Abstract
We cloned two glycoside hydrolase family 74 genes, the sav_1856 gene and the sav_2574 gene, from Streptomyces avermitilis NBRC14893 and characterized the resultant recombinant proteins. The sav_1856 gene product (SaGH74A) consisted of a catalytic domain and a family 2 carbohydrate-binding module at the C terminus, while the sav_2574 gene product (SaGH74B) consisted of only a catalytic domain. SaGH74A and SaGH74B were expressed successfully and had molecular masses of 92 and 78 kDa, respectively. Both recombinant proteins were xyloglucanases. SaGH74A had optimal activity at 60°C and pH 5.5, while SaGH74B had optimal activity at 55°C and pH 6.0. SaGH74A was stable over a broad pH range (pH 4.5 to 9.0), whereas SaGH74B was stable over a relatively narrow pH range (pH 6.0 to 6.5). Analysis of the hydrolysis products of tamarind xyloglucan and xyloglucan-derived oligosaccharides indicated that SaGH74A was endo-processive, while SaGH74B was a typical endo-enzyme. The C terminus of SaGH74A, which was annotated as a carbohydrate-binding module, bound to β-1,4-linked glucan-containing soluble polysaccharides such as hydroxyethyl cellulose, barley glucan, and xyloglucan.
INTRODUCTION
Plant cell walls consist of cellulose, hemicellulose, structural proteins, and phenolic compounds. Xyloglucan is the major hemicellulose in primary cell walls of flowering plants (6, 31). This complex polysaccharide has a highly branched cellulosic backbone linked by hydrogen bonding to cellulose microfibrils to form a cellulose-xyloglucan network. The cellulosic backbone comprises 1,4-linked β-glucopyranosyl (β-Glcp) residues, most of which are replaced at the C-6 position by α-xylopyranosyl (α-Xylp) residues that then attach to either galactopyranosyl (Galp), l-fucopyranosyl, or l-arabinofuranosyl residues. The side chains vary depending on the plant species. Fry et al. proposed a single-letter nomenclature for naming the xyloglucan side chain structures, in which the unsubstituted and substituted Glc residues are represented as G and X, respectively (11). Structural analyses suggest that most xyloglucans consist of repeating units of either XXXG (XXXG type) or XXGG (XXGG type) (30).
Glycoside hydrolases (GHs) are categorized into different families in the Carbohydrate-Active EnZymes (CAZy) database, according to the similarity between their amino acid sequences (5, 17). Xyloglucan-degrading enzymes are classified into families 5, 7, 12, 16, 44, and 74 (8, 14). The GH74 family includes endo-β-1,4-glucanase (EC 3.2.1.4), oligoxyloglucan reducing end-specific cellobiohydrolase (OXG-RCBH) (EC 3.2.1.150), and xyloglucanase (EC 3.2.1.151). All members of this family hydrolyze xyloglucan. According to the CAZy database, fungi have one or two GH74 genes. However, only one GH74 gene is present in Trichoderma reesei, a widely known cellulolytic filamentous fungus and the main industrial source of commercially available enzymes. The gene product has already been characterized as a xyloglucanase (7, 15). Geotrichum sp. M128 possesses two enzymes belonging to the GH74 family, i.e., xyloglucanase and OXG-RCBH (33, 34), which have distinct modes of action and substrate specificities. Xyloglucanase catalyzes the hydrolysis of xyloglucan in the endo mode, while OXG-RCBH recognizes the reducing end of various xyloglucan-derived oligosaccharides and releases two glycosyl residues from this site in the exo mode. In contrast, two or more genes classified as members of the GH74 family in the CAZy database are present in many bacteria. Streptomyces species are the most extensively studied of these bacteria and have been used as a source of hemicellulases (2, 4, 9, 12, 19, 20, 23, 28, 32). Streptomyces avermitilis possesses two GH74 genes, which are similar to those found in Geotrichum sp. M128. Although computational prediction of gene and protein functions is developing rapidly, experimental and observational data are required to determine the actual enzymatic activity.
To increase the amount of information available on GH74 enzymes, we cloned the two GH74 genes from S. avermitilis, the sav_1856 gene and the sav_2574 gene, and characterized the resulting recombinant proteins.
MATERIALS AND METHODS
Substrates.
Tamarind seed xyloglucan (TXG), carboxymethyl cellulose (CMC), curdlan, pachyman, konjac glucomannan, and ivory nut mannan were purchased from Megazyme International (Wicklow, Ireland). Oat spelt xylan, birch wood xylan, Avicel, locust bean gum, guar gum, β-1,3-β-1,4-glucan from barley, lichenan, and laminarin were purchased from Sigma Chemical Company (St. Louis, MO). Hydroxyethyl cellulose was purchased from Nacalai Tesque (Kyoto, Japan), and carboxymethyl curdlan (CM-curdlan) was purchased from Wako Pure Chemical Industries (Osaka, Japan). Pustulan, derived from Umbilicaria papullosa, was purchased from Calbiochem (Darmstadt, Germany). Celish microfibers were purchased from Daicel Chemical Industries, Ltd. (Tokyo, Japan), and chitin was purchased from Yaizu-Suisan (Yaizu, Japan). Various xyloglucan-derived oligosaccharides, such as XXX, XXXG, XLXG, XXLG, XLLG, GXXXG, XXXGXXXG, and XXXGXXXGXXXG, were prepared from TXG as previously described (33–35). G, X, and L refer to unbranched β-Glcp, α-Xylp-β-1,6-Glcp, and β-Galp-α-1,2-Xylp-β-1,6-Glcp residues, respectively.
Expression and purification of recombinant proteins.
S. avermitilis NBRC14893 was obtained from the National Institute of Technology and Evaluation (Kazusa, Japan). The full-length sav_1856 (GenBank accession number BAC69567) and sav_2574 (GenBank accession number BAC70285) genes were amplified from S. avermitilis genomic DNA by PCR using the primers listed in Table S1 in the supplemental material, as well as Phusion DNA polymerase (Finnzymes, Espoo, Finland). The sav_1856 and sav_2574 gene products were named SaGH74A and SaGH74B, respectively. The recombinant proteins were expressed as secreted proteins by use of a Streptomyces expression system as previously described (13). SaGH74A and SaGH74B were purified from culture filtrates by ammonium sulfate precipitation, ion-exchange chromatography using Q-Sepharose (GE Healthcare, UK, Ltd., Buckinghamshire, United Kingdom), and size exclusion chromatography using Sephacryl S-200 (GE Healthcare). The eluted proteins were verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and dialyzed against distilled water. The resulting preparations were used as the purified enzymes.
Enzyme assay and protein measurement.
Xyloglucanase activity was determined by the Somogyi-Nelson method (27). In brief, the enzyme assay mixture contained 200 μl of McIlvaine buffer (0.2 M Na2HPO4 and 0.1 M citric acid) at a pH of 5.5 (for the SaGH74A assay) or 6.0 (for the SaGH74B assay), 250 μl of 1% (wt/vol) TXG, and 50 μl of the enzyme preparation. The reactions were performed at 50°C. At regular time intervals, 50-μl aliquots of the reaction mixture were collected, and 50 μl of copper reagent was added to the reaction mixture. The reaction was terminated by heating at 100°C for 20 min. Nelson reagent (100 μl) was added to the solution and mixed until the precipitate dissolved. The solution was diluted by adding 800 μl of water. A blank solution was prepared by using water in the reaction mixture. The amount of reducing sugars was determined by measuring the increase in absorbance at 500 nm, and the assays were performed in duplicate. One unit of enzyme activity was defined as the amount of enzyme that released 1 μmol of glucose per minute from the substrate under these conditions. The substrate specificities of SaGH74A and SaGH74B for various polysaccharides were determined by the Somogyi-Nelson method as described above. The reactions were performed in McIlvaine buffer (pH 5.5 for SaGH74A and pH 6.0 for SaGH74B) containing 0.5% (wt/vol) substrate and 0.3 μM SaGH74A or 0.2 μM SaGH74B. The effects of pH and temperature on SaGH74A and SaGH74B activity and stability were investigated as previously described (18). The activity of the xyloglucanases was assayed under the conditions described above. The protein concentration was determined using a bicinchoninic acid (BCA) protein assay kit (Pierce, Rockford, IL) with bovine serum albumin (BSA) as the standard.
The hydrolysis products of TXG and xyloglucan oligosaccharides were analyzed by high-performance anion-exchange chromatography with a pulsed amperometric detection (HPAEC-PAD) system and a CarboPac PA1 column (4 × 250 mm) (Dionex Corp., Sunnyvale, CA). The samples were eluted with 0.1 M NaOH (0 to 5 min), followed by a linear gradient (5 to 35 min) of sodium acetate (0 to 0.2 M) at a flow rate of 1 ml/min. For hydrolysis of TXG, an aliquot of enzyme (0.1 μM) was incubated with 0.5% (wt/vol) substrate in McIlvaine buffer (pH 5.5 for SaGH74A and pH 6.0 for SaGH74B) for 30 min at 50°C. For hydrolysis of xyloglucan oligosaccharides such as XXXGXXXGXXXG and XXXGXXXG, a reaction mixture containing 10 μM substrate, enzyme, and 0.0003% (wt/vol) arabinose was incubated at 37°C, with arabinose as the internal standard. The reactions were terminated by heating at 100°C for 10 min.
Substrate-binding assay.
The affinity of carbohydrate-binding module family 2 (CBM2) of SaGH74A was determined using insoluble polysaccharides (Avicel, insoluble oat spelt xylan, insoluble birch wood xylan, lichenan, pachyman, ivory nut mannan, and chitin) as previously described (1). The assay mixture contained McIlvaine buffer (pH 5.5), substrate (0.3 mg), and SaGH74A (80 μg) in a final volume of 0.2 ml. Affinity gel electrophoresis was also used to investigate the affinity of SaGH74A for soluble polysaccharides. The method used various soluble polysaccharides at a concentration of 0.3% (wt/vol), as previously described (29). BSA was used as a nonbinding negative control.
RESULTS
Expression and characterization of recombinant GH74 proteins from S. avermitilis.
SignalP (http://www.cbs.dtu.dk/services/SignalP/) predicted that the N termini of both proteins (amino acids 1 to 26 of SaGH74A and amino acids 1 to 33 of SaGH74B) were signal sequences (see Fig. S1 in the supplemental material). The deduced amino acid sequence that corresponded to the catalytic domain of SaGH74A resembled the sequences of both bacterial and fungal enzymes (see Table S2). The catalytic domain of SaGH74A showed 42% identity and 55% similarity with that of SaGH74B. In contrast to SaGH74A, the sequence of SaGH74B showed relatively low similarity with the other GH74 enzymes. SaGH74A has a C-terminal region (amino acids 778 to 882) with similarity to CBM2. CBM2 of SaGH74A showed similarity with the sequences of CBMs from Cellulomonas fimi and Clostridium cellulovorans, whose three-dimensional structures had already been determined (10, 26) (see Table S2). The molecular masses of SaGH74A and SaGH74B, excluding the masses of the signal peptides, were predicted to be 89.5 and 75.2 kDa, respectively.
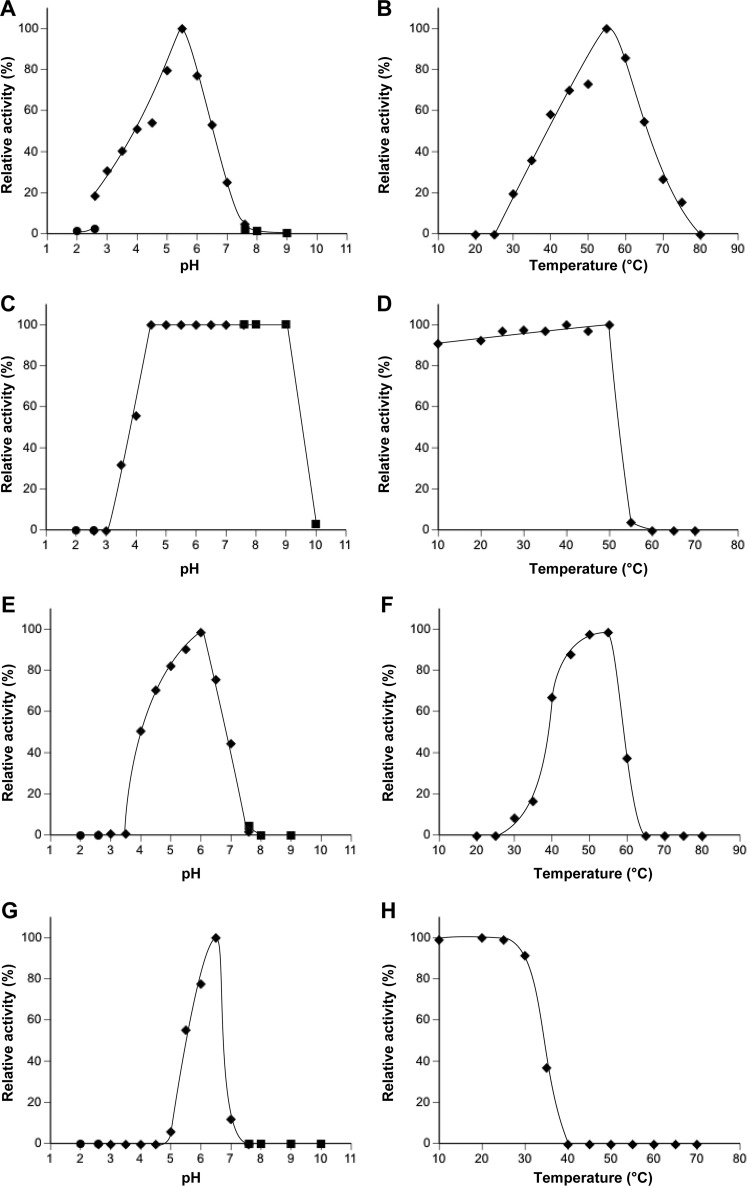
SaGH74A and SaGH74B were expressed successfully as secreted proteins by use of a Streptomyces expression system, whereas the catalytic domain of SaGH74A could not be expressed on its own. Therefore, all experiments were performed using SaGH74A. Recombinant expression yielded 280 mg of SaGH74A and 110 mg of SaGH74B per liter of culture. The molecular masses of SaGH74A and SaGH74B were estimated by SDS-PAGE to be 92 and 78 kDa, respectively (see Fig. S2 in the supplemental material), which correlated closely with the predicted molecular masses. When the proteins were incubated with TXG, both hydrolyzed the substrate. The optimal reaction conditions for hydrolysis of TXG were 60°C and a pH of 5.5 for SaGH74A and 55°C and a pH of 6.0 for SaGH74B (Fig. 1A, B, E, and F). SaGH74A was stable over a broad pH range, i.e., pH 4.5 to 9.0, at which more than 80% of enzyme activity was retained (Fig. 1C). In contrast, SaGH74B was stable over a relatively narrow pH range, i.e., pH 6.0 to 6.5, at which more than 80% of the activity was retained (Fig. 1G). SaGH74A retained more than 80% of activity below 50°C, while SaGH74B retained more than 80% of activity below 30°C (Fig. 1D and H). Under optimal reaction conditions, the specific activities of SaGH74A and SaGH74B were 1.6 and 2.4 units/mg, respectively.
Fig 1.
Enzymatic properties of SaGH74A and SaGH74B. Effects of pH and temperature on activity (A and B for SaGH74A, E and F for SaGH74B) and stability (C and D for SaGH74A, G and H for SaGH74B) were investigated. Symbols: ●, glycine-HCl buffer (pH 1 to 2.6); ◆, McIlvaine buffer (pH 2.6 to 7.6); ■, Atkins-Pantin buffer (pH 7.6 to 10).
Substrate specificities of the recombinant proteins.
The enzymes did not show any activity for Avicel (crystalline cellulose), Celish (microfibers), CMC (β-1,4-glucan), barley β-glycan (β-1,3-β-1,4-glucan), and lichenan (β-1,3-β-1,4-glucan), despite the main chain of TXG consisting of β-1,4-glucan linkages. Furthermore, the enzymes did not show any activity for other β-glucans, such as CM-curdlan (β-1,3-glucan), laminarin (β-1,3-β-1,6-glucan), and pustulan (β-1,6-glucan). In addition, the enzymes showed no activity for β-1,4-glycans, such as birch wood xylan, oat spelt xylan, guar gum, and locust bean gum. These results suggested that SaGH74A and SaGH74B were xyloglucanases.
Mode of action.
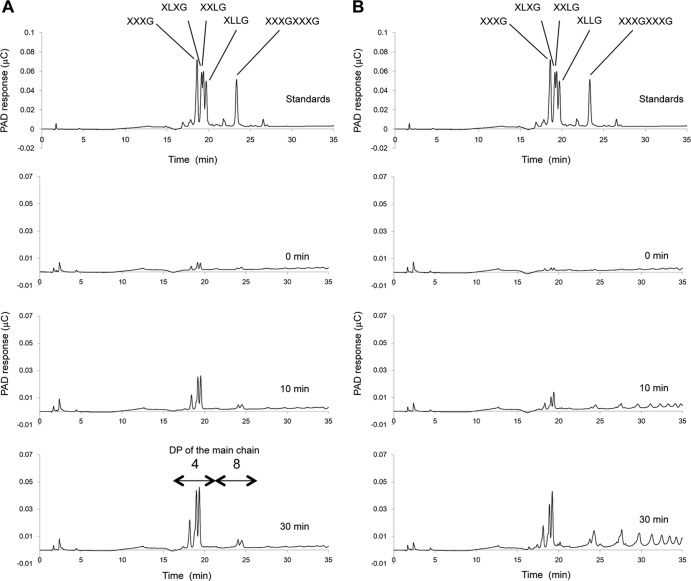
The hydrolysis products of TXG generated by SaGH74A and SaGH74B were analyzed by HPAEC-PAD (Fig. 2). Xyloglucan-derived oligosaccharides such as XXXG, XLXG, XXLG, XLLG, and XXXGXXXG were used as the standards (see Fig. S3 in the supplemental material). The enzymes released a mixture of oligosaccharides from TXG, indicating that both enzymes are endo-enzymes. However, the hydrolysis patterns were different. In the initial stage of the reaction, xyloglucan oligosaccharides, with a degree of polymerization (DP) of 4 in the main chain (retention time between 17 and 20 min), were generated as major products by SaGH74A (Fig. 2A). Small peaks were also detected that corresponded to a mixture of xyloglucan oligosaccharides, with a DP of 8 in the main chain (retention time between 23 and 25 min). There was an increase in the amount of major peaks as the reaction progressed, although there was almost no change in minor peaks (Fig. 2A). In contrast, several oligosaccharides with a DP of 4 or greater in the main chain were generated by SaGH74B, even at the initial stage of the reaction (Fig. 2B). The amounts of the corresponding peaks increased as the reaction progressed.
Fig 2.
HPAEC-PAD analysis of hydrolysis products of TXG generated by SaGH74A and SaGH74B. (A) Hydrolysis products of SaGH74A, with standards. (B) Hydrolysis products of SaGH74B, with standards. Each enzyme was incubated with 0.5% (wt/vol) TXG at 50°C for an appropriate time, and the samples were then subjected to HPAEC-PAD analysis.
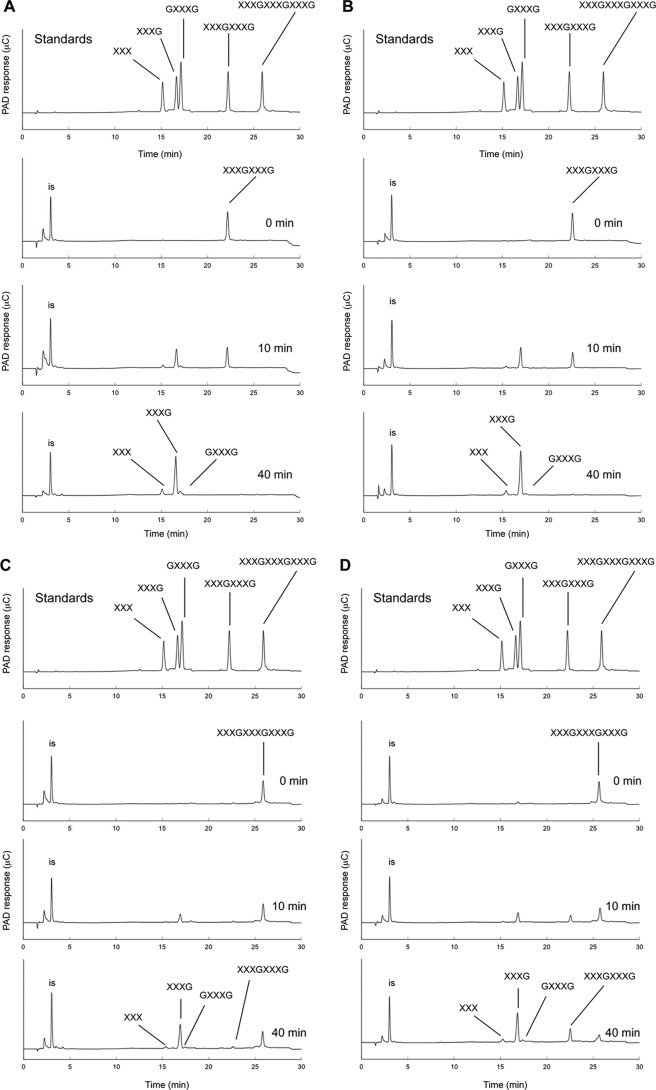
To investigate the hydrolysis pattern in detail, the hydrolysis products (xyloglucan oligosaccharides) generated by SaGH74A and SaGH74B were also analyzed. In the hydrolysis of XXXGXXXG, three types of products, represented as XXXG, XXX, and GXXXG, were generated by SaGH74A and SaGH74B (Fig. 3A and B). The results suggested that both enzymes cleave the glycosidic bonds of branched and unbranched Glc residues, although cleavage efficiency was slower for branched than unbranched Glc residues.
Fig 3.
HPAEC-PAD analysis of hydrolysis products of XXXGXXXG by SaGH74A (A) and SaGH74B (B) and of XXXGXXXGXXXG by SaGH74A (C) and SaGH74B (D). Each enzyme was incubated with 10 μM substrate at 37°C for an appropriate time, and the samples were then subjected to HPAEC-PAD analysis. Arabinose was used as the internal standard (is).
In the hydrolysis of XXXGXXXGXXXG, four types of products, represented as XXXG, XXX, GXXG, and XXXGXXXG, were generated by SaGH74A and SaGH74B. However, the amounts of XXXGXXXG formed by SaGH74A and SaGH74B were different (Fig. 3C and D). SaGH74A generated mainly XXXG, even at the initial stage of the reaction (Fig. 3C, 10 min). As the reaction progressed, the amount of XXXG increased, and both XXX and GXXXG were also detected (Fig. 3C, 40 min). However, XXXGXXXG was represented by an extremely small peak throughout the reaction. In contrast to the case with SaGH74A, XXXGXXXG and XXXG were generated by SaGH74B at the initial stage of the reaction (Fig. 3D, 10 min). As the reaction progressed, XXXG, XXX, GXXXG, and XXXGXXXG were detected (Fig. 3D, 40 min), and the amounts of XXXG and XXXGXXXG increased.
Affinity of C-terminal CBM2.
To analyze the function of the C-terminal CBM2 of SaGH74A, we attempted to express CBM2 as a His-tagged fusion protein. Because the recombinant protein could not be expressed, SaGH74A was used in several studies investigating the characteristics of CBM2. In the binding assay, SaGH74A did not show affinity for any insoluble polysaccharide substrates, such as Avicel, oat spelt xylan (β-1,4-xylan), birch wood xylan (β-1,4-xylan), chitin, lichenan (β-1,3-β-1,4-glucan), pachyman (β-1,3-glucan), and ivory nut mannan (β-1,4-mannan) (see Fig. S4 in the supplemental material). In subsequent affinity gel electrophoresis using soluble substrates, SaGH74A showed affinity for the following soluble substrates (in order of decreasing affinity): hydroxyethyl cellulose (β-1,4-glucan), barley β-glucan (β-1,3-β-1,4-glucan), TXG, konjak glucomannan, and locust bean gum (galactomannan) (Fig. 4B to F). SaGH74A did not show any affinity for oat spelt xylan (β-1,4-xylan), birch wood xylan (β-1,4-xylan), and laminarin (β-1,3-β-1,6-glucan) (data not shown). These results revealed that CBM2 of SaGH74A is a β-1,4-glucan-binding module.
Fig 4.
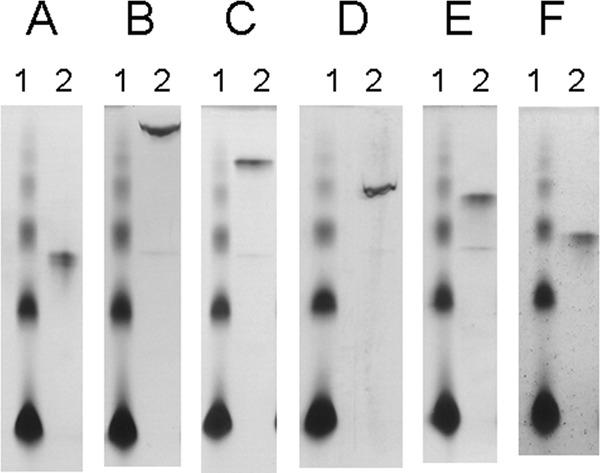
Affinity gel electrophoresis of SaGH74A. The affinity of SaGH74A for soluble polysaccharides was analyzed by native PAGE with gels containing no substrate (A), hydroxyethyl cellulose (β-1,4-glucan) (B), barley β-glucan (β-1,3-β-1,4-glucan) (C), TXG (D), konjak glucomannan (β-1,4-mannan) (E), and locust bean gum (β-1,4-mannan) (F). Lanes 1, BSA (6 μg); lanes 2, SaGH74A (6 μg).
DISCUSSION
GH74 enzymes that hydrolyze xyloglucans include endo-β-1,4-glucanase (EC 3.2.1.4), xyloglucanases (EC 3.2.1.151), and OXG-RCBH (EC 3.2.1.150). Both SaGH74A and SaGH74B also showed activity against xyloglucans and were therefore classified as xyloglucanases. SaGH74A and SaGH74B were slow xyloglucanases compared to other bacterial xyloglucanases (specific activity for that of Jonesia sp., 30 U/mg; for that of Geotrichum sp., 68 U/mg; for that of Clostridium thermocellum, 295 U/mg; and for that of Thermobifida fusca YX, 875 U/mg) (21, 25, 34, 36). The enzymes released a mixture of oligosaccharides from TXG, indicating that both enzymes are endo-enzymes. SaGH74B displayed a typical endo-type hydrolysis pattern, releasing a complete range of TXG fragment sizes. In contrast, oligosaccharides with a DP of 4 in the main chain were the major products formed by SaGH74A (Fig. 2A). Oligosaccharides with a DP of 8 in the main chain were also detected, although the amount of these products did not increase as the reaction progressed. When the enzyme hydrolyzed XXXGXXXGXXXG, the substrate was hydrolyzed first to XXXG and XXXGXXXG by SaGH74A (Fig. 3C). SaGH74A subsequently cleaved XXXGXXXG without releasing these initial products, with the result that the amount of XXXGXXXG did not increase as the reaction progressed (Fig. 3C). The hydrolysis pattern of TXG and xyloglucan oligosaccharides suggested that SaGH74A is an endo-processive enzyme. Endo-processive enzymes are found in Chrysosporium lucknowense, T. reesei (Hypocrea jecorina), Paenibacillus sp. KM21, and Phanerochaete chrysosporium (15, 22, 35). The Streptomyces coelicolor A(3) xyloglucanase Sco6545, belonging to the GH74 family, was recently cloned and characterized (9). The deduced amino acid sequence of Sco6545 was found to be similar to that of SaGH74A. Sco6545 is an endo-enzyme that also has a CBM2 at the C terminus, similar to SaGH74A. However, the method used to process the enzyme was not described in the report.
The CBM2 consists of a β-sandwich fold and is categorized as CBM2a or CBM2b, based on its structure and function (3). CBM2a is a type A CBM that binds to insoluble, highly crystalline cellulose and/or chitin, while CBM2b is a type B glycan chain-binding CBM that binds to soluble polysaccharides such as glucan, xylan, and mannan. CBM2a has an extra loop containing eight amino acids that is not found in CBM2b. The amino acid sequence of CBM2 of SaGH74A showed high similarity with that of CBM2a in C. fimi and C. cellulovorans (10, 24), and three Trp residues in CBM2a which are involved in substrate binding are completely conserved in CBM2 of SaGH74A (26). The results of the binding assay and affinity gel electrophoresis in our study showed that the CBM2 of SaGH74A had an affinity for soluble β-1,4-glucan chains but not for insoluble and crystalline substrates. These results suggest that the CBM2 of SaGH74A may be a novel CBM with unique properties compared to the characterized CBM2a or CBM2b. Some GH74 enzymes from fungi contain a CBM1 (15, 16, 22). In contrast, several bacterial GH74 enzymes contain a CBM2 and/or CBM3 (21, 25, 35). CBM1 and CBM3 as well as CBM2a are type A CBMs (3). The CBM2 of SaGH74A recognized different components of the plant cell wall compared to type A CBMs. This indicated that the localization of SaGH74A was different from that of other GH74 enzymes that harbored type A CBMs.
In conclusion, SaGH74A is an endo-processive xyloglucanase, while SaGH74B is an endo-xyloglucanase. SaGH74A differs from other GH74 enzymes in the substrate-binding ability of the CBM. Our results increase the variety of GH74 enzymes and may assist in research on enzymatic hydrolysis of plant cell walls.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by grants-in-aid (Development of Biomass Utilization Technologies for Revitalizing Rural Areas) from the Ministry of Agriculture, Forestry, and Fisheries of Japan and from the New Energy and Industrial Technology Development Organization (NEDO).
Footnotes
Published ahead of print 31 August 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Araki Y, Karita S, Tanaka A, Kondo M, Goto M. 2009. Characterization of family 17 and family 28 carbohydrate-binding modules from Clostridium josui Cel5A. Biosci. Biotechnol. Biochem. 73: 1028– 1032 [DOI] [PubMed] [Google Scholar]
- 2. Arcand N, Kluepfel D, Paradis FW, Morosoli R, Shareck F. 1993. Beta-mannanase of Streptomyces lividans 66: cloning and DNA sequence of the manA gene and characterization of the enzyme. Biochem. J. 290: 857– 863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382: 769– 781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Canals A, et al. 2003. Structure of xylanase Xys1delta from Streptomyces halstedii. Acta Crystallogr. D Biol. Crystallogr. 59: 1447– 1453 [DOI] [PubMed] [Google Scholar]
- 5. Cantarel BL, et al. 2009. The Carbohydrate-Active enZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37: D233– D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carpita NC, Gibeaut DM. 1993. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3: 1– 3 [DOI] [PubMed] [Google Scholar]
- 7. Desmet T, et al. 2007. An investigation of the substrate specificity of the xyloglucanase Cel74A from Hypocrea jecorina. FEBS J. 274: 356– 363 [DOI] [PubMed] [Google Scholar]
- 8. Eklöf JM, Ruda MC, Brumer H. 2012. Distinguishing xyloglucanase activity in endo-β(1→4)glucanases. Methods Enzymol. 510: 97– 120 [DOI] [PubMed] [Google Scholar]
- 9. Enkhbaatar B, et al. 2012. Identification and characterization of a xyloglucan-specific family 74 glycosyl hydrolase from Streptomyces coelicolor A3(2). Appl. Environ. Microbiol. 78: 607– 611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Foong FC, Doi RH. 1992. Characterization and comparison of Clostridium cellulovorans endoglucanases-xylanases EngB and EngD hyperexpressed in Escherichia coli. J. Bacteriol. 174: 1403– 1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fry SC, et al. 1993. An unambiguous nomenclature of xyloglucan-derived oligosaccharides. Physiol. Plant. 89: 1– 3 [Google Scholar]
- 12. Fujimoto Z, et al. 2000. Crystal structure of Streptomyces olivaceoviridis E-86 β-xylanase containing xylan-binding module. J. Mol. Biol. 300: 575– 585 [DOI] [PubMed] [Google Scholar]
- 13. Fujimoto Z, et al. 2009. Crystallization and preliminary crystallographic analysis of β-l-arabinopyranosidase from Streptomyces avermitilis NBRC14893. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65: 632– 634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gilbert HJ, Stålbrand H, Brumer H. 2008. How the walls come crumbling down: recent structural biochemistry of plant polysaccharide degradation. Curr. Opin. Plant Biol. 11: 338– 348 [DOI] [PubMed] [Google Scholar]
- 15. Grishutin SG, et al. 2004. Specific xyloglucanases as a new class of polysaccharide-degrading enzymes. Biochim. Biophys. Acta 1674: 268– 281 [DOI] [PubMed] [Google Scholar]
- 16. Hasper AA, Dekkers E, van Mil M, van de Vondervoort PJ, de Graaff LH. 2002. EglC, a new endoglucanase from Aspergillus niger with major activity towards xyloglucan. Appl. Environ. Microbiol. 68: 1556– 1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Henrissat B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280: 309– 316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ichinose H, et al. 2005. An exo-β-1,3-galactanase having a novel β-1,3-galactan-binding module from Phanerochaete chrysosporium. J. Biol. Chem. 280: 25820– 25829 [DOI] [PubMed] [Google Scholar]
- 19. Ichinose H, Yoshida M, Fujimoto Z, Kaneko S. 2008. Characterization of a modular enzyme of exo-1,5-α-l-arabinofuranosidase and arabinan binding module from Streptomyces avermitilis NBRC14893. Appl. Microbiol. Biotechnol. 80: 399– 408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ichinose H, et al. 2009. A β-l-arabinopyranosidase from Streptomyces avermitilis is a novel member of glycoside hydrolase family 27. J. Biol. Chem. 284: 25097– 25106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Irwin DC, Cheng M, Xiang B, Rose JK, Wilson DB. 2003. Cloning, expression and characterization of a family-74 xyloglucanase from Thermobifida fusca. Eur. J. Biochem. 270: 3083– 3091 [DOI] [PubMed] [Google Scholar]
- 22. Ishida T, Yaoi K, Hiyoshi A, Igarashi K, Samejima M. 2007. Substrate recognition by glycoside hydrolase family 74 xyloglucanase from the basidiomycete Phanerochaete chrysosporium. FEBS J. 274: 5727– 5736 [DOI] [PubMed] [Google Scholar]
- 23. Matsuo N, Kaneko S, Kuno A, Kobayashi H, Kusakabe I. 2000. Purification, characterization and gene cloning of two α-l-arabinofuranosidases from Streptomyces chartreusis GS901. Biochem. J. 346: 9– 15 [PMC free article] [PubMed] [Google Scholar]
- 24. Ong E, Gilkes NR, Miller RC, Jr, Warren RA, Kilburn DG. 1993. The cellulose-binding domain (CBDcex) of an exoglucanase from Cellulomonas fimi: production in Escherichia coli and characterization of the polypeptide. Biotechnol. Bioeng. 42: 401– 409 [DOI] [PubMed] [Google Scholar]
- 25. Sianidis G, et al. 2006. Functional large-scale production of a novel Jonesia sp. xyloglucanase by heterologous secretion from Streptomyces lividans. J. Biotechnol. 121: 498– 507 [DOI] [PubMed] [Google Scholar]
- 26. Simpson PJ, Xie H, Bolam DN, Gilbert HJ, Williamson MP. 2000. The structural basis for the ligand specificity of family 2 carbohydrate-binding modules. J. Biol. Chem. 275: 41137– 41142 [DOI] [PubMed] [Google Scholar]
- 27. Somogyi M. 1952. Notes on sugar determination. J. Biol. Chem. 195: 19– 23 [PubMed] [Google Scholar]
- 28. Sulzenbacher G, Shareck F, Morosoli R, Dupont C, Davies GJ. 1997. The Streptomyces lividans family 12 endoglucanase: construction of the catalytic cre, expression, and X-ray structure at 1.75 Å resolution. Biochemistry 36: 16032– 16039 [DOI] [PubMed] [Google Scholar]
- 29. Tomme P, Boraston A, Kormos JM, Warren RA, Kilburn DG. 2000. Affinity electrophoresis for the identification and characterization of soluble sugar binding by carbohydrate-binding modules. Enzyme Microb. Technol. 27: 453– 458 [DOI] [PubMed] [Google Scholar]
- 30. Vincken J-P, York WS, Beldman G, Voragen AGJ. 1997. Two general branching patterns of xyloglucan, XXXG and XXGG. Plant Physiol. 114: 9– 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vogel J. 2008. Unique aspects of the grass cell wall. Curr. Opin. Plant Biol. 11: 301– 307 [DOI] [PubMed] [Google Scholar]
- 32. Wouters J, et al. 2001. Crystallographic analysis of family 11 endo-β-1,4-xylanase Xyl1 from Streptomyces sp. S38. Acta Crystallogr. D Biol. Crystallogr. 57: 1813– 1819 [DOI] [PubMed] [Google Scholar]
- 33. Yaoi K, Mitsuishi Y. 2002. Purification, characterization, cloning, and expression of a novel xyloglucan-specific glycosidase, oligoxyloglucan reducing end-specific cellobiohydrolase. J. Biol. Chem. 277: 48276– 48281 [DOI] [PubMed] [Google Scholar]
- 34. Yaoi K, Mitsuishi Y. 2004. Purification, characterization, cDNA cloning, and expression of a xyloglucan endoglucanase from Geotrichum sp. M128. FEBS Lett. 560: 45– 50 [DOI] [PubMed] [Google Scholar]
- 35. Yaoi K, Nakai T, Kameda Y, Hiyoshi A, Mitsuishi Y. 2005. Cloning and characterization of two xyloglucanases from Paenibacillus sp. strain KM 21. Appl. Environ. Microbiol. 71: 7670– 7678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zverlov VV, Schantz N, Schmitt-Kopplin P, Schwarz WH. 2005. Two new major subunits in the cellulosome of Clostridium thermocellum: xyloglucanase Xgh74A and endoxylanase Xyn10D. Microbiology 151: 3395– 3401 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.