Abstract
We previously found that a short exposure of Staphylococcus aureus to subinhibitory (SI) doses of epigallocatechin gallate (EGCG) results in increased cell wall thickness, adaptation, and enhanced tolerance to cell-wall-targeted antibiotics. In this study, the response to EGCG of sigB and vraSR transcription factor mutants was characterized. We show that in contrast to the results observed for wild-type (WT) strains, an S. aureus 315 vraSR null mutant exposed to SI doses of EGCG did not exhibit increased tolerance to EGCG and oxacillin. A diminished increase in tolerance to ampicillin (from 16-fold to 4-fold) and no change in the magnitude of resistance to vancomycin were observed. Preexposure to EGCG enhanced the tolerance of wild-type and sigB null mutant cells to lysostaphin, but this enhancement was much weaker in the vraSR null mutant. Marked upregulation (about 60-fold) of vraR and upregulation of the peptidoglycan biosynthesis-associated genes murA, murF, and pbp2 (2-, 5-, and 6-fold, respectively) in response to SI doses of EGCG were determined by quantitative reverse transcription-PCR (qRT-PCR). EGCG also induced the promoter of sas016 (encoding a cell wall stress protein of unknown function which is not induced in vraSR null mutants) in a concentration-dependent manner, showing kinetics comparable to those of cell-wall-targeting antibiotics. Taken together, our results suggest that the two-component VraSR system is involved in modulating the cell response to SI doses of EGCG.
INTRODUCTION
Staphylococcus aureus causes a wide array of infectious diseases in humans, ranging from localized skin lesions to systemic infections such as osteomyelitis, endocarditis, pneumonia, bacteremia, and other life-threatening complications (25). Methicillin-resistant S. aureus (MRSA) has become a major nosocomial pathogen of hospital-acquired infections of surgical wounds and infections associated with indwelling medical devices (15). Hospital strains of S. aureus are usually resistant to a variety of different antibiotics, antiseptics, and disinfectants, leaving very limited therapeutic options for the treatment of staphylococcal infections. A few strains are resistant to all clinically useful antibiotics except for vancomycin; however, even vancomycin-resistant strains (VRSA) have now been reported (34).
The cell wall of Gram-positive bacteria is the first line of defense against environmental, physical, and chemical hazards, and its integrity and homeostasis are crucial for survival. It is thus no wonder that many clinically important antibiotics, from beta-lactams to vancomycin, have been developed over the years to target or interfere with cell wall biosynthesis to fight bacterial infections (20, 27). To maintain cell wall architecture and function under hostile conditions, Gram-positive bacteria are equipped with cell wall stress sensor systems that respond to alterations and dysfunctions by activating repair mechanisms (31). In S. aureus, the two-component system VraSR is strongly induced by a number of antibiotics that perturb cell wall synthesis, but not by general stresses such as heat, osmotic shock, or pH shifts (22). VraSR alters the expression of pbp2, encoding an essential penicillin-binding protein (PBP) crucial for peptidoglycan cross-linking, and murF, which encodes an enzyme catalyzing the last cytoplasmic step in peptidoglycan biosynthesis (12, 36). The intermembrane sensor VraS activates the response regulator VraR by phosphorylation, which in its active form binds VraR-responsive promoter sequences and controls the expression of target genes (2). In addition, rapid adaptation in response to environmental stresses in S. aureus has been shown to be mediated by the alternate transcription factor SigB (30). Moreover, sigB null mutants show alterations in cell-wall-associated proteins, and overexpression of sigB results in 20% thicker cell walls than those of the parent strain, with increased resistance to the lytic activity of lysostaphin and cell-wall-active antibiotics (28).
In contrast to antibiotics, which act selectively against specific cell targets, most plant extracts exert their antimicrobial effect on several sites within the cell simultaneously (6, 35). Epigallocatechin gallate (EGCG), the major polyphenol component of green tea extract, has been shown to possess several antibacterial activities, such as partitioning the lipid bilayer of the bacterial cytoplasmic membrane (7, 19, 37, 38) and specifically inhibiting the activities of bacterial FabG and FabI reductases (key enzymes in fatty acid synthesis) (42), DNA gyrase (16), and gelatinase (5). Moreover, EGCG has been shown to interfere with the integrity of the cell wall through direct binding to peptidoglycan (43). We previously reported that a short exposure of S. aureus to subinhibitory (SI) doses of EGCG causes a >2-fold increase in cell wall thickness. Furthermore, it increases resistance to antibiotics targeting the bacterial cell wall (4). We postulate that EGCG, like cell-wall-targeted antibiotics, acts as a cell wall stress signal that induces expression of the VraSR or SigB system, or both, which in turn enhances the transcription of genes encoding cell wall repair enzymes.
MATERIALS AND METHODS
Bacterial isolates, growth, and EGCG adaptation conditions.
S. aureus strains (Table 1) were cultured in Mueller-Hinton broth (MHB; Difco, Detroit, MI) at 37°C. For solid agar medium (MHA), Bacto agar (Difco) was added to 1.5% in MHB. For adaptation experiments, bacterial cultures were grown to late exponential phase in MHB and diluted 1,000-fold in fresh MHB, and growth was then continued until the culture reached a turbidity (optical density at 600 nm [OD600]) of 0.1 (approximately 107 CFU/ml). The culture was transferred to 30°C, and 20 μg/ml of EGCG was added for an additional 120 min of growth (adaptation), or growth was continued without supplemental EGCG (control).
Table 1.
Staphylococcus aureus strains used in this study
Antimicrobial assays.
MICs were determined by the broth dilution method as recommended by the Clinical and Laboratory Standards Institute (CLSI), using cation-adjusted MHB (Difco), with minor modifications. Bacterial cultures were grown for 2 h at 30°C in the presence or absence (control) of 20 μg/ml EGCG to an OD600 of 0.3 (approximately 108 CFU/ml) and were then washed twice with phosphate-buffered saline (PBS) by centrifugation, diluted in PBS to 5 × 105 CFU/ml, and used to inoculate 200 μl MHB containing 2-fold serial dilutions of EGCG, vancomycin, oxacillin, or ampicillin (Sigma, St. Louis, MO). The MIC was determined as the lowest concentration at which no visible growth occurred.
Lysostaphin lysis assay.
A lysostaphin lysis assay was performed to examine alterations in susceptibility to lysostaphin. EGCG-adapted and control cultures were grown to an OD600 of 1.0, washed, and resuspended in the same volume of MHB containing 200 μg/ml lysostaphin (Sigma). These suspensions were then incubated at 37°C with shaking, and aliquots were frequently removed for OD600 determination for 90 min. Data for autolysis and lysostaphin lysis were quantified as the proportion of absorbance remaining at each time point relative to the initial absorbance.
RNA isolation.
S. aureus strain Newman was grown in MHB to an OD600 of 0.1, and the culture was transferred to 30°C and supplemented, or not, with 20 μg/ml of EGCG. After 15 min, aliquots of suspended cells containing approximately 108 CFU were centrifuged (12,000 × g, 7 min, 4°C). The supernatant was decanted and the pellet immediately frozen in liquid nitrogen and stored at −80°C. Bacterial pellets were lysed in 1 ml of Tri reagent (Sigma) in a Fastprep24 tissue and cell homogenizer (MP Biomedicals, Solon, OH). Total RNA was isolated according to the manufacturer's protocol for Tri reagent. RNAs (150 ng) isolated from independent samples of bacteria were reverse transcribed into cDNAs by use of random hexamers and a high-capacity cDNA reverse transcription (RT) kit (Applied Biosystems, Foster City, CA). Verification of RNA integrity and concentration, as well as RNA purification and transcription into cDNA, was performed as previously described (17).
qRT-PCR.
Transcription levels of murA, murF, pbp2, vraR, and aaa were determined by quantitative real-time RT-PCR (qRT-PCR) analysis of S. aureus Newman grown in the presence of 20 μg/ml EGCG. The expression level of the 16S rRNA gene was used to normalize the expression data for the gene of interest, as this gene showed a constant low expression level throughout the treatment protocols. Primers were designed using Primer Express, version 2 (Applied Biosystems), to generate amplicons ranging from 50 to 150 bp in size (see Table 2 for primer sequences). Real-time PCR amplification was performed on an ABI 7300 detection system (Applied Biosystems), with two independent cultures amplified in triplicate in 20-μl reaction mixtures containing 10 μl Power SYBR green PCR master mix (Applied Biosystems), 1 μl of each primer (0.5 μM), and 5 ng cDNA. A reaction mixture without template was run as a control. Thermal cycling conditions were as follows: initial denaturation at 95°C for 15 min followed by 40 cycles of 94°C for 15 s, primer annealing at the optimal temperature for 30 s, and 72°C for 30 s. The 2−ΔΔCT method (24) was used to determine the relative expression levels of the investigated genes in cells exposed to EGCG compared to nonexposed (control) cells. The significance of the difference between EGCG-exposed and control cells was calculated by the t test (using GraphPad Prism, version 23) and deemed statistically significant if the P value was <0.05 or <0.01.
Table 2.
Primer sequences for quantitative RT-PCR
| Gene | Primer direction | Primer sequence (5′ → 3′) |
|---|---|---|
| murF | Forward | GTTCCATACGCATACCAGTTAAGCT |
| Reverse | TGCGGTTGGTCATGAATTAGG | |
| pbp2 | Forward | ACGTCGTTAACAGAAACCAAGCA |
| Reverse | TGCGGTTGGTCATGAATTAGG | |
| vraR | Forward | TGAGTCGTCGCTTCTACACCAT |
| Reverse | ATTGCCAAAGCCCATGAGTT | |
| aaa | Forward | TTTAATCGTCGTGCTGAAATTGG |
| Reverse | TGCTGCCGCTGCGTTAT | |
| 16S rRNA gene | Forward | CAACGAGCGCAACCCTTAAG |
| Reverse | TTTGTCACCGGCAGTCAACTT |
Luciferase assay.
For luciferase assay, an overnight culture of the lux reporter in S. aureus strain BB255 harboring the reporter plasmid psas016p-luc+ (8) was diluted 500-fold in fresh Luria-Bertani (LB) broth (Difco Laboratories), and its growth was continued to an OD600 of 0.5. Different concentrations of oxacillin (0 to 0.5 μg/ml) or EGCG (0 to 128 μg/ml) were added, 2-ml samples were collected after 1 h and centrifuged (20,000 × g, 2 min), and the pellets were frozen at −20°C. To measure luciferase activity, pellets were thawed briefly and resuspended in PBS (pH 7.4) to an OD600 of 1. Aliquots (50 μl) of each of the cell suspensions (for each oxacillin or EGCG concentration) were then transferred to wells of a 96-well flat-bottomed white polystyrene tissue culture plate (Costar). To each well, 50 μl of luciferase assay substrate (Promega) was added, and bioluminescence was measured for 10 s, after a delay of 3 s, on a Veritas microplate luminometer and expressed in relative light units (RLU).
RESULTS
Cross-resistance of staphylococci exposed to EGCG.
A short exposure of S. aureus to SI doses of EGCG leads to its adaptation and enhanced tolerance to cell-wall-targeted antibiotics (4). To verify the role of VraSR and SigB in the adaptation, acquisition of cross-protection, and response to EGCG, two wild-type (WT) staphylococcus strains and their corresponding vraSR or sigB null mutant were preexposed to SI doses of EGCG and challenged with vancomycin, oxacillin, ampicillin, and EGCG.
Preexposure to SI doses of EGCG led to an adaptive response to EGCG in both WT strains, as evidenced by a 2-fold increase in MIC value for S. aureus Newman and a 4-fold increase for S. aureus N315 (Table 3). Similar increases (2- to 16-fold) in MIC values of the cell-wall-targeting antibiotics vancomycin, oxacillin, and ampicillin were also observed for both strains following preexposure to EGCG. When the Newman sigB null mutant was preexposed to EGCG, no increase in tolerance to EGCG, oxacillin, or ampicillin was observed, but there was a 2-fold increase in the MIC value of vancomycin. Preexposure of the N315 vraSR null mutant to EGCG resulted in no increase in tolerance to EGCG or oxacillin but to increases in the MIC values of vancomycin and ampicillin (2- and 4-fold, respectively) (Table 3).
Table 3.
MICs of antibiotics and EGCG for EGCG-adapted and control staphylococcal strains
| S. aureus strain | Strain status | MIC (μg/ml) |
|||
|---|---|---|---|---|---|
| Vancomycin | Ampicillin | Oxacillin | EGCG | ||
| Newman | Control | 0.25 | 4 | 0.5 | 60 |
| Adapted | 0.75 | 64 | 4 | 120 | |
| Newman sigB null | Control | 0.25 | 0.5 | 0.25 | 60 |
| Adapted | 0.5 | 0.5 | 0.25 | 60 | |
| N315 | Control | 0.25 | 8 | 2 | 25 |
| Adapted | 0.5 | 128 | 8 | 100 | |
| KVR | Control | 0.125 | 4 | 0.5 | 50 |
| Adapted | 0.25 | 16 | 0.5 | 50 | |
Lysostaphin susceptibility.
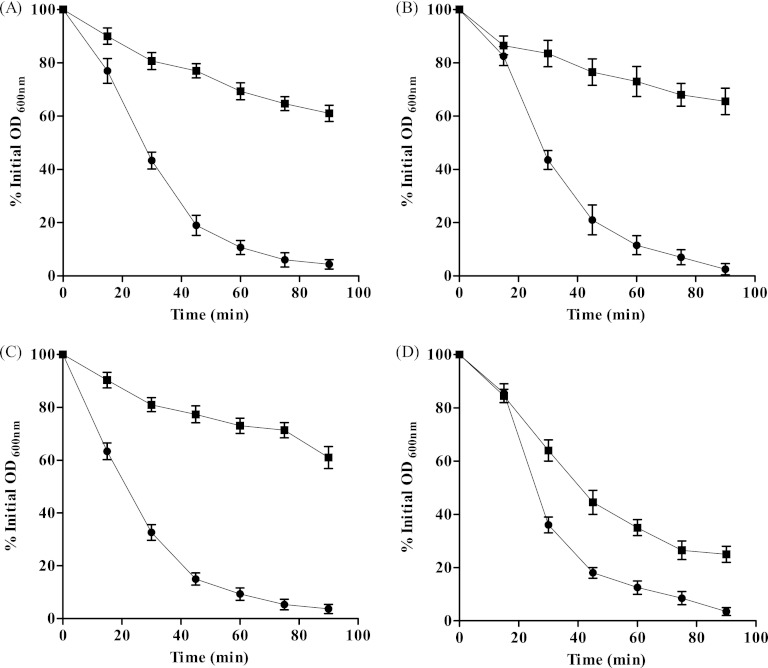
S. aureus exposure to SI doses of EGCG results in a significant increase in cell wall thickness (4). We suspected that this was due to EGCG modulating peptidoglycan cross-linking and causing alterations in the overall cell wall architecture. Therefore, the susceptibility of each WT strain and its isogenic mutant strain to the peptidoglycan-targeting endopeptidase lysostaphin, with and without adaptation to EGCG, was determined. Upon addition of lysostaphin, all WT and isogenic mutant strains reached 50% lysis within approximately 25 min, whereas preexposure to SI doses of EGCG resulted in a decrease in susceptibility, to approximately 30% lysis after as long as 90 min in all tested strains except for the N315 vraSR null mutant, which exhibited 50% lysis after 40 min (Fig. 1). Control reactions carried out in the absence of lysostaphin showed no significant changes in kinetics, indicating that the effects observed at the given incubation times were due to lysostaphin, not to autolysis. Based on these results, we concluded that preexposure of S. aureus to SI doses of EGCG results in increased resistance to lysostaphin. This increase was not achieved with the N315 vraSR null strain, probably as a direct result of the deletion of the VraSR genes.
Fig 1.
Effect of EGCG on lysostaphin sensitivity. The graphs indicate the susceptibilities of S. aureus strains Newman (A), N315 (B), Newman sigB null (C), and N315 vraSR null (D) and their respective EGCG-adapted strains (circles and squares, respectively). Results are percentages of the initial OD600. Experiments were repeated three times, and standard errors of the means (SEM) for all experiments are shown.
Gene expression in staphylococci exposed to EGCG.
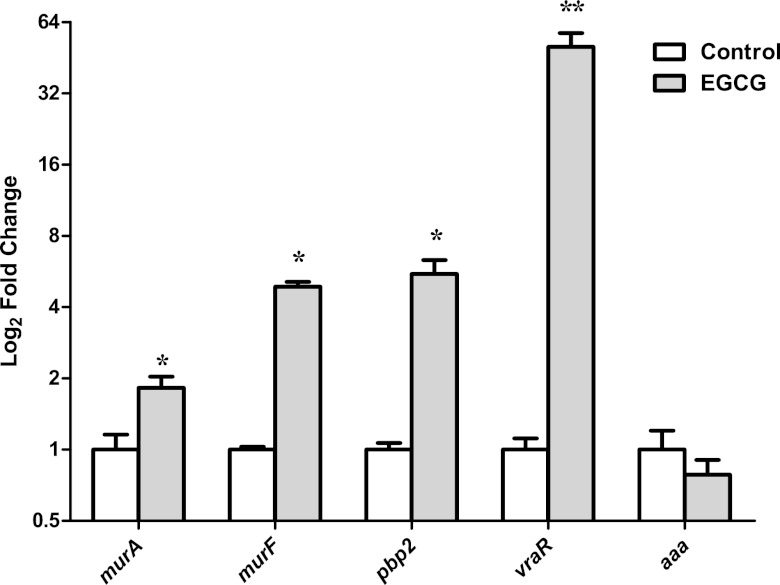
The attenuated response of the vraSR null mutant strain to EGCG established by the lysostaphin susceptibility assay, as opposed to the sigB null mutant's response, suggests that VraSR is a prime mediator of the cell response to EGCG. Therefore, the effects of an SI concentration of EGCG on transcriptional modulation of cell wall biosynthesis genes (murA, murF, and pbp2), the degradation gene aaa, and the regulator gene vraR from S. aureus strain Newman were assessed by qRT-PCR.
These analyses revealed a 30-fold increase in the relative expression (RE) of vraR after as little as 5 min (not shown) of exposure to EGCG, which increased to 60-fold after 15 min. We analyzed the RE of the cell wall biosynthesis- and degradation-associated genes after 15 min of exposure to EGCG. All three biosynthesis genes—murA, murF, and pbp2—were significantly upregulated (2-, 5-, and 6-fold, respectively) in response to EGCG. In contrast, there was no significant difference in the RE of aaa between EGCG-exposed and control cells (Fig. 2).
Fig 2.
Relative levels of murA, murF, pbp2, vraR, and aaa expression in S. aureus Newman as determined by qRT-PCR. Values are expressed as fold changes (log2) in EGCG-exposed cells (gray bars) compared to control cells (not exposed to EGCG) (white bars; values were standardized to 1). SEM for two independent cultures run in triplicate are shown. Significant differences (Student's t test) between EGCG-exposed and control cells are noted as follows: *, P < 0.05; **, P < 0.001.
Cell wall damage reporter assay.
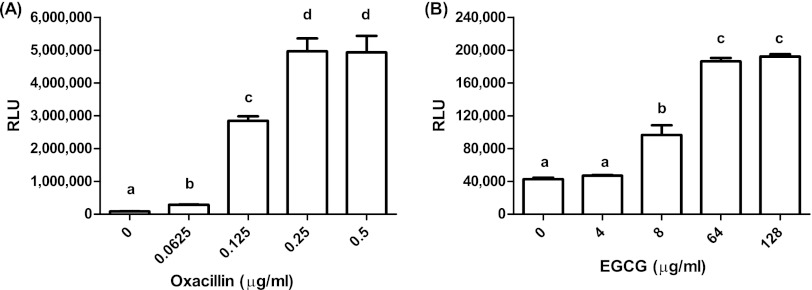
Transcription analysis of S. aureus exposed to cell-wall-targeting antibiotics revealed remarkable upregulation of a gene designated sas016, encoding a hypothetical protein (26, 32). S. aureus strain BB255, harboring a plasmid containing the sas016 promoter fused to the luciferase reporter gene, was used to test whether EGCG induces a reaction analogous to that caused by cell-wall-targeting antibiotics. EGCG and oxacillin at 1× MIC (64 μg/ml and 0.25 μg/ml, respectively) triggered an immediate response, with induction beginning within 10 min and maximal expression occurring at 60 min (not shown). The sas016 promoter was induced by EGCG and oxacillin in a concentration-dependent manner. Significant induction was obtained for oxacillin and EGCG at 0.25× MIC and 0.125× MIC, respectively, with maximal induction at 1× MIC for each of the antimicrobial compounds. The maximal induction level obtained by oxacillin was about 25-fold stronger than that with EGCG (Fig. 3).
Fig 3.
RLU measured upon induction of BB255/psas016p-luc+ after a 60-min exposure to different concentrations of oxacillin (A) or EGCG (B). The results of two induction experiments for oxacillin and EGCG, with SEM, are shown. Different letters indicate statistically significant differences in RLU (by Tukey's multiple-comparison test; P < 0.05).
DISCUSSION
We previously reported that a 2-h exposure of five Staphylococcus strains to an SI dose of EGCG significantly elevated their resistance to antibiotics targeting the bacterial cell wall and increased the cell wall thickness (4). In this study, our aim was to identify the cellular regulation system mediating this cross-resistance phenomenon.
In view of the specific role of the two-component VraSR signal transduction system in acquiring a certain level of tolerance to cell-wall-targeting antibiotics (1, 12) and in activating cell wall synthesis in vancomycin-resistant strains (9, 21), we suspected that EGCG might activate the VraSR system by inducing a cell wall stress signal. This hypothesis is supported by the fact that VraSR responds rapidly to several different cell wall antibiotics with different targets (12, 29, 36). Moreover, knockout of vraSR leads to a significant increase in the susceptibility of the S. aureus cell to antibiotics that inhibit cell wall synthesis (22). Indeed, in this work we observed that in contrast to the results observed for the WT strains, an S. aureus 315 vraSR null mutant exposed to SI doses of EGCG did not exhibit increased tolerance to EGCG and oxacillin. A diminished increase in tolerance to ampicillin (from 16-fold to 4-fold) and no change in the magnitude of resistance to vancomycin were observed. We postulate that EGCG binding to the cell wall or cell membrane (CM) moieties interrupts their integrity, inducing a stress response by an as yet unknown factor that mediates the upregulation of VraSR. In fact, binding of EGCG to peptidoglycan has been reported (18, 41, 43). Peptidoglycan homeostasis is maintained by very subtle and orchestrated enzymatic activities that might be modulated by EGCG. It has been proposed that damage inflicted to cell wall peptidoglycan is the main source of the stimuli to which VraR responds due to the tight control of VraS on the phosphorylation state of VraR (1). In addition, radiolabeling has shown that epicatechin gallate (ECG) binds predominantly to the CM, inducing an increase in its fluidity which results in uncoupling of some cell wall components that then cause delocalization of PBP2; as a consequence, the cells become more sensitive to beta-lactams and oxacillin (3). The structural similarity between ECG and EGCG and their common antimicrobial properties (33) tempt us to suggest that binding of EGCG to CM and relocation of PBP2 might be an additional mechanism conferring tolerance to beta-lactams.
We also showed that a mutant deleted in the transcription factor sigB is unable to respond to EGCG by adaptation and cross-protection, suggesting that this transcription factor is also involved in the S. aureus response to EGCG. We do not yet know if each of the investigated transcription factors operates separately or if there is cross talk between them. Moreover, we cannot exclude the possibility of another transcription factor playing a role in mediating the cellular response to EGCG.
To further demonstrate the involvement of these transcription factors in the phenotypic alteration induced by EGCG, lysostaphin susceptibility was determined for vraSR and sigB mutant strains and their corresponding isogenic WT strains. Exposure of all tested strains to an SI dose of EGCG resulted in increased tolerance to lysostaphin, with the exception of the vraSR null mutant (Fig. 1), emphasizing the central role of VraSR in mediating the cellular response to EGCG. The fact that the sigB null mutant did not show any notable differences in susceptibility to lysostaphin from its isogenic WT strain upon preexposure to EGCG but prevented the increase in tolerance to some of the antibiotics suggests more subtle changes in the cell wall architecture, with no substantial change in the peptidoglycan mass. Our results are supported by electron microscopic observations indicating no remarkable phenotypic difference between the sigB knockout mutant and its isogenic WT strain (28). We therefore suggest that SigB plays an important role in the EGCG-mediated increase in resistance to antibiotics that perturb cell wall synthesis.
The rapidity and magnitude of the increase in vraR expression in response to SI doses of EGCG (Fig. 2), in addition to the vraSR null mutant's attenuated response to EGCG, as evidenced by prevention of the increase in tolerance to cell-wall-targeting antibiotics and lysostaphin, emphasize the pivotal role of the VraSR system in mediating the cellular response to EGCG. This system is capable of rapidly sensing cell wall peptidoglycan damage and coordinating an immediate and massive change in transcription of a unique set of genes involved in cell wall biosynthesis and in conferral of resistance (1, 12, 22, 26, 36, 39). The increased expression of the cell wall biosynthesis-associated genes murA, murF, and pbp2 but not of the cell-wall-degrading gene aaa upon exposure to EGCG substantiates the modulation of VraSR in sensing EGCG and activating the transcription of genes associated with peptidoglycan biosynthesis. Morphological plasticity has been described as a survival strategy for staphylococci grown with different treatments, such as polyanionic substances (40), low concentrations of chloramphenicol (14), or various concentrations of penicillin (13). Our observations raise concerns over the potential of utilization of EGCG (and perhaps other natural antimicrobials) in therapy and as a preservative in that exposure to SI doses of the substance may contribute to the development and enhancement of microbial resistance mechanisms.
Aside from the upregulation of vraSR and genes encoding proteins involved in cell wall metabolism, genome-wide transcriptional profiling of S. aureus in the presence of cell-wall-active antibiotics has revealed distinct overexpression of stress response genes, including msrA, htrA, psrA, and hslO (39). Interestingly, an even higher magnitude of upregulation was induced in S. aureus challenged with SI doses of antimicrobial peptides, specifically in three genes/operons—vraDE, SA0205, and SAS016—encoding an ABC transporter, a putative membrane-bound lysostaphin-like peptidase, and a small functionally unknown protein, respectively (32). Construction of a plasmid carrying a luciferase reporter gene fused to sas016 enabled monitoring of the kinetics and detection of very subtle differences in expression of the cell wall stress signal (8). Here we show that EGCG induces this system in the same manner as cell-wall-active antibiotics, albeit at a lower magnitude than that with oxacillin. Interestingly, inactivation of vraR abolishes cell-wall-targeting antibiotic induction of sas016 expression (8). The results presented here corroborate our hypothesis that SI doses of EGCG, similar to SI doses of cell-wall-targeting antibiotics, induce a cell wall stress response in S. aureus that is modulated by the two-component VraSR system.
ACKNOWLEDGMENTS
We thank A. L. Cheung for providing the Newman sigB null strain, M. Kuroda for strains N315 and KVR, and N. McCallum for strain BB255/psas016p-luc+.
Footnotes
Published ahead of print 31 August 2012
REFERENCES
- 1. Belcheva A, Golemi-Kotra D. 2008. A close-up view of the VraSR two-component system. A mediator of Staphylococcus aureus response to cell wall damage. J. Biol. Chem. 283:12354–12364 [DOI] [PubMed] [Google Scholar]
- 2. Belcheva A, Verma V, Golemi-Kotra D. 2009. DNA-binding activity of the vancomycin resistance associated regulator protein VraR and the role of phosphorylation in transcriptional regulation of the vraSR operon. Biochemistry 48:5592–5601 [DOI] [PubMed] [Google Scholar]
- 3. Bernal P, et al. 2010. Insertion of epicatechin gallate into the cytoplasmic membrane of methicillin-resistant Staphylococcus aureus disrupts penicillin-binding protein (PBP) 2a-mediated beta-lactam resistance by delocalizing PBP2. J. Biol. Chem. 285:24055–24065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bikels-Goshen T, Landau E, Saguy S, Shapira R. 2010. Staphylococcal strains adapted to epigallocathechin gallate (EGCG) show reduced susceptibility to vancomycin, oxacillin and ampicillin, increased heat tolerance, and altered cell morphology. Int. J. Food Microbiol. 138:26–31 [DOI] [PubMed] [Google Scholar]
- 5. Blanco AR, et al. 2003. (−)Epigallocatechin-3-gallate inhibits gelatinase activity of some bacterial isolates from ocular infection, and limits their invasion through gelatine. Biochim. Biophys. Acta 1620:273–281 [DOI] [PubMed] [Google Scholar]
- 6. Burt S. 2004. Essential oils: their antibacterial properties and potential applications in foods—a review. Int. J. Food Microbiol. 94:223–253 [DOI] [PubMed] [Google Scholar]
- 7. Caturla N, Vera-Samper E, Villalain J, Mateo CR, Micol V. 2003. The relationship between the antioxidant and the antibacterial properties of galloylated catechins and the structure of phospholipid model membranes. Free Radic. Biol. Med. 34:648–662 [DOI] [PubMed] [Google Scholar]
- 8. Dengler V, Meier PS, Heusser R, Berger-Bachi B, McCallum N. 2011. Induction kinetics of the Staphylococcus aureus cell wall stress stimulon in response to different cell wall active antibiotics. BMC Microbiol. 11:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Doddangoudar VC, Boost MV, Tsang DN, O'Donoghue MM. 2011. Tracking changes in the vraSR and graSR two component regulatory systems during the development and loss of vancomycin non-susceptibility in a clinical isolate. Clin. Microbiol. Infect. 17:1268–1272 [DOI] [PubMed] [Google Scholar]
- 10. Donegan NP, Cheung AL. 2009. Regulation of the mazEF toxin-antitoxin module in Staphylococcus aureus and its impact on sigB expression. J. Bacteriol. 191:2795–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Duthie ES, Lorenz LL. 1952. Staphylococcal coagulase; mode of action and antigenicity. J. Gen. Microbiol. 6:95–107 [DOI] [PubMed] [Google Scholar]
- 12. Gardete S, Wu SW, Gill S, Tomasz A. 2006. Role of VraSR in antibiotic resistance and antibiotic-induced stress response in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:3424–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Giesbrecht P, et al. 1994. A novel, “hidden” penicillin-induced death of staphylococci at high drug concentration, occurring earlier than murosome-mediated killing processes. Arch. Microbiol. 161:370–383 [DOI] [PubMed] [Google Scholar]
- 14. Giesbrecht P, Wecke J, Reinicke B. 1976. On the morphogenesis of the cell wall of staphylococci. Int. Rev. Cytol. 44:225–318 [DOI] [PubMed] [Google Scholar]
- 15. Gould IM. 2007. MRSA bacteraemia. Int. J. Antimicrob. Agents 30(Suppl 1):S66–S70 [DOI] [PubMed] [Google Scholar]
- 16. Gradisar H, Pristovsek P, Plaper A, Jerala R. 2007. Green tea catechins inhibit bacterial DNA gyrase by interaction with its ATP binding site. J. Med. Chem. 50:264–271 [DOI] [PubMed] [Google Scholar]
- 17. Hew CM, Korakli M, Vogel RF. 2007. Expression of virulence-related genes by Enterococcus faecalis in response to different environments. Syst. Appl. Microbiol. 30:257–267 [DOI] [PubMed] [Google Scholar]
- 18. Hu ZQ, et al. 2002. Epigallocatechin gallate synergistically enhances the activity of carbapenems against methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:558–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ikigai H, Nakae T, Hara Y, Shimamura T. 1993. Bactericidal catechins damage the lipid bilayer. Biochim. Biophys. Acta 1147:132–136 [DOI] [PubMed] [Google Scholar]
- 20. Jordan S, Hutchings MI, Mascher T. 2008. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol. Rev. 32:107–146 [DOI] [PubMed] [Google Scholar]
- 21. Katayama Y, Murakami-Kuroda H, Cui L, Hiramatsu K. 2009. Selection of heterogeneous vancomycin-intermediate Staphylococcus aureus by imipenem. Antimicrob. Agents Chemother. 53:3190–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuroda M, et al. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807–821 [DOI] [PubMed] [Google Scholar]
- 23. Kuroda M, et al. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225–1240 [DOI] [PubMed] [Google Scholar]
- 24. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 25. Lowy FD. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520–532 [DOI] [PubMed] [Google Scholar]
- 26. McAleese F, et al. 2006. Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate-S. aureus-type resistance to vancomycin. J. Bacteriol. 188:1120–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCallum N, Berger-Bachi B, Senn MM. 2010. Regulation of antibiotic resistance in Staphylococcus aureus. Int. J. Med. Microbiol. 300:118–129 [DOI] [PubMed] [Google Scholar]
- 28. Morikawa K, et al. 2001. Overexpression of sigma factor, σB, urges Staphylococcus aureus to thicken the cell wall and to resist beta-lactams. Biochem. Biophys. Res. Commun. 288:385–389 [DOI] [PubMed] [Google Scholar]
- 29. Muthaiyan A, Silverman JA, Jayaswal RK, Wilkinson BJ. 2008. Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrob. Agents Chemother. 52:980–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nicholas RO, et al. 1999. Isolation and characterization of a sigB deletion mutant of Staphylococcus aureus. Infect. Immun. 67:3667–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parkinson JS. 1993. Signal transduction schemes of bacteria. Cell 73:857–871 [DOI] [PubMed] [Google Scholar]
- 32. Pietiainen M, et al. 2009. Transcriptome analysis of the responses of Staphylococcus aureus to antimicrobial peptides and characterization of the roles of vraDE and vraSR in antimicrobial resistance. BMC Genomics 10:429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sakanaka S, Aizawa M, Kim M, Yamamoto T. 1996. Inhibitory effects of green tea polyphenols on growth and cellular adherence of an oral bacterium, Porphyromonas gingivalis. Biosci. Biotechnol. Biochem. 60:745–749 [DOI] [PubMed] [Google Scholar]
- 34. Severin A, et al. 2004. High level oxacillin and vancomycin resistance and altered cell wall composition in Staphylococcus aureus carrying the staphylococcal mecA and the enterococcal vanA gene complex. J. Biol. Chem. 279:3398–3407 [DOI] [PubMed] [Google Scholar]
- 35. Shapira R, Mimran E. 2007. Isolation and characterization of Escherichia coli mutants exhibiting altered response to thymol. Microb. Drug Resist. 13:157–165 [DOI] [PubMed] [Google Scholar]
- 36. Sobral RG, et al. 2007. Extensive and genome-wide changes in the transcription profile of Staphylococcus aureus induced by modulating the transcription of the cell wall synthesis gene murF. J. Bacteriol. 189:2376–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tamba Y, Ohba S, Kubota M, Yoshioka H, Yamazaki M. 2007. Single GUV method reveals interaction of tea catechin (−)-epigallocatechin gallate with lipid membranes. Biophys. J. 92:3178–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsuchiya H. 1999. Effects of green tea catechins on membrane fluidity. Pharmacology 59:34–44 [DOI] [PubMed] [Google Scholar]
- 39. Utaida S, et al. 2003. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149:2719–2732 [DOI] [PubMed] [Google Scholar]
- 40. Wecke J, Lahav M, Ginsburg I, Kwa E, Giesbrecht P. 1986. Inhibition of wall autolysis of staphylococci by sodium polyanethole sulfonate “liquoid.” Arch. Microbiol. 144:110–115 [DOI] [PubMed] [Google Scholar]
- 41. Yoda Y, Hu ZQ, Zhao WH, Shimamura T. 2004. Different susceptibilities of Staphylococcus and Gram-negative rods to epigallocatechin gallate. J. Infect. Chemother. 10:55–58 [DOI] [PubMed] [Google Scholar]
- 42. Zhang YM, Rock CO. 2004. Evaluation of epigallocatechin gallate and related plant polyphenols as inhibitors of the FabG and FabI reductases of bacterial type II fatty-acid synthase. J. Biol. Chem. 279:30994–31001 [DOI] [PubMed] [Google Scholar]
- 43. Zhao WH, Hu ZQ, Okubo S, Hara Y, Shimamura T. 2001. Mechanism of synergy between epigallocatechin gallate and beta-lactams against methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1737–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]