Abstract
Cryptococcus neoformans is a human-pathogenic basidiomycete that commonly infects HIV/AIDS patients to cause meningoencephalitis (7, 19). C. neoformans grows as a budding yeast during vegetative growth or as hyphae during sexual reproduction. Pseudohyphal growth of C. neoformans has been observed rarely during murine and human infections but frequently during coculture with amoeba; however, the genetics underlying pseudohyphal growth are largely unknown. Our studies found that C. neoformans displays pseudohyphal growth under nitrogen-limiting conditions, especially when a small amount of ammonium is available as a sole nitrogen source. Pseudohyphal growth was observed with Cryptococcus neoformans serotypes A and D and Cryptococcus gattii. C. neoformans pseudohyphae bud to produce yeast cells and normal smooth hemispherical colonies when transferred to complete media, indicating that pseudohyphal growth is a conditional developmental stage. Subsequent analysis revealed that two ammonium permeases encoded by the AMT1 and AMT2 genes are required for pseudohyphal growth. Both amt1 and amt2 mutants are capable of forming pseudohyphae; however, amt1 amt2 double mutants do not form pseudohyphae. Interestingly, C. gattii pseudohypha formation is irreversible and involves a RAM pathway mutation that drives pseudohyphal development. We also found that pseudohyphal growth is related to the invasive growth into the medium. These results demonstrate that pseudohyphal growth is a common reversible growth pattern in C. neoformans but a mutational genetic event in C. gattii and provide new insights into understanding pseudohyphal growth of Cryptococcus.
INTRODUCTION
Cryptococcus neoformans is a human-pathogenic basidiomycete and a causative agent of fungal meningoencephalitis mainly in immunocompromised patients (7). Pathogenic cryptococci normally proliferate as unicellular budding yeast during vegetative growth or infection of animal hosts (reviewed in references 30 and 31). During mating, C. neoformans also forms true hyphae that are septate. Like other basidiomycetes, C. neoformans forms typical dikaryotic hyphae, which are followed by aerial hypha formation and basidium formation at the apex of the hyphae, and chains of spore progeny decorate the basidia (reviewed in references 28 and 31).
In addition to yeast and hyphal growth, C. neoformans can also exhibit pseudohyphal growth. When cocultured with amoeba, C. neoformans cells form pseudohyphae, where yeast C. neoformans cells are engulfed by amoebae and pseudohyphal C. neoformans cells are free from amoebae, either having not been engulfed or having escaped (4, 41). C. neoformans also forms atypical pseudohyphae during host infection, where pseudohyphal or hyphal growth has been observed in infected host tissues (11, 14, 15, 39, 48, 49, 54). These observations may imply that C. neoformans cells form pseudohyphae as a survival strategy to escape from natural predators or host defenses. Fries and her colleagues found that pseudohyphal growth changes C. neoformans colony morphology into a wrinkled form (12). A recent study by Walton and her colleagues found that RAM signaling pathway mutants undergo pseudohyphal morphogenesis and exhibit altered colony morphology (52).
Characteristics of pseudohyphal growth include an elongated cell shape with mother and daughter cells physically attached and lateral branch formation (22). Environmental cues and genetics of pseudohyphal growth in the model yeast Saccharomyces cerevisiae are well described. For example, nitrogen starvation conditions induce pseudohyphal growth of diploid, but not haploid, yeast strains of the Σ1278b and other wild-type (prototrophic) lineages (16). The cyclic AMP (cAMP) signaling pathway operating via protein kinase A (PKA) and the mitogen-activated protein kinase (MAPK) signaling pathway are responsible for pseudohyphal development via promoting the expression of the cell surface flocculin Flo11 (35). A series of previous studies revealed that the high-affinity ammonium permease Mep2 is required for pseudohyphal growth and plays a role in sensing ammonium (2, 37, 47). The upstream factor of the PKA pathway is the G-protein-coupled receptor Gpr1, which interacts with the alpha subunit of the heterotrimetic G protein Gpa2, which activates adenylyl cyclase, resulting in activation of the PKA pathway (23, 38). The MAPK pathway regulates the heteromeric Ste12-Tec1 transcription factor that contributes to induce expression of Flo11 to initiate pseudohyphal growth (13, 45).
In the human-pathogenic yeast Candida albicans, Cdc42 is a key regulator of hyphal and pseudohyphal growth. Deletions of the GTPase-activation protein genes RGA2 and BEM3 lead to hyphal growth under conditions that normally induce pseudohyphal growth (8). The Ume6 is known as a hyphal growth regulator from yeast or from pseudohyphae that operates via Hgc1-dependent inactivation of Rga2 (1, 5, 6).
In the plant-pathogenic fungus Ustilago maydis, the high-affinity ammonium permease Ump2 is required for invasive hyphal growth under low-ammonium conditions and, interestingly, the U. maydis UMP2 gene complements the pseudohyphal defect of S. cerevisiae mep1 mep2 mep3 triple mutants (37, 51). In another smut fungus, Microbotryum violaceum, two high-affinity ammonium permeases, mepA and mepC, were identified and mepA is also upregulated during the yeast-hyphal dimorphic transition during mating (20).
From classic studies, it is well established that C. neoformans exhibits pseudohyphal growth. Yet relatively little is known about the genetic and environmental factors that induce C. neoformans pseudohyphal growth. In this study, we found that nitrogen-limited conditions, especially when a small amount of ammonium serves as the sole nitrogen source, trigger pseudohyphal growth of both C. neoformans and C. gattii. We also found that two ammonium permease genes, AMT1 and AMT2, are required for pseudohyphal growth of C. neoformans, where wild-type cells and amt1 or amt2 single mutants are capable of forming pseudohyphae and wrinkled colonies, whereas amt1 amt2 double mutants fail to form pseudohyphae and exhibit a smooth colony morphology only under conditions of nitrogen-limited growth.
MATERIALS AND METHODS
Strains, media, and culture conditions.
C. neoformans and C. gattii strains used in this study are listed in Table 1. The strains were grown on yeast extract-peptone-dextrose (YPD) agar before inoculation. All media and conditions tested for hyphal and pseudohyphal growth formation in this study are listed in Table S1 in the supplemental material.
Table 1.
C. neoformans strains used in this study
| Species | Ploidy | Strain | Genotype | Reference |
|---|---|---|---|---|
| A | Haploid | H99 | MATα wild type | 44 |
| KN99α | MATα wild type | 42 | ||
| KN99a | MATa wild type | 42 | ||
| JR1 | MATα amt1::NEO | 46 | ||
| JR3 | MATα amt2::NAT | 46 | ||
| JR5 | MATα amt1::NEO amt2::NAT | 46 | ||
| JR11 | MATa amt1::NAT amt2::NAT | 46 | ||
| Diploid | MMRL743 | αAAα | 34 | |
| MMRL795 | αAAα | 34 | ||
| MMRL799 | αAAα | 34 | ||
| MMRL1088 | αAAα | 34 | ||
| MMRL1368 | αAAα | 34 | ||
| MMRL1370 | αAAα | 34 | ||
| MMRL2445 | αAAα | 34 | ||
| KN4B4-10 | αAAα | 42 | ||
| KN2B5-18 | αAAα | 42 | ||
| 102-14 | αAAα | 34 | ||
| 1054 | αAAα | 34 | ||
| 1035 | αAAα | 34 | ||
| 1052 | αAAα | 34 | ||
| XL1500 | αAAα | 33 | ||
| XL1501 | αAAα | 33 | ||
| D | Haploid | JEC21 | MATα wild type | 18 |
| JEC20 | MATa wild type | 18 |
To test the hypothesis that pseudohyphae can be revived, we transferred cells on the edge of colonies after 1 to 2 weeks of incubation on ¼ yeast nitrogen base (YNB) medium (1.7 g YNB without amino acids, 50 μM ammonium sulfate, 2% glucose per liter) to rich YPD media. We monitored the growth every 30 min for 4 h with a Zeiss Axioskop 2 Plus microscope with an AxioCam MRm camera (Carl Zeiss Inc., Thornwood, NY) at room temperature.
Mating and basidiospore dissection.
To assess association between the amt1 and amt2 mutations and pseudohyphal growth, we crossed amt1 amt2 double-mutant strain JR11 (MATa amt1::NEO amt2::NAT) with wild-type KN99α or JR5 (MATα amt1::NEO amt2::NAT) with wild-type KN99a. Two parental strains were cultured in YPD liquid media overnight and washed with sterile water. Then, mixtures of the two parental strains were spotted on MS media. After 2 weeks of incubation in the dark at room temperature, the spores from individual basidia were dissected as previously described (21). A total of 121 viable progeny were obtained from 12 basidia, and the genotypes of the progeny were determined based on drug markers. Representative progeny from a basidium in the cross of JR11 × KN99α are presented in Table S2 in the supplemental material.
Pseudohyphal growth of C. gattii.
We screened a panel of C. gattii strains (see Table S3 in the supplemental material) for pseudohyphal growth under nutrient-limiting conditions for 3 weeks on media that included V8 (pH 5.0) and filament agar in the absence of light. To test whether pseudohyphae represented mutants, we microdissected the pseudohyphae that were produced by VGIII strain 97/433 on filament agar and purified for single colonies on YPD media at 30°C twice following microdissection. Four individual colonies from each of the four microdissected pseudohyphae were screened for cell morphology using bright-field microscopy. Then, we tested for temperature sensitivity at 30°C and 37°C on YPD media.
Sequencing of the RAM pathway genes in the 97/433 isolate.
We sequenced the MOB2, CBK1, KIC1, TAO3, and SOG2 genes, including the ∼500-bp 5′ and 3′ flanking regions of wild-type and pseudohyphal mutant strains. The whole-genome sequence of C. gattii VGIII strain is not available. Therefore, we designed primers for the RAM pathway genes in strain 97/433 based on the conserved sequences of the genes in C. gattii VGI strain WM276 and VGII strain R265 (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans_b/MultiHome.html), for which whole genomes are available (9). The entire genes, including the ∼500-bp 5′ and 3′ flanking regions, were PCR amplified and subsequently sequenced. The primers used are listed in Table S4 in the supplemental material. The GenBank accession numbers for the genes are listed below.
Microscopy.
Colony morphology was observed using a Nikon Eclipse E400 microscope equipped with a Nikon DXM1200F camera. For detailed imaging, a Zeiss Axioskop 2 Plus with an AxioCam MRm camera (Carl Zeiss Inc., Thornwood, NY) was used.
For scanning electron microscopy (SEM), the edges of the colonies were washed with 0.1 M Na cacodylate buffer (pH 6.8), and 1-mm3 blocks of edge areas were excised and incubated in fixation buffer at 4°C. Samples were then rinsed in cold 0.1 M Na cacodylate buffer three times, postfixed in 2% osmium tetroxide–0.1 M Na cacodylate buffer for 2.5 h at 4°C, critical point dried, and sputter coated before being viewed by SEM.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the genes sequenced in this study are JX535489, JX535490, JX535491, JX535492, JX535493, and JX684012.
RESULTS
Nitrogen limitation triggers pseudohyphal growth of C. neoformans serotype A diploids.
Cryptococcus neoformans has two mating types, a and α, and mating occurs when two mating-type cells encounter and recognize each other (24–26). However, in natural and clinical populations, the α mating type is predominant and this limits opportunities for α-a sexual reproduction (3, 10, 17, 27). α-α unisexual reproduction has been previously described and genetically verified in C. neoformans var. neoformans (serotype D) (32), but C. neoformans var. grubii (serotype A) is not yet known to undergo unisexual mating under laboratory conditions, although serotype A is the more predominant type within the pathogenic Cryptococcus species. As a part of this study, we initially tested the possibility of same-sex reproduction of αAAα diploids of C. neoformans var. grubii (serotype A) under various conditions (see Table S1 in the supplemental material). Although we did not observe monokaryotic fruiting, we found that nitrogen limitation conditions trigger pseudohyphal growth. After 10 days of incubation on ¼ YNB media containing 9.5 mM ammonium sulfate, the αAAα diploid strains exhibited pseudohyphal growth at the edge of the colonies (Fig. 1). The ¼ YNB media contains a small amount of ammonium as the sole nitrogen source.
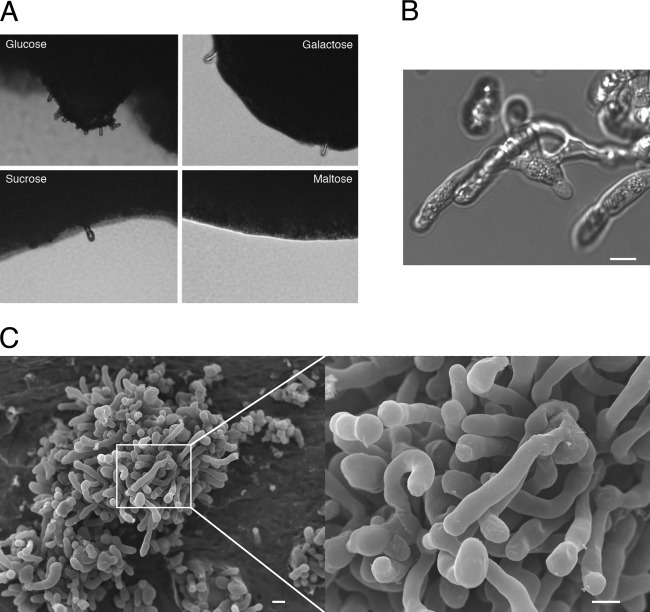
Fig 1.
Pseudohypha formation of an αAAα diploid in response to nitrogen limitation. (A) The αAAα diploid strain MMRL1368 produced pseudohyphae on the edge of colonies when glucose, galactose, or sucrose was used as the fermentable carbon source. However, when maltose was used as the carbon source, the strain did not form pseudohyphae. (B) Differential interference contrast (DIC) image of pseudohyphae of MMRL1368 displays branched pseudohyphae without the clamp connections observed during hypha formation. (C) Scanning electron microscopic images show details of pseudohyphae. Scale = 5 μm.
The MMRL1368 strain was further used to test whether carbon sources play a role in pseudohyphal growth. Under the conditions tested, galactose and sucrose did not change the strain's ability to form pseudohyphae. However, interestingly, when maltose was used as a carbon source, the strain failed to form pseudohyphae. When we supplemented ¼ YNB media with amino acids as a nitrogen source instead of ammonium, the αAAα diploid strain did not produce pseudohyphae (data not shown), indicating that ammonium may be important for the pseudohyphal morphogenesis. The phenotypes include lateral budding and branching (Fig. 1B). Some strains did not show pseudohyphal growth when tested under the same conditions (see Fig. S1 in the supplemental material). Scanning electron microscopy (SEM) analysis shows that the pseudohyphae produced by strain MMRL1368 are elongated cells with branching but do not have clamp connections or basidia decorated with spore progeny (Fig. 1C).
Haploid serotype A and D strains also undergo pseudohyphal growth under nitrogen limitation conditions.
We then tested whether pseudohyphal morphogenesis is also observed in haploid strains under the same nitrogen limitation conditions. Haploid strains of serotype A and D were grown on ¼ YNB media, and after 10 days of incubation at room temperature in the dark, we observed pseudohyphal growth (Fig. 2). The pseudohyphae formed from the H99 haploid strain (n = 10, 14.2 ± 2.4 μm) appear shorter than those from the MMRL1368 diploid strain (n = 10, 26.1 ± 5.6 μm) (Fig. 1; see also Fig. S2 in the supplemental material). However, when we tested pseudohyphal growth of an isogenic diploid of H99, the length of pseudohyphae was not significantly different from that of the haploid (data not shown), suggesting that longer pseudohyphae are not associated with ploidy. These results indicate that pseudohyphal growth of C. neoformans is a common phenotype throughout the different serotypes.
Fig 2.

Pseudohyphal growth in serotype A and serotype D haploids. Both mating types of both serotype A (H99α and KN99a) and serotype D (JEC21α and JEC20a) form pseudohyphae in ¼ YNB media. Scale = 10 μm.
Pseudohyphae revive as yeast on YPD-rich media.
We tested whether the pseudohyphae were alive and able to undergo cell division. The H99 haploid strain was grown on ¼ YNB media to induce pseudohyphal growth. After 2 weeks, pseudohyphal cells on the edge of the colonies were transferred onto YPD media. In a time course analysis, the pseudohyphae started producing daughter yeast cells from the apex of the pseudohyphae and went on to form smooth dome-shaped wild-type yeast colonies. This result indicates that pseudohyphal growth is not a dead end in development but a reversible developmental stage of C. neoformans involving both yeast-pseudohypha and pseudohypha-yeast dimorphic transitions (Fig. 3).
Fig 3.
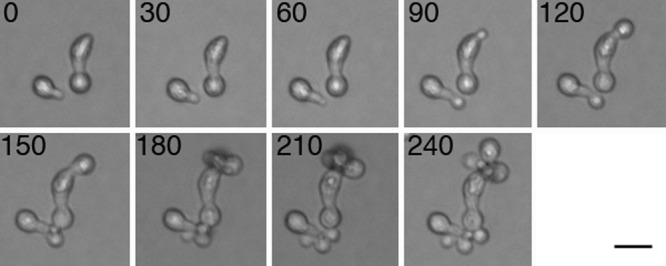
Daughter cells bud from pseudohyphae. When pseudohyphae formed on ¼ YNB media were transferred onto rich YPD media, yeast daughter cells budded from the apex of the pseudohyphae. Numbers represent minutes of growth. Scale = 5 μm.
Ammonium permeases Amt1 and Amt2 are required for pseudohyphal growth.
We observed pseudohyphal growth only in nitrogen-limited ¼ YNB media among the conditions tested, and in these media, ammonium was the only nitrogen source (see Table S1 in the supplemental material). This result prompted us to test whether ammonium permease mutants are able to produce pseudohyphae. C. neoformans has two ammonium permease genes: AMT1 encodes a low-affinity ammonium permease and is expressed constitutively and AMT2 encodes a high-affinity ammonium permease that is induced by nitrogen limitation conditions (46). We tested whether amt1 or amt2 single mutants are able to form pseudohyphae in the ¼ YNB media. Both the amt1 mutants and the amt2 mutants exhibited pseudohyphal growth (Fig. 4) under the given conditions. However, interestingly, the independent amt1 amt2 double mutants lacked pseudohyphal growth. To further verify the genetic linkage between the pseudohyphal phenotype and the two ammonium permease genes, we crossed the double-mutant JR11 strain (amt1::NEO amt2::NAT) with the wild-type KN99a strain and another double-mutant JR5 strain (amt1::NEO amt2::NAT) with the wild-type KN99α strain. A total of 121 progeny from 12 basidia were analyzed; from those, we obtained 22 wild-type, 31 amt1 mutant, 36 amt2 mutant, and 32 amt1 amt2 double-mutant progeny based on segregation of the integrated drug resistance selectable markers. When all progeny were grown on ¼ YNB media, wild-type, amt1 mutant, and amt2 mutant progeny all produced pseudohyphae, whereas the amt1 amt2 double-mutant progeny all lacked pseudohyphal morphogenesis, providing further evidence that both ammonium permeases are required for pseudohyphal growth. The colony morphology of amt1 amt2 also differed: wild-type cells and amt1 and amt2 single mutants formed wrinkled colonies with rough edges, whereas amt1 amt2 double mutants formed smooth colonies (Fig. 4). When washed with tap water and gently rubbed with a finger, wild-type cells and amt1 and amt2 single mutants showed more invasive growth into the solid media than amt1 amt2 double mutants, indicating that pseudohyphae are involved in invasive growth in C. neoformans. Data determined from a representative set of progeny from a single basidium (JR11 × KN99α) are presented in Table S3 in the supplemental material.
Fig 4.
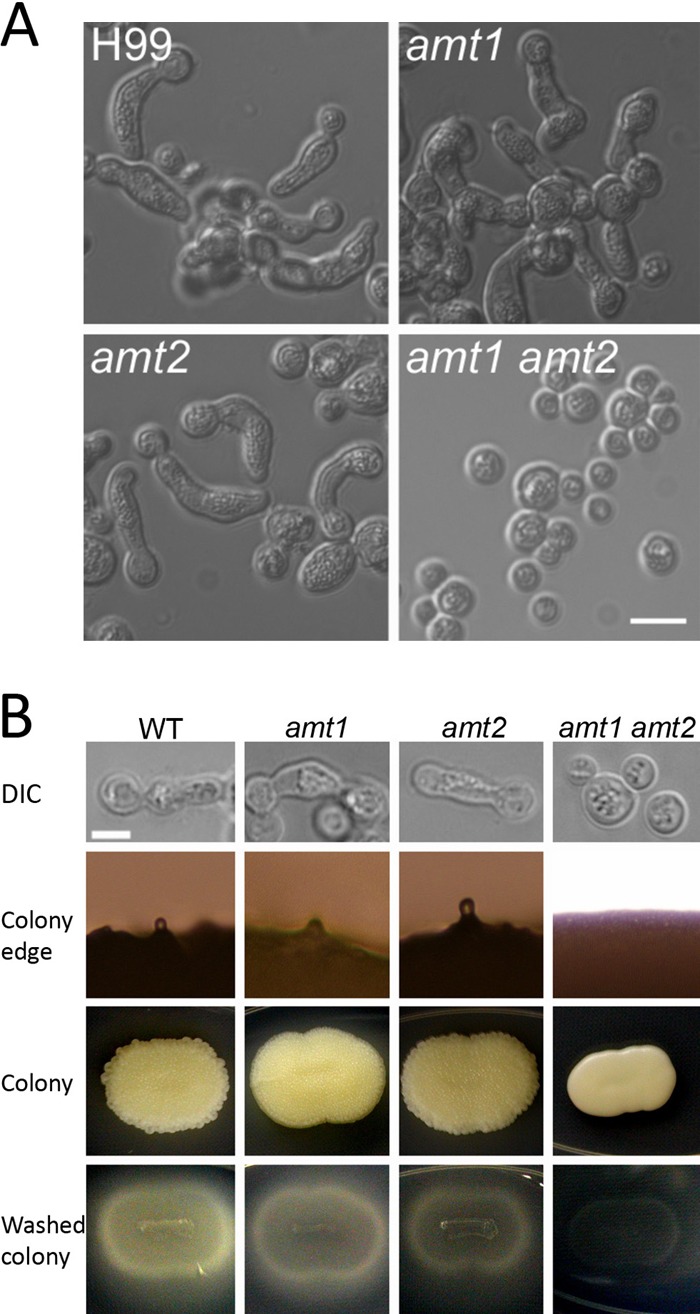
Requirement of both ammonium permeases to form pseudohyphae in C. neoformans. (A) Wild-type cells and amt1 and amt2 single mutants were able to exhibit pseudohyphal growth on ¼ YNB media; however, amt1 amt2 double mutants were not able to undergo pseudohyphal growth. Scale = 10 μm. (B) Wild-type (WT), amt1, amt2, and amt1 amt2 progeny were obtained from a cross between congenic strains JR11 (amt1 amt2) and KN99a (wild type). Wild-type, amt1, and amt2 progeny exhibited pseudohyphal growth on ¼ YNB media, showing rough-edged, wrinkled colonies and invasive growth into the agar, whereas the amt1 amt2 double mutant formed smooth colonies with no pseudohyphae and less invasive growth into the agar. Scale = 5 μm.
Pseudohypha formation by C. gattii under nitrogen limitation conditions.
We also observed pseudohyphal growth of C. gattii, and strains 97/433 and ICB108 formed pseudohyphae on filamentation agar (1.7 g YNB without ammonium sulfate and without amino acids, 0.5% glucose) (Fig. 5). A few other strains and mating mixtures also occasionally but less reliably formed pseudohyphae (see Table S1 in the supplemental material and Fig. 5A) on filamentation agar.
Fig 5.
Pseudohyphal growth of C. gattii. (A) The C. gattii 97/433 strain exhibited pseudohyphal growth on filamentation agar. (B) Microdissected pseudohyphae of C. gattii cells continued to grow as pseudohyphae, unlike those of C. neoformans, indicating that inheritable genetic change(s) had occurred that promoted pseudohyphal growth. Scale = 10 μm. (C) The mutants always displayed pseudohyphal growth, even on rich YPD media, whereas wild-type cells grew as yeast. When washed, the pseudohyphae showed more invasive growth into the agar than wild-type cells on YPD media. The pseudohypha-forming mutants also exhibited temperature-sensitive growth at 37°C and were unable to proliferate.
C. neoformans formed pseudohyphae uniformly around the colonies, and they regrew as yeast when transferred onto rich media. In contrast, C. gattii pseudohyphae emerged only from certain sectors of the colonies, which might indicate that a spontaneous mutation occurred that resulted in pseudohyphal morphogenesis. To test whether the pseudohyphal morphogenetic trait in this case was caused by inheritable mutation(s), we dissected pseudohyphae from four different sectors from one colony and transferred them onto rich nutrient media. We found that all microdissected pseudohyphae from strain 97/433 showed a phenotype typical of cell morphology mutants (Fig. 5), i.e., four independent colonies grown on YPD media from each of the microdissected pseudohyphae maintained an attached-cell morphology at 30°C in solid and liquid media and did not return to a wild-type yeast form. This suggests that the differentiation into pseudohyphae was not reversible and that the microdissected isolates were mutants. As observed in a previous study on RAM pathway mutants in C. neoformans (52), our mutants showed a dry/rough colony morphology as opposed to the slimy/wetter morphology of wild-type 97/433. Similar to the RAM pathway mutants, we also found these pseudohypha-forming mutants were (i) temperature sensitive at 37°C, and more sensitive to the calcineurin inhibitor FK506, and (ii) more invasive into the agar than the parental 97/433 strain (see Fig. S3 in the supplemental material and Fig. 5C). However, C. neoformans formed pseudohyphae even at 37°C, and progeny from pseudohyphae were not temperature sensitive (see Fig. S4 in the supplemental material).
Based on sequence analysis of the RAM pathway genes (including MOB2, CBK1, KIC1, TAO3, and SOG2), we found there was a mutation (C to T at position 70) in the MOB2 gene of four isolates of pseudohyphal cells from an isolated colony, resulting in the introduction of a premature stop codon. The sequences of the other RAM pathway genes were found to be wild type in the pseudohyphal mutant strains. We sought to test whether the mob2 mutation cosegregates with the mutant phenotype; however, interestingly, the pseudohyphal mutants were unable to complete mating with the wild type; they formed mating hyphae and basidia, but no sporulation was observed (data not shown).
DISCUSSION
Fungi change morphology in response to various environmental conditions. These morphogenetic traits include yeast growth, hyphal growth, and pseudohyphal differentiation. Like most fungi, Cryptococcus neoformans changes growth morphology: in rich nutrients, round yeast form growth predominates, whereas during mating, C. neoformans forms dikaryotic hyphae. Several decades ago, pseudohyphal or hyphal morphogenesis of C. neoformans was reported in infected animal tissues and during interaction with amoebae (11, 14, 15, 39, 48, 49, 54). Recently, Magditch and colleagues found that RAM pathway mutants form pseudohyphae, which survive better during coculture with amoeba (40). Conditions inducing yeast and mating hyphal growth in C. neoformans are well known. However, although pseudohyphal growth is not uncommon, the conditions and genes involved have been largely uncharacterized.
In the model yeast S. cerevisiae, nitrogen limitation conditions with fermentable carbon sources induce pseudohyphal growth. The MAPK and PKA pathways orchestrate pseudohyphal morphogenesis in S. cerevisiae. The high-affinity ammonium permease Mep2 both transports and senses ammonium, and thus, under nitrogen limitation conditions, Mep2 plays a central role in pseudohyphal morphogenesis (2, 37, 47). In C. neoformans, the high-affinity ammonium permease Amt2 is required for initiating mating under low-ammonium conditions, implying a role for ammonium and this ammonium permease during mating (46). It is intriguing that C. neoformans exhibits pseudohyphal growth under nitrogen limitation conditions such as in the ¼ YNB media tested here (see Table S1 in the supplemental material and Fig. 1). Furthermore, that ammonium permeases play an essential role for pseudohyphal growth is a key feature shared by C. neoformans and S. cerevisiae. Interestingly, the ammonium permeases Amt1 and Amt2 are both required for cryptococcal pseudohyphal growth (Fig. 4), whereas in S. cerevisiae, only the high-affinity ammonium permease Mep2 is required for pseudohyphal growth (37). This is the first finding that both low- and high-affinity ammonium permeases are required for a morphogenic switch in C. neoformans. This intriguing observation indicates that C. neoformans regulates nitrogen level more precisely, for example, for hyphal morphogenesis during mating, where only the high-affinity ammonium permease Amt2 is required (46). Although nitrogen limitation conditions induce pseudohyphal growth, we found that too low a concentration of ammonium, such as in SLAD (synthetic low ammonium dextrose) media (50 μM ammonium sulfate in SLAD versus 9.5 mM ammonium sulfate in ¼ YNB medium) (see Table S1 in the supplemental material), results in the absence of induction of pseudohyphal growth. This may explain why C. neoformans requires both Amt1 and Amt2 for pseudohyphal growth: balanced activity between the two ammonium permeases at an appropriate ammonium concentration may be required to induce pseudohyphal growth.
Pseudohyphae of C. neoformans divide to produce yeast when transferred to rich nutrient conditions (Fig. 3). This finding suggests that pseudohyphae represent an intermediate developmental stage. For example, in natural habitats, cryptococcal pseudohyphae may serve as a way to escape from natural predators. Neilson et al. observed that when Cryptococcus cells were cocultured with Acanthamoeba polyphaga, the soil amoebae phagocytosed and killed yeast Cryptococcus cells, whereas the yeast cells that developed pseudohyphae survived (41). Previous studies found that C. neoformans also forms pseudohyphae during host infection, albeit rarely (reviewed in reference 30). Morphotype transition is linked with virulence in C. neoformans (53), and pseudohyphae may also serve as part of an escape strategy; for example, similarly to giant/titan cells of C. neoformans (43, 55), pseudohyphae might be resistant to phygocytic engulfment by macrophages. Alternatively, phagocytosed yeast cells might form pseudohyphae to kill macrophages, as observed in other pathogenic fungi, including C. albicans and the zygomycete pathogen Mucor circinelloides (29, 36); however, interestingly, amt1 and amt2 double mutants were found to be as virulent as wild-type cells in a murine inhalation model (46).
Walton and colleagues found that RAM pathway mutants exhibit pseudohyphal growth (52). The phenotype of these mutants differs from those that arose under the nitrogen limitation conditions we tested: the RAM mutants continued to grow as pseudohyphae, whereas pseudohyphae formed during nitrogen limitation were fully reversible to yeast on rich media, and more similar to those observed in histopathology samples from tissues infected by C. neoformans, and the pseudohyphae did not grow further as elongated cells; instead, daughter cells budded out of the apex of pseudohyphae when grown in complete media. Interestingly, C. gattii pseudohyphae are strikingly similar to the RAM mutants, for instance, continuing to grow as pseudohyphae. Moreover, the pseudohyphae formed by C. gattii did not bud to produce yeast on rich media, indicating that a spontaneous mutation might have occurred in the RAM pathway or related genes. Indeed, the C. gattii pseudohyphal mutant carries a mutation in the MOB2 gene generating a premature stop codon, indicating that the RAM pathway in C. gattii is also involved in pseudohyphal differentiation, as seen in C. neoformans (40, 52). It is also possible that C. gattii may undergo a reversible morphological switch between yeast and pseudohyphae under certain conditions, or in a strain-specific fashion, as in the case of C. neoformans.
Establishing laboratory conditions to induce pseudohyphal growth is a key advance of this study. Although there are implications that pseudohyphal growth may enable escape from host defenses as discussed above, this remains to be experimentally validated in cultured immune cells or animal models. In addition, signaling pathways for pseudohyphal development in S. cerevisiae are well understood, but in C. neoformans, no genetic studies had been reported beyond the analyses of RAM mutants (40, 52). The C. albicans pescadillo homolog Pes1 is required for the hypha-yeast dimorphic transition (50). Therefore, future research to test interactions between pseudohyphae and innate immune systems and immune cells, to identify molecular genetics of pseudohypha formation, and to identify genes involved in the pseudohypha-yeast transition is a promising venue to pursue.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alex Idnurm for valuable discussions and communication of results on RAM pathway mutants forming pseudohyphae prior to publication. We are indebted to Anna Averette and Alicia Li for technical support. We are also grateful to Valerie Knowlton and the NC State EM core laboratory for SEM analysis.
S.C.L. is supported by the NIH Molecular Mycology and Pathogenesis Training Program (AI52080). This work was supported by NIH/NIAID R37 grant AI39115-14 to J.H.
Footnotes
Published ahead of print 21 September 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Bastidas RJ, Heitman J. 2009. Trimorphic stepping stones pave the way to fungal virulence. Proc. Natl. Acad. Sci. U. S. A. 106:351–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boeckstaens M, André B, Marini AM. 2007. The yeast ammonium transport protein Mep2 and its positive regulator, the Npr1 kinase, play an important role in normal and pseudohyphal growth on various nitrogen media through retrieval of excreted ammonium. Mol. Microbiol. 64:534–546 [DOI] [PubMed] [Google Scholar]
- 3. Brandt ME, et al. 1996. Molecular subtype distribution of Cryptococcus neoformans in four areas of the United States. Cryptococcal Disease Active Surveillance Group. J. Clin. Microbiol. 34:912–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bunting LA, Neilson JB, Bulmer GS. 1979. Cryptococcus neoformans: gastronomic delight of a soil amoeba. Sabouraudia 17:225–232 [DOI] [PubMed] [Google Scholar]
- 5. Carlisle PL, et al. 2009. Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc. Natl. Acad. Sci. U. S. A. 106:599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carlisle PL, Kadosh D. 2010. Candida albicans Ume6, a filament-specific transcriptional regulator, directs hyphal growth via a pathway involving Hgc1 cyclin-related protein. Eukaryot. Cell 9:1320–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Casadevall A, Perfect JR. 1998. Cryptococcus neoformans. ASM Press, Washington, DC [Google Scholar]
- 8. Court H, Sudbery P. 2007. Regulation of Cdc42 GTPase activity in the formation of hyphae in Candida albicans. Mol. Biol. Cell 18:265–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. D'Souza CA, et al. 2011. Genome variation in Cryptococcus gattii, an emerging pathogen of immunocompetent hosts. mBio 2:e00342–10 doi:10.1128/mBio.00342-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Franzot SP, Hamdan JS, Currie BP, Casadevall A. 1997. Molecular epidemiology of Cryptococcus neoformans in Brazil and the United States: evidence for both local genetic differences and a global clonal population structure. J. Clin. Microbiol. 35:2243–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Freed ER, Duma RJ, Shadomy HJ, Utz JP. 1971. Meningoencephalitis due to hyphae-forming Cryptococcus neoformans. Am. J. Clin. Pathol. 55:30–33 [DOI] [PubMed] [Google Scholar]
- 12. Fries BC, Goldman DL, Cherniak R, Ju R, Casadevall A. 1999. Phenotypic switching in Cryptococcus neoformans results in changes in cellular morphology and glucuronoxylomannan structure. Infect. Immun. 67:6076–6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gavrias V, Andrianopoulos A, Gimeno CJ, Timberlake WE. 1996. Saccharomyces cerevisiae TEC1 is required for pseudohyphal growth. Mol. Microbiol. 19:1255–1263 [DOI] [PubMed] [Google Scholar]
- 14. Gazzoni AF, Fd, et al. 2010. Unusual morphologies of Cryptococcus spp. in tissue specimens: report of 10 cases. Rev. Inst. Med. Trop. Sao Paulo 52:145–149 [DOI] [PubMed] [Google Scholar]
- 15. Gazzoni AF, Severo CB, Barra MB, Severo LC. 2009. Atypical micromorphology and uncommon location of cryptococcosis: a histopathologic study using special histochemical techniques (one case report). Mycopathologia 167:197–202 [DOI] [PubMed] [Google Scholar]
- 16. Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077–1090 [DOI] [PubMed] [Google Scholar]
- 17. Halliday CL, Carter DA. 2003. Clonal reproduction and limited dispersal in an environmental population of Cryptococcus neoformans var gattii isolates from Australia. J. Clin. Microbiol. 41:703–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heitman J, Allen B, Alspaugh JA, Kwon-Chung KJ. 1999. On the origins of congenic MATα and MATa strains of the pathogenic yeast Cryptococcus neoformans. Fungal Genet. Biol. 28:1–5 [DOI] [PubMed] [Google Scholar]
- 19. Heitman J, Kozel TR, Kwon-Chung JK, Perfect JR, Casadevall A. 2011. Cryptococcus: from human pathogen to model yeast. ASM Press, Washington, DC [Google Scholar]
- 20. Hughes CF, Perlin MH. 2005. Differential expression of mepA, mepC and smtE during growth and development of Microbotryum violaceum. Mycologia 97:605–611 [DOI] [PubMed] [Google Scholar]
- 21. Idnurm A. 2010. A tetrad analysis of the basidiomycete fungus Cryptococcus neoformans. Genetics 185:153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kron SJ, Styles CA, Fink GR. 1994. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 5:1003–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kübler E, Mosch H-U, Rupp S, Lisanti MP. 1997. Gpa2p, a G-protein α-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J. Biol. Chem. 272:20321–20323 [DOI] [PubMed] [Google Scholar]
- 24. Kwon-Chung KJ. 1976. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia 68:821–833 [PubMed] [Google Scholar]
- 25. Kwon-Chung KJ. 1975. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia 67:1197–1200 [PubMed] [Google Scholar]
- 26. Kwon-Chung KJ. 1976. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia 68:943–946 [PubMed] [Google Scholar]
- 27. Kwon-Chung KJ, Bennett JE. 1978. Distribution of α and a mating types of Cryptococcus neoformans among natural and clinical isolates. Am. J. Epidemiol. 108:337–340 [DOI] [PubMed] [Google Scholar]
- 28. Lee SC, Ni M, Li W, Shertz C, Heitman J. 2010. The evolution of sex: a perspective from the fungal kingdom. Microbiol. Mol. Biol. Rev. 74:298–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li CH, et al. 2011. Sporangiospore size dimorphism is linked to virulence of Mucor circinelloides. PLoS Pathog. 7:e1002086 doi:10.1371/journal.ppat.1002086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lin X. 2009. Cryptococcus neoformans: morphogenesis, infection, and evolution. Infect. Genet. Evol. 9:401–416 [DOI] [PubMed] [Google Scholar]
- 31. Lin X, Heitman J. 2006. The biology of the Cryptococcus neoformans species complex. Annu. Rev. Microbiol. 60:69–105 [DOI] [PubMed] [Google Scholar]
- 32. Lin X, Hull CM, Heitman J. 2005. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature 434:1017–1021 [DOI] [PubMed] [Google Scholar]
- 33. Lin X, Nielsen K, Patel S, Heitman J. 2008. Impact of mating type, serotype, and ploidy on the virulence of Cryptococcus neoformans. Infect. Immun. 76:2923–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin X, et al. 2009. Diploids in the Cryptococcus neoformans serotype A population homozygous for the α mating type originate via unisexual mating. PLoS Pathog. 5:e1000283 doi:10.1371/journal.ppat.1000283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lo W-S, Dranginis AM. 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9:161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lorenz MC, Bender JA, Fink GR. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 3:1076–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lorenz MC, Heitman J. 1998. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 17:1236–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lorenz MC, Heitman J. 1997. Yeast pseudohyphal growth is regulated by GPA2, a G protein α homolog. EMBO J. 16:7008–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lurie HI, Shadomy HJ. 1971. Morphological variations of a hypha-forming strain of Cryptococcus neoformans (Coward strain) in tissues of mice. Sabouraudia 9:10–14 [DOI] [PubMed] [Google Scholar]
- 40. Magditch DA, Liu T-B, Xue C, Idnurm A. DNA mutations mediate microevolution between host-adapted forms of the pathogenic fungus Cryptococcus neoformans. PLoS Pathog., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Neilson JB, Ivey MH, Bulmer GS. 1978. Cryptococcus neoformans: pseudohyphal forms surviving culture with Acanthamoeba polyphaga. Infect. Immun. 20:262–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nielsen K, et al. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect. Immun. 71:4831–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Okagaki LH, et al. 2010. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 6:e1000953 doi:10.1371/journal.ppat.1000953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Perfect JR, LSD, Durack DT. 1980. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am. J. Pathol. 101:177–194 [PMC free article] [PubMed] [Google Scholar]
- 45. Rupp S, Summers E, Lo H-J, Madhani H, Fink G. 1999. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 18:1257–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rutherford J, Lin X, Nielsen K, Heitman J. 2008. The Amt2 permease is required to induce ammonium responsive invasive growth and mating in Cryptococcus neoformans. Eukaryot. Cell 7:237–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rutherford JC, Chua G, Hughes T, Cardenas ME, Heitman J. 2008. A Mep2-dependent transcriptional profile links permease function to gene expression during pseudohyphal growth in Saccharomyces cerevisiae. Mol. Biol. Cell 19:3028–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shadomy HJ, Lurie HI. 1971. Histopathological observations in experimental cryptococcosis caused by a hypha-producing strain of Cryptococcus neoformans (Coward strain) in mice. Sabouraudia 9:6–9 [DOI] [PubMed] [Google Scholar]
- 49. Shadomy HJ, Utz JP. 1966. Preliminary studies on a hypha-forming mutant of Cryptococcus neoformans. Mycologia 58:383–390 [PubMed] [Google Scholar]
- 50. Shen J, Cowen LE, Griffin AM, Chan L, Koehler JR. 2008. The Candida albicans pescadillo homolog is required for normal hypha-to-yeast morphogenesis and yeast proliferation. Proc. Natl. Acad. Sci. U. S. A. 105:20918–20923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Smith DG, Garcia-Pedrajas MD, Gold SE, Perlin MH. 2003. Isolation and characterization from pathogenic fungi of genes encoding ammonium permeases and their roles in dimorphism. Mol. Microbiol. 50:259–275 [DOI] [PubMed] [Google Scholar]
- 52. Walton FJ, Heitman J, Idnurm A. 2006. Conserved elements of the RAM signaling pathway establish cell polarity in the basidiomycete Cryptococcus neoformans in a divergent fashion from other fungi. Mol. Biol. Cell 17:3768–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang L, Zhai B, Lin X. 2012. The link between morphotype transition and virulence in Cryptococcus neoformans. PLoS Pathog. 8:e1002765 doi:10.1371/journal.ppat.1002765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Williamson JD, Silverman JF, Mallak CT, Christie JD. 1996. Atypical cytomorphologic appearance of Cryptococcus neoformans: a report of five cases. Acta Cytol. 40:363–370 [DOI] [PubMed] [Google Scholar]
- 55. Zaragoza O, et al. 2010. Fungal cell gigantism during mammalian infection. PLoS Pathog. 6:e1000945 doi:10.1371/journal.ppat.1000945 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.