubMed] [Google Scholar]
- 21. Lenz O, Friedrich B. 1998. A novel multicomponent regulatory system mediates H2 sensing in Alcaligenes eutrophus. Proc. Natl. Acad. Sci. U. S. A. 95:12474–12479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lenz O, Schwartz E, Dernedde J, Eitinger M, Friedrich B. 1994. The Alcaligenes eutrophus H16 hoxX gene participates in hydrogenase regulation. J. Bacteriol. 176:4385–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lenz O, Strack A, Tran-Betcke A, Friedrich B. 1997. A hydrogen-sensing system in transcriptional regulation of hydrogenase gene expression in Alcaligenes species. J. Bacteriol. 179:1655–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin K, Elbahloul Y, Steinbüchel A. 2012. Physiological conditions conducive to high cell density and high cyanophycin content in Ralstonia eutropha strain H16 possessing a KDPG aldolase gene-dependent addiction system. Appl. Microbiol. Biotechnol. 93:1885–1894 [DOI] [PubMed] [Google Scholar]
- 25. Mackerras AH, de Chazal NM, Smith GD. 1990. Transient accumulations of cyanophycin in Anabaena cylindrica and Synechocystis 6308. J. Gen. Microbiol. 136:2057–2065 [Google Scholar]
- 26. Marx CJ, Lidstrom ME. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147:2065–2075 [DOI] [PubMed] [Google Scholar]
- 27. Meussen BJ, Weusthuis RA, Sanders JP, Graaff LH. 2012. Production of cyanophycin in Rhizopus oryzae through the expression of a cyanophycin synthetase encoding gene. Appl. Microbiol. Biotechnol. 93:1167–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mooibroek H, et al. 2007. Assessment of technological options and economical feasibility for cyanophycin biopolymer and high-value amino acid production. Appl. Microbiol. Biotechnol. 77:257–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Neumann K, et al. 2005. Production of cyanophycin, a suitable source for the biodegradable polymer polyaspartate, in transgenic plants. Plant Biotechnol. J. 3:249–258 [DOI] [PubMed] [Google Scholar]
- 30. Ong SE, et al. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics 1:376–386 [DOI] [PubMed] [Google Scholar]
- 31. Ong SE, Kratchmarova I, Mann M. 2003. Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC). J. Proteome Res. 2:173–181 [DOI] [PubMed] [Google Scholar]
- 32. Peplinski K, et al. 2010. Genome-wide transcriptome analyses of the ‘Knallgas’ bacterium Ralstonia eutropha H16 with regard to polyhydroxyalkanoate metabolism. Microbiology 156:2136–2152 [DOI] [PubMed] [Google Scholar]
- 33. Peplinski K, Ehrenreich A, Döring C, Bömeke M, Steinbüchel A. 2010. Investigations on the microbial catabolism of the organic sulfur compounds TDP and DTDP in Ralstonia eutropha H16 employing DNA microarrays. Appl. Microbiol. Biotechnol. 88:1145–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pohlmann A, Cramm R, Schmelz K, Friedrich B. 2000. A novel NO-responding regulator controls the reduction of nitric oxide in Ralstonia eutropha. Mol. Microbiol. 38:626–638 [DOI] [PubMed] [Google Scholar]
- 35. Pohlmann A, et al. 2006. Genome sequence of the bioplastic-producing “Knallgas” bacterium Ralstonia eutropha H16. Nat. Biotechnol. 24:1257–1262 [DOI] [PubMed] [Google Scholar]
- 36. Raberg M, et al. 2011. Proteomic and transcriptomic elucidation of the mutant ralstonia eutropha G+1 with regard to glucose utilization. Appl. Environ. Microbiol. 77:2058–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reinecke F, Steinbüchel A. 2009. Ralstonia eutropha strain H16 as model organism for PHA metabolism and for biotechnological production of technically interesting biopolymers. J. Mol. Microbiol. Biotechnol. 16:91–108 [DOI] [PubMed] [Google Scholar]
- 38. Schwarze A, Kopczak MJ, Rogner M, Lenz O. 2010. Requirements for construction of a functional hybrid complex of photosystem I and [NiFe]-hydrogenase. Appl. Environ. Microbiol. 76:2641–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology 1:784–791 [Google Scholar]
- 40. Simon RD. 1976. The biosynthesis of multi-L-arginyl-poly(L-aspartic acid) in the filamentous cyanobacterium Anabaena cylindrica. Biochim. Biophys. Acta 422:407–418 [DOI] [PubMed] [Google Scholar]
- 41. Steinle A, Oppermann-Sanio FB, Reichelt R, Steinbüchel A. 2008. Synthesis and accumulation of cyanophycin in transgenic strains of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 74:3410–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Steinle A, Witthoff S, Krause JP, Steinbüchel A. 2010. Establishment of cyanophycin biosynthesis in Pichia pastoris and optimization by use of engineered cyanophycin synthetases. Appl. Environ. Microbiol. 76:1062–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Szyperski T, et al. 1999. NMR structure of the chimeric hybrid duplex r(gcaguggc).r(gcca)d(CTGC) comprising the tRNA-DNA junction formed during initiation of HIV-1 reverse transcription. J. Biomol. NMR 13:343–355 [DOI] [PubMed] [Google Scholar]
- 44. Voss I, Diniz SC, Aboulmagd E, Steinbüchel A. 2004. Identification of the Anabaena sp. strain PCC7120 cyanophycin synthetase as suitable enzyme for production of cyanophycin in gram-negative bacteria like Pseudomonas putida and Ralstonia eutropha. Biomacromolecules 5:1588–1595 [DOI] [PubMed] [Google Scholar]
- 45. Voss I, Steinbüchel A. 2006. Application of a KDPG-aldolase gene-dependent addiction system for enhanced production of cyanophycin in Ralstonia eutropha strain H16. Metab. Eng. 8:66–78 [DOI] [PubMed] [Google Scholar]
- 46. Ziegler K, Deutzmann R, Lockau W. 2002. Cyanophycin synthetase-like enzymes of non-cyanobacterial eubacteria: characterization of the polymer produced by a recombinant synthetase of Desulfitobacterium hafniense. Z. Naturforsch. C 57:522–529 [DOI] [PubMed] [Google Scholar]
- 47. Ziegler K, et al. 1998. Molecular characterization of cyanophycin synthetase, the enzyme catalyzing the biosynthesis of the cyanobacterial reserve material multi-L-arginyl-poly-L-aspartate (cyanophycin). Eur. J. Biochem. 254:154–159 [DOI] [PubMed] [Google Scholar]