Recent studies have identified cytoskeletal elements in bacteria which play important roles in cellular morphology, cell division, DNA segregation and the establishment of cell polarity. However, our understanding of the contribution the bacterial cytoskeleton makes toward virulence is lacking. The MreB protein is a bacterial homolog of eukaryotic actin and interacts intimately with MreC and MreD. We provide evidence that the Mre-based cytoskeleton directly influences pathogenicity in Salmonella. Disruption of MreC and MreD caused the downregulation of the Salmonella pathogenicity island 1 (SPI-1) type 3 secretion system (T3SS) essential for host cell-invasion, and also flagella-mediated motility. These phenotypic effects were mediated by RcsC, the sensor kinase of the Rcs phosphorelay two-component system and a major envelope stress regulator. Curiously, the SPI-2 T3SS remained viable. Our results demonstrate the importance of the integrity of the bacterial cytoskeleton for virulence, highlighting the impact of associated global regulatory mechanisms on pathogenicity.
Structural homologs of all three major eukaryotic cytoskeletal protein classes are now known to exist in bacteria. These homologs include MreB and ParM (actin), crescentin (intermediate filaments) and FtsZ (microtubules). Ongoing research seeks to understand the mechanisms by which the bacterial cytoskeleton and associated proteins control cell shape, through their control of synthesis of the stress-bearing peptidoglycan cell wall (Shih and Rothfield, Microbiol Mol Biol Rev 2006).
To date six essential cell shape-determinant proteins have been identified in non-spherical bacteria: MreB, MreC, MreD, penicillin binding protein 2 (PBP2), RodA and RodZ. The chemical or genetic inactivation of any of these proteins causes a loss of rod-shape and, in the absence of compensatory mutations, eventual cell-lysis. The cell shape-determinant proteins are thought to form a large multi-protein complex together with the peptidoglycan synthetic enzymes, to coordinate longitudinal cell wall synthesis and hence cell shape (Bendezú and de Boer, J Bacteriol 2008; van den Ent et al., EMBO 2010). The cytoskeletal MreB protein was believed to form long helical scaffold-like filaments along the length of the cell, directing the positioning of the peptidoglycan synthetic enzymes to spatially regulate cell wall synthesis. However, recent studies that used a sensitive total internal reflection fluorescence microscopy technique (TIRFM) to study MreB filament dynamics suggested that MreB and associated proteins in fact form short dynamic filaments that move around the cell perpendicularly to its long-axis (Domínguez-Escobar et al., Science 2011; Garner et al., Science 2011).
Bacterial cytoskeletal proteins are recognized for their roles in defining cellular morphology, division and polarity (Shih and Rothfield, Microbiol Mol Biol Rev 2006). However, we have little knowledge on how the bacterial cytoskeleton, peptidoglycan-synthetic complexes and cell shape may influence pathogenicity. Bacterial pathogens employ many crucial cell envelope-associated virulence factors, since this forms the initial contact with the host. Such factors include macromolecular cell wall-spanning organelles such as flagella or type 3 secretion systems (T3SSs) and multi-protein needle complexes that actively inject effector proteins into the host cell cytoplasm (Cornelis, Nat Rev Microbiol 2006; Stecher et al., Infect Immun 2004). It is reasonable to suggest that the bacterial cytoskeleton itself, or the peptidoglycan sacculus, may function in directing the assembly, localization and ultimately function of cell wall-spanning virulence organelles.
Many cell wall-spanning virulence organelles exhibit specific localization patterns, which may be influenced by cytoskeletal elements. MreB has been implicated in regulating the polar or side-wall localization of virulence-related proteins, including secretion system complexes, type IV pili and motility proteins (Shih and Rothfield, Microbiol Mol Biol Rev 2006; Mauriello et al., EMBO 2010; Cowles and Gitai, Mol Microbiol 2010).
The extent to which the bacterial cytoskeleton is involved in the assembly, function and localization of virulence organelles remains to be examined. We characterized the effects of disrupting the bacterial cytoskeleton on the pathogenicity of Salmonella enterica serovar Typhimurium (S. Typhimurium), investigating the effects on major virulence factors (Bulmer et al., PLoS Pathogens 2012).
Salmonella enterica serovars remain major human pathogens, causing disease ranging from gastroenteritis to life-threatening systemic typhoid fever (Haraga et al., Nat Rev Microbiol 2008). Challenges associated with Salmonella include the spread of multidrug-resistant strains and the emergence of serious systemic “non-typhoidal salmonellosis,” associated with immunocompromised patients (Chuang et al., Epidemiol Infect 2009; Gordon, J Infect 2008). Salmonella infections are acquired upon ingestion of contaminated food or drink. Virulence is mediated by a variety of factors including two pathogenicity island-encoded T3SSs: “SPI-1” and “SPI-2.” The SPI-1 T3SS is required for the initial invasion of intestinal epithelial cells and macrophages; SPI-2 T3SS is essential for intracellular survival, enabling Salmonella to subvert host cell defense mechanisms and replicate within modified “Salmonella-containing vacuoles” (SCVs) (Haraga et al. Nat Rev Microbiol 2008). Flagella/chemotaxis systems also assist invasion (Asten and van Dijk, FEMS Immunol Med Mic 2005).
The MreB, MreC and MreD cell shape-determining proteins are encoded contiguously within the mre operon in Salmonella. The cytoplasmic membrane proteins, MreC and MreD, may function to bridge MreB and the peptidoglycan synthesis machinery. Evidence suggests that MreC directly interacts with both MreB and MreD (Kruse et al., Mol Microbiol 2005; van den Ent et al., EMBO 2010). Furthermore, the localization patterns of MreB, MreC and MreD were shown to be similar and interdependent, and the inactivation of MreB, MreC or MreD results in the formation of phenotypically indistinguishable spherical cells (Divakaruni et al., Proc Natl Acad Sci USA 2005; Bendezú and de Boer, J Bacteriol 2008; Bulmer et al., PLoS Pathogens 2012). These observations demonstrate the relation and interdependency in function between the Mre proteins, suggesting that inactivation of any one protein disrupts both the cytoskeleton and longitudinal peptidoglycan synthesis.
In many bacteria mreB appears to be essential and viable ΔmreB mutants can only be generated through acquiring compensatory mutations (Kruse et al., Mol Microbiol 2005; Shih and Rothfield, Microbiol Mol Biol Rev 2006). To study the role of the Mre proteins in Salmonella pathogenicity, precise knockout mutants of both ΔmreC and ΔmreD were generated in S. Typhimurium and maintained by expressing the mre operon in trans from an inducible promoter. Spherical cells were formed upon the depletion of mre expression, as expected. These and all other observed mutant phenotypes were fully complementable upon restoration of MreC and MreD expression (Fig. 1). We were unable to construct a viable ΔmreB mutant. However, the chemical inactivation of MreB caused wild-type S. Typhimurium to form spherical cells that were phenotypically identical to the ΔmreC and ΔmreD mutants. The chemical disruption of MreB was achieved using A22, a drug that specifically binds MreB, inhibiting polymerization (Bean et al., Biochemistry 2009).
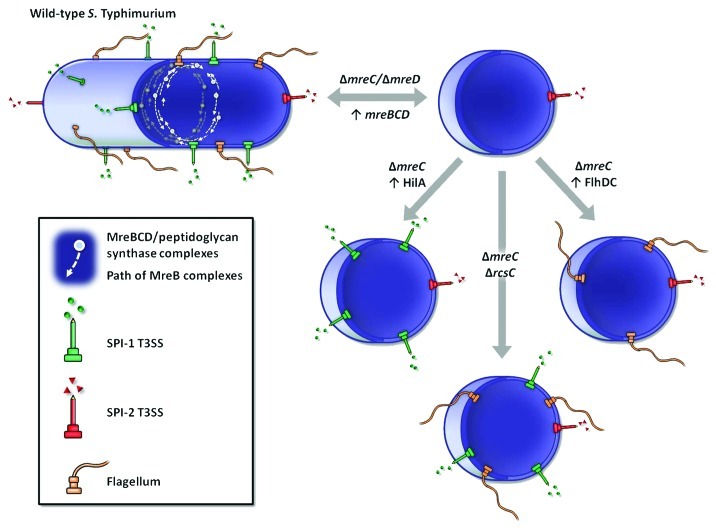
Figure 1. Generation and phenotypic characterization of ΔmreC mutants of S. Typhimurium. Schematic representation of wild-type S. Typhimurium expressing the invasion-associated SPI-1 T3SS, flagella for motility and the SPI-2 T3SS essential for intracellular survival. Wild-type Salmonella cells express 1–2 polar SPI-2 needles and approximately 6–8 flagella and SPI-1 needles, distributed predominantly along the lateral cell wall (Bulmer et al., PLoS Pathog 2012). Putative peptidoglycan synthetic complexes comprising the MreBCD cytoskeletal proteins, cell shape-determinants and peptidoglycan synthetic enzymes, are shown as short dynamic complexes moving perpendicularly to the long-axis of the cell. Upon MreC or MreD depletion S. Typhimurium formed spherical cells, with a loss of SPI-1 T3SS and flagella expression; SPI-2 expression remained active. SPI-1 T3SS and flagella expression were restored upon recovery of either mre, hilA (SPI-1 master regulator) or flhDC (flagella master regulator) expression respectively, or with the inactivation of rcsC, sensor kinase of the Rcs phosphorelay envelope stress response.
Phenotypic screening of S. Typhimurium ΔmreC and ΔmreD demonstrated that both motility and the SPI-1 T3SS were almost completely downregulated in these mutants; cells were non-motile and did not express the structural or secreted proteins of either the flagella or SPI-1 organelles (Fig. 1). In addition, transepithelial resistance (TER) assays showed that the ability of ΔmreC cells to disrupt the tight junctions between polarized Caco-2 cells, a feature which is dependent upon SPI-1 functionality (Boyle, Cell Microbiol 2006), was impaired. Fundamentally, expression of both flagella and SPI-1 genes was repressed at the level of the master-regulators, flhDC and hilA respectively (Apel and Surette, Biochim Biophys Acta 2008; Ellermeier and Slauch, Curr Opin Microbiol 2007).
Few studies have documented or explored virulence defects resulting from the inactivation of cell shape-determinants or the bacterial cytoskeleton. E. coli ΔrodZ cells were previously noted to be non-motile, the genes encoding the flagella structural proteins and flagellin being downregulated. flhDC expression, however, remained unaffected (Niba et al., FEMS Microbiol Lett 2010). Another study reported that flhDC expression was decreased in E. coli cells treated with mecillinam, an antibiotic that specifically inhibits the longitudinal-transpeptidase penicillin-binding protein 2 (PBP2) (Laubacher and Ades, J Bacteriol 2008). However, little is known about the mechanisms responsible for the downregulation of motility in spherical E. coli or Salmonella.
Surprisingly, the SPI-2 T3SS was expressed and appeared functional in round-cell ΔmreC and ΔmreD mutants. SseB is a SPI-2-secreted protein that forms part of the SPI-2 T3SS “translocon” at the needle tip (Nikolaus et al., J Bacteriol 2001). This protein was detected in membrane fractions and visualized at the cell surface, in both wild-type and ΔmreC cells. These results were reflected in vivo; the ΔmreC mutant was inoculated intravenously into C57/BL6 mice, to bypass the SPI-1-dependent stages of infection. Although clearly attenuated compared with the wild-type, the ΔmreC mutant was able to survive and replicate within the host, demonstrating the retention of some virulence and a functional SPI-2 T3SS.
Several hypotheses were formulated to explain mechanistically the observed virulence phenotypes in ΔmreC and ΔmreD mutants. For instance, the bacterial cytoskeleton or peptidoglycan layer may physically support multi-protein wall-spanning organelles. Thus an inability of Δmre cells to correctly assemble and localize cell wall-spanning organelles, either directly from the loss of a functional cytoskeleton or through resulting peptidoglycan structural defects, may cause the downregulation of motility and SPI-1. In this case, retention of SPI-2 functionality could relate to the different spatial organization of this T3SS, its localization potentially being controlled by different factors. Alternatively, the activation of stress response pathways resulting from cytoskeletal or cell wall defects, may lead indirectly to the downregulation of virulence factors whose expression is controlled by global stress regulators, or may cause the cell to specifically repress non-essential energetically expensive processes.
In order to determine whether the flagella and SPI-1 defects were due, at least in part, to the physical inability of ΔmreC mutants to assemble and localize such structures, we attempted to recover SPI-1 and flagella expression in these cells. flhDC and hilA were therefore each individually expressed in trans under the control of an inducible promoter, releasing the flagella and SPI-1 genes from upstream regulatory mechanisms, and causing the recovery of flagella or SPI-1 gene and protein expression. Surface expression of both the SPI-1 T3SS and flagella organelles was observed in ΔmreC cells when hilA or flhDC were constitutively expressed, respectively (Fig. 1). Furthermore, TER experiments and motility assays demonstrated the functionality of these respective organelles. Our results demonstrate that round-cell mutants lacking a functional cytoskeleton, and/or harboring significant cell wall defects, retain the ability to support functional cell wall-spanning organelles.
These insights led us to examine the regulatory mechanisms responsible for the observed ΔmreC and ΔmreD virulence phenotypes. Many global regulators responding to environmental stress in Salmonella comprise two-component systems. These systems consist of an integral membrane histidine kinase coupled to a response regulator, which upon activation by phosphorylation, directly regulates transcription (Stock et al., Annu Rev Biochem 2000). Upon generating and screening a panel of mutants of major two-component systems in S. Typhimurium ΔmreC, we identified a ΔmreC ΔrcsC mutant in which significant motility and SPI-1 effector protein expression and functionality were recovered. The restoration of rcsC expression in trans in S. Typhimurium ΔrcsC ΔmreC resulted in a subsequent loss of flagella and SPI-1 expression.
Thus RcsC is an important regulator of virulence in response to the inactivation of mreC. The Rcs phosphorelay regulates gene expression when activated by envelope-associated stress. The sensor kinase RcsC activates the response regulator RcsB via the RcsD protein (Laubacher and Ades, J Bacteriol 2008; Huang et al., Res Microbiol 2006). This complex phosphorelay is well known to activate colanic acid biofilm synthesis. However, it also regulates virulence, repressing motility and SPI-1, and activating SPI-2 expression in Salmonella (Arricau et al., Mol Microbiol 1998; Garcia-Calderon et al., J Bacteriol 2007). Interestingly, mecillinam treatment of wild-type E. coli also resulted in a reduction in flhDC expression and the activation of several stress response pathways, including the Rcs phosphorelay (Laubacher and Ades, J Bacteriol 2008).
On identifying RcsC as a candidate global regulator of SPI-1 and motility in ΔmreC mutants, ongoing work aimed to characterize the roles of other members of the Rcs phosphorelay. However, SPI-1 functionality and motility were not recovered in ΔrcsB, ΔrcsD, ΔrcsDB and ΔrcsF double mutants of S. Typhimurium ΔmreC. This was surprising since the inactivation of any of these genes would be expected to disrupt the system, preventing RcsB activation and the RcsB-dependent repression of motility and SPI-1 T3SS. RcsF, an outer membrane lipoprotein that activates RcsC in response to peptidoglycan defects, would also be expected to specifically activate RcsC in ΔmreC mutants.
That a ΔrcsCBD mutant was unable to restore motility or SPI-1 T3S in the ΔmreC mutant suggests that the presence of RcsB and/or RcsD is essential for the restoration of their expression, in the absence of RcsC. An explanation for these results could relate to the complex nature of the RcsC protein, which possesses both kinase and phosphatase activity, and is therefore capable of activating or deactivating RcsB. Unlike ΔrcsD or ΔrcsB mutants, ΔrcsC mutants may retain residual Rcs phosphorelay activity (García-Calderón et al., Microbiology 2005; Girgis et al., PLoS Genet 2007). Hypothetically, low-level RcsB activation may be necessary for the downstream recovery of flhDC and hilA expression, while higher levels of activated RcsB lead to their repression. However, in E. coli the inactivation of rcsB, rcsD or rcsF was shown to restore motility in non-motile mutants, while rcsC inactivation did not restore motility as effectively (Girgis et al., PLoS Genet 2007).
It appears that the inactivation of the bacterial cytoskeleton brings about the RcsF-independent activation of RcsC. However, the downstream regulatory mechanisms that repress SPI-1 T3SS, motility and activate SPI-2 T3SS are potentially complex and atypical. The regulation of SPI-1 T3SS and flagella expression in ΔmreC mutants is potentially multifaceted, involving several global regulators to provide tighter control over virulence—especially given that the inactivation of rcsC alone was not sufficient to completely restore wild-type levels of motility or SPI-1 T3SS activity.
Furthermore, it cannot be excluded that cell wall defects may also directly influence motility/SPI-1 functionality in these mutants; hence the reduced motility in ΔmreC ΔrcsC cells (compared with wild-type cells) may be caused at least partially by some dependency of these organelles upon the structural integrity of the cell envelope/peptidoglycan layer. The extent to which cell shape and the robustness of the peptidoglycan cell wall directly affects the assembly and function of the SPI-1 and motility systems is unclear. Studies in Gram-negative bacteria, including Salmonella, have demonstrated that mutations in the peptidoglycan-binding regions of structural proteins of both flagella and various secretion system organelles reduced their function (Scheurwater and Burrows, FEMS Microbiol Lett 2011; Pucciarelli and García-del Portillo, Mol Microbiol 2003). Peptidoglycan structural changes also affected virulence in Salmonella, causing a reduction in the number of SPI-1 T3SS needles per cell (Pucciarelli and García-del Portillo, Mol Microbiol 2003). Together these studies suggest that alterations to cell shape and peptidoglycan structural integrity may both directly and significantly affect virulence in ΔmreC mutants.
The activation or overexpression of envelope stress response regulators would be expected in ΔmreC mutants, where the peptidoglycan structural integrity may be significantly compromised. Surprisingly, within a panel of two-component system mutants including mutants of two major envelope stress response pathways, ΔcpxAR and ΔbaeSR, only ΔrcsC mutants restored SPI-1 T3S or motility to the ΔmreC cells (Raivio, Mol Microbiol 2005). It is possible that these regulators only play a minor regulatory role, or that some redundancy exists in the regulatory mechanisms. However, only the Rcs phosphorelay has so far been recognized to respond to peptidoglycan-associated stress specifically (Laubacher and Ades, J Bacteriol 2008).
We have characterized the role of the bacterial cytoskeleton in S. Typhimurium, investigating the effects that inactivation of the Mre cytoskeleton had upon the assembly and function of major cell wall-spanning virulence organelles (Bulmer et al., PLoS Pathog 2012). While the SPI-1 T3SS and flagella systems were downregulated in round-cell ΔmreC mutants, the SPI-2 T3SS remained functional. Interestingly, this phenotype is not due to an inherent inability of round-cell mutants to express and assemble functional wall-spanning virulence organelles. This suggests that the bacterial cytoskeleton does not play an essential role in directing the assembly and localization of such organelles, but affects the expression of these systems. Furthermore, expression of the flhDC and hilA master regulators in trans in the ΔmreC mutants, resulted in expression and assembly of flagella and SPI-1 T3S needles, respectively.
The Rcs phosphorelay, a global envelope stress-response pathway, appears to be largely responsible for the repression of the SPI-1 T3SS and motility in response to the inactivation of the bacterial cytoskeleton. However, it remains unknown what role RcsB and RcsD play in this regulation. The precise nature of the signal(s) responsible for activating the Rcs phosphorelay in round-cell mutants also remain(s) to be identified. The bacterial cytoskeleton itself may be involved in activating global regulators; it has been suggested that the integrity of the bacterial cytoskeleton is utilized for monitoring the bacterial cell’s physiological state, as is observed in eukaryotic cells (Chiu et al., Appl Environ Microbiol 2008). Future work aims to further elucidate of the roles of the Rcs phosphorelay, alternative global stress regulators and post-transcriptional regulators, in controlling virulence in Salmonella upon disruption of the bacterial cytoskeleton and longitudinal cell wall synthesis.
It is now clear that the integrity of the bacterial cytoskeleton can attenuate the virulence of S. Typhimurium, by downregulating the expression of important virulence genes. This regulation is mediated by the Rcs two-component system. The broader importance of the cytoskeleton in virulence needs to be investigated in greater detail in a variety of other bacterial pathogens. These observations highlight the potential of the bacterial cytoskeleton serving as a novel target for a new generation of antimicrobial therapeutics.
Acknowledgments
This work was supported by Medical Research Council UK grant (G0801212) to C.M.A.K. A.C.D. was supported by a Medical Research Council (UK) PhD studentship with C.M.A.K. L.K. was supported by a Ford Foundation of America PhD studentship with C.M.A.K.
Footnotes
Previously published online: www.landesbioscience.com/journals/virulence/article/20993