Abstract
Water utilities in parts of the U.S. control microbial regrowth in drinking water distribution systems (DWDS) by alternating postdisinfection methods between chlorination and chloramination. To examine how this strategy influences drinking water microbial communities, an urban DWDS (population ≅ 40,000) with groundwater as the source water was studied for approximately 2 years. Water samples were collected at five locations in the network at different seasons and analyzed for their chemical and physical characteristics and for their microbial community composition and structure by examining the 16S rRNA gene via terminal restriction fragment length polymorphism and DNA pyrosequencing technology. Nonmetric multidimension scaling and canonical correspondence analysis of microbial community profiles could explain >57% of the variation. Clustering of samples based on disinfection types (free chlorine versus combined chlorine) and sampling time was observed to correlate to the shifts in microbial communities. Sampling location and water age (<21.2 h) had no apparent effects on the microbial compositions of samples from most time points. Microbial community analysis revealed that among major core populations, Cyanobacteria, Methylobacteriaceae, Sphingomonadaceae, and Xanthomonadaceae were more abundant in chlorinated water, and Methylophilaceae, Methylococcaceae, and Pseudomonadaceae were more abundant in chloraminated water. No correlation was observed with minor populations that were detected frequently (<0.1% of total pyrosequences), which were likely present in source water and survived through the treatment process. Transient microbial populations including Flavobacteriaceae and Clostridiaceae were also observed. Overall, reversible shifts in microbial communities were especially pronounced with chloramination, suggesting stronger selection of microbial populations from chloramines than chlorine.
INTRODUCTION
Drinking water is one of the most closely monitored and strictly regulated resources. To produce drinking water, water utilities select a combination of treatment processes most appropriate to treat the contaminants found in the raw water used by the system. These processes in general include a sequential treatment that employs procedures such as coagulation, flocculation, filtration, and disinfection (Fig. 1A). The final disinfection step typically involves the addition of chlorine and chloramines to ensure pathogen removal and reduce cell numbers. After disinfection treatment, however, the remaining bacteria released into the drinking water distribution systems (DWDS) may interact with microbial populations in the distribution network, where they carry out microbially mediated processes such as biofilm growth, nitrification, biocorrosion, and pathogen persistence (28). A constant disinfectant residual concentration is thus required for limiting regrowth of bacteria in the DWDS.
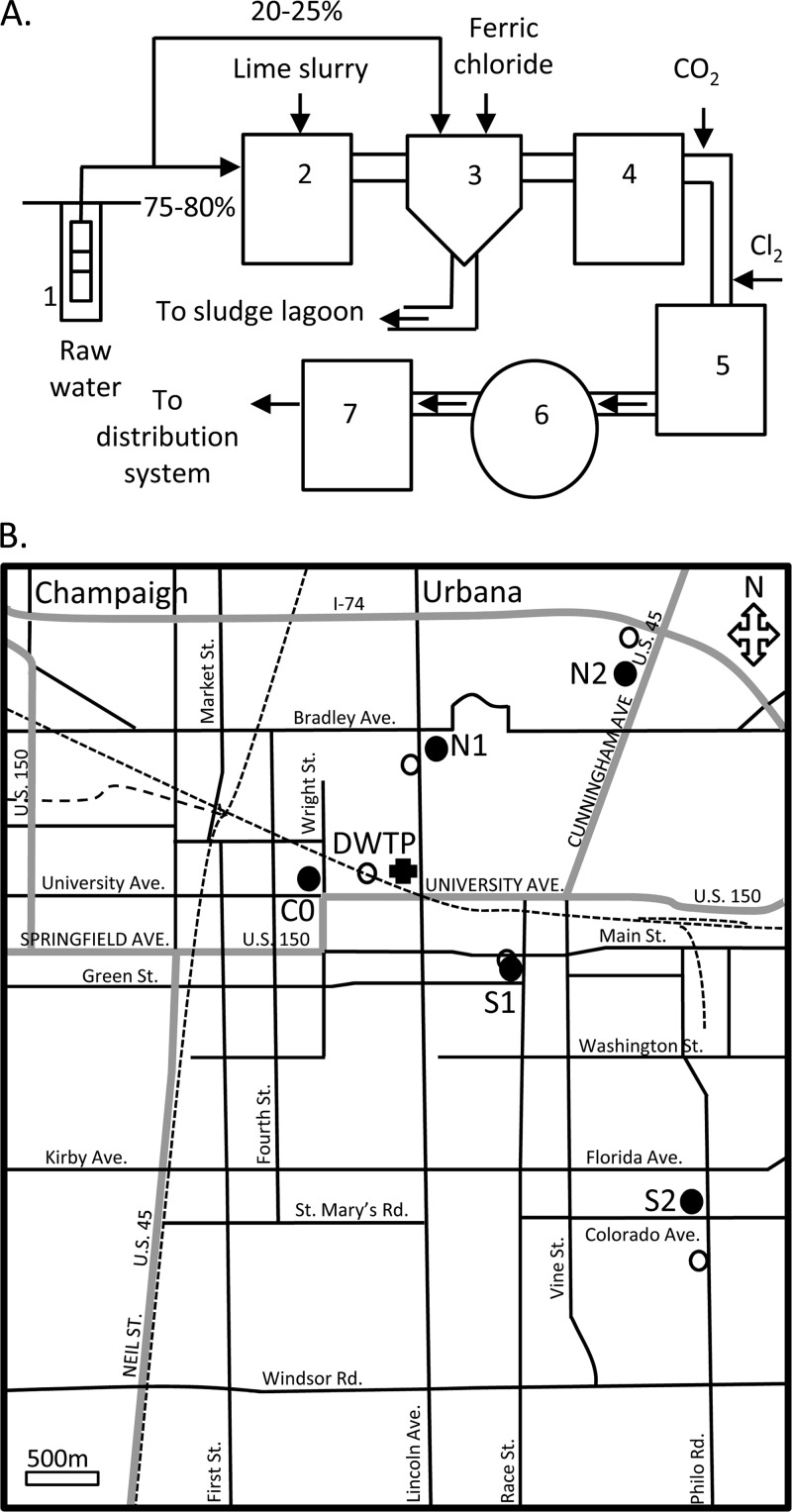
Fig 1.
(A) Schematic diagram of the drinking water treatment process at the water utility in Urbana, IL. Numbered sites: 1, water well; 2, primary basin; 3, mixing basin; 4, secondary basin; 5, filter unit; 6, clarifier; 7, clear well. (B) Map of the five water sampling sites for the present study (closed circles) and the closest sites for water chemistry measurements conducted by the water utility (open circles). The sites are labeled according to their relative position to the water utility (DWTP, cross symbol): one in the center (C0, immediate from the water treatment plant), two in the north (N1 and N2) and two in the south (S1 and S2). Water from S1 is collected for our study and the water utility.
Nevertheless, bacterial regrowth can still occur and be heavily influenced by disinfection practices (4, 19, 29). Bacterial regrowth has been observed under the diminished presence of disinfectant residuals (i.e., chlorine and chloramine), which is influenced by the complex interactions between chemical and physical parameters including but not limited to temperature, alkalinity, pH, organic matter/humic substances, phosphate, and pipe materials, etc. (5, 20, 27, 42). The chemical disinfection reaction with organic matter can also lead to an increase in assimilable organic carbon (AOC) (10, 22, 33) and other chemicals termed disinfection by-products (DBPs) (32). AOC can serve as a carbon source to microorganisms and subsequently affect the biological stability of water. In chloraminated systems, the ammonia-oxidizing bacteria (AOB) can be directly or indirectly involved in promoting biological nitrification and regrowth via consumption of ammonia (1) and the degradation of trihalomethanes, the most common DBP (39). The presence of secondary disinfectants, on the other hand, can select for drinking water biofilms that are resistant to disinfection (19, 41).
It is very important for water utilities in the United States to monitor microbial regrowth in distribution systems to remain in compliance with U.S. Environmental Protection Agency (EPA) drinking water regulations. Water utilities are required to use cultivable microbial indicators (e.g., coliforms and Escherichia coli) to assess the biological effectiveness of the treatment processes and quality of finished drinking water. Cultivation-based methods are also used to evaluate the impact of disinfection on the behavioral response in drinking water isolates (21, 25, 35) and biofilms (7, 26). However, cultivation-based methods do not capture a complete view of drinking water bacterial diversity since only 1% of the known microbes can be cultivated under laboratory settings (36). Recent developments in molecular techniques have allowed the transition from cultivation-based studies of microbes in the DWDS to the use of 16S rRNA gene as the biomarker for bacterial identification. Much research has then been conducted on the impact of chlorination or chloramination, but not a combination of both on bacterial communities in pilot-scale or model DWDS. Williams et al. (40) showed that Alphaproteobacteria populations predominated in both chlorinated and chloraminated water and suggested that Betaproteobacteria populations were more sensitive to disinfectant exposure than Alphaproteobacteria. Episodic chlorination was also shown to induce resilience in certain proteobacterial populations (24). In biofilms of various ages, the different disinfection methods used also led to the development of different bacterial communities (33). These studies showed that monitoring the changes in the microbial community in DWDS is important to further understand the mechanisms of microbial regrowth. Although bacterial numbers are indicators of microbial regrowth, the information alone could not provide the reasons behind the phenomenon. Therefore, we sought to identify the microbial populations present in the DWDS water and to correlate the microbial populations to the measured water parameters under different disinfectant treatments or nutrient availability.
Unlike the aforementioned laboratory-based studies, the present study assessed the structure and composition of microbial communities in the DWDS of the city of Urbana, IL, using terminal restriction fragment length polymorphism (T-RFLP) and 454 pyrosequencing of 16S rRNA gene based on nucleic acids extracted from a set of drinking water samples over a 2-year period. In the past few decades, the DWDS in Urbana has been treated with chloramines; however, in recent years, the system has been subjected to periodic transitions from chloramines to free chlorine in order to control nitrification and maintain optimum chlorine residual levels. The free chlorine treatment in the distribution system can range from 1 to 5 months or more per year. This provided an opportunity to compare the microbial community profiles generated from T-RFLP and 454 pyrosequencing and to determine the influence of environmental factors such as the hydraulic retention time or water age, seasonal changes, and periodic switches in disinfection methods to the changes in the drinking water microbial communities.
MATERIALS AND METHODS
Water treatment process.
A schematic diagram of the water treatment facility for the city of Urbana (population ≅ 40,000) is shown in Fig. 1A. Raw water drawn from two aquifers is combined into the raw water piping before entering the treatment basins. The water utilities routinely monitor raw water quality by measuring temperature, pH, alkalinity, fluoride, iron, total and calcium hardness, total dissolved solids (TDS), ammonia, and bacterial count. Approximately 70 to 75% of the water is added to the primary basin and treated with lime (240-ppm dose) to raise the pH to 11 for the removal of calcium and magnesium. The water is then transported to the mixing basin where ferric chloride (1.2 ppm) is added to the lime to help further remove calcium, magnesium, and other particulates. At this point, the remaining 25 to 30% of the raw water is added to the basin to raise the hardness and decrease the pH to around 10 to allow for iron removal. The water enters the secondary basin to settle the lime sludge and other particles to the bottom of the water, and the clear water is drawn from the top. As the water leaves the secondary basin, carbon dioxide is bubbled into the water to lower the pH to 8.8. After carbon dioxide addition and just before the water reaches the filtration system, chlorine is added to the water. The water is further filtered through dual-medium filters and sent to the clear wells for storage and contact time before being introduced into the distribution system (Fig. 1A). To maintain a disinfectant residual, the water utility switches between chloramination and chlorination. Chloramine is formed by reacting chlorine with the naturally occurring ammonia in the source water with a goal of eliminating free ammonia. During chlorination, additional chlorine is added to react with the naturally occurring ammonia to result in chlorine residual.
Site description.
The water utility performs periodical water sampling for the presence of coliform bacteria, residual chlorine, and other water chemistry analyses to monitor water quality. Typically, the water is collected from fire departments, hospitals, hotels, supermarkets, and other public location throughout the district. For our study, the water sampling locations were selected based on ease of water access, e.g., without the need for consideration of security issues that may be required at certain locations. We considered the water sampling sites based on their position in relation to the location of the water utility and then by matching to the sampling sites used by the water utility. We narrowed our water sampling sites to five locations (Fig. 1B), which included three gas stations, one apartment complex, and one public library. Since these locations are situated on major streets, their position also indicated that the water is received from the main pipeline, thus allowing us to make a comparable analysis. Sampling site C0 denotes a gas station located immediately next to the water utility; the two sampling sites north of the water utility, N1 and N2, are the apartment complex and another gas station, respectively; and the two sampling sites south of the water utility, S1 and S2, are the public library and the third gas station, respectively (Fig. 1B).
Sample collection, DNA extraction, and water chemistry analysis.
Water samples were collected in two consecutive years, 2010 and 2011 during the winter (March 2010 and January 2011), summer (May 2010 and July 2011), and fall (month of October in both years) seasons. These sampling dates coincided with the periods during which transitions from chloramines to free chlorine was applied by the water utility. Each sample was named according to the season, year, and the site at which it was collected, for example, “W10_C0” denotes winter 2010 sample collected at site C0, etc. The tap water was run for extensive time to ensure that the water was coming through the main pipe and not stagnant water in order to obtain representative samples from the distribution system. Water was then collected in sterile 10-liter plastic carboys and processed within 4 h of collection. Bacterial biomass was collected by filtering 10-liter water samples through 0.22-μm-pore-size nitrocellulose membrane filters (Millipore, MA). DNA was extracted using Schmidt's protocol (34) and was stored at −80°C. The protocol was selected based on a previous publication that evaluated the different protocols for DNA extraction of DWDS samples (16).
On site, the samples were measured for pH, temperature, and dissolved oxygen (DO), as well as chlorine and total chlorine concentrations, which were measured using a N,N-diethyl-p-phenylenediamine (DPD) chlorine test kit (Hach, CO). Other water chemistry measured at the laboratory included inductively coupled plasma (ICP) metals, orthophosphate, anions (nitrate and sulfate), ammonia, nitrite, nonvolatile organic carbon (NVOC), alkalinity, and TDS. These chemical analyses were conducted by the Illinois State Water Survey in accordance with EPA-based methods (see Table S1 in the supplemental material). The data for hydraulic retention time (24 day average/year) was provided by the water utility according to a hydrological model developed internally.
T-RFLP and 454 pyrosequencing analysis.
For T-RFLP analysis, 16S rRNA gene was PCR-amplified using universal bacterial primer pair 47F (6 FAM-CYTAACACATGCAAGTCG) and 927r (ACCGCTTGTGCGGGCCC) as described previously (16). T-RFLP profiles were generated based on peak area and normalized similar to the method of Lukow et al. (23), with individual T-RF data expressed as the relative abundance. For pyrosequencing analysis, the extracted DNA was amplified with the following bacterial specific forward 515F (5′-Fusion A-Barcode-CA linker-GTGYCAGCMGCCGCGGTA-3′) and reverse 909R (5′-Fusion B-TC linker-CCCCGYCAATTCMTTTRAGT-3′) primers as described previously (37). 454 pyrosequencing was carried out on the Titanium platform (Roche/454 Life Sciences) at the W. M. Keck Center, part of the Roy J. Carver Biotechnology Center at the University of Illinois at Urbana-Champaign. Sequences generated from pyrosequencing analysis of 16S rRNA gene amplicons were processed using the Quantitative Insights Into Microbial Ecology program (QIIME v1.1 [3]) pipeline with the default settings. Briefly, the flow diagrams were denoised and the UCLUST algorithm was used for operational taxonomic unit (OTU) assignment (97% pairwise identity). Representative sequences from each OTU were selected for alignment to the Greengenes imputed core reference alignment using PyNAST with default settings. Chimiera Slayer was used to identify chimeras, which were then removed from the reference set. The alignment was then filtered to remove gaps and a maximum-likelihood tree was constructed from the filtered sequences using FastTree. Taxonomy assignment was performed with an RDP classifier with data set from Greengenes OTUs at a 0.8 minimum confidence level. An unweighted UniFrc distance matrix was constructed from the phylogenetic tree and visualized using principal coordinates analysis (PCoA; as implemented in QIIME). Samples S11_S2 and F11_S1 did not yield quality sequences and were not included in subsequent analysis.
Statistical analysis.
Analysis of variance (ANOVA) was performed using Origin (OriginLab Corp., Northampton, MA) with the Tukey and Fisher tests to determine inter treatment differences. Nonmetric multidimensional scaling (NMDS) was performed with the Bray-Curtis coefficient using Primer 6 version 1.0.3 (PRIMER-E, Ltd., Ivy Bridge, United Kingdom) to ordinate the similarity data as previously described (30) for both sets of square-root transformed T-RF and pyrosequencing OTU data. Bray-Curtis coefficient has been suggested to be the most suitable coefficient for construction of similarity matrices (30). Canonical correspondence analysis (CCA) performed by CANOCO v.4.5 (Microcomputer Power, Ithaca, NY) was used to correlate the microbial community profiles (T-RF and pyrosequencing OTU data) with different variables, including water chemistry, water temperature, water age, and sampling time. Detrended correspondence analysis was performed to determine whether a linear or unimodal model should be used to calculate the distribution of species across environmental gradients. Manual forward selection with Monte Carlo permutation tests was then performed to determine the significance of the potential explanatory (environmental) variables with 499 permutations under the full model as described by ter Braak and Šmilauer (38). CCA was performed with no transformation of species data and biplot scaling with focusing on interspecies difference. Each CCA was then plotted with the set of potential explanatory variables.
RESULTS
Water chemistry analysis.
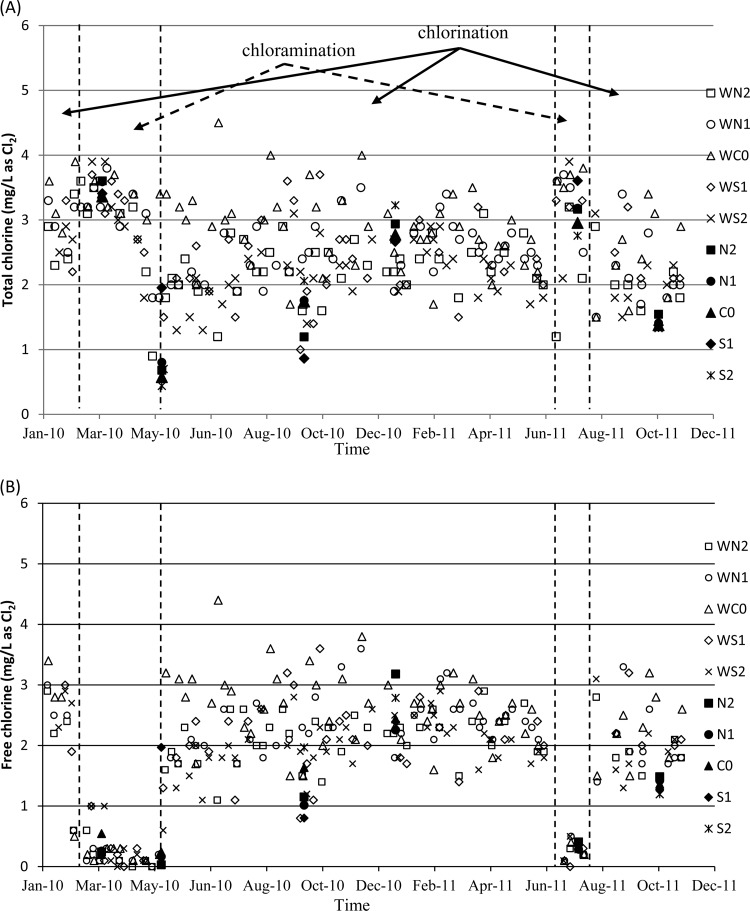
Both chlorination and chloramination were used as the disinfection methods in the distribution system studied. Figure 2 indicates the concentrations of free chlorine and total chlorine for samples taken at different locations during a period of approximately 2 years. As shown in panel A, the total residual chlorine measured by the utility varied from 1.0 to 4.0 mg/liter as Cl2 (average = 2.63 ± 0.65), and during the study period, only one measurement at WC0 in July 2010 was observed to exceed 4.0 mg/liter. Panel B indicated the free available residual chlorine or free chlorine during three different chlorination periods (i.e., before 31 January 2010, between 6 May 2011 and 1 July 2011, and after 15 August 2011). It fluctuated between 1 and 4 mg/liter as Cl2, which was consistently below the maximum regulated residual concentration (4.0 mg/liter), except one measurement in WC0 in July 2010. As expected, the free chlorine was low during the chloramination periods, and the combined available residual chlorine (or combined chlorine) could be calculated by subtracting the total chlorine with the free chlorine. In the present study, water samples were taken at five locations close to the sampling locations used by the utility at six different time points (i.e., W10, Su10, F10, W11, Su11, and F11). The concentrations of total chlorine and free chlorine were within the range measured by the utility, except the time point on 8 May 2010. Four of the five samples still contained low free chlorine during the transition from chloramination to chlorination from 6 May. The observed low free chlorine concentrations were likely due to breakpoint reaction as the residuals converted to free chlorine. It should be noted that measurements on ammonia were conducted by the water utilities only during chloramination, as completion reaction of ammonia with free chlorine is anticipated during chlorination.
Fig 2.
On-site measurements of the total chlorine concentration over the 2-year study period (solid arrows and dashed arrows indicate periods of chlorination and chloramination, respectively) (A) and the free chlorine concentration (B). Open symbols indicate measurements conducted by the water utility; closed symbols indicate measurements conducted from water sampling sites for the present study.
Furthermore, temperature varied season to season, with the colder temperatures measured in the winters and warmer temperatures measured during fall and summer seasons, ranging from approximately 7 to 23°C. The measured pH (8.2 to 9.0) was relatively constant. Values for other water chemistry including ICP metals (e.g., B, Ba, Cu, and Sr), orthophosphate, anions (nitrite, nitrate, and sulfate), ammonia, nonvolatile organic carbon (NVOC), alkalinity, and total dissolved solids were averaged and presented in see Table S1 in the supplemental material. The average hydraulic retention times (estimated water age in hours) for the sites were as follows: N2, 21.24 h; N1, 3.60; h C0, 4.18 h; S1, 3.73 h; and S2, 17.86 h.
Microbial diversity and composition.
The microbial diversity in the water sample was determined using 16S pyrosequencing with a 3% cutoff to define an OTU. Based on the rarefaction curves (see Fig. S1 in the supplemental material) generated, most of the samples achieved saturated (flat) curves, except for the winter 2010 samples and W11_C0 sample, which suggested that more sequences may be required for these samples. The total number of OTUs varied from 26 to 256 based on the minimum number of sequences analyzed (i.e., 1,206) (see Fig. S2A in the supplemental material). The steepness of the curves for these samples also reflected species richness (estimated by Chao1 and phylogenetic diversity metric). The highest species richness was observed with the winter 2010 samples, and samples W11_C0 and F11_N2 (see Fig. S2B and C in the supplemental material). The Chao1 diversity index varied from 40 to 500, and the phylogenetic diversity metric (PD) from 3.0 to 32.6. In general, samples from W10 and F11 seemed to have higher species richness than samples taken at other time points. ANOVA test indicated that significant variation of microbial diversity was observed among different sampling points (P < 0.05), but the differences between any two given sampling points were not correlated to the different disinfection treatments.
Pyrosequencing analysis of 16S rRNA gene was used to identify drinking water microbial community composition. There were some unclassified sequences, ranging from 0.06 to 4.6%, in some of the samples (see Fig. S3 in the supplemental material). At the phylum level, the majority of the sequences were assigned to Proteobacteria (37.5 to 99.5%), Bacteroidetes (0 to 53.4%), Cyanobacteria (0 to 35.8%), and Firmicutes (0 to 51.8%) (see Fig. S3 in the supplemental material). Analysis of the phylum Proteobacteria at the class level showed that most samples were dominated by Gammaproteobacteria, followed by Alphaproteobacteria (except for samples F10_S2, F11_N1, and F11_S2) and Betaproteobacteria (see Fig. S4 in the supplemental material). Furthermore, the majority of Bacteroidetes were especially abundant during winter 2010. The Cyanobacteria were mainly detected throughout fall of 2010 and fall 2011 (see Fig. S3 in the supplemental material), and the Firmicutes were very abundant in F11 samples. In addition, sequences from five other known or candidate bacterial phyla were found. For archaeal populations, sequences primarily related to Methanobacteriaceae and Methanosaetaceae were detected in several samples with the highest abundance (3.4%) observed in F11_N2. As described below, the differences between the samples became more evident at a more refined phylogenetic resolution such as the family level (see Fig. S3 in the supplemental material).
Microbial population dynamics based on T-RFLP and pyrosequencing community profile.
T-RFLP fingerprinting patterns and pyrosequencing results revealed changes or shifts of microbial populations among those samples taken at different time points. Certain T-RFs were detected at higher relative abundance during periods of chloramination or chlorination (see Fig. S5 in the supplemental material). To depict this observation, representative populations observed from site C0 was selected (Fig. 3A). T-RFs 405 and 415 were present at higher relative abundance during chloramination than chlorination, while the opposite was observed for T-RFs 395 and 452. From pyrosequencing results, the differences among the samples were already evident at the family level even though the observed sequences could be classified at the genus-level. Methylococcaceae-like sequences were observed at higher relative abundance during chloramination than chlorination while the opposite was observed for Cyanobacteria and Xanthomonadaceae-like sequences (Fig. 3B). The pyrosequencing microbial community profile of each sample also reflected different effects of the two different disinfection treatments on the drinking water microbial composition. For instance, the Methylococcaceae and Xanthomonadaceae were more abundant during chloramination and chlorination, respectively (see Fig. S3 in the supplemental material). Likewise, the Sphingomonadaceae, Enterobacteriaceae, and Cyanobacteria-like sequences were more frequently detected during chlorination.
Fig 3.
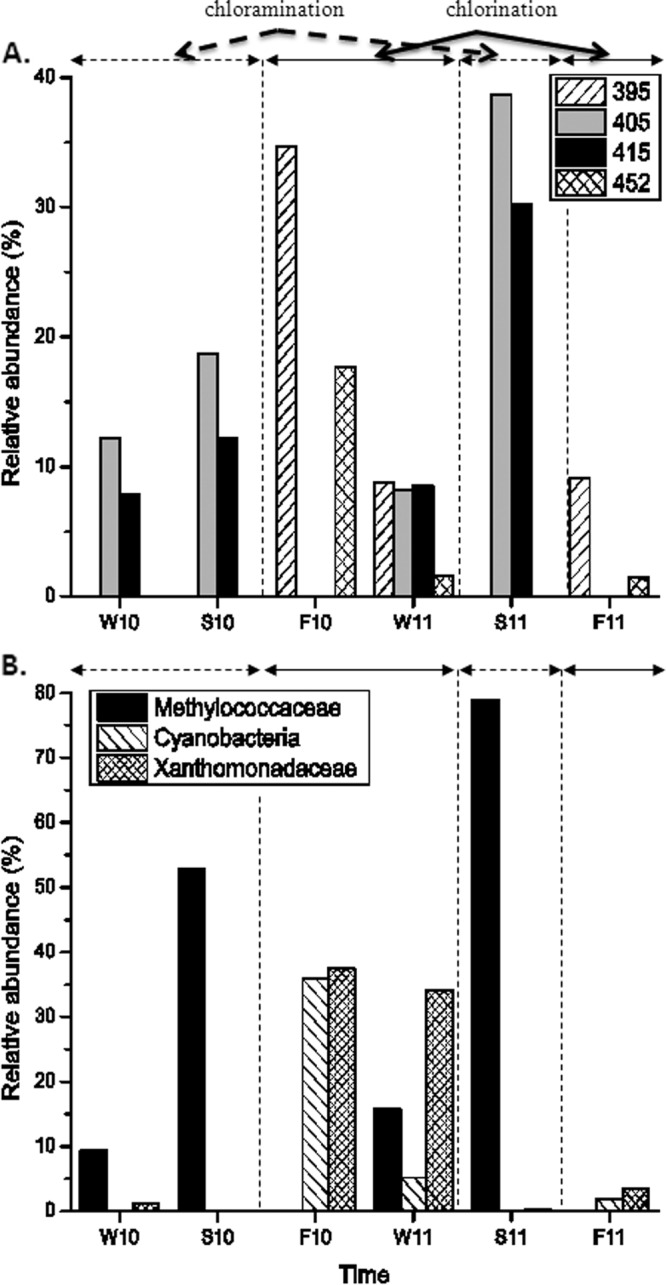
Changes in selected T-RFs (A) and family-level microbial populations (B) throughout the sampling period. Solid arrows and dashed arrows indicate periods of chlorination and chloramination, respectively.
Furthermore, several bacterial groups were observed to predominate in a specific sampling time, and their abundances had no direct relation with the type of disinfection treatments. For example, Flavobacteriaceae was observed to dominate only in samples primarily from winter 2010, Methylobacteriaceae in sample F10_S2, and Clostridiaceae in the fall 2011 samples. It is possible that other factors besides disinfection could also play significant roles in affecting the dominance of specific populations in the drinking water. Similar phenomena were observed with the occurrence of some T-RFs (see Fig. S3 in the supplemental material).
NMDS analysis.
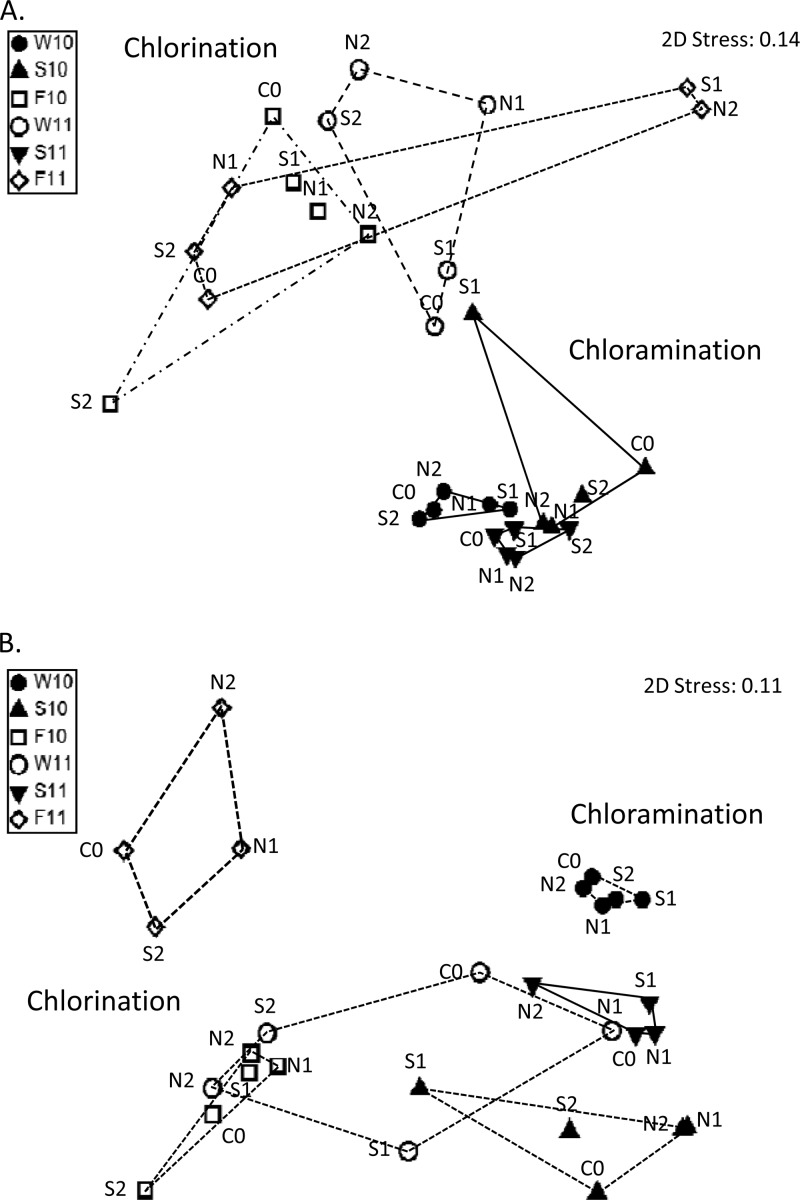
The shift of the drinking water microbial communities throughout the 2-year sampling period was analyzed through nonmetric multidimensional scaling (NMDS) analysis, which correlates the differences of microbial communities between two given samples through the distance on a two-dimensional ordination plot (i.e., a shorter distance indicates a higher similarity in microbial community structure). Figure 4 showed that the two-dimensional stress value of the NMDS analysis was below 0.2 for results obtained based on T-RFLP and pyrosequencing, respectively, suggesting that the microbial community variations were significant. NMDS analysis of T-RFLP microbial community profile (Fig. 4A) clearly revealed shifts of microbial community structures due to the type of disinfection treatments used. Under chloramination, water samples taken in W10 were closely clustered among each other, and this cluster was very closely related to the other two clusters containing samples taken in S10 and S11, except S10_S1, which contained a high free chlorine concentration and was clustered distantly away from other samples. Spatial location or hydraulic retention time showed no apparent effect on the microbial structure. For water samples taken from F10, W11, and F11 under chlorination, they did not form distinct and individual clusters but exhibited some degree of overlapping among them. The clustering patterns were different from those observed with samples taken under chloramination. By examining all clusters formed at those six time points, a reversible shift in microbial community structure between chloramination and chlorination was observed.
Fig 4.
Ordination plots of NMDS results, showing distribution of samples collected during chloramination (solid symbols) and chlorination (open symbols) in T-RFs (A) and pyrosequencing microbial community profiles (B).
NMDS analysis of pyrosequencing results also gave different clustering among the samples taken at different time points (Fig. 4B). Water samples from W10, S10, F10, S11, and F11 were observed to form relatively smaller and distinct clusters than samples from W11. A noticeable separation among W10, Su10, and Su11, and between F10 and F11 further suggested some degree of effect due to temporal change or microbial variations in source water. Clusters W10, S10, and S11 and clusters F10 and F11 were observed to situate at different sides of Fig. 4B, suggesting correlation to different disinfection methods used. Samples from W11 and F11 gave a relatively large area of clusters, and this indicated that other factors (e.g., location, water age, and source water) alone or in combination might also have effects on the microbial community structure of samples.
Different sample clustering patterns between T-RFLP and pyrosequencing results were also observed. We suspected that the difference could be due to the higher resolution of pyrosequencing data in identifying individual sequences down to possibly a genus level, compared to T-RFLP, where each T-RF may represent one or more bacterial populations from different genera or higher phylogenetic levels.
CCA.
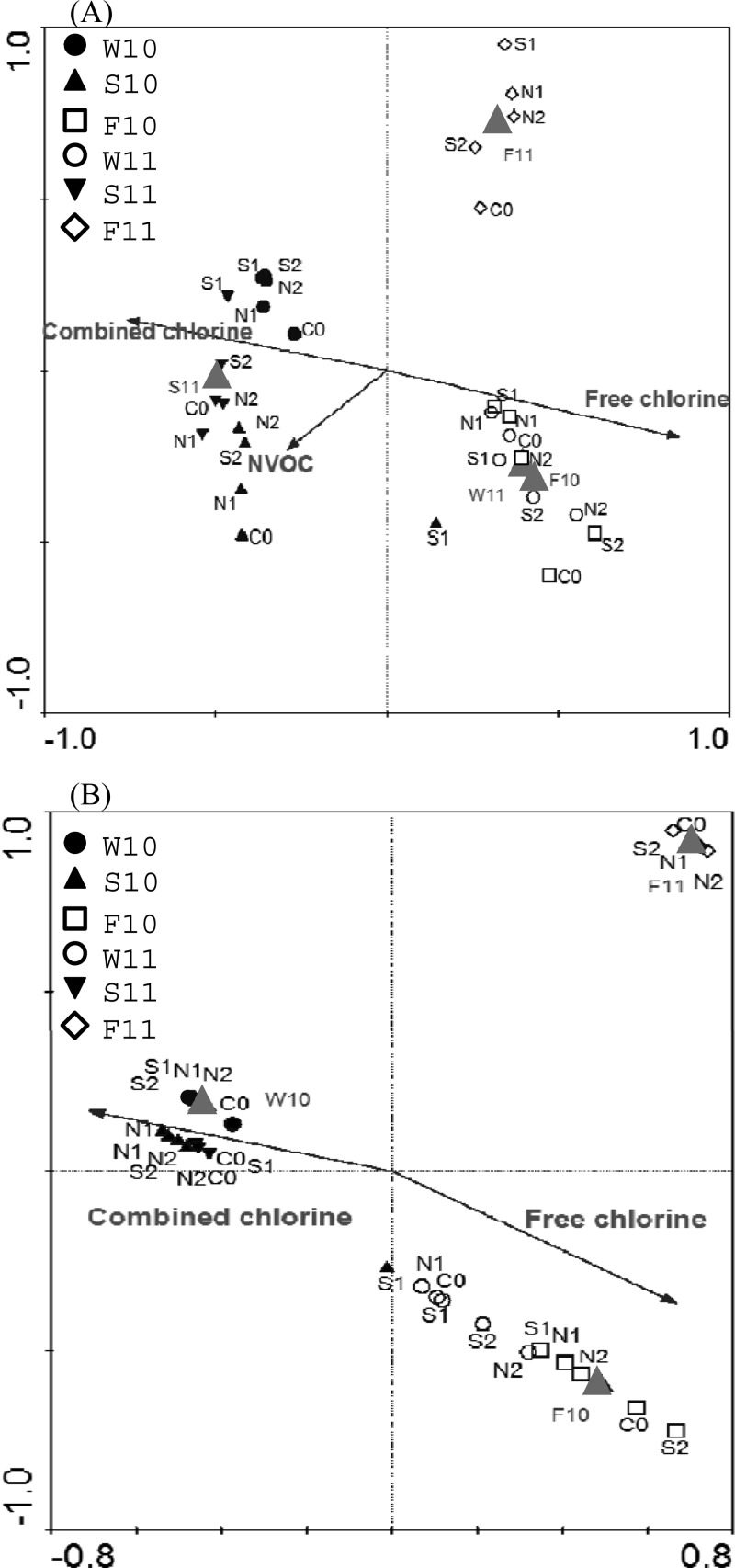
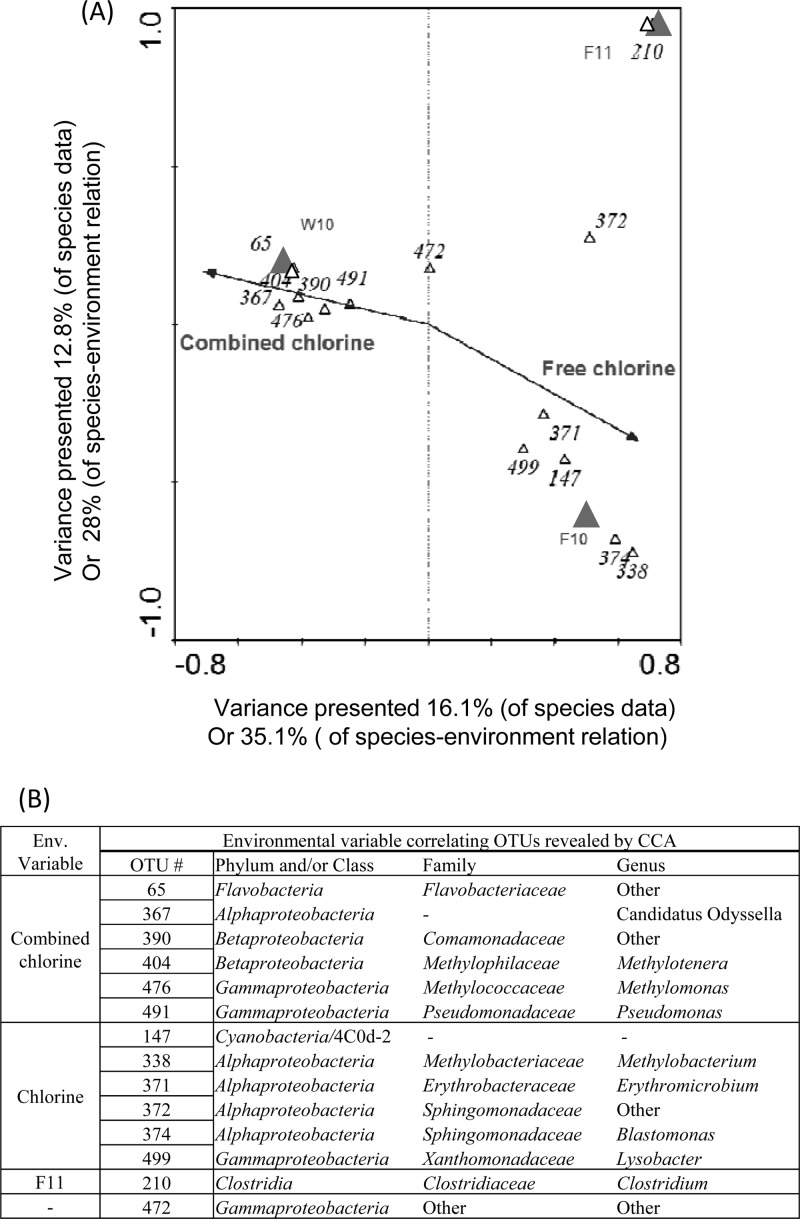
Canonical correspondence analysis (CCA) was used to correlate the effect of environmental variables on the microbial community structure and composition. Figure 5 indicates that both T-RFLP and 454 pyrosequencing could explain similar amount of variation, 63.1 and 57.7%, respectively, within the species-environment relationship across the first two canonical axes (P = 0.01; Fig. 5). As indicated by the length of the environmental variable arrows in the CCA plot, the strongest determinant (indicated by longer arrows) for microbial community composition was free chlorine and combined chlorine concentrations from both T-RFLP and pyrosequencing analysis. It was obvious that the samples receiving chlorination and chloramination were divided into two sides with the exception of sample S10_S1, which contained high free chlorine under the treatment of chloramination. Samples taken at fall 2010 (season 3) and fall 2011 (season 6) also had strong effects on the microbial community structure. Furthermore, NVOC and seasons 4 and 5 were also observed to have influence on sample distribution in the T-RFLP data (Fig. 5A), and season 1 had an influence on sample distribution in the pyrosequencing data (Fig. 5B). Likewise, this difference was due to the difference in the resolution for differentiating microbial populations.
Fig 5.
Ordination plots of CCA results for T-RFLP (A) and pyrosequencing microbial community profiles (B).
The microbial populations present in samples that are strongly affected by chlorination or chloramination were further compared (Fig. 5B). Eleven and thirteen samples associated with free chlorine and combined chlorine, respectively, were selected. Within each sample, the total number of microbial populations or OTU was determined by excluding 16S pyrosequences that are singletons or doubletons. In total, 216 OTUs were detected among all samples, with 69 observed only chlorinated water samples, 49 in chloraminated water samples, and 98 common to both chlorinated and chloraminated water samples.
Figure 6 further revealed major microbial populations or OTU (>5% in total abundance) that were influenced by the environmental variables. In total, 14 different OTUs had strong correlation with nine identified down to the genus level. Among them, six OTUs were strongly correlated to combined chlorine and season 1 (winter 2010), five to free chlorine and season 3 (fall 2010), and one (OTU = 210) to season 6 (fall 2011). OTU 472 was plotted close to the origin of the axes, which implies that its appearance could not be explained by the environmental variables tested.
Fig 6.
(A) Ordination plots were provided to interpret sample-environment correspondence and species-environment correspondence. OTUs with weight > 5% were shown on the species-environment plots. (B) Abundant OTUs (weight > 5%) correlated with free or combined chlorine disinfection and their phylogenetic affiliation.
DISCUSSION
Microorganisms have been suggested to be good indicators of ecosystem stability as they are sensitive to changes in the environment. In the DWDS, microbial community fingerprints have also been suggested for use as indicators of stability in the drinking water microflora (6). In the last several years, many water utilities have started to switch from using chlorine for disinfection to chloramines due to the rising concern of DBP formed during chlorination. In the Urbana distribution system, the disinfectant residual is maintained by periodic use of chlorination and chloramination. During chlorination, breakpoint addition is conducted, and free chlorine is produced to control biofilm growth. While studies have examined the impact of chlorination and chloramination on microbial communities, to our knowledge, the present study is the first to use molecular methods for community composition analysis and to determine the influence of transitory switches between these two disinfection methods on drinking water microbial communities in a full-scale DWDS.
This study used two different approaches, T-RFLP and 454 pyrosequencing, to characterize the drinking water microflora. The results suggested that the free chlorine concentration was an obvious factor in explaining variations in drinking water microflora. NMDS analysis indicated that there was a temporal change in the microbial community, and further analysis revealed that the change was a result of the different disinfectant strategy conducted by the water utility during each season, which influenced the free chlorine concentration in the DWDS. Similar results were obtained by CCA, with some degree of effects due to seasonal or temporal change. It is known that chlorination and chloramination exhibited different degrees of effectiveness in shaping the microbial composition through chemically oxidizing bacterial cells and suppressing bacterial growth (12). In our study, 49 or 69 OTUs were mainly detected in chloraminated or chlorinated water, respectively. The different dominant bacterial groups detected in each cluster could represent differences in planktonic cells that were damaged but not lysed, survived and possibly proliferate, or dislodged from distribution systems surfaces during the disinfection treatments.
Previous studies have reported the predominance of Alphaproteobacteria or Betaproteobacteria or both in the drinking water (40). In contrast, our results showed the predominance of the Gammaproteobacteria population over the Alpha- and Betaproteobacteria populations in most of the samples. This could be attributed to the differences in the source water quality (groundwater versus surface water), physicochemical variables of the water, and efficiency of disinfection treatment (28). Studies have also demonstrated the influence of disinfectants on the Proteobacteria populations where Alphaproteobacteria predominated in both chloraminated and chlorinated water (40), whereas Betaproteobacteria and Gammaproteobacteria in drinking water biofilms were favored with increased chlorination (24).
Although we did not observe the same trend on the overall proteobacterial populations (see Fig. S4 in the supplemental material), our results did show the influence of chlorination and chloramination on certain major bacterial groups (≥5% abundance) or core microbial populations at the family level of classification. Under chloramination, Methylophilaceae, Methylococcaceae, and Pseudomonadaceae were the dominant families detected, whereas, under chlorination, Cyanobacteria, Methylobacteriaceae, Sphingomonadaceae, and Xanthomonadaceae were dominant. The presence of Methylophilaceae, Methylococcaceae, and Methylobacteriaceae were possibly influenced by dissolved methane, which has been detected in the groundwater that serves as the source water in Urbana and Champaign area (18). However, we do not have sufficient information to explain the presence of other bacterial families detected, their resistance to chlorine and chloramine and the microbial ecology roles they played. For example, Blastomonas sp. was the main population detected in Sphingomonadaceae, which comprise of a group of bacteria that have a versatile metabolism and are ubiquitous in soil and water habitats. Although the close relatives of Blastomonas sp. are common inhabitants of chlorinated water (11), its resistance to chlorination remains unclear. Lysobacter species, a common soil and freshwater bacterium linked to the production of the musty-earthy odor detected in water (8), were the most abundant members found in Xanthomonadaceae and were also detected from a biofilm microbial community a water meter collected from the same distribution system (15). The possibility for Lysobacter spp. to be sloughed from the biofilms should be further confirmed. Other studies have demonstrated that not only are Cyanobacteria present in chlorinated DWDS but some are also potentially active members of the DWDS microbial community (17, 31), suggesting that they could survive from different physical and chemical treatments and enter the DWDS (13). We think the use of cultivation-based methods together with metagenomics approaches should be pursued to further understand the ecological roles of predominant populations in DWDS.
Among minor core populations (<0.1% of total pyrosequences), correlation between their abundances and disinfection practice, and sampling time and location was not obvious. These included, for example, metal reducers (e.g., Gallionellaceae, Geobacteraceae, and Shewanellaceae), methanogens (e.g., Methanobacteriaceae, Methanosaetaceae, and Methanosarcinaceae), anaerobic syntrophs (e.g., Syntrophaceae and Syntrophobacteraceae), and nitrifying populations (e.g., Nitrosomonadaceae, Nitrospinaceae, Nitrospirales, and Nitrosopumilus). Most of these populations were likely those present in source water and have survived through the water treatment processes or perhaps were those that have established themselves in the biofilm and were detected in the planktonic phase upon biofilm detachment. Legionella spp. (<0.006 of total sequences obtained for a given sample) and Mycobacterium spp. (<0.008) were also detectable in some of the samples. However, due to the short 16S pyrosequence reads obtained, we could not precisely determine the identity and confirm the presence of potential pathogenic organisms. It can be anticipated that some of these populations could serve as inocula and proliferate in the DWDS if optimal growth conditions are provided.
We were also able to identify microbial populations that exhibited transient responses. Flavobacteriaceae and Clostridiaceae were two major populations observed during winter 2010 and fall 2011 (i.e., season 6), respectively, and Methylobacteriaceae was only detected in sample F10_S2. In particular, most of the sequences in Clostridiaceae were associated with Clostridium species, which are known spore-forming organisms and have strong resistance to disinfection treatments. Their appearances could likely be due to compromises or perturbations in treatment barriers or to changes in source water microbial composition. Thus, it will be useful to find out the correlation between the transient microbial populations and causes and use the information to monitor the integrity of system operation in the future.
No spatial patterns in the microbial communities were observed during most sampling time points (Fig. 4 and 5), suggesting that the water age at the sampling sites might not be long enough to allow the development of distinct local drinking water microflora. This finding is good for water utilities, since almost the same water quality produced at the water treatment plant can be provided to individual households. It was further observed that the microbial community clustering between the sites was less pronounced in samples collected during chlorination than chloramination. One possible explanation is that free chlorine is a stronger disinfectant than chloramines and can rapidly react with microbial cells. As a result, the cells could be lysed, and microbial community structures in the water phase and biofilms could be altered to an extent greater than that observed between two given sampling points. Alternately, chlorine can cause an increase in assimilable organic carbon (AOC) due to the reaction of free chlorine with the dissolved organic carbon present in natural water (10, 22). If the AOC generated could serve as substrates for growth of heterotrophic bacteria, although unlikely under a short water age, shifts in the microbial communities could be expected.
Biodiversity is a subject of interest in ecological studies and is usually used as an indicator of ecosystem stability to cope with perturbations. With this notion, biodiversity could be used as an indicator of distribution system compromise (e.g., inefficient treatment, pipe leak, contamination, corrosion, etc.). However, in our study, the bacterial diversity estimations based on 16S pyrosequencing were insensitive for detecting the influences of disinfectant strategies. It is possible that diversity measurements may exhibit less variability than species composition in response to environmental factors because changes in some taxonomic groups can be compensated for by others, and thus the changes observed in microbial communities are not necessarily reflected in microbial diversity (9). This is in agreement with previous soil microbial ecological studies reporting that microbial community composition and structures were important for environmental monitoring rather than microbial diversity measurement (2, 14). Overall, we characterized here the drinking water microflora of a DWDS that receives both chlorination and chloramination treatments over a 2-year period. Although our study is based on DNA-based analysis that is useful for identifying present members, unlike RNA-based analysis that could provide information to active members, our study nonetheless demonstrated that chlorine and chloramines caused microbial community shifts and that certain populations are particularly affected by the different treatments (i.e., a reduction in cell numbers or reduced sensitivity to the treatments). Chlorination or chloramination treatments are reported to exert a strong selection process in the microflora in the drinking water that is consumed at the tap (6, 28, 40). Our findings revealed that the reversible shifts in microbial communities were especially pronounced with chloramination, which perhaps suggested a stronger selection pressure of microbial populations exerted from chloramines compared to chlorine. This impact of different disinfection methods on the overall drinking water microflora and detailed analysis of microbial communities in the distribution system can provide insight to the selection of disinfection strategies used by water utilities that provide potable water to the end users.
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff at Illinois American Waters for obtaining the water meters and Sang Liao for assistance in T-RFLP analysis.
This study was supported by the Water Research Foundation.
Footnotes
Published ahead of print 31 August 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Berry D, Xi C, Raskin L. 2006. Microbial ecology of drinking water distribution systems. Curr. Opin. Biotechnol. 17: 297–302 [DOI] [PubMed] [Google Scholar]
- 2. Cao Y, et al. 2006. Relationships between sediment microbial communities and pollutants in two California salt marshes. Microb. Ecol. 52: 619–633 [DOI] [PubMed] [Google Scholar]
- 3. Caporaso JG, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7: 335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Codony F, Morato J, Mas J. 2005. Role of discontinuous chlorination on microbial production by drinking water biofilms. Water Res. 39: 1896–1906 [DOI] [PubMed] [Google Scholar]
- 5. Cordoba M, Del Coco VF, Minvielle MC, Basualdo JA. 2010. Influencing factors in the occurrence of injured coliforms in the drinking water distribution system in the city of La Plata, Argentina. J. Water Health 8: 2205–2211 [DOI] [PubMed] [Google Scholar]
- 6. Eichler S, et al. 2006. Composition and dynamics of bacterial communities of a drinking water supply system as assessed by RNA- and DNA-based 16S rRNA gene fingerprinting. Appl. Environ. Microbiol. 72: 1858–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ellis BD, Butterfield P, Jones WL, McFeters GA, Camper AK. 1999. Effects of carbon source, carbon concentration, and chlorination on growth related parameters of heterotrophic biofilm bacteria. Microb. Ecol. 38: 330–347 [DOI] [PubMed] [Google Scholar]
- 8. Emde KME, Best N, Hrudey SE. 1992. Production of the potent odor agent, isopropyl methoxypyrazine, by Lysobacter enzymogenes. Environ. Technol. 13: 201–206 [Google Scholar]
- 9. Ernest SKM, Brown JH. 2001. Homeostasis and compensation: the role of species and resources in ecosystem stability. Ecology 82: 2118–2132 [Google Scholar]
- 10. Fass S, et al. 2003. Release of organic matter in a discontinuously chlorinated drinking water network. Water Res. 37: 493–500 [DOI] [PubMed] [Google Scholar]
- 11. Furuhata K, et al. 2006. Isolation and identification of Methylobacterium species from the tap water in hospitals in Japan and their antibiotic susceptibility. Microbiol. Immunol. 50: 11–17 [DOI] [PubMed] [Google Scholar]
- 12. Gagnon GA, et al. 2004. Comparative analysis of chlorine dioxide, free chlorine and chloramines on bacterial water quality in model distribution systems. J. Environ. Eng. 130: 1269–1279 [Google Scholar]
- 13. Gomez-Alvarez V, Revetta RP, Santo Domingo JW. 2012. Metagenomic analysis of drinking water receiving different disinfection treatments. Appl. Environ. Microbiol. 78: 6095–6102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hartmann M, Widmer F. 2006. Community structure analyses are more sensitive to differences in soil bacterial communities than anonymous diversity indices. Appl. Environ. Microbiol. 72: 7804–7812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hong PY, et al. 2010. Pyrosequencing analysis of bacterial biofilm communities in water meters of a drinking water distribution system. Appl. Environ. Microbiol. 76: 5631–5635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hwang C, Ling F, Andersen GL, Lechevallier MW, Liu WT. 2012. Evaluation of methods for the extraction of DNA from drinking water distribution system biofilms. Microbes Environ. 27: 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kahlisch L, et al. 2010. Molecular analysis of the bacterial drinking water community with respect to live/dead status. Water Sci. Technol. 61: 9–14 [DOI] [PubMed] [Google Scholar]
- 18. Kirk MF, et al. 2004. Bacterial sulfate reduction limits natural arsenic contamination in groundwater. Geology 32: 953–956 [Google Scholar]
- 19. LeChevallier MW, Cawthon CD, Lee RG. 1988. Factors promoting survival of bacteria in chlorinated water supplies. Appl. Environ. Microbiol. 54: 649–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. LeChevallier MW, Welch NJ, Smith DB. 1996. Full-scale studies of factors related to coliform regrowth in drinking water. Appl. Environ. Microbiol. 62: 2201–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Le Dantec C, et al. 2002. Occurrence of mycobacteria in water treatment lines and in water distribution systems. Appl. Environ. Microbiol. 68: 5318–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu W, et al. 2002. Investigation of assimilable organic carbon (AOC) and bacterial regrowth in drinking water distribution system. Water Res. 36: 891–898 [DOI] [PubMed] [Google Scholar]
- 23. Lukow T, Dunfield PF, Liesack W. 2000. Use of the T-RFLP technique to assess spatial and temporal changes in the bacterial community structure within an agricultural soil planted with transgenic and non-transgenic potato plants. FEMS Microbiol. Ecol. 32: 241–247 [DOI] [PubMed] [Google Scholar]
- 24. Mathieu L, Bouteleux C, Fass S, Angel E, Block JC. 2009. Reversible shift in the alpha-, beta-, and gammaproteobacteria populations of drinking water biofilms during discontinuous chlorination. Water Res. 43: 3375–3386 [DOI] [PubMed] [Google Scholar]
- 25. Mir J, Morato J, Ribas F. 1997. Resistance to chlorine of freshwater bacterial strains. J. Appl. Microbiol. 82: 7–18 [DOI] [PubMed] [Google Scholar]
- 26. Ndiongue S, Huck PM, Slawson RM. 2005. Effects of temperature and biodegradable organic matter on control of biofilms by free chlorine in a model drinking water distribution system. Water Res. 39: 953–964 [DOI] [PubMed] [Google Scholar]
- 27. Niquette P, Servais P, Savoir R. 2001. Bacterial dynamics in the drinking water distribution system of Brussels. Water Res. 35: 675–682 [DOI] [PubMed] [Google Scholar]
- 28. Poitelon JB, et al. 2010. Variations of bacterial 16S rDNA phylotypes prior to and after chlorination for drinking water production from two surface water treatment plants. J. Ind. Microbiol. Biotechnol. 37: 117–128 [DOI] [PubMed] [Google Scholar]
- 29. Propato M, Uber JG. 2004. Vulnerability of water distribution systems to pathogen intrusion: how effective is a disinfectant residual? Environ. Sci. Technol. 38: 3713–3722 [DOI] [PubMed] [Google Scholar]
- 30. Rees GN, Baldwin DS, Watson GO, Perryman S, Nielsen DL. 2004. Ordination and significance testing of microbial community composition derived from terminal restriction fragment length polymorphisms: application of multivariate statistics. Antonie Van Leeuwenhoek 86: 339–347 [DOI] [PubMed] [Google Scholar]
- 31. Revetta RP, Matlib RS, Santo Domingo JW. 2011. 16S rRNA gene sequence analysis of drinking water using RNA and DNA extracts as targets for clone library development. Curr. Microbiol. 63: 50–59 [DOI] [PubMed] [Google Scholar]
- 32. Richardson SD. 2003. Disinfection by-products and other emerging contaminants in drinking water. Trends Analyt. Chem. 22: 666–684 [Google Scholar]
- 33. Roeder RS, et al. 2010. Long-term effects of disinfectants on the community composition of drinking water biofilms. Int. J. Hyg. Environ. Health 213: 183–189 [DOI] [PubMed] [Google Scholar]
- 34. Schmidt TM, Delong EF, Pace NR. 1991. Analysis of a marine picoplankton community by 16S ribosomal-RNA gene cloning and sequencing. J. Bacteriol. 173: 4371–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shrivastava R, et al. 2004. Suboptimal chlorine treatment of drinking water leads to selection of multidrug-resistant Pseudomonas aeruginosa. Ecotoxicol. Environ. Saf. 58: 277–283 [DOI] [PubMed] [Google Scholar]
- 36. Staley JT, Konopka A. 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39: 321–346 [DOI] [PubMed] [Google Scholar]
- 37. Tamaki H, et al. 2011. Analysis of 16S rRNA amplicon sequencing options on the Roche/454 next-generation titanium sequencing platform. PLoS One 6: e25263 doi:10.1371/journal.pone.0025263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. ter Braak CJ, Šmilauer L. 1998. CANOCO reference manual and user's guide to CANOCO for Windows. Microcomputer Power, Ithaca, NY [Google Scholar]
- 39. Wahman DG, Kirisits MJ, Katz LE, Speitel GE., Jr 2011. Ammonia-oxidizing bacteria in biofilters removing trihalomethanes are related to Nitrosomonas oligotropha. Appl. Environ. Microbiol. 77: 2537–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Williams MM, Domingo JW, Meckes MC, Kelty CA, Rochon HS. 2004. Phylogenetic diversity of drinking water bacteria in a distribution system simulator. J. Appl. Microbiol. 96: 954–964 [DOI] [PubMed] [Google Scholar]
- 41. Wolfe RL, Ward NR, Olson BH. 1985. Inactivation of heterotrophic bacterial-populations in finished drinking-water by chlorine and chloramines. Water Res. 19: 1393–1403 [Google Scholar]
- 42. Zhang WD, DiGiano FA. 2002. Comparison of bacterial regrowth in distribution systems using free chlorine and chloramine: a statistical study of causative factors. Water Res. 36: 1469–1482 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.