Abstract
Heterotrimeric G proteins (G proteins) govern growth, development, and secondary metabolism in various fungi. Here, we characterized ricA, which encodes a putative GDP/GTP exchange factor for G proteins in the model fungus Aspergillus nidulans and the opportunistic human pathogen Aspergillus fumigatus. In both species, ricA mRNA accumulates during vegetative growth and early developmental phases, but it is not present in spores. The deletion of ricA results in severely impaired colony growth and the total (for A. nidulans) or near (for A. fumigatus) absence of asexual sporulation (conidiation). The overexpression (OE) of the A. fumigatus ricA gene (AfricA) restores growth and conidiation in the ΔAnricA mutant to some extent, indicating partial conservation of RicA function in Aspergillus. A series of double mutant analyses revealed that the removal of RgsA (an RGS protein of the GanB Gα subunit), but not sfgA, flbA, rgsB, or rgsC, restored vegetative growth and conidiation in ΔAnricA. Furthermore, we found that RicA can physically interact with GanB in yeast and in vitro. Moreover, the presence of two copies or OE of pkaA suppresses the profound defects caused by ΔAnricA, indicating that RicA-mediated growth and developmental signaling is primarily through GanB and PkaA in A. nidulans. Despite the lack of conidiation, brlA and vosA mRNAs accumulated to normal levels in the ΔricA mutant. In addition, mutants overexpressing fluG or brlA (OEfluG or OEbrlA) failed to restore development in the ΔAnricA mutant. These findings suggest that the commencement of asexual development requires unknown RicA-mediated signaling input in A. nidulans.
INTRODUCTION
Heterotrimeric G proteins (G proteins) are conserved in all eukaryotes and are involved in almost all biological processes (22, 43, 58). Basic units of the heterotrimeric G protein signaling system include a G protein-coupled receptor (GPCR), a G protein, composed of α, β, and γ subunits, and a variety of effectors, which relay the signal into cells to elicit appropriate physiological and biochemical responses (6).
G proteins are regarded as biological switches that oscillate between on and off states (7). Under nonstimulated conditions, the inactive Gα-GDP::Gβγ trimeric complex prevails in the cell membrane, and the signaling pathway remains off. Typically, G proteins are turned on by the guanine nucleotide exchange caused by ligand-bound (sensitized) GPCRs, causing the dissociation of the GTP-bound Gα subunit and the Gβγ heterodimer, which then transduce signals by interacting with various effectors, including adenylyl cyclase-protein kinase A (PKA), phospholipase C, ionic channels, and mitogen-activated protein kinases (18, 45). The signal is turned off when Gα-GTP is hydrolyzed back to Gα-GDP by the intrinsic GTPase activity of the Gα subunit, forming the inactive trimeric complex. One key element facilitating inactivation is the regulator of G protein signaling (RGS), which accelerates GTP hydrolysis catalyzed by the Gα subunit (15). While GPCR-mediated signaling accounts for the majority of G protein-regulated cellular control mechanisms, the evolutionarily conserved RIC-8 (resistance to inhibitors of cholinesterase 8) protein is a proven critical guanine nucleotide exchange factor (GEF) that activates a subset of Gα subunits (24). Ric-8 interacts with monomeric Gα-GDP, stimulates the release of GDP, forms a stable nucleotide-free transition-state complex with the Gα subunit, and catalyzes the exchange of GDP for GTP (Fig. 1A) (5, 24).
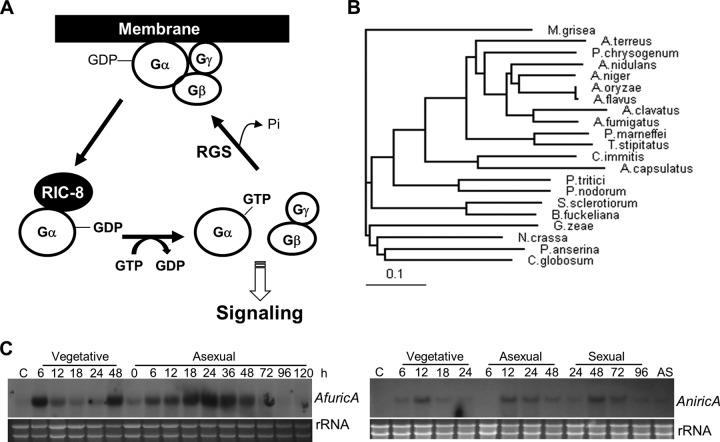
Fig 1.
Summary of the ricA genes. (A) Proposed Ric-8-mediated G protein signaling. (B) Phylogenetic tree of the putative RicA proteins identified in various fungal species (from top to bottom: Magnaporthe grisea, A. terreus, P. chrysogenum, A. nidulans, A. niger, A. oryzae, A. flavus, A. clavatus, A. fumigatus, Penicillium marneffei, Talaromyces stipitatus, Coccidioides immitis, Ajellomyces capsulatus, P. tritici, Phaeosphaeria nodorum, Sclerotinia sclerotiorum, G. zeae, Botryotinia fuckeliana, Podospora anserine, and Chaetomium globosum). (C) mRNA levels of ricA during the life cycles of A. fumigatus and A. nidulans. C, conidia (asexual spores); AS, ascospores (sexual spores in A. nidulans). Numbers indicate the time (in hours) of incubation in liquid MMG or MMG plus 0.1% YE (vegetative) or after transfer onto solid MMG glucose under conditions favoring asexual or sexual development. Equal loading of total RNA was evaluated by ethidium bromide staining of rRNA.
In fungi, G protein signaling governs cell growth, morphogenesis, sexual/asexual development, mating, pathogenicity, secondary metabolism, and many more processes (33, 34, 69, 73). The model filamentous fungus Aspergillus nidulans contains three Gα subunits (FadA, GanB, and GanA) (10, 69, 74), one Gβ subunit (SfaD) (48), and one Gγ subunit (GpgA) (53). Genetic studies have revealed that both FadA (Gα) and SfaD::GpgA (Gβγ) mediate signaling that promotes vegetative growth while inhibiting development and biosynthesis of the carcinogenic mycotoxin sterigmatocystin (ST) (23, 48, 73, 74). Further studies have shown that FadA signaling is in part transduced via the cyclic AMP (cAMP)-dependent protein kinase PkaA (56). This FadA→PkaA-mediated signaling in turn inhibits asexual development (conidiation), which is activated by the FluG→BrlA pathway and completed by VosA (2, 44, 69, 70). FlbA is the cognate RGS protein, whose primary role is to negatively control FadA-mediated vegetative growth signaling (31, 74). Both ΔflbA and constitutively active FadA mutations (G42R, R178C, and Q204L, resulting in defective intrinsic GTPase) produce the fluffy autolytic phenotype (64, 74). Importantly, this FadA-mediated signaling for vegetative growth, development, and toxigenesis is conserved in the aflatoxin-producing fungi Aspergillus parasiticus and Aspergillus flavus (23, 49) and the opportunistic human pathogen Aspergillus fumigatus (38, 69).
The GanB Gα subunit negatively regulates conidiation and plays a positive role in the germination of conidia, whereas GanA's role is not yet understood (10). Additional studies have revealed that GanB and SfaD::GpgA constitute a functional heterotrimer controlling cAMP-PKA signaling and conidial germination in response to glucose, where GanB is the primary signaling element and SfaD::GpgA functions for proper activation of GanB (30). Among the three additional RGS proteins, RgsA, RgsB, and RgsC (69), RgsA acts as the negative regulator of GanB signaling in A. nidulans (21). The lack of RgsA results in phenotypes similar to those caused by constitutive activation of GanB (Q208L), i.e., germination of conidia in the absence of an external C source and an enhanced stress response (21). Furthermore, the overexpression of rgsA causes elaboration of asexual developmental structures (conidiophores) in liquid submerged cultures, as observed in ΔganB or GanBG207R mutants (10, 21). In A. fumigatus, signaling mediated by GpaB (GanB homolog) is associated with the activation of the predominant PKA catalytic subunit PkaC1, which governs hyphal growth and development (36, 37).
Despite such a pivotal role of G proteins in many aspects of Aspergillus biology, upstream mechanisms of signal activation remain to be understood. While at least 16 putative GPCRs have been identified in the genome of A. nidulans (69), none has been proven to specifically activate FadA- or GanB-mediated signaling. In an effort to understand the upstream activation of G protein signaling in Aspergillus, we identified and characterized the Ric-8 ortholog RicA in A. nidulans and A. fumigatus. Functional studies of the ricA gene revealed that it plays a crucial (or essential) role in vegetative growth and development in both species, with a partially conserved function. Genetic and biochemical studies further indicated that RicA primarily activates the GanB→PkaA signaling cascade in A. nidulans. Finally, as normal or elevated expression of key developmental activators fails to trigger conidiation in the absence of RicA, it is proposed that an unknown RicA-mediated signal input, independent of the FluG→BrlA→VosA pathway, is required for asexual development in A. nidulans.
MATERIALS AND METHODS
Strains and culture conditions.
A. nidulans and A. fumigatus strains used in this study are listed in Table 1. Glucose minimal medium (MMG) and MMG with 0.5% (wt/vol) yeast extract (YE) with appropriate supplements were used for general culture of A. nidulans strains (26, 47). For A. fumigatus pyrimidine and arginine auxotrophic mutant strains (AF293.1 and AF293.6 [67]), MMG plus 0.1% YE was supplemented with 5 mM uridine, 10 mM uracil (for pyrG1), and 0.1% arginine (for argB1). Minimal medium with 100 mM threonine as a sole carbon source (MMT) with 0.5% YE was used for alcAp-mediated overexpression. To check the phenotype of the overexpression strains under control of the alcA promoter (40, 63) in A. nidulans and A. fumigatus, wild-type (WT) and overexpression strains were inoculated on MMG and MMT plus 0.5% YE solid media and incubated at 37°C for 5 days. Effects of overexpression of the target genes under the niiA promoter (4) in A. nidulans were examined by growing the strains in both MM with 0.2% (wt/vol) ammonium tartrate (MM plus AT; noninducing) and also MMG (containing 0.6% [wt/vol] sodium nitrate; inducing). For Northern blot assays to confirm overexpression by the alcA promoter, strains were cultured in liquid MMG at 37°C, 220 rpm, for 12 h, and the mycelial aggregates were collected, rinsed with liquid MMT, transferred into liquid MMT, and further induced at 37°C, 220 rpm, for 6 h. Overexpression under niiA(p) was performed by culturing the strains in liquid MMG for 16 h at 37°C, 220 rpm. The Saccharomyces cerevisiae L40 strain (Clontech) was used to check the protein-protein interactions between the RicA-fused DNA binding domain and Gα subunits FadA, GanA, and GanB with the activation domain in a yeast two-hybrid assay. The L40 strain was grown in synthetic dropout minimal medium (SD) with the necessary supplements (10 g/liter leucine, 2 g/liter tryptophan, and 2 g/liter histidine) (55) and incubated at 30°C for 2 to 3 days. Escherichia coli DH5α and DH10B were grown in the Luria-Bertani (LB) medium with ampicillin (50 μg/ml; Sigma) or zeocin (20 μg/ml; Invitrogen) for plasmid amplification and construction. The oligonucleotides used in this study are listed in Table S1 of the supplemental material.
Table 1.
Aspergillus strains used in this study
| Species and strain | Genotype | Source |
|---|---|---|
| A. nidulans strains | ||
| FGSC4 | veA+ (wild type) | FGSCa |
| RJMP1.59 | pyrG89 pyroA4 veA+ | 54 |
| TNJ21 | pyrG89 pyroA4 ΔricA::AfpyrG+ veA+ | This study |
| TNJ36 | pyrG89 AfpyrG+ pyroA4 veA+ | 28 |
| TNJ42 | pyrG89 ΔflbA::AnpyroA+ pyroA4 veA+ | This study |
| TNJ49 | pyrG89 ΔflbA::AnpyroA+ pyroA4 ΔricA::AfpyrG+ veA+ | This study |
| TNJ57 | pyrG89 ΔsfgA::AfpyrG+ pyroA4 veA+ | This study |
| TNJ58 | pyrG89 ΔsfgA::AfpyrG+ pyroA4 ΔricA::AnpyroA+ veA+ | This study |
| TNJ59b | pyrG89 pyroA4 niiA(p)::fluG::Flag::3/4pyroA+ veA+ | This study |
| TNJ60b | pyrG89 pyroA4 niiA(p)::fluG::Flag::3/4pyroA+ ΔricA::AnpyroA+ veA+ | This study |
| TNJ61 | pyrG89 pyroA4 ΔrgsA::AnpyroA+ veA+ | This study |
| TNJ62 | pyrG89 pyroA4 ΔrgsA::AnpyroA+ ΔricA::AfpyrG+ veA+ | This study |
| TNJ63 | pyrG89 ΔrgsB::AfpyrG+ pyroA4 veA+ | This study |
| TNJ64 | pyrG89 ΔrgsB::AfpyrG+ pyroA4 ΔricA::AnpyroA+ veA+ | This study |
| TNJ65 | pyrG89 pyroA4 ΔrgsC::AfpyrG+ veA+ | This study |
| TNJ66 | pyrG89 pyroA4 ΔricA::AnpyroA+ ΔrgsC::AfpyrG+ veA+ | This study |
| TNJ68 | pyrG89 pyroA4 ganBQ208L::AfpyrG+ veA+ | This study |
| TNJ69 | pyrG89 pyroA4 ΔricA::AnpyroA+ ganBQ208L::AfpyrG+ veA+ | This study |
| TNJ85b | pyrG89 pyroA4 niiA(p)::brlA::Flag::3/4pyroA+ veA+ | This study |
| TNJ86b | pyrG89 pyroA4 niiA(p)::brlA::Flag::3/4pyroA+ ΔricA::AnpyroA+ veA+ | This study |
| TNJ87b | pyrG89 pyroA4 ricA(p)::ricA::ricA(t)::3/4pyroA+ ΔricA::AfpyrG+ veA+ | This study |
| TNJ89b | pyrG89 pyroA4 niiA(p)::pkaA::3/4pyroA+ veA+ | This study |
| TNJ90b | pyrG89 pyroA4 niiA(p)::pkaA::3/4pyroA+ ΔricA::AfpyrG+ veA+ | This study |
| TNJ94b | pyrG89 pyroA4 alcA(p)::AfricA::3/4pyroA+AfveA+ | This study |
| TNJ95b | pyrG89 pyroA4 alcA(p)::AfricA::3/4pyroA+ ΔricA::AfpyrG+ veA+ | This study |
| A. fumigatus strains | ||
| AF293 | Wild type | 9 |
| AF293.1 | pyrG1 | 67 |
| AF293.6 | pyrG1 argB1 | 67 |
| FNJ12 | pyrG1 ΔricA::AnpyrG+ | This study |
| FNJ13c | pyrG1 ricA(p)::ricA::ricA(t)::3/4pyrG+ ΔricA::AnargB+ argB1 | This study |
FGSC, Fungal Genetics Stock Center.
The 3/4 pyroA marker in pHS causes targeted integration at the pyroA locus.
The 3/4 AfpyrG marker in pNJ25 causes targeted integration at the pyrG locus.
Database analyses, nucleic acid isolation, and manipulation.
The putative RicA proteins were retrieved from an NCBI BLASTX (http://blast.ncbi.nlm.nih.gov/Blast.cgi) search based on A. nidulans ricA. A phylogenetic tree of the 21 putative RicA proteins was created by using information in EMBL-EBI (http://www.ebi.ac.uk/Tools/clustalw2/index.html). The construction and analysis of the phylogenetic tree were carried out within EMBL-EBI (http://www.ebi.ac.uk/Tools/clustalw2/help.html#tree). The AnricA and AfricA genes were PCR amplified from A. nidulans (FGSC4) and A. fumigatus (AF293) genomic DNA. cDNA of AnricA was isolated from an A. nidulans cDNA library (provided by K. Y. Jahng, Chonbuk National University, Jeonju, Korea) with the primer pairs oNK-39 and oNK-394. AfricA cDNA was isolated from reverse transcriptase-treated total RNA and primers oNK-391 and oNK-392. Isolation of genomic DNA and total RNA for Northern blot analysis was carried out as described previously (38, 72). About 10 μg of total RNA isolated from each sample was separated by electrophoresis, using a 1.1% (wt/vol) agarose gel containing 3% (wt/vol) formaldehyde and ethidium bromide, then transferred onto a Hybond-N+ membrane (Amersham). The probes for brlA (1) and vosA (44) in A. nidulans and brlA, abaA, and wetA in A. fumigatus for Northern blot analyses were prepared by PCR amplification with primer pairs oNK-556/oNK-557 (AnbrlA), oNK-14/oNK-15 (AnvosA), oNK-594/oNK-595 (AfbrlA), oHS-382/oHS-383 (AfabaA), and oTL-7/oTL-8 (AfwetA), respectively (see Table S1 of the supplemental material).
Construction of deletion, complementation, and overexpression strains.
The ricA deletion (ΔricA) mutants for A. nidulans and A. fumigatus were generated by double-joint PCR (DJ-PCR) as described previously (72). The flanking regions of each ricA gene were amplified by PCR with primer pairs oNK-352/oNK-353 (An5′ with AfpyrG tail), oNK-354/oNK-355 (An3′ with AfpyrG tail), oNK-352/oNK-474 (An5′ with AnpyroA tail), oNK-475/oNK-355 (An3′ with AfpyroA tail), oNK-358/oNK-359 (Af5′ with AnpyrG tail), oNK-360/oNK-361 (Af3′ with AnpyrG tail), oNK-358/oNK-933 (Af5′ with AnargB tail), and oNK-934/oNK-361 (Af3′ with AnargB tail) from both genomic DNAs, respectively. The AnpyrG, AnpyroA, AnargB, and AfpyrG markers were amplified with the primer pairs oBS-08/oBS-09, oNK-395/oNK-396, oNK-104/oNK-105, and oJH-83/oJH-86, respectively. The final deletion constructs were amplified with the nested primer set oNK-356/oNK-357 (A. nidulans) or oNK-362/oNK-363 (A. fumigatus), respectively. The final PCR products were introduced into RJMP1.59 (N. P. Keller; veA+) and RNIW3 (M. Ni; veA1) for A. nidulans AF293.1 or AF293.6 for A. fumigatus (67) using the Vinoflow FCE lysing enzyme (Novo Nordisk) (59). For the deletion mutants of flbA, sfgA, rgsA, rgsB, and rgsC in A. nidulans, each flanking region was PCR amplified using primer pairs oNK-412/oNK-413 (5′ flbA with AnpyroA tail), oNK-414/oNK-415 (3′ flbA with AnpyroA tail), oNK-397/oNK-398 (5′ sfgA with AfpyroA tail), oNK-399/oNK-400 (3′ sfgA with AfpyroA tail), oNK-540/oNK-541 (5′ rgsA with AfpyroA tail), oNK-542/oNK-543 (3′ rgsA with AfpyroA tail), oNK-562/oNK-563 (5′ rgsB with AfpyroA tail), oNK-564/oNK-567 (3′ rgsB with AfpyroA tail), oNK-568/oNK-569 (5′ rgsC with AfpyroA tail), and oNK-603/oNK-604 (3′ rgsC with AfpyroA tail), respectively. The final deletion constructs were amplified with oNK-416/oNK-417 (AnflbA), oNK-401/oNK-402 (AnsfgA), oNK-544/oNK-545 (AnrgsA), oNK-605/oNK-606 (AnrgsB), and oNK-607/oNK-608 (AnrgsC). These deletion mutants were used to generate double-deletion mutants with ΔAnricA by subsequent transformation.
To generate the complemented strains, genomic DNA fragments of AnricA and AfricA were PCR amplified using the primer pairs oNK-870/oNK-871 and oNK-868/oNK-869 from each genomic DNA, then digested with BamHI (followed by treatment with Klenow fragment) and NotI, and cloned between PvuII and NotI of pHS3 (28) containing the ¾AnpyroA (46) marker and pNJ25 (29) containing the ¾AfpyrG (14) marker with the alcA promoter (19), FLAG tag (DYKDDDDK), and the trpC terminator (68), respectively. Each construct was introduced into the recipient ΔAnricA and ΔAfricA strains, where preferentially a single copy is inserted into the AnpyroA or AfpyrG locus, respectively. The complemented strains were confirmed by PCR amplification using the primers of each vector from the genomic DNA of transformants.
To generate the ricA overexpression mutant, the ricA genes were amplified by primer pairs, oNK-393/oNK-394 (AnricA) and oNK-391/oNK-392 (AfricA) from each genomic DNA. The amplified genes were digested with restriction enzymes EcoRI and NotI and ligated between the alcA promoter and the trpC terminator in pHS3 and pNJ25, respectively. The final plasmids were used to transform TNJ36 and AF293.1, and single integration at pyroA in A. nidulans and pyrG locus in A. fumigatus was confirmed.
Autolysis and cell death assays.
The alamarBlue (AB) assay to assess the cell viability based on the percent reduction of alamarBlue was used as described previously (57). A total of 106 conidia of the WT and ΔAfricA strains were cultured in MMG plus 0.1% YE liquid medium at 37°C for 7 days. Aliquots (0.5 ml) of cultures according to time (days), including the mycelial aggregates and liquid medium, were transferred into 24-well plates (Nunc), 1 ml of fresh liquid medium containing 150 μl of alamarBlue (AbD Serotec) was added to each sample, and then the cultures were incubated for a further 6 h at 37°C. The solution samples were transferred into 96-well plates excluding mycelial aggregates, and absorbance was read at 570 and 600 nm. The percent alamarBlue reduction was detected with a Synergy HT apparatus (Bio-Tek) and the KC4 v3.1 software and was calculated using the following formula: [(117,216 × the A570 of sample) − (80,586 × the A600 of the sample)]/[(155,677 × the A600 of the medium) − (14,652 × the A570 of the medium)] × 100 (39), as described by Shin et al. (57)
Yeast two-hybrid assay.
Open reading frames (ORFs) of RicA (oNK-521/oNK-394), FadA (oNK-507/oNK-508), GanA (oNK-509/oNK-510), and GanB (oNK-511/oNK-512) were PCR amplified from the cDNA library of A. nidulans. The ricA ORF was cloned into the pTLexA vector (provided by S. K. Chae, Paichai University, Daejeon, Republic of Korea) (11) carrying the LexA DNA binding domain (DBD) (8), which was generated by modifying pHybLex/Zeo (Invitrogen) via insertion of the TRP1 marker from pGBT9 (Clontech). The cDNA-derived ORFs of fadA, ganA, and ganB were each fused under the activation domain of pGAD424 (Clontech). Plasmids were sequence verified and cointroduced into S. cerevisiae L40 by lithium acetate-polyethylene glycol-mediated yeast transformation (25). The yeast transformants were selected on SD medium in the absence of uracil, tryptophan, and leucine (-UWL). To test the reporters β-galactosidase and histidine (H) by the interaction of RicA with Gα subunits in yeast, the transformants were inoculated on the -UWL medium containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 40 μg/ml; Sigma) and the -UHWL medium, and the growth and color of the colonies were examined. To confirm and quantify the trans-activation activity, five transformants per each test set were tested for β-galactosidase activity (32) with a yeast β-galactosidase assay kit (Pierce).
GST pulldown assay.
The AnricA ORF was cloned between EcoRI and NotI sites into pGEX-5X-1 (GE Healthcare) and then introduced into E. coli BL21(DE3) to express the glutathione S-transferase (GST)–AnRiA fusion protein. The E. coli strain was grown to an optical density at 600 nm of 0.5 to 0.6 at 37°C, 220 rpm, and 1 mM IPTG (isopropyl β-d-1-thiogalactopyranoside; Sigma) was added to induce fusion protein expression. The culture was then further incubated at 30°C, 220 rpm, for 3 h. Subsequently, cells were lysed by sonication in ice-cold E. coli lysis buffer (0.1% Triton X-100, 0.1 mM EDTA, 500 mM NaCl, with 1 protease inhibitor cocktail tablet [Roche] per 50 ml added before use). Then, the cell lysates were cleared of cellular debris by centrifugation, and the supernatant was collected and incubated with glutathione-Sepharose 4B beads (GE Healthcare) on a mixer at 4°C overnight. The beads were washed with the lysis buffer three times and resuspended in 500 μl lysis buffer. In addition, ORFs for A. nidulans fadA, ganA, ganB, and ricA were each cloned into pcDNA3 (Invitrogen) to translate in vitro by using the TNT T7 quick-coupled transcription/translation system (Promega). One-microgram aliquots of individual plasmids were incubated with 20 μCi of [35S]methionine (PerkinElmer) in TNT mixture for 90 min at 30°C. Equal amounts of in vitro-translated proteins were added to glutathione bead-GST-RicA or glutathione bead-GST (control) suspensions. The mixtures were incubated on a mixer at 4°C overnight. After washing with E. coli lysis buffer five times, the samples were mixed with Laemmli sample buffer (Bio-Rad) and loaded onto SDS-PAGE gels. After electrophoresis, the gels were dried under a vacuum onto three layers of Whatman 3MM filter paper. Autoradiography was performed at −80°C with Fuji SuperRX film.
Microscopy.
The colony photographs were taken using a Sony DSC-T30 digital camera. Photomicrographs were taken using a Zeiss M2Bio microscope equipped with AxioCam and AxioVision digital imaging software.
Nucleotide sequence accession numbers.
Our newly determined AnRicA and AfRicA sequences and annotations have been deposited in GenBank under the accession numbers JN410838 and JN582330, respectively.
RESULTS
Identification of RicA in A. nidulans and A. fumigatus.
A genome search with the Caenorhabditis elegans RIC-8 protein (GenBank accession number AF288812.1) resulted in the identification one putative ortholog in each Aspergillus species: Afu4g08820 (AfRicA; score of 134 and e-value of 2.5e−05) and AN1661 (AnRicA; score of 131 and e-value of 6.5e−05). To verify the corresponding ORFs, we isolated and analyzed the ricA cDNA from the A. fumigatus and A. nidulans cells. Briefly, the AnRicA protein is composed of 466 amino acids (aa; ORF of 1,401 bp with 6 introns), and AfRicA is composed of 461 aa (ORF of 1,386 bp with 6 introns), with predicted masses of 51.7 kDa and 51.2 kDa, respectively. Employing these protein sequences, we further identified putative RicA orthologs in other fungi and carried out alignments (see Fig. S1 in the supplemental materal) and phylogenetic analyses (Fig. 1B). The RIC8 ortholog is absent in the genomes of S. cerevisiae and plants (65). As presented, the A. nidulans RicA is close to that of A. niger, A. oryzae, and A. flavus, while the A. fumigatus RicA is close to the A. clavatus RicA. To characterize the ricA gene, levels of ricA mRNA at different time points in the life cycle of the two species were examined. As shown in Fig. 1C, in both species the ricA transcript (each ∼2.5 kb) was detectable in a somewhat undulant manner during growth and developmental phases, but not in conidia. It appears that AnricA mRNA is present at low levels in sexual spores (ascospores [Fig. 1C]).
Characterization of ricA in A. fumigatus.
To understand the function of RicA in A. fumigatus, we generated the AfricA deletion mutant (ΔAfricA) by replacing its coding region with AnpyrG+ (for AF293.1) or AnargB+ (for AF293.6; for further complementation information, see Table 1). Subsequently, by using a ΔAfricA::AnargB+ pyrG1 strain as a transformation host, we further generated AfricA-complemented strains by introducing the AfricA WT allele into the pyrG locus, via pNJ25 (57). We then examined the phenotypes of ΔAfricA (FNJ12), WT (AF293), and complemented (FNJ13) strains (Table 1). Most markedly, the ΔAfricA mutant exhibited highly restricted colony growth (about 20% of WT) and produced very few and abnormal conidia at the center of colonies (Fig. 2A). Conidiation at this point was less than 1% that of the WT at 5 days after point inoculation on solid MMG with YE. To test whether the ΔAfricA abnormal conidia were viable, we tested the rates of conidial germination on solid MMG plus 0.1% YE. As shown in Fig. 2B, WT conidia showed germination rates of 90% ± 1.78% (mean ± standard deviation) at 6 h and 100% at 12 h postinoculation. On the contrary, the ΔAfricA conidia displayed extremely delayed and defective germination: 0%, 2.9% ± 0.18%, and 34.5% ± 2.8% of conidia germinated at 6 h, 12 h, and 18 h, respectively (Fig. 2B), whereas only ∼35% of conidia germinated eventually (data not shown). These results indicate that AfRicA is necessary for the functionality (germination) of conidia.
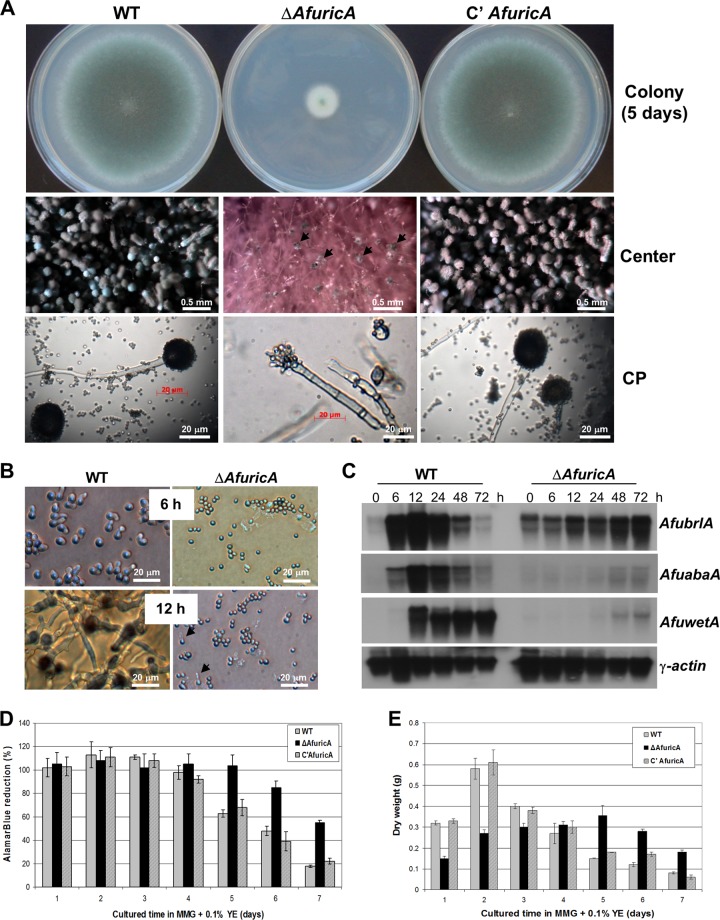
Fig 2.
Phenotypes caused by ΔAfricA. (A) WT (AF293), ΔAfricA (FNJ12), and complemented (C′ AfricA; FNJ13) strains were point inoculated on solid MMG plus 0.1% YE and incubated at 37°C for 5 days. Photographs of colony sizes and close-up views of the centers of the colonies and conidiophores are shown. Black arrows indicate abnormal conidiophores at the center of the ΔAfricA colony. (B) Germination of WT (AF293) and ΔAfricA (FNJ12) conidia inoculated on MMG plus 0.1% YE plates and incubated for 6 and 12 h. Black arrows in the image for ΔAfricA indicate germinated conidia at 12 h. (C) Accumulation of brlA, abaA, wetA, and γ-actin mRNA post-asexual developmental induction of WT (AF293) and ΔAfricA (FNJ12) strains. Development at 0 h indicates vegetative growth in MMG plus 0.1% YE liquid for 18 h. The A. fumigatus γ-actin gene (16, 29) was used as a control. (D) alamarBlue reduction data, indicating relative cell death rates. The mycelial aggregates of WT, ΔAfricA, and complemented strains were mixed with the AB reagent to check the cell viability for 7 days. (E) Dry weights of WT, ΔAfricA, and the complemented strain in MMG plus 0.1% YE submerged cultures were quantified for 7 days at 37°C, 220 rpm.
Aspergillus conidiophore formation requires sequential activities of the central regulatory components brlA, abaA, and wetA (2, 61, 70). As shown in Fig. 2A (middle and bottom panels), the ΔAfricA mutant produced abnormal conidiophores (black arrowheads) showing improper septation in stalks and incomplete formation of phialides and conidia. As shown by Northern blot analysis, the ΔAfricA mutant exhibited severely delayed and reduced levels of AfbrlA, AfabaA, and AfwetA mRNA during the progression of conidiation (Fig. 2C). In the WT, AfbrlA mRNA levels increased at 6 h, peaked at 12 h, began to decrease at 24 h, and became almost undetectable at 72 h post-developmental induction. Accumulation of the AfabaA and AfwetA transcripts followed the AfbrlA mRNA accumulation pattern. However, in the ΔAfricA mutant, AfbrlA mRNA accumulated at low levels at 0 h (vegetative growth at 16 h) and gradually increased until 72 h, i.e., there was reduced, delayed, and uncontrolled accumulation of AfbrlA. Moreover, transcripts of AfabaA and AfwetA accumulated at low levels even at 48 and 72 h post-developmental induction. These results indicate that AfRicA is necessary for proper expression and regulation of key developmental regulators that coordinate the formation, integrity, and vitality of spores.
Finally, we tested whether the deletion of AfricA affects cell death and autolysis in A. fumigatus by using AB reduction (57) and dry weight assays, respectively. As shown in Fig. 2D and E, the absence of AfRicA resulted in delayed cell death and autolysis. Whereas both WT and complemented strains exhibited reduced AB reduction rates at day 4 and levels of only 20% at day 7, the ΔAfricA mutant exhibited 100% AB reduction at day 5, a decreased rate from day 6 (to about 82%), and retained ∼58% AB reduction even at day 7 (Fig. 2D). Similarly, while WT and complemented strains exhibited maximum dry weight at day 2 and reduced dry weights from day 3, the ΔAfricA mutant showed a peak dry weight at day 5 and gradually reduced weight at days 6 and 7 (Fig. 2E). These findings suggest that AfRicA is necessary for proper vegetative proliferation and normal progression of cell death and autolysis in A. fumigatus.
Characterization of AnricA and cross-species complementation.
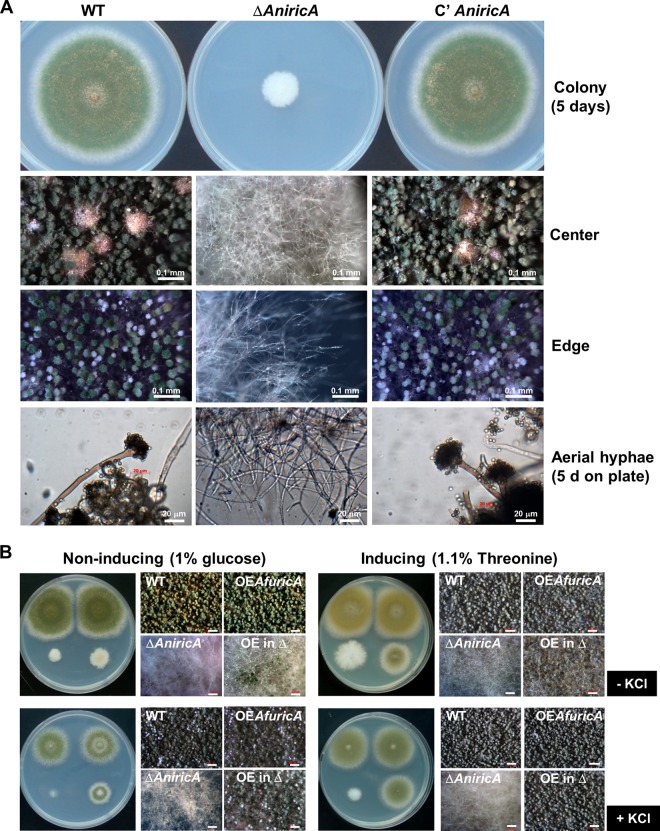
The deletion of AnricA resulted in severely impaired growth and a complete lack of asexual and sexual development in A. nidulans. As clearly noticeable from the colonies of WT (TNJ36), ΔAnricA (TNJ21), and complemented (TNJ87) strains grown on solid MMG at 37°C for 5 days, the ΔAnricA mutant formed a very small colony (∼30% of the WT colony diameter) composed of hyphae without any conidiophores, conidia, cleistothecia (sexual fruiting bodies), or Hülle cells (Fig. 3A). These growth and developmental defects could not be alleviated by changing the growth conditions, e.g., high salt (0.6 M KCl or 0.8 M NaCl), the lack of carbon source, sexual induction, or rich nutrient (0.5% YE) (data not shown). Thus, all further experiments employing the ΔAnricA mutant were done by collecting and inoculating hyphal fragments from a high number of air-exposed mutant colonies. In submerged shake cultures following inoculation of the ΔAnricA aerial hyphae, the ΔAnricA mutant exhibited extremely slow hyphal proliferation compared to the WT, and the size of ΔAnricA mycelial aggregates was only 15% of that of WT and complemented strains at 3 days (data not shown). We then asked whether introduction and/or overexpression of the AfricA WT allele could restore growth and development in the ΔAnricA mutant. We found that the presence of a genomic fragment of AfricA, including its own promoter, coding region, and terminator, was not sufficient to restore growth and development in the ΔAnricA mutant (data not shown). However, as shown in Fig. 3B, the overexpression of AfricA under the control of the AnalcA promoter partially enhanced growth, and there was fully restored conidiation in the ΔAnricA mutant under the inducing condition (MMT plus 0.5% YE). Supplementation with 0.6 M KCl enhanced the developmental restoration by AfricA in the ΔAnricA mutant grown on MMT plus 0.5% YE, and even on MMG (noninducing) (Fig. 3B, bottom, +KCl). These results indicate that AnRicA plays a crucial and essential role for growth and development in A. nidulans, and AfRicA can partially replace AnRicA.
Fig 3.
Requirement of RicA in A. nidulans growth and development. (A) A. nidulans WT (TNJ36), ΔAnricA (TNJ21), and complemented (C′ AnricA; TNJ87) strains were inoculated on MMG and cultured for 5 days at 37°C. Colonies and conidiophores of the 5-day-old culture on solid MMG were observed under a stereomicroscope. (B) Partial complementation by AfricA overexpression in ΔAnricA. WT (TNJ36), OEAfricA (TNJ94), ΔAnricA (TNJ21), and OEAfricA ΔAnricA (TNJ95; OE in Δ) were point inoculated on solid MMG (noninducing), MMT plus 0.5% YE (induction by the alcA promoter), MMG without KCl, or MMT plus 0.5% YE with 0.6 M KCl and incubated for 3 days 37°C. Bar, 200 μm.
Suppression of ΔricA by ΔrgsA in A. nidulans.
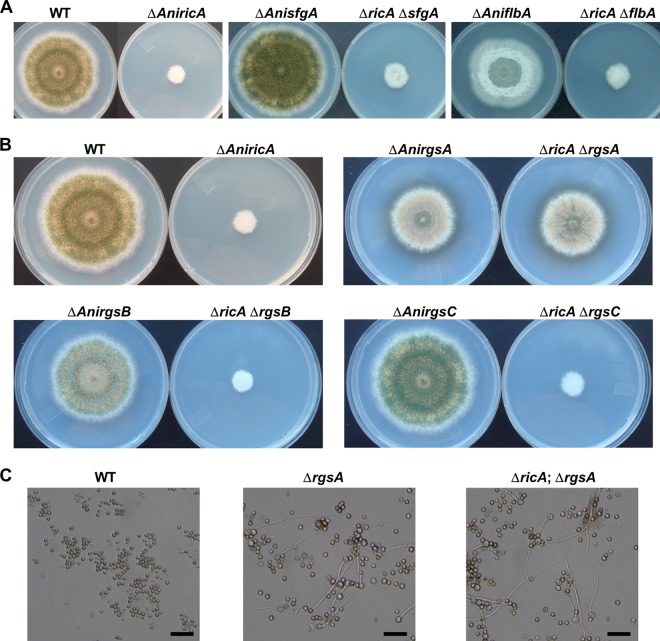
If RicA is an ortholog of Ric-8 in animals, it likely functions in activation of heterotrimeric G protein signaling (24). As RicA is clearly needed for growth and development, we carried out a series of double-mutant analyses to examine the genetic interactions of RicA with other key regulators in A. nidulans. The flbA gene encodes an RGS protein required for the attenuation of vegetative proliferation signaling mediated by FadA and activation of conidiophore development in Aspergillus (23, 31, 38, 74). SfgA (suppressor of fluG) is a negative regulator of conidiation, functioning downstream of FluG but upstream of other key developmental activators, including FlbD, FlbC, FlbB, and BrlA (52). As shown in Fig. 4A, the ΔricA ΔflbA and ΔricA ΔsfgA double mutants exhibited phenotypes identical to the ΔricA single mutant. These findings indicate that the removal of the key negative regulators of growth (FlbA) or conidiation (SfgA) could not alleviate the growth and developmental defects caused by ΔricA and that RicA functions either upstream or independently of the FlbA/FadA growth and FluG/SfgA developmental control pathways. We then asked whether RicA is associated with activation of other G protein pathways. RgsA is an RGS protein that inhibits GanB-mediated signaling for germination/developmental control and stress response (10, 21). RgsB and RgsC are putative RGS proteins that remain to be characterized (21). As shown in Fig. 4B, while the deletion of rgsB or rgsC failed to suppress ΔricA, the absence of rgsA restored growth and development to the ΔrgsA level in the ΔricA mutant. Furthermore, as found in the ΔrgsA mutant (21), the ΔricA ΔrgsA mutant conidia germinated proficiently in liquid medium lacking an external carbon source, whereas WT conidia did not show any sign of germination (Fig. 4C). These results indicate that RicA mediates signaling for growth and development, primarily through the RgsA/GanB signaling pathway. It might do so by activating GanB in the absence of the (unidentified) corresponding GPCR(s).
Fig 4.
Double mutant analyses in A. nidulans. (A) WT (TNJ36), ΔAnricA (TNJ21), ΔAnsfgA (TNJ57), ΔAnsfgA ΔAnricA (TNJ58) ΔAnflbA (TNJ42), and ΔAnflbA ΔAnricA (TNJ49) strains grown on solid MMG for 5 days at 37°C. (B) WT (TNJ36), ΔricA (TNJ21), ΔrgsA (TNJ61), ΔricA ΔrgsA (TNJ62), ΔrgsB (TNJ63), ΔricA ΔrgsB (TNJ64), ΔrgsC (TNJ65), and ΔricA ΔrgsC (TNJ66) were inoculated on MMG and incubated at 37°C for 5 days. (C) Germination of WT (TNJ36), ΔrgsA (TNJ61), and ΔricA ΔrgsA (TNJ62) conidia in the absence of an external carbon source. Photographs were taken after inoculating 1 × 106 conidia into liquid MM without an external C source and cultured for 16 h at 37°C and 220 rpm. Bar, 25 μm.
Physical interaction between RicA and GanB.
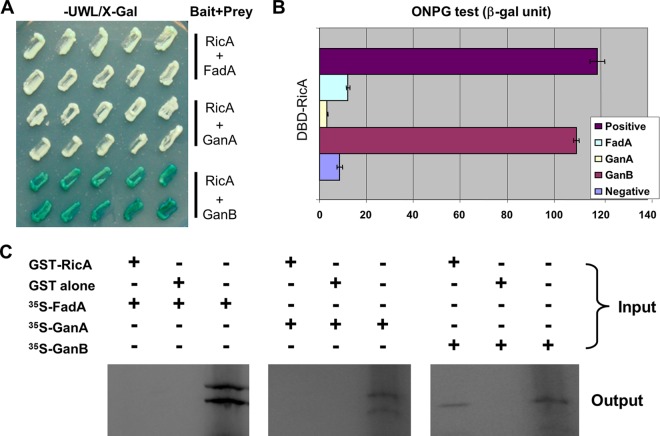
To further test the hypothesis that the primary target of AnRicA is AnGanB, we first tested the physical interaction between AnRicA and individual Gα subunits by employing a yeast two-hybrid assay. The AnricA ORF PCR fragment derived from cDNA was fused with the LexA DBD in the pTLexA vector (11), and each Gα ORF, AnFadA, AnGanA, and AnGanB, was fused with the Gal4 activation domain (Gal4 AD) in the pGAD424 vector (Clontech). Individual pairs of plasmids were introduced into the yeast and examined for levels of β-galactosidase reporter activity. As shown in Fig. 5A, only the AnRicA-AnGanB pair exhibited a blue color on the X-Gal SD medium lacking uracil, tryptophan, and leucine. Quantification of the β-galactosidase activity of each pair in yeast by using o-nitrophenyl-galactosidase (ONPG) further demonstrated that only the AnRicA-AnGanB pair resulted in high levels of reporter expression in yeast (Fig. 5B), i.e., about 90% of that of the well-known transcriptional activator AnAflR (positive control) (44, 71). To map the critical interacting domains, we further tested the physical interaction between the truncated AnRicA (aa 1 to 398 or 51 to 466, of the full-length 466 aa) and AnGanB (aa 1 to 325 or 35 to 356 of the full-length 356 aa), and we found that the full-length AnRicA and AnGanB are necessary for the interaction in yeast (data not shown). The physical interaction of AnRicA and AnGanB was further tested in vitro in a GST-pulldown assay (Fig. 5C). The AnricA ORF was fused with GST in the pGEX 5X-1 vector (GE Healthcare), and the AnRicA protein was expressed and purified in Escherichia coli. The ORF regions of AnfadA, AnganA, and AnganB were cloned under the T7 promoter of pCDNA3, and each Gα subunit was translated in vitro and labeled with 35S. An equal amount of in vitro-translated proteins was added to glutathione bead-GST-AnRicA or glutathione bead-GST (control) suspensions and subjected to pulldown. As shown in Fig. 5C, 35S-labeled AnGanB, but not AnFadA or AnGanA, could be copurified with GST-AnRicA specifically, indicating that AnRicA directly binds to AnGanB in vitro. Collectively, these data suggest that GanB is a primary target of RicA-mediated signaling in A. nidulans, and the full-length AnRicA and AnGanB are necessary for their physical interaction.
Fig 5.
RicA and GanB physically interact in yeast and in vitro. (A) Colony photographs of yeast strains expressing both the LexA DNA binding domain fused the AnRicA protein and the Gal4 activation domain fused to Gα proteins, AnFadA, AnGanA, and AnGanB on X-Gal medium without uracil, tryptophan, or leucine (-UWL/X-Gal). (B) Quantitative analyses of β-galactosidase activities, using ONPG in yeast strains, including the positive control (AflR) (71) and negative control (pTLex vector; LexA DNA binding domain alone), shown on the left. (C) GST pulldown assay for GST-AnRicA and in vitro-translated [35S]Gα proteins. The right lane of each panel shows the in vitro-translated FadA, GanA, and GanB proteins, respectively (10 μl from 50-μl translation reaction volumes). The in vitro-translated proteins were divided into two parts (each 20 μl) and mixed with the GST-AnRicA protein (left lane) or GST alone (middle lane). The expected protein sizes of FadA, GanA, and GanB were about 39, 40, and 39 kDa, respectively.
The RicA→GanB→PkaA signaling cascade.
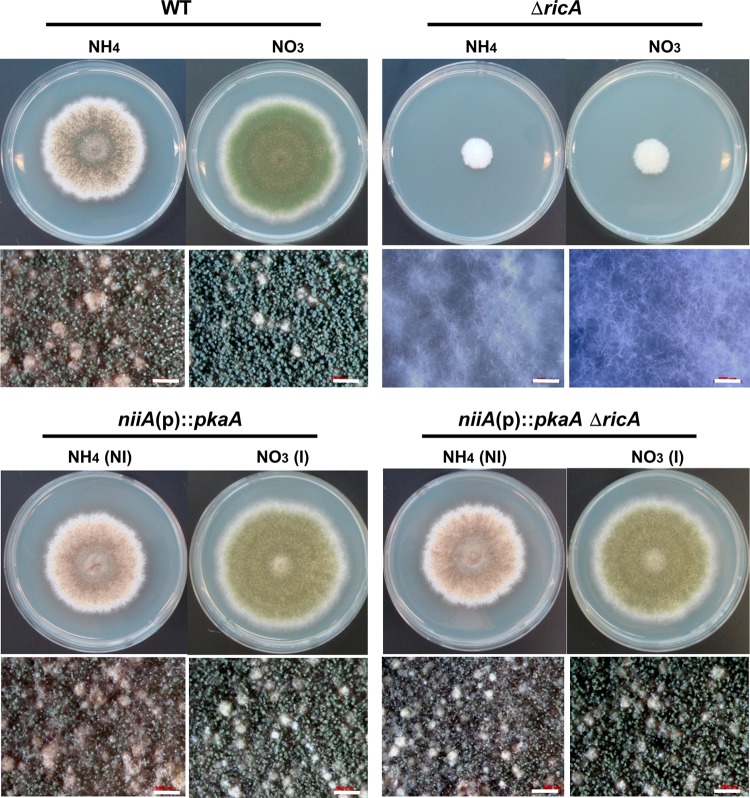
In A. nidulans, early events of conidial germination in response to sensing carbon sources are controlled by GanB (Gα) and SfaD::GpgA (Gβγ) and the cAMP-dependent protein kinase PkaA (17, 30, 56). As GanB has been proven to be a primary target of RicA, we asked whether the elevated expression of pkaA could restore growth and development in the ricA deletion mutant. While supplementation of exogenous (up to 10 mM) cAMP and dibutyryl cAMP failed to restore growth and development (data not shown), two copies (one native and an ectopic) or the overexpression of pkaA was sufficient to restore growth and development in the ΔricA mutant. As shown in Fig. 6, the niiA(p)::pkaA single and ΔricA niiA(p)::pkaA double mutants exhibited identical phenotypes on solid medium with 0.2% ammonium tartrate (noninducing) or 0.6% sodium nitrate (inducing) as a nitrogen source. The overexpression of pkaA regardless of the presence or absence of RicA resulted in enhanced production of aerial hyphae and a reduced density of conidia per unit area on inducing medium, as described by Shimizu and Keller (56). These results suggest that RicA likely mediates signaling through GanB→PkaA in A. nidulans.
Fig 6.
Suppression of ΔAnricA by an ectopic copy or overexpression of pkaA. Colony photographs (top) and close-up views (bottom) of A. nidulans WT (TNJ36), ΔAnricA (TNJ21), OEpkaA (TNJ89), and OEpkaA ΔAnricA (TNJ90) strains are shown. These strains were point inoculated on noninducing medium (NI; MMG containing 0.2% ammonium tartrate as a nitrogen source) or inducing medium (I; MMG containing 0.6% sodium nitrate as a nitrogen source) and incubated for 5 days at 37°C.
Unknown role of RicA-mediated signaling in A. nidulans development.
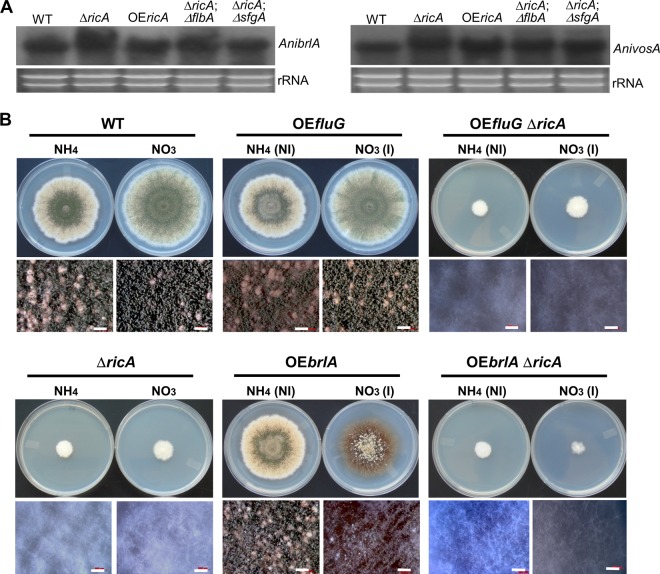
Finally, we checked whether the developmental defect caused by ΔricA is due to the defective expression of the key regulators brlA (1) and vosA (44). The mycelial mats of WT, ΔAnricA, niiA(p)::ricA (OEricA), ΔflbA, ΔflbA ΔricA, ΔsfgA, and ΔsfgA ΔricA strains grown on the surface of liquid stationary MMG were collected and subjected to Northern blot analyses. It is important to note that the mutant strains containing ΔricA were cultured for 3 days in order to generate enough hyphal mass on the surface of liquid MMG. Despite the complete lack of conidia, all mutants containing ΔricA showed comparable levels of brlA and vosA mRNAs (Fig. 7A), suggesting that expression of brlA is not sufficient to activate conidiation in the absence of RicA and that RicA-mediated signaling may provide a critical input for conidiophore development. This was further tested by overexpressing fluG and brlA in the absence of ricA. Neither OEfluG nor OEbrlA could restore conidiation in the ΔricA mutant on solid medium (Fig. 7B). Moreover, while OEbrlA ricA+ caused the formation of conidia at the hyphal tip in liquid submerged culture, the OEbrlA ΔricA double mutant showed only reduced mycelial growth without forming conidia (data not shown). These results suggest that expression of brlA is not sufficient for conidiation, and unknown RicA-mediated developmental signaling, likely independent of the fluG →brlA pathway, is necessary for conidiophore development in A. nidulans (Fig. 8).
Fig 7.
Requirement of AnricA in conidiation independent of fluG and brlA. (A) mRNA levels of brlA and vosA in WT (TNJ36), ΔricA (TNJ21), OEricA (TNJ88), ΔflbA ΔricA (TNJ49), and ΔsfgA ΔricA (TNJ58) cells collected from MMG liquid surface culture. Note the strains, including ΔricA, were cultured for 3 days on the surface of liquid MMG to obtain equal amounts of the hyphal aggregates. (B) WT, ΔricA (TNJ21), OEfluG (TNJ59), OEfluG ΔricA (TNJ60), OEbrlA (TNJ85), and OEbrlA ΔricA (TNJ86) strains were point inoculated on noninducing (NI; MMG with 0.2% ammonium tartrate) or inducing (I; MMG with 0.6% sodium nitrate) solid medium and incubated at 37°C for 5 days.
Fig 8.
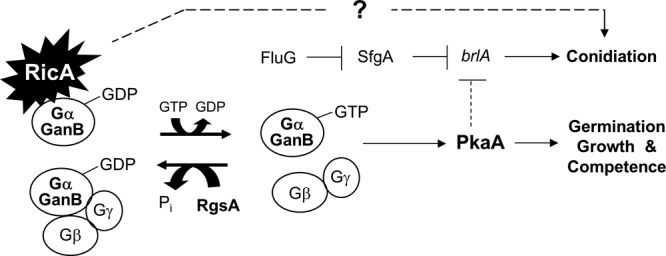
Model for how RicA governs A. nidulans growth and development (see the text). The putative GEF RicA governs upstream signaling for spore germination, vegetative growth, and development in Aspergillus, primarily through the GanB→PkaA signaling cascade. RicA is also (indirectly) required for conidiophore development, which may involve the acquisition of developmental competence via modulating GanB→PkaA signals. In addition, a potential direct role of RicA in activating conidiation, independent of FluG/BrlA, is indicated. It is further speculated that brlA is downregulated by GanB and PkaA during early phases of vegetative growth in A. nidulans.
DISCUSSION
During the past decade, novel families of proteins that can modulate the on-off state of G proteins have been identified, raising the complexity of the regulation of signal transduction (50). Among the newly identified components that may function as positive modulators of G proteins is Ric-8 (also known as synembryn [24]). Ric-8 is a cytoplasmic protein that was initially identified by a genetic screening of C. elegans mutants that are resistant to the cholinesterase inhibitor aldicarb (41) and by a yeast two-hybrid screen searching for interacting partners of mammalian Gα subunits (27, 60). Later, it was revealed that RIC-8 is involved in the asymmetric division of C. elegans embryos (3, 12, 42) and Drosophila melanogaster neuroblasts (13, 20, 62). The RIC-8 orthologs are present in genomes of animals and filamentous fungi, but not in baker's yeast or plants. Whereas the C. elegans and Drosophila genomes contain a single Ric-8 gene, mammals possess two Ric-8 orthologs, Ric-8A and Ric-8B (24). Unlike GPCRs, Ric-8 proteins cannot stimulate the guanine nucleotide exchange activity of the Gα subunit associated with Gβγ, i.e., the inactive heterotrimeric complex. Thus, Ric-8 proteins can only function on dissociated (free) monomeric Gα-GDP (Fig. 1A) (24, 60). It has been hypothesized that RIC-8 was acquired by animals and certain fungi after G proteins and GPCRs evolved and is therefore a fairly recent addition to G protein regulatory pathways (65).
In this report, we present experimental evidence that the Ric-8 ortholog (RicA) plays a crucial role in governing vegetative growth and development in two Aspergillus species. The lack of AnRicA function results in profound defects in hyphal proliferation and asexual/sexual fruiting. In particular, the AnricA null mutants were severely impaired in hyphal growth and unable to form conidia or ascospores in A. nidulans. Thus, the ΔAnricA cultures were derived from the mutant hyphal fragments, and all double mutants were generated by deleting AnricA from individual single mutants through transformation. Likewise, the ΔAfricA mutant produces defective conidia, many of which are unable to germinate, making it extremely difficult to study RicA function in both species. Whereas the primary structure of the two Aspergillus RicA proteins is highly conserved (75% identity, 89% similarity, and 0% gaps [http://blast.ncbi.nlm.nih.gov/Blast.cgi]), the overexpression of AfRicA only partially restored growth and development in A. nidulans. We found here that RicA interacts with the Gα subunit GanB, but not with FadA or GanA, in A. nidulans. Furthermore, RicA is incapable of forming a homodimer or a multimer (Fig. 5). Han et al. reported that the deletion of ganB, but not fadA or ganA, suppressed the developmental and metabolic defects caused by ΔrgsA, and they concluded that RgsA negatively regulates GanB-mediated signaling in A. nidulans (21). We found that RicA physically interacts with GanB, and only ΔrgsA suppresses the defective growth and development phenotypes caused by ΔricA; the ΔrgsA ΔricA double mutant exhibited a phenotype identical to the ΔrgsA single mutant (Fig. 4B and C). All these findings suggest that RicA-mediated signaling is transduced primarily via GanB in A. nidulans (Fig. 8).
Studies of the Ric-8 ortholog in Neurospora crassa (66) and Magnaporthe oryzae (35) have revealed that RIC-8 plays a highly conserved role in filamentous fungi. As found in Aspergillus, the N. crassa ric-8 (Ncric-8) deletion mutant shows severe defects in growth and development. The phenotypes caused by Ncric-8 are similar to those observed in the mutant lacking the Gα genes gna-1 and gna-3. Moreover, ΔNcric-8 results in greatly reduced levels of all three Gα subunits and one Gβ subunit (GNB-1) (66). Further studies have revealed that NcRIC8 positively regulates GNA-1 and GNA-3 and physically interacts with and acts as a GEF for GNA-1 (equivalent to AnFadA) and GNA-3 (AnGanB) in vitro, with the strongest effect on GNA-3. The rice blast fungus M. oryzae RIC-8 (MoRIC-8) is known to be a novel component of G protein signaling during infection into plants (35). The deletion of MoRIC-8 results in nonpathogenicity and impairment in cellular differentiation associated with sporulation, sexual development, and plant infection. MoRic-8 is highly expressed in the appressorium, a specialized plant tissue-invading structure, and physically interacts with MagB (AnFadA) but not MagA (AnGanB) in the yeast two-hybrid system. Collectively, these results indicate that Ric-8/RicA plays a crucial role in upstream activation of various signaling pathways in filamentous fungi and that the primary targets of RicA vary.
Studies in these three fungi (Aspergillus, Neurospora, and Magnaporthe) indicate that RIC-8-mediated signaling likely involves cAMP-dependent protein kinase signaling and might affect G protein levels. In N. crassa, ΔNcric-8 results in low levels of adenylyl cyclase protein. Moreover, ΔNcric-8 can be suppressed by a mutation in the PKA regulatory subunit (66). MoRic-8 is thought to act upstream of the cyclic AMP response pathway that is necessary for appressorium morphogenesis. In accordance with these concepts, we have shown that two copies and/or overexpression of the primary catalytic subunit of the cAMP-dependent protein kinase PkaA suppresses the profound defects caused by the lack of RicA function. Previous studies have also revealed that RIC-8 is also required for normal levels of various G proteins in Neurospora, Drosophila, C. elegans, and mammalian cells. Likewise, the deletion of MoRIC-8 causes the downregulation of Gα subunits, MAGA, MAGB, and MAGC, as well as the Gγ (MGG1) subunit, but not the Gβ subunit (MGB1) (35). Taken together, these results suggest that maintenance of normal levels of G proteins and adenylyl cyclase (and perhaps other yet-unknown regulatory components) are important in the conserved function of the RIC-8 protein (66) and that RicA-mediated signaling is transduced primarily through the GanB→PkaA pathway in Aspergillus (Fig. 8).
While the absence of RicA function essentially abolished conidiation in both species, mRNA of the key developmental activator brlA accumulated to some extent in both A. nidulans and A. fumigatus. Moreover, while abaA and wetA mRNA levels were very low in the A. fumigatus ricA null mutant, AnvosA mRNA accumulated in hyphal cells. Furthermore, neither the removal of the key repressor of conidiation SfgA (51, 52) nor the overexpression of the key activators FluG/BrlA rescued the developmental defects caused by the lack of RicA. This suggests that, in addition to the activation of brlA expression, an unknown RicA-mediated signaling input is essential for conidiation in A. nidulans. As the deletion of rgsA and the overexpression of PkaA restored the production of conidia in the ricA null mutant, we speculate that the RicA→GanB→PkaA signaling input is somehow (indirectly) necessary for the activation of conidiophore development (Fig. 8). As a possible explanation, we propose that RicA-mediated activation of GanB and PkaA signaling is necessary for the acquisition of developmental competence (reference 2 and references therein). As previously reported, conidiation does not usually occur in A. nidulans until cells have gone through a defined period of vegetative growth (reference 2 and references therein), supporting the hypothesis that the early aspect of Aspergillus conidiophore development occurs as an integral part of the life cycle rather than as a response to unfavorable environmental conditions. We speculate that the absence of ricA function abolishes transduction of signals for carbon source sensing, spore germination, vegetative proliferation, and thereby the acquisition of developmental competence; all are thought to be primarily mediated by the GanB→PkaA pathway (Fig. 8). In a previous model, it was proposed that GanB and PkaA signaling inhibited expression of brlA (10, 56). We further speculate that GanB and PkaA play a role in downregulating brlA expression in order to prevent precocious conidiation and to confer vegetative growth for a certain period of time during the life cycle of A. nidulans. Further investigation of RicA-mediated signaling and the PkaA downstream components that are associated with the developmental regulation/competence needs to be carried out in Aspergillus species.
Supplementary Material
ACKNOWLEDGMENTS
We thank our lab members for helpful discussions and Ellin Doyle for critically reviewing the manuscript.
This work was supported by National Science Foundation (IOS-0950850), USDA CSREES Hatch (WIS01195), and UW Food Research Institute grants to J.-H.Y. The work at Konkuk University was supported by a National Research Foundation of Korea grant funded by the Korean Government (NRF-2011-619-E0002) to S.J. The work at the Korea Advanced Institute of Science and Technology was supported in part by the Intelligent Synthetic Biology Center of Global Frontier Project (2011-0031955), funded by the Ministry of Education, Science and Technology, Republic of Korea, to S.C.K.
Footnotes
Published ahead of print 21 September 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Adams TH, Boylan MT, Timberlake WE. 1988. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54:353–362 [DOI] [PubMed] [Google Scholar]
- 2. Adams TH, Wieser JK, Yu J-H. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62:35–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Afshar K, et al. 2004. RIC-8 is required for GPR-1/2-dependent Gα function during asymmetric division of C. elegans embryos. Cell 119:219–230 [DOI] [PubMed] [Google Scholar]
- 4. Arst HN, Jr, Rand KN, Bailey CR. 1979. Do the tightly linked structural genes for nitrate and nitrite reductases in Aspergillus nidulans form an operon? Evidence from an insertional translocation which separates them. Mol. Gen. Genet. 174:89–100 [DOI] [PubMed] [Google Scholar]
- 5. Bastiani C, Mendel J. 2006. Heterotrimeric G proteins in C. elegans. WormBook (13 October 2006), p 1–25 The C. elegans Research Community, http://www.wormbook.org. doi:10.1895/wormbook.1.75.1 [DOI] [PMC free article] [PubMed]
- 6. Birnbaumer L. 1990. G proteins in signal transduction. Annu. Rev. Pharmacol. Toxicol. 30:675–705 [DOI] [PubMed] [Google Scholar]
- 7. Birnbaumer L. 2007. Expansion of signal transduction by G proteins. The second 15 years or so: from 3 to 16 α subunits plus βγ dimers. Biochim. Biophys. Acta 1768:772–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brent R, Ptashne M. 1985. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell 43:729–736 [DOI] [PubMed] [Google Scholar]
- 9. Brookman JL, Denning DW. 2000. Molecular genetics in Aspergillus fumigatus. Curr. Opin. Microbiol. 3:468–474 [DOI] [PubMed] [Google Scholar]
- 10. Chang M-H, Chae K-S, Han D-M, Jahng K-Y. 2004. The GanB Gα-protein negatively regulates asexual sporulation and plays a positive role in conidial germination in Aspergillus nidulans. Genetics 167:1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cho J-H, et al. 2003. Identification and cloning of jipA encoding a polypeptide that interacts with a homolog of yeast Rad6, UVSJ in Aspergillus nidulans. J. Microbiol. 41:46–51 [Google Scholar]
- 12. Couwenbergs C, Spilker AC, Gotta M. 2004. Control of embryonic spindle positioning and Gα activity by C. elegans RIC-8. Curr. Biol. 14:1871–1876 [DOI] [PubMed] [Google Scholar]
- 13. David NB, et al. 2005. Drosophila Ric-8 regulates Galphai cortical localization to promote Gαi-dependent planar orientation of the mitotic spindle during asymmetric cell division. Nat. Cell Biol. 7:1083–1090 [DOI] [PubMed] [Google Scholar]
- 14. d'Enfert C. 1996. Selection of multiple disruption events in Aspergillus fumigatus using the orotidine-5′-decarboxylase gene, pyrG, as a unique transformation marker. Curr. Genet. 30:76–82 [DOI] [PubMed] [Google Scholar]
- 15. De Vries L, Zheng B, Fischer T, Elenko E, Farquhar MG. 2000. The regulator of G protein signaling family. Annu. Rev. Pharmacol. Toxicol. 40:235–271 [DOI] [PubMed] [Google Scholar]
- 16. Fidel S, Doonan JH, Morris NR. 1988. Aspergillus nidulans contains a single actin gene which has unique intron locations and encodes a gamma-actin. Gene 70:283–293 [DOI] [PubMed] [Google Scholar]
- 17. Fillinger S, Chaveroche MK, Shimizu K, Keller N, d'Enfert C. 2002. cAMP and ras signalling independently control spore germination in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 44:1001–1016 [DOI] [PubMed] [Google Scholar]
- 18. Goldsmith ZG, Dhanasekaran DN. 2007. G protein regulation of MAPK networks. Oncogene 26:3122–3142 [DOI] [PubMed] [Google Scholar]
- 19. Gwynne DI, et al. 1987. Comparison of the cis-acting control regions of two coordinately controlled genes involved in ethanol utilization in Aspergillus nidulans. Gene 51:205–216 [DOI] [PubMed] [Google Scholar]
- 20. Hampoelz B, Hoeller O, Bowman SK, Dunican D, Knoblich JA. 2005. Drosophila Ric-8 is essential for plasma-membrane localization of heterotrimeric G proteins. Nat. Cell Biol. 7:1099–1105 [DOI] [PubMed] [Google Scholar]
- 21. Han K-H, Seo J-A, Yu J-H. 2004. Regulators of G-protein signalling in Aspergillus nidulans: RgsA downregulates stress response and stimulates asexual sporulation through attenuation of GanB (Gα) signalling. Mol. Microbiol. 53:529–540 [DOI] [PubMed] [Google Scholar]
- 22. Harnett MM, Klaus GG. 1988. G protein regulation of receptor signalling. Immunol. Today 9:315–320 [DOI] [PubMed] [Google Scholar]
- 23. Hicks JK, Yu J-H, Keller NP, Adams TH. 1997. Aspergillus sporulation and mycotoxin production both require inactivation of the FadA G alpha protein-dependent signaling pathway. EMBO J. 16:4916–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hinrichs MV, Torrejon M, Montecino M, Olate J. 2012. Ric-8: Different cellular roles for a heterotrimeric G-protein GEF. J. Cell Biochem. 113:2797–2805 [DOI] [PubMed] [Google Scholar]
- 25. Ito H, Fukuda Y, Murata K, Kimura A. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kafer E. 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19:33–131 [DOI] [PubMed] [Google Scholar]
- 27. Klattenhoff C, et al. 2003. Human brain synembryn interacts with Gsα and Gqα and is translocated to the plasma membrane in response to isoproterenol and carbachol. J. Cell Physiol. 195:151–157 [DOI] [PubMed] [Google Scholar]
- 28. Kwon N-J, Garzia A, Espeso EA, Ugalde U, Yu J-H. 2010. FlbC is a putative nuclear C2H2 transcription factor regulating development in Aspergillus nidulans. Mol. Microbiol. 77:1203–1219 [DOI] [PubMed] [Google Scholar]
- 29. Kwon N-J, Shin K-S, Yu J-H. 2010. Characterization of the developmental regulator FlbE in Aspergillus fumigatus and Aspergillus nidulans. Fungal Genet. Biol. 47:981–993 [DOI] [PubMed] [Google Scholar]
- 30. Lafon A, Seo J-A, Han K-H, Yu J-H, d'Enfert C. 2005. The heterotrimeric G-protein GanBα-SfaDβ-GpgAγ is a carbon source sensor involved in early cAMP-dependent germination in Aspergillus nidulans. Genetics 171:71–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee BN, Adams TH. 1994. Overexpression of flbA, an early regulator of Aspergillus asexual sporulation, leads to activation of brlA and premature initiation of development. Mol. Microbiol. 14:323–334 [DOI] [PubMed] [Google Scholar]
- 32. Lee KH, Na DS, Kim JW. 1999. Calcium-dependent interaction of annexin I with annexin II and mapping of the interaction sites. FEBS Lett. 442:143–146 [DOI] [PubMed] [Google Scholar]
- 33. Lengeler KB, et al. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li L, Wright SJ, Krystofova S, Park G, Borkovich KA. 2007. Heterotrimeric G protein signaling in filamentous fungi. Annu. Rev. Microbiol. 61:423–452 [DOI] [PubMed] [Google Scholar]
- 35. Li Y, et al. 2010. MoRic8 is a novel component of G-protein signaling during plant infection by the rice blast fungus Magnaporthe oryzae. Mol. Plant Microbe Interact. 23:317–331 [DOI] [PubMed] [Google Scholar]
- 36. Liebmann B, Gattung S, Jahn B, Brakhage AA. 2003. cAMP signaling in Aspergillus fumigatus is involved in the regulation of the virulence gene pksP and in defense against killing by macrophages. Mol. Genet. Genomics 269:420–435 [DOI] [PubMed] [Google Scholar]
- 37. Liebmann B, Muller M, Braun A, Brakhage AA. 2004. The cyclic AMP-dependent protein kinase a network regulates development and virulence in Aspergillus fumigatus. Infect. Immun. 72:5193–5203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mah J-H, Yu J-H. 2006. Upstream and downstream regulation of asexual development in Aspergillus fumigatus. Eukaryot. Cell 5:1585–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McBride J, Ingram PR, Henriquez FL, Roberts CW. 2005. Development of colorimetric microtiter plate assay for assessment of antimicrobials against Acanthamoeba. J. Clin. Microbiol. 43:629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McKnight GL, et al. 1985. Identification and molecular analysis of a third Aspergillus nidulans alcohol dehydrogenase gene. EMBO J. 4:2093–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miller KG, et al. 1996. A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc. Natl. Acad. Sci. U. S. A. 93:12593–12598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miller KG, Rand JB. 2000. A role for RIC-8 (Synembryn) and GOA-1 (Goα) in regulating a subset of centrosome movements during early embryogenesis in Caenorhabditis elegans. Genetics 156:1649–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Neves SR, Ram PT, Iyengar R. 2002. G protein pathways. Science 296:1636–1639 [DOI] [PubMed] [Google Scholar]
- 44. Ni M, Yu J-H. 2007. A novel regulator couples sporogenesis and trehalose biogenesis in Aspergillus nidulans. PLoS One 2:e970 doi:10.1371/journal.pone.0000970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oldham WM, Hamm HE. 2008. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 9:60–71 [DOI] [PubMed] [Google Scholar]
- 46. Osmani AH, May GS, Osmani SA. 1999. The extremely conserved pyroA gene of Aspergillus nidulans is required for pyridoxine synthesis and is required indirectly for resistance to photosensitizers. J. Biol. Chem. 274:23565–23569 [DOI] [PubMed] [Google Scholar]
- 47. Pontecorvo G, Roper JA, Hemmons LM, Macdonald KD, Bufton AW. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141–238 [DOI] [PubMed] [Google Scholar]
- 48. Rosen S, Yu J-H, Adams TH. 1999. The Aspergillus nidulans sfaD gene encodes a G protein beta subunit that is required for normal growth and repression of sporulation. EMBO J. 18:5592–5600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Roze LV, Beaudry RM, Keller NP, Linz JE. 2004. Regulation of aflatoxin synthesis by FadA/cAMP/protein kinase A signaling in Aspergillus parasiticus. Mycopathologia 158:219–232 [DOI] [PubMed] [Google Scholar]
- 50. Sato M, Blumer JB, Simon V, Lanier SM. 2006. Accessory proteins for G proteins: partners in signaling. Annu. Rev. Pharmacol. Toxicol. 46:151–187 [DOI] [PubMed] [Google Scholar]
- 51. Seo J-A, Guan Y, Yu J-H. 2003. Suppressor mutations bypass the requirement of fluG for asexual sporulation and sterigmatocystin production in Aspergillus nidulans. Genetics 165:1083–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Seo J-A, Guan Y, Yu J-H. 2006. FluG-dependent asexual development in Aspergillus nidulans occurs via derepression. Genetics 172:1535–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Seo J-A, Han K-H, Yu J-H. 2005. Multiple roles of a heterotrimeric G-protein gamma-subunit in governing growth and development of Aspergillus nidulans. Genetics 171:81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shaaban MI, Bok JW, Lauer C, Keller NP. 2010. Suppressor mutagenesis identifies a velvet complex remediator of Aspergillus nidulans secondary metabolism. Eukaryot. Cell 9:1816–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sherman F. 1991. Getting started with yeast. Methods Enzymol. 194:3–21 [DOI] [PubMed] [Google Scholar]
- 56. Shimizu K, Keller NP. 2001. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157:591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shin K-S, et al. 2009. Differential roles of the ChiB chitinase in autolysis and cell death of Aspergillus nidulans. Eukaryot. Cell 8:738–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Simon MI, Strathmann MP, Gautam N. 1991. Diversity of G proteins in signal transduction. Science 252:802–808 [DOI] [PubMed] [Google Scholar]
- 59. Szewczyk E, et al. 2006. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1:3111–3120 [DOI] [PubMed] [Google Scholar]
- 60. Tall GG, Krumins AM, Gilman AG. 2003. Mammalian Ric-8A (synembryn) is a heterotrimeric Gα protein guanine nucleotide exchange factor. J. Biol. Chem. 278:8356–8362 [DOI] [PubMed] [Google Scholar]
- 61. Tao L, Yu J-H. 2011. AbaA and WetA govern distinct stages of Aspergillus fumigatus development. Microbiology 157:313–326 [DOI] [PubMed] [Google Scholar]
- 62. Wang H, et al. 2005. Ric-8 controls Drosophila neural progenitor asymmetric division by regulating heterotrimeric G proteins. Nat. Cell Biol. 7:1091–1098 [DOI] [PubMed] [Google Scholar]
- 63. Waring RB, May GS, Morris NR. 1989. Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin-coding genes. Gene 79:119–130 [DOI] [PubMed] [Google Scholar]
- 64. Wieser J, Yu J-H, Adams TH. 1997. Dominant mutations affecting both sporulation and sterigmatocystin biosynthesis in Aspergillus nidulans. Curr. Genet. 32:218–224 [DOI] [PubMed] [Google Scholar]
- 65. Wilkie TM, Kinch L. 2005. New roles for Gα and RGS proteins: communication continues despite pulling sisters apart. Curr. Biol. 15:R843–R854 [DOI] [PubMed] [Google Scholar]
- 66. Wright SJ, Inchausti R, Eaton CJ, Krystofova S, Borkovich KA. 2011. RIC8 is a guanine-nucleotide exchange factor for Gα subunits that regulates growth and development in Neurospora crassa. Genetics 189:165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Xue T, Nguyen CK, Romans A, Kontoyiannis DP, May GS. 2004. Isogenic auxotrophic mutant strains in the Aspergillus fumigatus genome reference strain AF293. Arch. Microbiol. 182:346–353 [DOI] [PubMed] [Google Scholar]
- 68. Yelton MM, Hamer JE, de Souza ER, Mullaney EJ, Timberlake WE. 1983. Developmental regulation of the Aspergillus nidulans trpC gene. Proc. Natl. Acad. Sci. U. S. A. 80:7576–7580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yu J-H. 2006. Heterotrimeric G protein signaling and RGSs in Aspergillus nidulans. J. Microbiol. 44:145–154 [PubMed] [Google Scholar]
- 70. Yu J-H. 2010. Regulation of development in Aspergillus nidulans and Aspergillus fumigatus. Mycobiology 38:229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yu J-H, et al. 1996. Conservation of structure and function of the aflatoxin regulatory gene aflR from Aspergillus nidulans and A. flavus. Curr. Genet. 29:549–555 [DOI] [PubMed] [Google Scholar]
- 72. Yu J-H, et al. 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41:973–981 [DOI] [PubMed] [Google Scholar]
- 73. Yu J-H, Keller N. 2005. Regulation of secondary metabolism in filamentous fungi. Annu. Rev. Phytopathol. 43:437–458 [DOI] [PubMed] [Google Scholar]
- 74. Yu J-H, Wieser J, Adams TH. 1996. The Aspergillus FlbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J. 15:5184–5190 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.