Abstract
The alkane hydroxylase system of Pseudomonas putida GPo1 allows it to use alkanes as the sole source of carbon and energy. Bacterial alkane hydroxylases have tremendous potential as biocatalysts for the stereo- and regioselective transformation of a wide range of chemically inert unreactive alkanes into valuable reactive chemical precursors. We have produced and characterized the first 2-dimensional crystals of the integral membrane component of the P. putida alkane hydroxylase system, the nonheme di-iron alkane monooxygenase AlkB. Our analysis reveals for the first time that AlkB reconstituted into a lipid bilayer forms trimers. Addition of detergents that do not disrupt the AlkB oligomeric state (decyl maltose neopentyl glycol [DMNG], lauryl maltose neopentyl glycol [LMNG], and octaethylene glycol monododecyl ether [C12E8]) preserved its activity at a level close to that of the detergent-free control sample. In contrast, the monomeric form of AlkB produced by purification in n-decyl-β-d-maltopyranoside (DM), n-dodecyl-β-d-maltopyranoside (DDM), octyl glucose neopentyl glycol (OGNG), and n-dodecyl-N,N-dimethylamine-N-oxide (LDAO) was largely inactive. This is the first indication that the physiologically active form of membrane-embedded AlkB may be a multimer. We present for the first time experimental evidence that 1-octyne acts as a mechanism-based inhibitor of AlkB. Therefore, despite the lack of any significant full-length sequence similarity with members of other monooxygenase classes that catalyze the terminal oxidation of alkanes, AlkB is likely to share a similar catalytic mechanism.
INTRODUCTION
Alkane hydroxylases are versatile biocatalysts that insert an oxygen atom derived from O2 into otherwise inert alkanes, often with great regio- and stereoselectivity. This reaction allows many bacteria to use alkanes as the sole source of carbon and energy, making them able to grow and survive in oil-contaminated environments (34). The selective oxidation activity of alkane hydroxylases under mild conditions offers tremendous potential to produce a large number of commercially valuable precursors for the chemical, pharmaceutical, and fragrance industries using green chemical approaches (1, 26). Therefore, there is a strong incentive to study in detail the structure/function relationships of the alkane hydroxylase systems.
Different alkane hydroxylases can be grouped together according to the chain lengths of the alkanes they oxidize: C1 to C4 (methane to butane; oxidized by two biochemically and structurally distinct monooxygenase classes, soluble methane monooxygenase [sMMO] and particulate methane monooxygenase [pMMO]), C5 to C16 (pentane to hexadecane; oxidized by integral membrane nonheme iron or cytochrome P450 enzymes), and C17+ (long-chain alkane monooxygenases) (10, 28). The integral membrane nonheme di-iron alkane hydroxylase from Pseudomonas putida GPo1 (AlkB) is one of the more unusual alkane hydroxylases, as its n-alkane substrate range is limited to C3 to C12 (gasoline-range alkanes), while many other related medium-chain alkane hydroxylases typically oxidize alkanes longer than C10 (28). The alkane hydroxylase system of P. putida GPo1 comprises three protein components: AlkB, soluble NADH-rubredoxin reductase (RR), and soluble electron transfer protein rubredoxin (Rd). AlkB transfers one oxygen atom from O2 to the alkane molecule, while the other oxygen is reduced to H2O using the electrons provided by RR (8) via Rd (9, 15). In addition to the terminal hydroxylation of linear alkanes (6, 31), P. putida AlkB catalyzes the hydroxylation of branched alkanes and alicyclic and alkylaromatic compounds, oxidation of terminal alcohols to the corresponding aldehydes, demethylation of branched methyl ethers, sulfoxidation of thioethers, and epoxidation of terminal olefins and allyl alcohol derivatives (17). The combination of a wide substrate range with a high regioselectivity makes this system useful for the production of fine chemicals such as fatty acids, alcohols, epoxides, and sulfoxides. Examples of industrial applications include the following: production of 1,2-epoxyoctane; specific epoxidation of 4-(2-methoxyethyl)phenylallyl ether, an intermediate in the production of β-blockers; regioselective oxidation of ethyl-substituted aromatic compounds to produce hydroxyethyl-substituted heterocyclic compounds; and synthesis of 1-alkanols from linear alkanes using alcohol dehydrogenase-deficient P. putida strains (17).
Although the biotechnological potential of AlkB is well recognized, very little is known about its 3-dimensional (3D) structure and the locations of the catalytic residues, the substrate binding pocket and the docking sites for its redox partners, RR and Rd. The topology of P. putida AlkB was studied by van Beilen et al. (29) using protein fusions to alkaline phosphatase and β-galactosidase. The protein is likely to have six transmembrane (TM) segments, with the amino terminus, two hydrophilic loops, and a large carboxy-terminal domain located in the cytoplasm. More recently, site-directed mutagenesis studies have confirmed the essential role of eight conserved histidine residues in enzymatic activity (21), and a single amino acid residue was found to determine the maximum length of the n-alkane substrates that can bind within the active site (30). Little is known about the amino acids lining the substrate binding pocket, the arrangement of the transmembrane helices within the lipid bilayer, or the conformation of the C-terminal domain.
The main obstacle in the structural characterization of AlkB has been the difficulty in isolating this integral membrane protein in a stable and active form. Consequently, all previous studies of AlkB used whole cells, cell extracts, or partially purified enzyme (6, 7, 13, 14, 18, 20, 23, 31). We have recently reported an improved protocol that overcomes this limitation and allows the isolation of milligram amounts of recombinant, detergent-solubilized P. putida GPo1 AlkB in a folded, catalytically active form to purity levels of above 90% (35).
In this paper, we report the first 2-dimensional crystallization of AlkB by reconstitution of detergent-purified enzyme into a lipid bilayer. Analysis of these crystals revealed an unexpected trimeric structure of AlkB. In addition, we report the effects of different detergents on its enzymatic activity and the kinetic characterization of 1-octyne as a likely mechanism-based inactivator of AlkB.
MATERIALS AND METHODS
Reagents and bacterial strains.
Escherichia coli BL21(DE3), C41(DE3), Rosetta-2(DE3), pLysS, and BL21-CondonPlus(DE3)-RIPL were purchased from Novagen. n-Dodecyl-β-d-maltopyranoside (DDM), n-decyl-β-d-maltopyranoside (DM), n-octyl-β-d-glucoside (OG), n-dodecyl-N,N-dimethylamine-N-oxide (LDAO), octaethylene glycol monododecyl ether (C12E8), lauryl maltose neopentyl glycol (LMNG), decyl maltose neopentyl glycol (DMNG), and octyl glucose neopentyl glycol (OGNG) were purchased from Anatrace. E. coli polar lipids and 1,2-ditetradecanoyl-sn-glycero-3-phosphocholine (DMPC) were purchased from Avanti Polar Lipids. Desthiobiotin, n-octane, and 1-octyne were from Sigma and NADH from Roche. The StrepTactin-alkaline phosphatase (AP) conjugate was from Bio-Rad and the chemiluminescent substrate CDP-Star from Invitrogen. The pGEc47 plasmid, containing the AlkBAC operon and the AlkR locus (3), was purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH.
Gene cloning and protein expression and purification.
The gene encoding P. putida AlkB with a C-terminal StrepII tag was constructed as previously described (35) and inserted into the pET22-b(+) expression vector to give the plasmid pET22-PpAlkB-SII. The gene encoding P. putida AlkT (RR) was amplified from the pGEc47 plasmid using the primers 5′-AGGAGATATACATATGGCAATCGTTGTTG-3′ and 5′-GATCTGAGCTCTAATCAGGTAATTTTATAC-3′, incorporating unique NdeI and SacI restriction sites (underlined), and cloned into pET22-b(+) to produce pET22-PpAlkT. All plasmids were confirmed by sequencing.
P. putida Rd was expressed and purified as previously described (9). For the expression of P. putida RR, the pET22-PpAlkT plasmid was transformed into E. coli BL21(DE3) cells, which were then grown in LB medium containing 100 μg/ml ampicillin at 18°C until an optical density at 600 nm (OD600) of 0.5 was reached, at which point overexpression of RR was induced by adding 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and growth was continued for a further 16 h. The protein was purified following a modified protocol described by Lee et al. (8). In brief, the cells were harvested, resuspended in buffer A (20 mM sodium phosphate [pH 7.4] and 20% glycerol), and lysed using a high-pressure homogenizer (Avestin). The clarified extract was applied to a DEAE (DE-52) column (GE Healthcare) preequilibrated in buffer A. The column was then washed with 3 column volumes (cv) of buffer A, and the protein was eluted with a linear gradient of 0 to 0.1 M KCl in buffer A. Fractions containing RR were pooled and dialyzed against buffer A. The sample was then loaded into a Cibracon Blue column (GE Healthcare) preequilibrated in buffer A. After the column was washed with 10 cv of buffer A, RR was eluted with a linear gradient of 0 to 0.1 M KCl in buffer A, dialyzed against buffer A, concentrated by ultrafiltration to 18.3 μM, and stored at −80°C. The concentration of the purified P. putida RR was determined using its extinction coefficient at 450 nm (11,100 M−1 cm−1 [24]).
Expression of P. putida AlkB (PpAlkB-SII) from the pET22-PpAlkB-SII plasmid was tested in E. coli strains BL21(DE3), C41(DE3), Rosetta-2(DE3), pLysS, and BL21-CondonPlus(DE3)-RIPL under different growth conditions. The highest expression levels as judged by Western blot analysis of SDS-polyacrylamide gels were achieved in C41(DE3) cells grown at 30°C to an OD600 of ∼0.5 and then induced with IPTG for 4 to 5 h. The cells were harvested by centrifugation, resuspended in buffer B (20 mM sodium phosphate [pH 7.4], 150 mM NaCl, 4 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride [PMSF]), and lysed using the Avestin homogenizer. Unbroken cells and cell debris were removed by a 30-min centrifugation at 15,000 × g. The supernatant was then subjected to overnight ultracentrifugation at 100,000 × g at 4°C to pellet the membranes containing PpAlkB-SII.
To purify PpAlkB-SII without detergent we followed a modified protocol described by Shanklin et al. (20). In brief, the membrane fraction was resuspended by sonication in 50 mM HEPES (pH 7.5) and urea was added to a final concentration of 1 M, after which the sample was spun down at 290,000 × g for 1 h. The pellet was then resuspended by sonication in buffer C (25 mM Tris [pH 8.0], 10% glycerol, 150 mM NaCl, and 1 mM PMSF) and applied to a 5-ml StrepTactin MacroPrep column (IBA). After washing with 5 cv of buffer C, the sample was eluted with buffer C supplemented with 5 mM desthiobiotin.
For the purification of PpAlkB-SII in the presence of detergent, the membrane fraction was resuspended by sonication in buffer D (20 mM Tris [pH 8.0], 150 mM NaCl, 15% glycerol, 4 mM β-mercaptoethanol, and 1 mM PMSF), after which detergent was added to approximately 20 times its critical micelle concentration (20 CMC) (1.75% DM, 0.17% DDM, 1.16% OGNG, 0.07% DMNG, 0.02% LMNG, 0.10% C12E8, or 0.46% LDAO) and the sample was incubated in a rotator mixer at 4°C for 1 h. The undissolved material was removed by ultracentrifugation at 100,000 × g for 30 min and the clarified sample loaded into the StrepTactin MacroPrep column. The column was washed with 5 cv of buffer D containing 4 CMC of detergent (0.35% DM, 0.035% DDM, 0.23% OGNG, 0.014% DMNG, 0.004% LMNG, 0.02% C12E8, or 0.10% LDAO), and PpAlkB-SII was eluted using buffer D supplemented with 4 CMC of detergent and 5 mM desthiobiotin. The protein was then concentrated by ultrafiltration using a 100-kDa cutoff filter and loaded onto a size exclusion Superdex 200 10/300 GL column (GE Healthcare) preequilibrated in buffer E (20 mM Tris [pH 7.5], 250 mM NaCl, 15% glycerol, 4 mM β-mercaptoethanol, and 1 mM PMSF) supplemented with 4 CMC detergent. The peak fractions were pooled, concentrated, and used immediately.
SDS-PAGE and Western blot analysis.
Proteins were separated on 12% (vol/vol) SDS-PAGE gels and visualized by Coomassie blue staining or transferred onto a nitrocellulose membrane for Western blot analysis. The StrepII tag in PpAlkB-SII was detected using StrepTactin-AP conjugate and the chemiluminescent substrate CDP-Star.
Enzymatic activity.
The hydroxylation activity of purified PpAlkB-SII with its natural substrate n-octane was measured using a previously described spectrophotometric assay (13, 35). The reaction buffer contained 20 mM Tris (pH 8.0), 15% glycerol, and 50 mM NaCl. A 50 mM n-octane stock solution was prepared in acetone. A typical reaction mixture contained 3 μM Rd, 0.6 μM RR, 0.7 to 2.2 μM PpAlkB-SII, 200 μM NADH, and 0.8 mM n-octane. The rate of NADH consumption was followed by measuring the change in absorbance at 340 nm at room temperature using a Varian 50-Bio UV-visible spectrophotometer. Background activity levels determined by the addition of acetone, the solvent of n-octane, were subtracted from the n-octane hydroxylation reaction rates. The rate of NADH consumption (absorbance units/min) was converted into AlkB specific hydroxylase activity (μmol n-octane/min/mg protein) using the extinction coefficient of NADH (ε340 = 6,220 M−1 cm−1) and the amount of protein determined using the Bradford assay. Heat inactivation of PpAlkB-SII was achieved by incubating the enzyme at 65°C for 15 min.
The effect of the potential mechanism-based inhibitor 1-octyne was studied by preincubating PpAlkB-SII with the alkyne in the reaction buffer containing Rd and RR in both the presence and absence of NADH and then measuring the remaining n-octane hydroxylation activity. Detergent-free PpAlkB-SII (0.7 μM) was preincubated with different concentrations of 1-octyne (50, 100, 250, and 500 μM) under turnover conditions (reaction buffer including 3 μM Rd, 0.6 μM RR, and 200 μM NADH [optionally]) for 2 min. The remaining activity was then tested by adding 0.8 mM n-octane and 200 μM NADH and measuring the change in absorbance at 340 nm. The inhibitor binding affinity (KI) and inactivation rate (kinact) were determined as described by Maurer and Fung (12) by fitting the equation −ln(E/E0)/t = (kinact × [I])/([I] + KI), where E0 represents the maximum enzyme activity following preincubation in the absence of inhibitor and E is the activity following preincubation with the inhibitor over time t.
The effects of different detergents on PpAlkB-SII activity were measured by preincubating the detergent-free purified enzyme with 4 CMC of OG (2.12%), DM (0.35%), DDM (0.035%), OGNG (0.23%), DMNG (0.014%), LMNG (0.004%), C12E8 (0.02%), or LDAO (0.10%) for 60 min at room temperature and measuring the remaining activity as described above.
CD analysis.
Far-UV circular dichroism (CD) spectra were recorded for PpAlkB-SII purified in the presence of C12E8 and DDM using a JASCO J600 spectropolarimeter. The 0.1-mg/ml protein samples were prepared in a buffer containing 20 mM sodium phosphate (pH 7.5), 250 mM NaCl, 15% glycerol, and 4 CMC detergent (0.02% C12E8 or 0.035% DDM). The spectra were collected over the wavelength range from 190 to 260 nm in a quartz cuvette with a 2-mm path length at a scan rate of 20 nm min−1, and the results were averaged over 5 accumulated spectra. The percentage of secondary structure was calculated using the DichroWeb server and the K2D program (32, 33).
2D crystallization.
PpAlkB-SII in C12E8 or DDM at a final concentration of 1 mg/ml was mixed with one of two lipids (1,2-dimyristoyl-sn-glycero-3-phosphocholine [DMPC] solubilized in DM or E. coli polar lipid extract solubilized in DM or DDM) at a lipid-to-protein ratio (LPR) range of 0.1 to 1.0, transferred into dialysis buttons (Hampton Research) and dialyzed against detergent-free buffer A (10 mM HEPES [pH 7.0], 10 mM MgCl2, 150 mM NaCl, 0.01% sodium azide) or buffer B (10 mM MES [morpholineethanesulfonic acid] [pH 6.5], 10 mM MgCl2, 150 mM NaCl, 0.01% sodium azide) at 37°C. The dialysis buffers were renewed every 7 days and supplemented with 10 mM CaCl2 after 3 weeks. The samples were first inspected at 4 weeks after dialysis commencement.
Transmission electron microscopy (TEM) imaging and data processing.
PpAlkB-SII purified with or without detergent was diluted in buffer E supplemented with 4 CMC detergent or buffer C, respectively, to a final concentration of 0.005 mg/ml, adsorbed onto carbon-coated copper grids, washed with deionized water, negatively stained with 2% uranyl acetate, and air dried. A 2D crystal suspension extracted from the dialysis buttons was applied to the grids without dilution and negatively stained as described above. The analysis of the grids was carried out using a Hitachi H7500 TEM 120-kV microscope fitted with a digital Gatan camera. The untilted images of the negatively stained crystals were processed using the 2dx software package (4, 5).
RESULTS
Expression and purification of P. putida AlkB.
A C-terminally StrepII-tagged version of the P. putida alkB product was expressed in E. coli BL21(DE3). We extracted and purified PpAlkB-SII in seven different detergents (DM, DDM, OGNG, DMNG, LMNG, C12E8, or LDAO) using StrepTactin affinity chromatography followed by size exclusion chromatography (SEC). PpAlkB was isolated in each detergent to purity levels of greater than 90% as judged by SDS-PAGE (Fig. 1A shows an example of C12E8 purification). Approximately 2 mg of pure protein was obtained from 1 liter of bacterial culture, a 20-fold yield improvement over our previously published protocol (35).
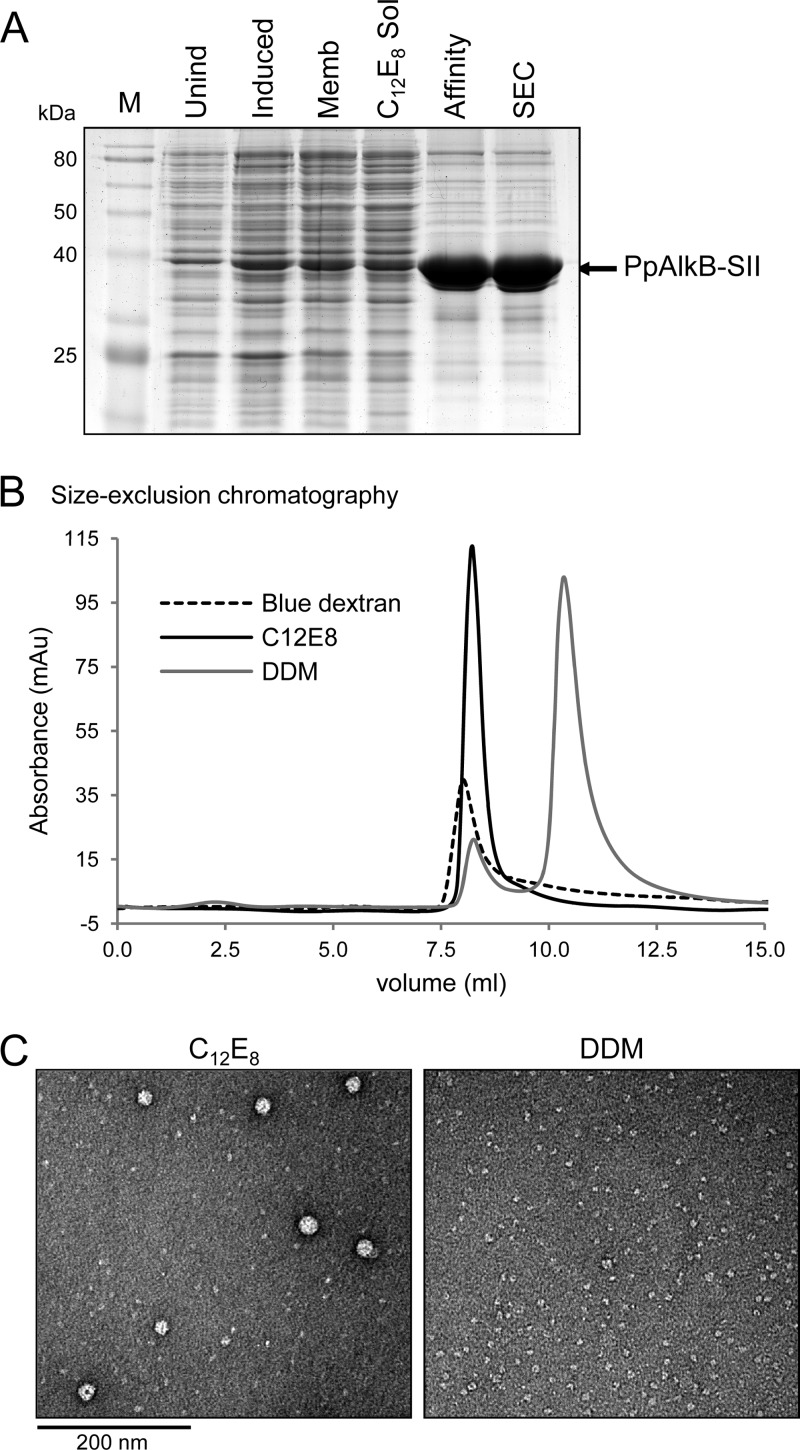
Fig 1.
Purification of PpAlkB-SII. (A) SDS-PAGE analysis of the purification process: M, marker; Unind, uninduced total cell lysate sample; Induced, induced sample; Memb, membrane fraction of the induced sample; C12E8 Sol, C12E8-solubilized fraction; Affinity, eluate from the MacroPrep StrepTactin column; SEC, pooled peak fraction from the size exclusion chromatography step. (B) Size exclusion chromatography elution profiles of blue dextran, and C12E8- and DDM-purified PpAlkB-SII. (C) TEM images of negatively stained DDM- and C12E8-purified PpAlkB-SII (for DDM, the second [main] peak from the size exclusion chromatography step was used for this analysis).
Two significantly different elution patterns were observed during the SEC step in the presence of different detergents (Fig. 1B). While the major peak of the samples purified with DM, DDM, and OGNG eluted at ∼11 ml, the C12E8, DMNG, LMNG, and LDAO samples eluted at ∼8.8 ml, close to the void volume of the column (8 ml). The association of a PpAlkB-SII monomer (molecular mass, 46 kDa) with detergent (for example, C12E8 micelle [∼66 kDa] and DDM micelle [∼72 kDa]) was estimated to produce a complex with a molecular mass in excess of 100 kDa. The observed value of ∼8.8 ml for the elution volume of PpAlkB-SII in C12E8, DMNG, LMNG, and LDAO corresponded to a much larger complex with a molecular mass in excess of 440 kDa. In order to rule out protein aggregation, the samples in DDM (11-ml elution volume) and C12E8 (8.8-ml elution volume) were further studied by electron microscopy (Fig. 1C). The sample in DDM was monodispersed, with particle sizes averaging 60 Å in diameter. Assuming a particle density of about 1.35 g cm−3 (average protein density), a 90-kDa spherical particle would have a diameter of ≈ 60 Å, where Na is the Avogadro number. This analysis suggests that purification in DDM produces micelle-bound PpAlkB-SII monomers. In contrast, the sample in C12E8 was a combination of both small and large particles. The larger protein clusters, ranging in size from 150 to 300 Å, appeared to have a distinctive ultrastructure indicative of protein multimers rather than aggregates.
Detergent-free preparation of membrane fraction enriched in PpAlkB-SII by means of StrepTactin affinity chromatography produced samples with purity levels of above 80% and a yield of ∼0.2 mg of PpAlkB-SII per 1 liter of bacterial culture (Fig. 2A). Electron microscopy imaging showed that PpAlkB-SII in these samples formed vesicular structures of ∼33 nm in diameter (Fig. 2B), which is consistent with the results of previous characterization of P. putida AlkB purified in the absence of detergent (20).
Fig 2.
Detergent-free purification of PpAlkB-SII. (A) SDS-PAGE analysis of the purification process: M, marker; Total, total membrane fraction; Urea, soluble protein after urea treatment; Unb, unbound (flowthrough) fraction from the MacroPrep StrepTactin column; Purif, final purified PpAlkB-SII protein. (B) TEM image of negatively stained detergent-free purified PpAlkB-SII.
Secondary structure of detergent-purified AlkB.
CD analysis of PpAlkB-SII isolated in DDM and C12E8 (Fig. 3) demonstrated that these samples had similar secondary structure despite their different SEC elution profile. The estimated α-helical and β-sheet contents (DDM, 48% α and 21% β; C12E8, 44% α and 17% β) were very close to those previously reported for the LDAO-purified AlkB (44% α and 19% β) (35) and to those predicted using JPred server sequence analysis (51% α and 14% β) (2). These results indicated that both the DDM- and C12E8-solubilized proteins were folded and that the observed difference in their quaternary structure was not likely to be the result of a large secondary structure rearrangement.
Fig 3.
Circular dichroism spectra of DDM- and C12E8-purified PpAlkB-SII at a concentration of 0.1 mg/ml. The secondary structure content calculated from these data using the DichroWeb server and the K2D program is shown in the table below the spectra.
Effects of detergents and alkynes on the enzymatic activity of alkane hydroxylase.
In line with previous reports (20, 21, 27), the n-octane hydroxylation activity of the detergent-free purified PpAlkB-SII was ∼2 μmol/min/mg (Fig. 4). The purification process resulted in a 40-fold increase in activity from the initial ∼0.05 μmol/min/mg of total protein in the original whole-cell lysate (Fig. 4A). The hydroxylase activity of purified PpAlkB-SII was significantly reduced after preincubation with the AlkB-specific iron-chelating inhibitor 8-hydroxyquinoline (13) and after heat treatment (Fig. 4A).
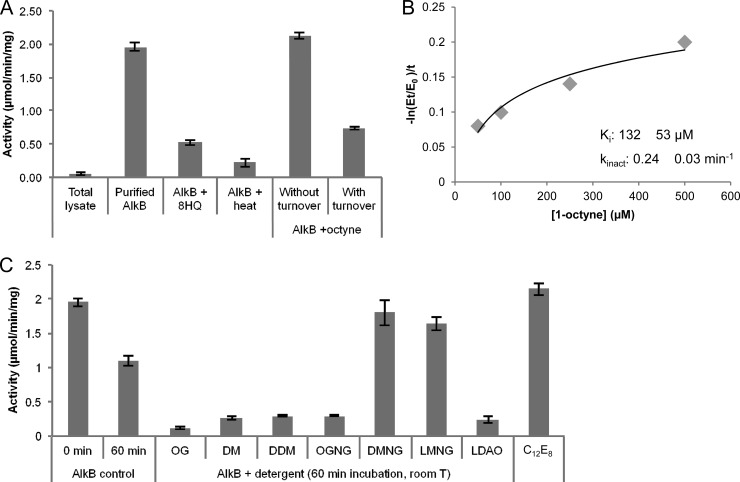
Fig 4.
Functional characterization of PpAlkB-SII. The hydroxylation of n-octane by the PpAlkB-SII/Rd/RR system was followed spectrophotometrically by measuring the coupled NADH consumption. Results are the averages from three independent replicates, and error bars show the standard deviations. (A) The initial n-octane hydroxylase activity of the whole-cell lysate (0.05 ± 0.02 μmol/min/mg total protein) (total lysate) increased ∼40-fold after protein purification (1.95 ± 0.05 μmol/min/mg purified PpAlkB-SII) (purified AlkB). Preincubation of PpAlkB-SII with a 1 mM concentration of the inhibitor 8-hydroxyquinoline (AlkB + 8HQ) or PpAlkB-SII inactivation by heat pretreatment at 65°C for 15 min (AlkB + heat) resulted in decreased activity. The effect of 1-octyne on the activity of the PpAlkB-SII/Rd/RR system was studied by adding 0.8 mM 1-octyne under turnover conditions (0.72 μM PpAlkB-SII, 3 μM Rd, 0.6 μM RR, and 200 μM NADH) (with turnover) or without turnover (no NADH). 1-Octyne behaved as a mechanism-based inactivator, inhibiting PpAlkB-SII only under turnover conditions. (B) Determination of the 1-octyne binding affinity (KI) and inactivation rate (kinact) by measuring the remaining activity after preincubation of PpAlkB-SII with different concentrations of 1-octyne under turnover conditions. (C) The effects of different detergents on the activity of PpAlkB-SII were tested by preincubating the detergent-free PpAlkB-SII with 4 CMC of OG, DM, DDM, OGNG, DMNG, LMNG, LDAO, or C12E8 at room temperature for 60 min prior to measuring its n-octane hydroxylase activity in the presence of Rd, RR, and NADH. DMNG, LMNG, and C12E8 preserved activity at a level close to that of the control sample at time zero, thus stabilizing the PpAlkB-SII protein compared to the detergent-free control.
To further characterize PpAlkB-SII, we tested the effect of 1-octyne on enzyme activity. Alkynes have been shown to act as mechanism-based inhibitors of monooxygenases (16, 36, 37). They bind to the oxygenase active site and are oxidized to an oxirene equivalent that rapidly rearranges to a reactive ketene that covalently acylates an active-site amino acid, causing a concurrent loss of enzyme activity. AlkB was reported to be inhibited by 1,7-octadiyne in previous studies (7); however, no relevant kinetic parameters were shown. Therefore, we decided to perform the kinetic characterization of 1-octyne as a potential suicide inhibitor of PpAlkB-SII. Incubation of detergent-free PpAlkB-SII with 1-octyne under turnover conditions resulted in inactivation of the enzyme. However, no inhibition was observed if NADH was excluded from the incubation solution (Fig. 4A). This result suggests that 1-octyne is a mechanism-based inactivator of PpAlkB-SII, since it is effective only under conditions of catalysis. This view is further supported by the observation that when no NADH is present, 1-octyne can be readily displaced by the addition of excess substrate n-octane and AlkB remains active (Fig. 4A). We measured the binding affinity KI and inactivation rate kinact of 1-octyne as 132 ± 53 μM and 0.24 ± 0.03 min−1, respectively (Fig. 4B).
To evaluate the effects of different detergents on PpAlkB-SII activity, detergent-free PpAlkB-SII was preincubated with 4 CMC of OG, DM, DDM, OGNG, DMNG, LMNG, C12E8, or LDAO for 1 h before measuring the rate of NADH oxidation by the PpAlkB-SII/Rd/RR system (Fig. 4C). Although the detergent-free purified enzyme was found to be stable at 4°C and resistant to freeze-thaw cycles as previously reported (20), incubation at room temperature for 60 min resulted in an ∼50% activity loss. Interestingly, incubation of PpAlkB-SII in the presence of detergents resulted in two very different outcomes. While significant loss of activity (>85%) was observed for OG, DM, DDM, OGNG, and LDAO, incubation with DMNG, LMNG, and C12E8 preserved activity at levels similar to that for the control sample at time zero. These results suggest that while membrane-bound PpAlkB-SII is sensitive to treatment with some detergents, the presence of the low-CMC detergents DMNG, LMNG, and C12E8 increase the stability of the protein at room temperature.
2D crystallization.
We performed 2D crystallization trials by reconstituting the detergent-purified PpAlkB-SII into a lipid bilayer during slow detergent dialysis for several weeks at 37°C, with weekly inspections after 1 month. Although it would be desirable to work with active PpAlkB-SII for the purpose of structural characterization, we note that the low CMC values of the detergents found to preserve its activity (DMNG, 0.0034%; LMNG, 0.001%; and C12E8, 0.0048%) make it difficult to remove them by dialysis. For this reason we selected two detergents for crystallization trials: C12E8, which has the highest CMC of the three detergents that allow isolation of an active, multimeric enzyme, and DDM, which possess a higher CMC (0.0087%) and produces monodisperse, likely monomeric (albeit inactive) protein.
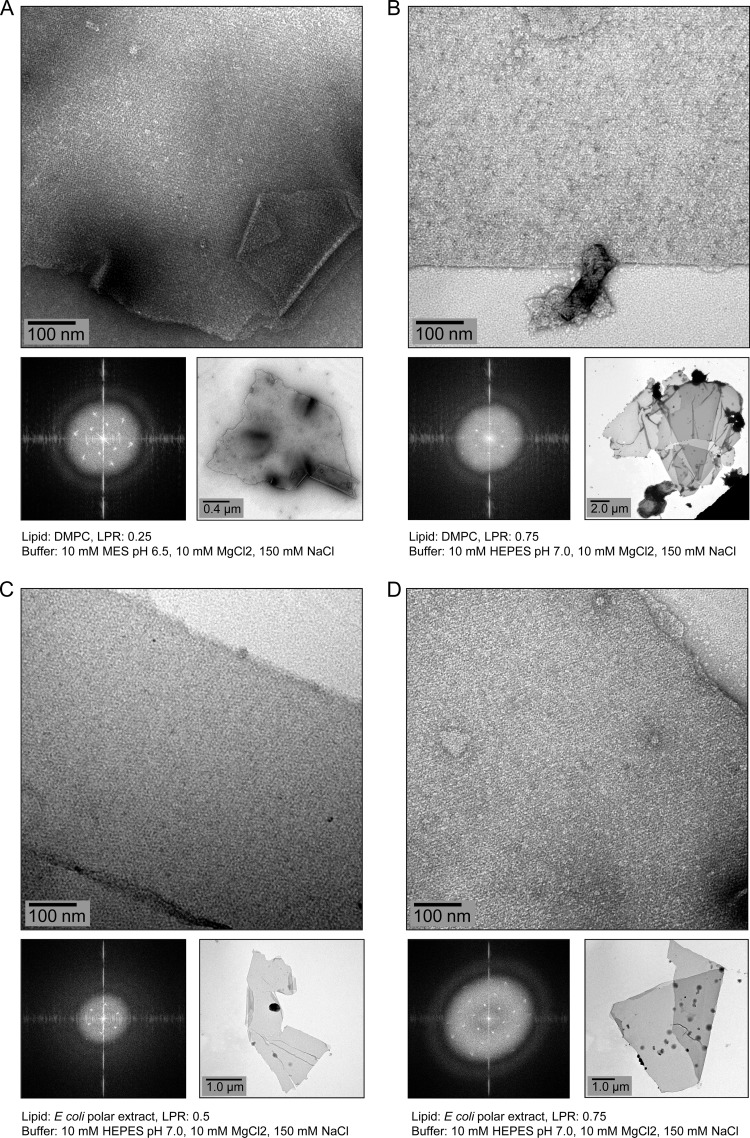
Reconstitution of C12E8-puified PpAlkB-SII in lipids did not produce any crystals, probably due to the difficulty of removing this low-CMC detergent by dialysis. In contrast, several 2D crystals of 1 μm or more were obtained for the DDM-purified enzyme using both E. coli polar lipids and DMPC under several conditions after 4 weeks of dialysis (Fig. 5). Transmission electron microscopy images of all negatively stained crystals showed two layers at the edges (clearly seen in Fig. 5A and D). Optical diffraction of these crystals showed single spots rather than twinning, indicating that these are double-layered crystals rather than the stacks of two, roughly parallel, separate single-layered crystals (“pseudocrystals”) (Fig. 5). The calculated power spectrum of these images showed clear diffraction spots up to ∼2.5-nm resolution. We were unable to detect n-octane hydroxylase activity in the reconstituted samples and cannot exclude the possibility that the crystals represent an inactive state of PpAlkB-SII.
Fig 5.
Electron micrographs of negatively stained 2D crystals of membrane-reconstituted PpAlkB-SII obtained after 4 weeks of dialysis of the DDM-purified protein containing DMPC (A and B) or E. coli polar lipids (C and D) against the indicated buffers.
Projection map of the AlkB protein.
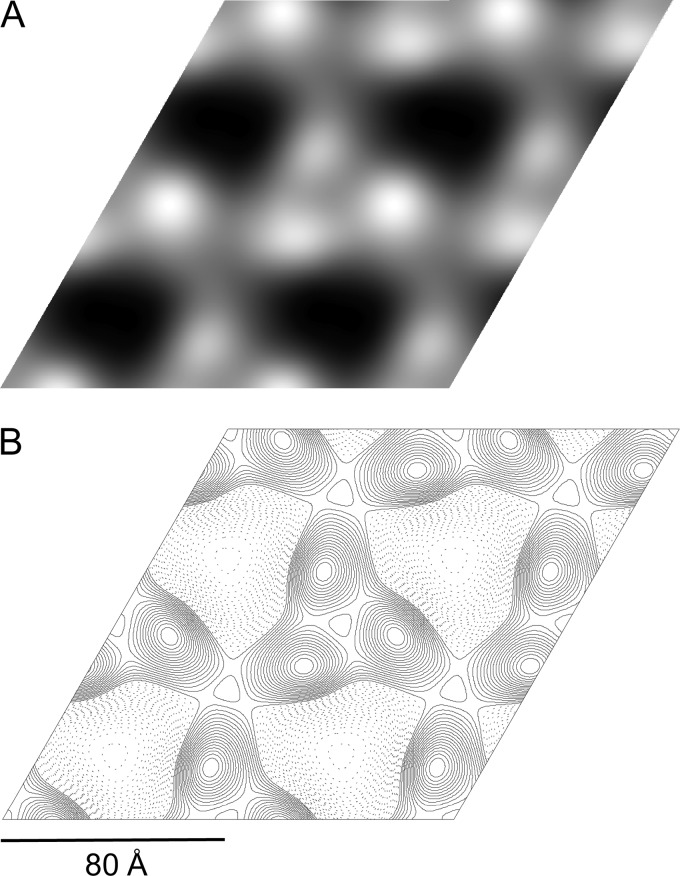
Merging and processing 5 images from different regions of a single negatively stained crystal obtained in E. coli polar lipids (LPR, 0.5) allowed us to generate the projection map shown in Fig. 6. Analysis of phase residuals using the ALLSPACE program (25) indicated p3 symmetry with the unit cell dimensions a = b = 80 Å and γ = 120° and a maximum resolution of 16 Å. This result is in agreement with analysis of the nonsymmetrized merged map (Fig. 6A), which shows a trimer with a 3-fold rotational symmetry, where each monomer has an elliptical shape with approximate dimensions of 40 by 30 Å.
Fig 6.
Reconstruction of the 2D PpAlkB-SII crystal. (A) Merged map showing 2-by-2 unit cells (protein is seen as white over a dark background). (B) Reconstruction map after application of p3 symmetry.
DISCUSSION
In this study, we have presented the functional and structural characterization of P. putida AlkB, an alkane hydroxylase enzyme that allows bacteria to grow in oil-rich environments. We have evaluated the effects of different detergents on the hydroxylation activity of the purified enzyme with its natural substrate n-octane, characterized the inhibitory effect of 1-octyne, and obtained the first projection image of a 2D crystal of membrane-reconstituted P. putida AlkB.
Out of all the detergents tested, only the ones with the lowest CMCs (DMNG, LMNG, and C12E8) preserved the PpAlkB-SII hydroxylase activity after purification. Furthermore, the same subset of detergents was found to have an activity-stabilizing effect on a detergent-free sample (Fig. 4C), presumably compensating for the lost lipid-protein interactions yet not delipidating the protein.
Interestingly, PpAlkB-SII purified in this subset of low-CMC detergents eluted as high-molecular-weight complexes during size exclusion chromatography. Electron microscopy analysis of the C12E8 sample, for example, revealed multimers of 150 to 300 Å in size (Fig. 1C). On the other hand, none of the samples that appeared to be monomeric (i.e., those purified in the higher-CMC detergents DM, DDM, and OGNG) showed any detectable specific activity. An implication of this observation is that AlkB likely functions as an oligomer rather than a monomer, and only the low-CMC detergents that preserve this multimeric state result in the isolation of active enzyme. The significant loss of enzymatic activity observed in other detergents likely resulted from the disruption of protein-protein interactions stabilizing the oligomer, partial delipidation of the enzyme, or both. Indeed, it has been previously shown that P. putida AlkB preparations from which most of the phospholipid was removed exhibited reduced activity unless supplemented with a phospholipid fraction of P. putida or with dilauroylglyceryl-3-phosphorylcholine (18).
We have presented for the first time experimental evidence that 1-octyne acts as a mechanism-based inactivator of PpAlkB-SII. This compound met the following kinetic criteria for a mechanism-based inhibitor (22): (i) the inactivation of PpAlkB-SII with 1-octyne was time dependent, (ii) the substrate n-octane protected the enzyme against inactivation, (iii) the inactivation was irreversible, and (iv) the inactivation occurred only under conditions of catalysis (Fig. 4A and B). These results suggest that 1-octyne needs to be activated by PpAlkB-SII via a normal catalytic mechanism in order to exert its inhibitory effect, which is the main feature of mechanism-based inhibitors (22). In line with what has previously been reported for other monooxygenases (16, 36, 37), enzymatic oxidation of the triple bond in 1-octyne is thought to produce a reactive species that forms a covalent bond with a PpAlkB-SII active-site residue, rendering the enzyme inactive.
We have also presented for the first time the generation and preliminary characterization of 2D crystals of lipid-reconstituted PpAlkB-SII. Analysis of the projection map obtained from negatively stained crystals points to p3 symmetry with unit cell parameters of a = b = 80 Å and γ = 120°. The protein, shown in white over a dark background in Fig. 6A, forms trimers, with each monomer represented by an elliptical shape of 40 by 30 Å (Fig. 6B). Until now, only a topological model of P. putida AlkB constructed by van Beilen et al. (29) was available. Using gene fusions with alkaline phosphatase and β-galactosidase, they predicted that AlkB is likely to have 6 TM helices in a hexagonal arrangement. The dimensions of each monomer in our projection map are similar to those of rhodopsin, which forms a 37- by 33-Å bundle of seven TM helices oriented mostly perpendicular to the membrane plane in 2D crystals (19), suggesting that the TM helices of PpAlkB-SII might adopt a similar compact arrangement. Although the trimeric structure of PpAlkB was not anticipated, it is not the first time that an integral membrane monooxygenase that catalyzes the terminal oxidation of alkanes has been shown to adopt such an arrangement. The heterologous protein particulate methane monooxygenase (pMMO) from Methylococcus capsulatus (Bath), composed of three subunits (α, β, and γ), was found to form a cylindrical α3β3γ3 trimer (11). The trimeric arrangement observed in 2D crystals of PpAlkB-SII and the sensitivity of PpAlkB-SII activity to detergents provide the first indication that the physiologically active form of membrane-embedded AlkB may be a trimer.
ACKNOWLEDGMENTS
This work was supported by the Australia-India Strategic Research Fund of the Department of Innovation, Industry, Science and Research (grant BF040030). A.R. is an Australian Research Council Research Fellow.
We thank Andreas Schenk (Harvard Medical School) for providing help with 2D crystallization and Ben Hankamer (University of Queensland) for technical advice on electron microscopy.
Footnotes
Published ahead of print 31 August 2012
REFERENCES
- 1. Ayala M, Torres E. 2004. Enzymatic activation of alkanes: constraints and prospective. Appl. Catal. A 272: 1– 13 [Google Scholar]
- 2. Cuff JA, Barton GJ. 2000. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins 40: 502– 511 [DOI] [PubMed] [Google Scholar]
- 3. Eggink G, Lageveen RG, Altenburg B, Witholt B. 1987. Controlled and functional expression of the Pseudomonas oleovorans alkane utilizing system in Pseudomonas putida and Escherichia coli. J. Biol. Chem. 262: 17712– 17718 [PubMed] [Google Scholar]
- 4. Gipson B, Zeng X, Stahlberg H. 2007. 2dx_merge: data management and merging for 2D crystal images. J. Struct. Biol. 160: 375– 384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gipson B, Zeng X, Zhang ZY, Stahlberg H. 2007. 2dx—user-friendly image processing for 2D crystals. J. Struct. Biol. 157: 64– 72 [DOI] [PubMed] [Google Scholar]
- 6. Johnson EL, Hyman MR. 2006. Propane and n-butane oxidation by Pseudomonas putida GPo1. Appl. Environ. Microbiol. 72: 950– 952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Katopodis AG, Wimalasena K, Lee J, May SW. 1984. Mechanistic studies on non-heme iron Monooxygenase catalysis—epoxidation, aldehyde formation, and demethylation by the omega-hydroxylation system of Pseudomonas oleovorans. J. Am. Chem. Soc. 106: 7928– 7935 [Google Scholar]
- 8. Lee HJ, Basran J, Scrutton NS. 1998. Electron transfer from flavin to iron in the Pseudomonas oleovorans rubredoxin reductase-rubredoxin electron transfer complex. Biochemistry 37: 15513– 15522 [DOI] [PubMed] [Google Scholar]
- 9. Lee HJ, Lian LY, Scrutton NS. 1997. Recombinant two-iron rubredoxin of Pseudomonas oleovorans: overexpression, purification and characterization by optical, CD and 113Cd NMR spectroscopies. Biochem. J. 328: 131– 136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li L, et al. 2008. Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: unveiling the long-chain alkane hydroxylase. J. Mol. Biol. 376: 453– 465 [DOI] [PubMed] [Google Scholar]
- 11. Lieberman RL, Rosenzweig AC. 2005. Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature 434: 177– 182 [DOI] [PubMed] [Google Scholar]
- 12. Maurer T, Fung HL. 2000. Comparison of methods for analyzing kinetic data from mechanism-based enzyme inactivation: application to nitric oxide synthase. AAPS PharmSci. 2: E8 doi:10.1208ps/020108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McKenna EJ, Coon MJ. 1970. Enzymatic omega-oxidation. IV. Purification and properties of the omega-hydroxylase of Pseudomonas oleovorans. J. Biol. Chem. 245:3882– 3889 [PubMed] [Google Scholar]
- 14. Nieboer M, Kingma J, Witholt B. 1993. The alkane oxidation system of Pseudomonas oleovorans: induction of the alk genes in Escherichia coli W3110 (pGEc47) affects membrane biogenesis and results in overexpression of alkane hydroxylase in a distinct cytoplasmic membrane subfraction. Mol. Microbiol. 8: 1039– 1051 [DOI] [PubMed] [Google Scholar]
- 15. Perry A, Lian LY, Scrutton NS. 2001. Two-iron rubredoxin of Pseudomonas oleovorans: production, stability and characterization of the individual iron-binding domains by optical, CD and NMR spectroscopies. Biochem. J. 354: 89– 98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Prior S. 1985. Acetylene as a suicide substrate and active site probe for methane monooxygenase from Methylococcus capsulatus (Bath). FEMS Microbiol. Lett. 29: 105– 109 [Google Scholar]
- 17. Ramos J-L, Levesque RC. 2004. Pseudomonas v. 3. Kluwer Academic/Plenum Springer, Boston, MA: [Google Scholar]
- 18. Ruettinger RT, Olson ST, Boyer RF, Coon MJ. 1974. Identification of the omega-hydroxylase of Pseudomonas oleovorans as a nonheme iron protein requiring phospholipid for catalytic activity. Biochem. Biophys. Res. Commun. 57: 1011– 1017 [DOI] [PubMed] [Google Scholar]
- 19. Schertler GF, Villa C, Henderson R. 1993. Projection structure of rhodopsin. Nature 362: 770– 772 [DOI] [PubMed] [Google Scholar]
- 20. Shanklin J, Achim C, Schmidt H, Fox BG, Münck E. 1997. Mössbauer studies of alkane omega-hydroxylase: evidence for a diiron cluster in an integral-membrane enzyme. Proc. Natl. Acad. Sci. U. S. A. 94: 2981– 2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shanklin J, Whittle E. 2003. Evidence linking the Pseudomonas oleovorans alkane omega-hydroxylase, an integral membrane diiron enzyme, and the fatty acid desaturase family. FEBS Lett. 545: 188– 192 [DOI] [PubMed] [Google Scholar]
- 22. Silverman RB. 1995. Mechanism-based enzyme inactivators. Methods in enzymology. 249: 240– 283 [DOI] [PubMed] [Google Scholar]
- 23. Smits THM, Balada SB, Witholt B, van Beilen JB. 2002. Functional analysis of alkane hydroxylases from gram-negative and gram-positive bacteria. J. Bacteriol. 184: 1733– 1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ueda T, Coon MJ. 1972. Enzymatic oxidation. VII. Reduced diphosphopyridine nucleotide-rubredoxin reductase: properties and function as an electron carrier in hydroxylation. J. Biol. Chem. 247: 5010– 5016 [PubMed] [Google Scholar]
- 25. Valpuesta JM, Carrascosa JL, Henderson R. 1994. Analysis of electron microscope images and electron diffraction patterns of thin crystals of phi 29 connectors in ice. J. Mol. Biol. 240: 281– 287 [DOI] [PubMed] [Google Scholar]
- 26. van Beilen J, Duetz W, Schmid A, Witholt B. 2003. Practical issues in the application of oxygenases. Trends Biotechnol. 21: 170– 177 [DOI] [PubMed] [Google Scholar]
- 27. van Beilen J, Kingma J, Witholt B. 1994. Substrate-specificity of the alkane hydroxylase system of Pseudomonas-oleovorans GPo1. Enzyme Microb. Technol. 16: 904– 911 [Google Scholar]
- 28. van Beilen JB, Funhoff EG. 2007. Alkane hydroxylases involved in microbial alkane degradation. Appl. Microbiol. Biotechnol. 74: 13– 21 [DOI] [PubMed] [Google Scholar]
- 29. van Beilen JB, Penninga D, Witholt B. 1992. Topology of the membrane-bound alkane hydroxylase of Pseudomonas oleovorans. J. Biol. Chem. 267: 9194– 9201 [PubMed] [Google Scholar]
- 30. van Beilen JB, et al. 2005. Identification of an amino acid position that determines the substrate range of integral membrane alkane hydroxylases. J. Bacteriol. 187: 85– 91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. van Beilen JB, Wubbolts MG, Witholt B. 1994. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation 5: 161– 174 [DOI] [PubMed] [Google Scholar]
- 32. Whitmore L, Wallace BA. 2004. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32: W668– W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Whitmore L, Wallace BA. 2008. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 89: 392– 400 [DOI] [PubMed] [Google Scholar]
- 34. Whyte LG, et al. 2002. Prevalence of alkane monooxygenase genes in Arctic and Antarctic hydrocarbon-contaminated and pristine soils. FEMS Microbiol. Ecol. 41: 141– 150 [DOI] [PubMed] [Google Scholar]
- 35. Xie M, Alonso H, Roujeinikova A. 2011. An improved procedure for the purification of catalytically active alkane hydroxylase from Pseudomonas putida GPo1. Appl. Biochem. Biotechnol. 165: 823– 831 [DOI] [PubMed] [Google Scholar]
- 36. Yeager CM, Bottomley PJ, Arp DJ, Hyman MR. 1999. Inactivation of toluene 2-monooxygenase in Burkholderia cepacia G4 by alkynes. Appl. Environ. Microbiol. 65: 632– 639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zahn JA, DiSpirito AA. 1996. Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath). J. Bacteriol. 178: 1018– 1029 [DOI] [PMC free article] [PubMed] [Google Scholar]