Abstract
The eukaryotic microbes known as oomycetes are common inhabitants of terrestrial and aquatic environments and include saprophytes and pathogens. Lifestyles of the pathogens extend from biotrophy to necrotrophy, obligate to facultative pathogenesis, and narrow to broad host ranges on plants or animals. Sequencing of several pathogens has revealed striking variation in genome size and content, a plastic set of genes related to pathogenesis, and adaptations associated with obligate biotrophy. Features of genome evolution include repeat-driven expansions, deletions, gene fusions, and horizontal gene transfer in a landscape organized into gene-dense and gene-sparse sectors and influenced by transposable elements. Gene expression profiles are also highly dynamic throughout oomycete life cycles, with transcriptional polymorphisms as well as differences in protein sequence contributing to variation. The genome projects have set the foundation for functional studies and should spur the sequencing of additional species, including more diverse pathogens and nonpathogens.
INTRODUCTION
Oomycetes are best known for their plant pathogens but also include saprophytes and colonizers of insects, vertebrates, and microbes. Oomycetes outwardly resemble fungi since both exhibit hyphal growth and heterotrophic absorptive nutrition and they reside in similar ecological niches. Oomycetes were misclassified as Mycota until the last part of the 20th century but are now accepted to have a distinct evolutionary history and to belong to the kingdom Stramenopila, which also includes brown algae and diatoms (7). One theory joins stramenopiles with other protists into the Chromalveolata superkingdom, linked by an ancestral endosymbiosis of a photosynthetic alga (44).
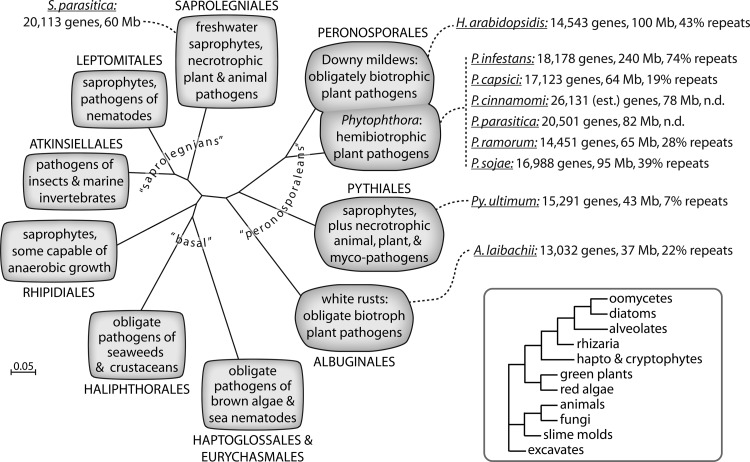
Of the hundreds of known oomycetes, those affecting agriculture and natural ecosystems are the most studied. Most fall into two large orders, the Peronosporales and Saprolegniales (Fig. 1). Most notorious is the potato and tomato late blight agent Phytophthora infestans, a peronosporalean that triggered the Irish Famine in the 1840s and still has major impact worldwide (22). Phytophthora contains over 100 species, all plant pathogens (48). Many are relatively recent discoveries, such as the Sudden Oak Death agent Phytophthora ramorum that has severely damaged woodlands in North America and Europe (29). No Phytophthora species is known to persist as a saprophyte, in contrast to its sister genus Pythium, in which most members are saprophytic and opportunistic phytopathogens. Mycoparasites and an animal pathogen also populate Pythium. Saprolegnian genera, like those in the Peronosporales, also include saprophytes and plant pathogens, but there is a greater tendency to an aquatic lifestyle. Most species within Achlya and Saprolegnia, for example, are usually innocuous inhabitants of freshwater ponds and streams, in which they grow on dead vegetable matter. They can, however, also parasitize a range of aquatic animals, particularly when immune systems are stressed. Saprolegnia parasitica, in particular, has become a growing problem in commercial aquaculture, in which high fish density and imperfect water quality increase stress and favor disease (88).
Fig 1.
Evolutionary history of oomycetes, lifestyles, and genome sequencing overview. The main part of the figure portrays phylogenetic relationships between the major orders based on 28S rRNA sequences. Orders comprising the peronosporalean group (∼1,000 species), saprolegnians (∼500 species), and basal clades are noted (8). While Phytophthora was traditionally considered part of the Pythiales, it is now recognized to belong to the Peronosporales along with the downy mildews (70). Species with publicly released genome data are listed along with their predicted gene number, genome size, and repetitive DNA content if known; values are based on the most recent publication or data on websites. n.d., the percentage of repeats was not determined. The phylogram in the lower right shows the position of oomycetes compared to other eukaryotes.
Much diversity in the nature of pathogenesis is displayed by different oomycetes. Species such as P. infestans and the soybean pest Phytophthora sojae colonize only a limited number of hosts. Other Phytophthora spp., such as P. ramorum, which infects over 60 trees and shrubs, and Phytophthora cinnamomi, which has over 1,000 hosts spanning both herbaceous and woody plants, are more cosmopolitan (19). Broad host range is also typical of most species of Pythium and Saprolegnia; the latter, for example, infects both fish and crustaceans. In contrast, with a few exceptions, each downy mildew and white rust colonizes only a single type of host. The white rust Albugo laibachii and the downy mildew Hyaloperonospora arabidopsidis infect only Arabidopsis thaliana, for example. Besides diversity in host range, pathogenic interactions involving oomycetes are also varied, ranging from biotrophy to necrotrophy. Pythium is largely necrotrophic, extracting resources from dying cells. At the other extreme, white rusts and downy mildews feed on living cells and do not directly kill their hosts; one Albugo species is even reported as an asymptomatic endophyte (64). Phytophthora are hemibiotrophs, which are biotrophic initially but shift to necrotrophy at the end of the disease cycle.
Genome sequences are now available for several oomycetes and are valuable for resolving long-standing issues about their biology and evolution. Questions that can be addressed include the following. What accounts for the diversity of oomycete lifestyles? How do the genes that oomycetes use to infect hosts compare to those of pathogens in other taxa? To what extent has horizontal gene transfer affected the pathogenic, metabolic, or regulatory characteristics of oomycetes? Do oomycete genomes have features that help oomycetes evolve to survive environmental changes or defeat efforts to control them?
Genome size and topography.
Oomycete genomes are estimated to range in size from a low of about 37 Mb, as in the cases of Pythium sylvaticum and A. laibachii, to a high of 280 Mb, as in some Phytophthora spp. Repetitive DNA, mostly in the form of transposable elements (TEs), is responsible for the bulk of variation (45, 55, 65). Assemblies annotated with gene models are now available publicly for nine plant pathogens, including six Phytophthora spp. (Phytophthora capsici, P. cinnamomi, P. infestans, Phytophthora parasitica, P. ramorum, and P. sojae), Pythium ultimum, H. arabidopsidis, and A. laibachii (6, 31, 45, 50, 51, 86). Data are also released for one animal pathogen, S. parasitica, but not for an exclusively saprophytic species. Predicted gene contents range from about 13 thousand to 26 thousand genes (Fig. 1). A striking feature is the wide variation in repetitive DNA content, which ranges from 7% for P. ultimum to 74% for P. infestans. Some correlation exists between gene number and genome size in P. infestans, P. ramorum, and P. sojae, but the trend loosens when considering other species. The recently released sequence of P. cinnamomi, for example, includes 26,131 predicted genes within a 78-Mb genome, compared to 16,988 genes for the 95-Mb genome of P. sojae. Initial gene counts are often overestimates; however, it is intriguing to consider that the large number of genes in P. cinnamomi relates to its ability to infect more than a thousand plant species.
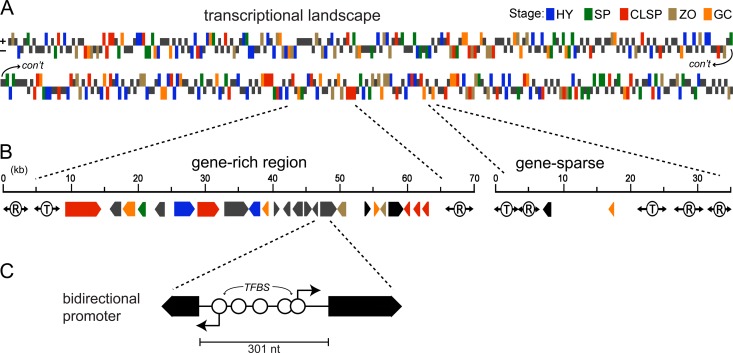
Genome-wide ortholog mapping suggests that a core set of about 8,000 to 9,500 genes is conserved between species (31, 75). The variable component thus represents nearly half of some proteomes. Interspecific differences are largely attributable to family expansion and/or gene loss, which, to some extent, relate to the novel partitioning of oomycete genomes into gene-dense and gene-sparse regions. In P. infestans, for example, about 80% of genes reside in clusters of tightly spaced genes, with the rest distant to each other (∼2 or more kb). This is illustrated in Fig. 2B for a portion of the P. infestans genome, which contains a gene-rich cluster of 24 genes with small intergenic distances near a gene-sparse region with larger intergenic distances. Insight toward the diversity of pathogenic lifestyles has come from studying the gene-sparse regions, as these are enriched for loci encoding host-modulating “effectors” (31, 66). Many genes in such regions have evolved faster than average in sequence and copy number, possibly aided by the nearby repeated DNA that fosters illegitimate recombination and a low gene density that minimizes lethal events.
Fig 2.
Structural and transcriptional landscape of the P. infestans genome. (A) Expression patterns of genes in a portion of a chromosome. Genes with >2-fold-higher mRNAs in one life stage than on average are colored based on when expression is highest according to the key in the upper right (HY, hyphae; SP, sporangia; CLSP, sporangia cleaving into zoospores; ZO, swimming zoospores; GC, zoospore cysts germinating and making infection structures, e.g., appressoria). Genes not changing are in dark gray at half height. Note that the scaffold is split into two portions and that the horizontal axis represents gene order and not distance. (B) Representative gene-dense and gene-sparse regions. Shown are gene orientations, expression patterns as in panel A, and flanking DNA transposon (T) or retroelement-like (R) sequences. The two regions are separated by 450 kb, which is also gene sparse. (C) Example of closely spaced opposing promoters from genes with strong EST support from panel B. The 301-nt intergenic region includes a combined 142 nt of 5′ untranslated regions (UTRs) and has five predicted transcription factor binding sites (TFBS). The genes have a similar configuration in P. sojae.
Some clusters of repeated genes can nevertheless be found in gene-dense regions, including some protein kinases and genes involved in sexual development (14, 34). P. infestans encodes 354 proteins with classic eukaryotic protein kinase domains, with other members of the genus having similar numbers. As with some other gene families, unequal crossing over appears to have contributed to its expansion, as about 20% of kinase loci reside in clusters of 2 to 13 genes. The same clusters tend to occur in other members of the genus, indicating their presence in the last common ancestor of Phytophthora, and a few are shared with H. arabidopsidis. The number of kinases in different oomycetes varies dramatically, ranging from less than 100 in P. ultimum to about 500 in S. parasitica.
Transcriptional landscape.
A typical oomycete life cycle involves vegetative filamentous growth followed by sporulation, spore germination, and, for pathogens, the formation of structures used to enter and feed from host tissues (37). Most species also form both sexual and asexual spores. mRNA profiles exhibit dynamic changes throughout these developmental transitions, with many genes expressed specifically in spore or infection stages. In P. infestans, mRNA levels for nearly one-half of genes change >2-fold during life stage changes, with about 14% being expressed exclusively in one stage or another (35). Studies in P. infestans, the lima bean pathogen Phytophthora phaseoli, and the cucumber downy mildew agent Pseudoperonospora cubensis also documented changes during different stages of growth (31, 49). Although P. infestans and P. cubensis have different lifestyles (hemibiotrophic and obligately biotrophic, respectively), most orthologs had similar expression patterns, suggesting that the nonorthologous genes may play roles in determining the mode of pathogenesis (71).
Many differentially expressed genes have functions that are logically required at only some stages of the life cycle. Examples include structural components of asexual or sexual spores, cell cycle proteins that may act to establish spore dormancy, and pathogenicity factors. Others, however, such as new isoforms of metabolic enzymes, have roles that are more challenging to discern. Many transcription factors also show strong differential expression, especially those in the bZIP and Myb families which have patterns suggestive of a transcription factor cascade (35, 71, 92).
Most oomycete genes are tightly spaced, with median intergenic distances being only 435 nucleotides (nt) within the gene-dense regions of P. infestans. In comparison, median intergenic regions in Saccharomyces cerevisiae, Arabidopsis thaliana, and Homo sapiens are 0.45, 1.5, and 35 kb, respectively (13, 47, 94). It is remarkable that despite the close spacing of genes in Phytophthora, neighboring genes typically lack similar patterns of developmental regulation. This can be seen in Fig. 2A, in which each color indicates upregulation in a particular developmental stage, with dark gray representing constitutive expression. Tandemly duplicated genes are frequently coexpressed, as illustrated in Fig. 2B, in which the right border of the gene-rich interval contains a tandemly arrayed coexpressed cluster. In contrast, other adjacent genes are not usually upregulated in the same stage. The transcriptional independence of most neighboring genes in Phytophthora contrasts with that of many eukaryotes, in which genes are often organized into coexpressed domains (40, 85, 94). The main mechanism for regulating transcription during oomycete development may therefore not involve chromatin level effects, which in yeasts and metazoans can influence genes within a 5- to 50-kb region (23).
One effect of high gene density in oomycetes appears to be a bias in gene orientation. Within the gene-dense regions of P. infestans, only about 40% of loci are transcribed divergently, i.e., head to head; this is much less than expected by random chance and less than in yeasts and plants. An example of genes in this orientation, for which the intergenic region is only 301 nt, is shown in Fig. 2C. About 10% of such closely spaced genes show some degree of correlated or anticorrelated expression (69). Oomycete promoters therefore not only are compact but may overlap and possibly antagonize their neighbors.
Intergenic distances are also small between the 3′ ends of genes transcribed toward each other, averaging 255 nt within the gene-dense regions of P. infestans (69). Such regions must include nearby transcriptional terminators, a configuration which may be mechanistically efficient. This also may generate overlapping transcripts for epigenetic regulation. A role for the latter is demonstrated in fungi and metazoans (16), and it would be interesting to see if this also holds true for oomycetes.
The tight spacing of genes may also help stabilize genomes. Insertion of a transposable element or any illegitimate recombination event within such a region would likely be lethal and eliminated quickly from the population. In contrast, if intergenic distances were large, then insertions would be tolerable in the short-term but might foster rearrangements that would be catastrophic during sexual reproduction. The selection for genome stability may counterbalance the normal propensity of transposable elements to insert within regions of high transcriptional activity, in which a higher fraction of DNA is uncoiled and exposed (21). The same principle likely underlies the expansion of the gene-poor regions of oomycete genomes, as these more than the gene-dense regions can tolerate transposon-mediated expansion.
The small size of most intergenic regions can also be helpful for identifying regulatory motifs through mutagenesis or bioinformatics (1, 69, 81, 93). For example, by searching just upstream of open reading frames for overrepresented motifs, over 100 putative transcription factor binding sites were able to be identified from Phytophthora, of which many were verified by functional assays (69). Most were within 200 nt of the transcription start, consistent with the compactness and potential simplicity of oomycete promoters. Nevertheless, sequenced Phytophthora spp. are predicted to encode between 543 and 669 transcription factors, similar to the number for fungal phytopathogens such as Fusarium graminearum and Magnaporthe oryzae (659 and 481, respectively [63]). H. arabidopsidis, however, has fewer predicted transcription factors at 369. This may reflect its simplified life cycle compared to that of Phytophthora spp.; while the latter form sporangia that release zoospores that encyst and then form germ tubes, H. arabidopsidis conidia germinate directly and lack the zoospore stage.
Most oomycete genes have few introns, although intron-rich genes are common. Means of about 1.6 per gene are predicted for Phytophthora and Pythium, although a lower number was reported for the subset of P. sojae genes with expressed sequence tag (EST) support (51, 77, 86). Many cases of “missplicing,” in which unexcised introns or alternate junctions may lead to premature translational termination, exist (77). There is, however, only one credible report of alternate splicing generating a new biological function. This comes from P. cubensis, for which the novel product had a new transporter activity (72).
Novel gene fusions.
Oomycetes are unusually rich in proteins with novel combinations of domains (59, 74). These have several predicted functions, which often involve signaling. Two notable examples are separate families of phosphatidylinositol-4-phosphate kinases and aspartic proteases with transmembrane domains characteristic of G protein-coupled receptors (43, 58). While their structures are mostly unconfirmed by cDNA or proteomic analysis, most appear conserved throughout Phytophthora. Protein kinases also have evolved domain combinations that are oomycete specific or seen only in closely related groups. One example found in Phytophthora, A. laibachii, H. arabidopsidis, and S. parasitica is a mitogen-activated protein (MAP) kinase with an N-terminal PAS signal-sensing domain (34). This combination does not appear in other eukaryotes, including diatoms or chromalveolates. Another unusual protein joins an N-terminal cyclin-dependent kinase with a C-terminal cyclin, which in other taxa are on separate proteins. This unusual configuration is detected in most oomycetes but otherwise appears only in apicomplexans and ciliates, which are chromalveolates.
Due to the tight spacing between oomycete genes, viable fusions can easily result from the loss of a stop codon in an upstream gene. The diploidy of oomycetes provides a setting in which innovative combinations of domains can persist for long periods during which their benefits can be tested or be eliminated by gene conversion during asexual growth or replacement with the wild-type allele during sexual reproduction.
Genes for pathogenesis and host colonization.
After an oomycete attaches to a plant or animal host, several events are required for successful infection. The invader must degrade host barriers such as cell walls, usually by secreting hydrolytic enzymes. Nutrients must be acquired, which, for necrotrophs such as P. ultimum, involves taking molecules leaked from damaged host cells. In contrast, most biotrophs and hemibiotrophs are assumed to acquire much of their nutrients through haustoria, which are specialized hyphae that insert into a host cell while remaining external to its plasma membrane. Movement through haustoria is presumably bidirectional; not only are nutrients probably moved from the plant, but oomycete proteins move across the interface to alter the physiology of the host, including its immune system. It should be noted that data on the role of oomycete haustoria in nutrient acquisition are limited (3), although infection-specific transporters can be identified from microarray data.
Analyses of the Phytophthora and P. ultimum genomes indicate that they are largely autotrophic and can use diverse molecules as substrates for metabolism. As described later, obligate biotrophs are deficient in several pathways. Haustorial oomycetes (A. laibachii, H. arabidopsidis, Phytophthora) have lost the gene for thiamine biosynthesis. The gene is present in P. ultimum and S. parasitica, which, as necrotrophs, are not thought to employ haustoria (84). While it seems that haustorial oomycetes have chosen to take this vitamin from their host, this is not the case for rust fungi such as Uromyces, which makes its own thiamine in haustoria (78).
Most plant-pathogenic oomycetes cannot make sterols due to the absence of the relevant biosynthetic genes. An exception is the legume root pathogen Aphanomyces euteiches, in which a biosynthetic pathway was predicted from an expressed sequence tag (EST) project and confirmed biochemically (53). The status of the sterol pathway has been a topic of long-standing interest, since such compounds are needed for sexual reproduction. Also, while the agrochemical industry has found sterol biosynthetic enzymes to be frequent fungicide targets, this has not been the case for oomycetes. It follows that a practical application of genome sequencing is the identification of targets for chemicals to defend against oomycete pathogens.
Plant defenses against oomycetes include producing enzymes that degrade the pathogen's cell wall. Oomycete proteins inhibiting these were first identified from EST projects and later catalogued from whole genomes. P. infestans, for example, encodes 38 secreted Kazal-like and cystatin protease inhibitors, some of which have been shown to suppress host apoplastic serine and cysteine proteases, respectively. These inhibitor-like genes are present in moderately reduced numbers in the other Phytophthora spp. and P. ultimum (51). The phytopathogenic oomycetes also make inhibitors of plant β-1,3-glucanases (15).
Oomycetes themselves use proteases and other host-degrading enzymes for offense, similar to other phytopathogens. Many, such as cysteine proteases, which average 37 in Phytophthora and 42 in P. ultimum, have similar numbers in Phytophthora and P. ultimum. Others, such as cutinases and pectin esterases, which have about 7 to 12 copies in each Phytophthora species but none in P. ultimum, vary widely. This may be because the latter infects primarily young, nonsuberized tissue while Phytophthora can colonize plant parts with thick cuticles. In contrast, lipases and subtilisin-related serine and aspartyl proteases are more common in P. ultimum. As expected, the opportunistic fish pathogen S. parasitica is largely deficient in enzymes for degrading plant cell walls, although its genome is annotated with two polygalacturonases. The latter might be maintained for saprophytic growth on plant debris.
Whether an oomycete consumes plants or animals, host attachment is required for infection; binding to a growth substrate must also be useful for saprophytes. Oomycetes express several factors that may act in this role, including glue-like proteins released by zoospores, mucins, thrombospondin repeat proteins, jacalin domain proteins, and cellulose-binding proteins, including CBELs, which genome data indicate are made throughout oomycetes (25, 31, 68, 84). Interestingly, CBELs contain a PAN/apple domain also used by parasitic apicomplexans (also chromalveolates) to bind their hosts. Recent discoveries in the P. ultimum and Phytophthora genomes also include cadherins, which play adhesion and secretory roles in animals (51). Except for CBELs, most of these lack proved roles in pathogenesis, and it should be noted that many oomycetes form adherent gametangia during sexual development which might also use such proteins (37).
Fast-evolving families of effectors.
Fungal, bacterial, and oomycete pathogens secrete proteins collectively known as effectors that modify their hosts. Some proteins described above are often also categorized as effectors, but much recent work has focused on families unique to oomycetes, especially the RXLRs and Crinklers/CRNs (41). The CRNs are named after a “crinkling” or necrotic phenotype that many cause when overexpressed in plants, and RXLR is named after an N-terminal motif (X is any amino acid) involved in uptake by host cells. Many RXLRs suppress plant immunity, such as Avr3a and AvrBlb2, which, respectively, stabilize an E3 ubiquitin ligase used in plant defense and block transport of a plant protease (10). The names of these two effectors indicate that they sometimes act as avirulence factors, which means that some plants encode resistance or R proteins that recognize them and respond with a cell death response (33). These proteins, but not necessarily all RXLRs, also contribute to pathogenesis against plant genotypes lacking the cognate R genes. The reader is directed to recent reviews for more details of their activities (10, 79).
RXLRs in particular represent a success story for bioinformatics. The family was recognized because of a short N-terminal motif shared between cloned avirulence proteins, as the rest of the proteins are very dissimilar. A conventional N-terminal signal peptide is required, and a nearby dEER motif often associates with RXLR. Early studies used that information to identify 563, 374, and 396 such proteins in P. infestans, P. ramorum, and P. sojae, respectively, and then 134 proteins in H. arabidopsidis. It was later observed that the original RXLR consensus may be too restrictive. For example, P. cubensis employs RXLQ and A. laibachii employs both RXLR and RXLQ (45, 82).
Identification of the RXLR motif stimulated searches of secretomes for conserved motifs and other hallmarks of host-translocated effectors. The CRNs were shown to have an LXLFLAK motif, Albugo spp. contained a family defined by a CHXC, and P. ultimum was predicted to encode a group with YXSL[RK] (45, 51, 73). Some of these, such as the CHXCs, which have 29 members in A. laibachii but much fewer in the other oomycetes, are not widely distributed. It should be stressed that outside A. laibachii and P. infestans, in which functional testing of the CHXCs and CRNs was done, these are just candidate effectors.
When examined at a genome-wide level, it is clear that effectors are quite variable within and between species. As noted before, their genes tend to reside in gene-poor regions in which DNA repeats may contribute to changes in family size, copy number, and sequence. Some RXLR genes, for example, exist in tandem arrays of near-identical copies, which vary in number in different strains (18). Comparisons of P. infestans with four close relatives that infect different plants found that RXLR genes and CRN genes were highly diversified, often through recombination within C-terminal domains which may confer new activities for adaptation to their hosts (65). Examinations of genome or EST data from other species have also identified many nonsynonymous changes that suggest an “arms race” between the pathogens and host resistance proteins that interact with the effectors (4, 45, 65, 73).
Not all oomycete pathogens employ RXLRs. While found in most sequenced peronosporalean genomes, RXLRs appear absent from P. ultimum. This probably relates to its necrotrophic lifestyle (in contrast, biotrophs and hemibiotrophs need effectors as part of stealthy infection strategies) and its broad host range (distinct RXLRs are probably needed for different hosts). Only a few candidates were found in the S. parasitica genome and in ESTs from the mycoparasite Pythium oligandrum, and none were found in ESTs of the basal oomycete and algal pathogen Eurychasma dicksonii and the legume root pathogen A. euteiches (25, 28, 32). However, inferences from EST studies are not definitive. For example, sterol carrier proteins known as elicitins are predicted from the S. parasitica genome sequence but were not detected in ESTs (84).
Another effector group of note is the NLP family, which encodes a phytotoxin active against dicots (26). Like those described above, it is diversified in copy number and sequence throughout the Oomycota. Unlike the others, NLPs represent an ancient family with members in bacteria and fungi. Counterintuitively, the necrotroph P. ultimum has about one-sixth the number of NLP genes of the hemibiotrophic Phytophthora spp., 7 versus about 45, respectively, excluding pseudogenes. A recent study in P. sojae, however, showed that only half of its NLP genes were expressed and only eight had necrosis-inducing activity (17). This means that a reduction in gene number does not necessarily translate to reduced protein levels, a finding which is an important warning to those making inferences from genome analyses without functional data.
Adaptations for obligate parasitism.
Downy mildews and white rusts are not culturable on artificial medium and seem to have evolved to be wholly dependent on their hosts. This occurred multiple times in oomycete history, and genome studies suggest an association with defects in metabolism (6, 45). Both A. laibachii and H. arabidopsidis lack nitrite and nitrate reductase and thus are limited in their use of inorganic nitrogen. A. laibachii, but not H. arabidopsidis, has also lost genes for making the molybdenum cofactor used by nitrate reductase and another cofactor-dependent enzyme, sulfite oxidase, which generates ATP from sulfite. Both also lack sulfite reductase. These enzymes are found in the other sequenced oomycetes, such as Phytophthora, but interestingly are also missing in Plasmodium, an obligately pathogenic chromalveolate (24). Knowing these defects might lead to the artificial culture of downy mildews and white rusts; however, this may be an oversimplification since the species may have also evolved to use plant signals to regulate their metabolism and development.
A. laibachii and H. arabidopsidis also encode reduced numbers of many proteins involved in pathogenesis, including enzymes for degrading plant cell walls. Pectate lyase, for example, is encoded by 30 genes in P. infestans but only 8 genes and 1 gene in A. laibachii and H. arabidopsidis, respectively. Reductions are less dramatic for other wall hydrolases, which may be used to form haustoria or remodel the pathogen's own walls.
The two species also have a reduced complement of CRNs and RXLRs. A. laibachii and H. arabidopsidis encode only 3 and 20 CRNs, respectively, compared to 196 for P. infestans. CRNs induce host necrosis, so this is consistent with a greater reliance on biotrophy. Genes for RXLR/Q proteins, which are encoded by >300 genes in each Phytophthora species, number only 49 in A. laibachii and 115 in H. arabidopsidis (note that A. laibachii also has 29 of the CHXC proteins). The reduction may be explained by the fact that these species need to interact with just one type of host. It is interesting to speculate whether the loss of effectors was an adaptation to a narrowed host range or its cause. It is also possible that since these species inflict less damage to host cells than do hemibiotrophs, fewer defenses against the plant immune response are required.
Gene content is also reduced in H. arabidopsidis due to loss of the zoospore pathway. This includes at least 200 genes encoding structural components of flagella along with >300 others with roles in zoospore assembly, chemotaxis, and encystment. Fossilized remnants of many of these genes can be detected within the genome, but most appear to have disappeared through slow mutation and genome contraction through recombination. Albugo has, however, maintained the pathway, as have many other downy mildews. Curiously, genes for flagella are retained in P. ultimum despite the apparent rarity of that stage in nature (51).
TEs and their impacts.
Oomycetes contain diverse populations of retrotransposon and DNA transposon-like sequences. Their abilities to self-replicate and form recombination-stimulating repeats shape the structure of genomes and induce variation. As noted above, TEs appear to have particularly influenced the gene-sparse regions of oomycetes. Within gene-dense regions in which gene order is fairly conserved between species, sequences resembling TEs often reside where microsynteny is disrupted.
As in many eukaryotes, the most abundant oomycete retrotransposons belong to the long terminal repeat (LTR)-containing gypsy and copia families, with non-LTR long interspersed elements (LINEs) also present. Most are degenerate and incapable of movement. Genome size in Phytophthora species correlates with the number of retroelements. P. infestans contains about 10,000 gypsy and 4,000 copia-like elements which comprise 37% of the genome, while P. sojae and P. ramorum contain only 3,000 and 2,400 in total, respectively (31). In contrast, in A. laibachii, these represent only 9% of the genome and copia outnumbers gypsy by 3:1, and the two make up only 2% of P. ultimum chromosomes (45, 51).
The ratio of DNA transposons to retroelements is strikingly different between species. For example, these comprise 10% and 37% of the P. infestans genome, respectively, and 12% and 9% of A. laibachii, respectively. A massive expansion of retrotransposons and not DNA elements thus primarily accounts for the larger Phytophthora genomes. Nevertheless, a diverse population of DNA transposons exists in Phytophthora, including those that replicate by cut-and-paste, rolling circle, and self-synthesizing mechanisms. Listed in declining order of copy number in P. infestans, ranging from about 2,000 to 8, these represent piggyBac, helitron, hAT, crypton, Tc1/mariner/pogo, MuDR/foldback, Sola, Maverick, and PIF/harbinger families (20, 31, 87). Most, and particularly piggyBac and helitrons, are more abundant in P. infestans. P. infestans has 273 helitrons, of which 13 appear intact, a number which is more than that for other eukaryotes. P. ramorum and P. sojae contain about one-tenth that number, with none appearing to be functional.
Most of the DNA transposons have representatives in A. laibachii, H. arabidopsidis, and P. ultimum, which suggests that they are ancient members of the oomycete lineage. Some, such as hAT and Tc1/mariner, may have been acquired after stramenopiles diverged from other eukaryotes, as they are reportedly absent from the diatom Thalassiosira pseudonana. The history of the cryptons is also interesting. Two of the six major subfamilies described in eukaryotes, F and S, are found throughout oomycetes, while diatoms contain only crypton S. Crypton F, in contrast, is in Phytophthora but not A. laibachii, H. arabidopsidis, or P. ultimum. Outside Phytophthora, crypton F has been found only in ascomycetes, which suggests horizontal transfer (46).
There is evidence for some movement of TEs. One P. infestans gene was found to contain an inserted gypsy element, including host target site duplications (31). hAT, helitron, and PIF elements may also have “captured” Phytophthora genes and copied them through the genome, based on the detection of their inverted terminal repeats with target site duplications flanking adjoining transposase/replicase and host genes (87). The latter encode SET domain proteins and AdoMet-dependent methyltransferases, which interestingly are both involved in epigenetic regulation, an ABC transporter, cysteine protease, and transglutaminase.
TEs, epigenetics, and transcriptional polymorphisms.
Although few TEs are active in oomycetes, many are transcribed (35). Others are silenced based on the detection in P. infestans of small interfering RNAs (siRNAs) (90). That oomycetes have an active epigenetic system was also shown by RNA interference studies, in which silencing was associated with siRNA and heterochromatinization that spread from the target locus (2, 39, 89). In nonoomycetes, TEs influence nearby genes by recruiting heterochromatin modifications that repress transcription or by insulating genes from such effects (52). Since a TE is within 2 kb of over half of the effector genes in P. infestans, their influence on expression should be considered.
In fact, certain P. infestans and P. sojae RXLR genes are known to be transcriptionally inactive, with virulent and avirulent alleles distinguished by differences in expression rather than protein sequence (18, 27). Some alleles also vary quantitatively in mRNA level (76). Promoter mutations were associated with one case of RXLR silencing, but others may involve epigenetic events. Both TEs and epigenetics may relate to the phenotypic variability observed within Phytophthora spp., including reports of strains with unstable avirulence phenotypes or losing pathogenicity. In P. ramorum, isolates with unusual colony morphologies and an early senescence phenotype were observed to have higher levels of expression of several copia elements (42). Although causality was not established, this may be a sign of epigenetic flux within their genomes. The contribution of expression level polymorphisms to oomycete biology, due to promoter differences or epialleles, is just starting to be appreciated.
It should be remembered that mobile elements and epigenetic phenomena are not the only mechanisms driving change within oomycete genomes. Changes in ploidy, gene conversion, and loss of heterozygosity are reported in several species (11, 30, 50). Unstable supernumerary chromosomes have also been described in Pythium (56).
HGT.
Prior to genome sequencing, it was known that some fungal and oomycete genes, such as the wall-degrading polygalacturonases, were similar, suggesting convergent evolution or horizontal gene transfer (HGT) (83). As noted above, the NLP phytotoxins were also seen as HGT candidates (26). Careful phylogenetic analyses using whole-genome data now provide strong support for HGT. Transfers from fungi to oomycetes appear to have involved about 20 types of genes (67). About two-thirds, including lipases, carbohydrate-depolymerizing enzymes, sugar and nitrogenous base transporters, and enzymes to degrade plant defense compounds, were predicted to be secreted and/or have potential roles in pathogenesis. Since transfers from fungi to oomycetes were much more common than the reverse, it was suggested that plant pathogenesis developed in oomycetes more recently than in fungi. The same study also reported very few cases of HGT for any gene in A. laibachii or S. parasitica (67), and it is interesting to speculate that the larger TE-rich Phytophthora genomes are more tolerant of recombination with foreign DNA.
Oomycetes also have obtained genes not related directly to pathogenesis by HGT, including several involved in cofactor metabolism and amino acid or lipopolysaccharide biosynthesis (91). Glucokinases are another example, with different genes coming from plants and bacteroidales (38, 91). The retention of similar genes from two origins is not very common in eukaryotes, but these may function in distinct cellular locations.
Uncertain support for the chromalveolate hypothesis.
In one evolutionary model, the Chromalveolata are a major eukaryotic group derived from a biflagellated cell with a red algal endosymbiont (44). Chromalveolates are proposed to include about half of known protists and algae, mainly cryptophytes, alveolates, haptophytes, and stramenopiles (with the latter including various classes of golden-brown or brownish-green algae, diatoms, and oomycetes). The diatom and algal lineages are thought to have retained the red alga as a photosynthetic plastid, with oomycetes losing the endosymbiont but retaining some genes (54). Studies of the first sequenced oomycetes, P. ramorum and P. sojae, reported 855 genes of likely red algal origin based on sequence similarity (86). Combined with the finding that chromalveolate and oomycete proteins share some novel domain combinations, this was taken by some to support the model (44, 59). However, endosymbiotic acquisition is hard to distinguish from other events, including HGT.
More-recent analyses may be leading to a consensus for a polyphyletic origin or falsification of the Chromalveolata. Diatom and oomycete genes having affinity with red alga have limited overlap, so some researchers suggested that separate endosymbiotic events occurred before and after they diverged from their common ancestor (5, 57, 62, 80). Support for endosymbiosis in the diatom lineage appears solid, but its occurrence prior to their divergence from oomycetes was challenged by a study that argued that other evolutionary events provide better explanations for the presence of red alga-like genes in oomycetes (80).
Regardless of the validity of the chromalveolate hypothesis, several data, including a recent analysis of 108 concatenated nuclear proteins, indicate that stramenopiles have a monophyletic origin (5). Oomycetes and diatoms also clearly contain common sets of genes of plant (“green genes”) and bacterial origin (9, 61). These include many membrane transporters associated with nutrient acquisition or metabolism with roles at the environmental interface or in organelles. Examples include transporters for taking up nitrate, phosphate, or sulfate and moving ADP/ATP across mitochondrial membranes (12).
CONCLUSIONS AND OUTLOOK
Genomics has provided insight into the biology of oomycetes and paved the way for future experimentation, but it also raises new questions. The species sequenced so far include agents of globally important diseases, such as Phytophthora and Pythium, and pathogens of a model plant. Phytophthora is also a good subject since it is amenable to transformation, enabling functional studies of genes (36). However, data from more species need to be made available to help researchers understand the foundations of pathogenesis and relationships between orders. These should include both nonpathogens and pathogens from additional clades. These need not be limited to species that are culturable in the lab, based on the success with the obligate pathogens A. laibachii and H. arabidopsidis, for which spores were taken from infected leaves for analysis. This should also be feasible for the graminicolous (grass- and grain-infecting) downy mildews which are of major economic importance. Basal genera such as Eurychasma and Haptoglossa, which include algal and nematode pathogens, are also obligate pathogens but grow intracellularly with few external structures (8). Perhaps sufficient DNA can be obtained from their zoospores or pathogen sequences can be separated from host DNA in silico.
The extant data indicate that oomycete genomes have had dynamic histories colored by expansions/contractions, birth/death events, gene fusions, and HGT. The acquisition of bacterial, fungal, and organellar genes has given oomycetes unique capabilities, enabling them to occupy multiple environmental niches. Variation within species is also just starting to be understood; one must wonder what pressures are imposed by environmental changes and agronomic practices such as agrochemical treatment, which may select for alterations in coding and regulatory sequences and may also have epigenetic effects. Several Phytophthora spp. appear to be recent interspecific hybrids (48), perhaps fostered by man's movement of plant material, and it is of interest to see how their genomes evolve.
Evidence of adaptations is evident in the genomes of the obligately biotrophic oomycetes, which are partially condensed. They have not undergone the dramatic reductions of obligate pathogens like Giardia and Plasmodium (24, 60), perhaps since plants provide a less stable environment than an animal host. The benefits of genome reduction in H. arabidopsidis due to loss of the zoospore might seem obvious, but neither Albugo nor P. ultimum (which rarely, if ever, makes zoospores) has chosen this path. While much remains to be learned about the ecological and environmental pressures that influence the evolution of oomycetes, it is clear that nature provides niches for both streamlined and expanded genomes.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institute of Food and Agriculture of the United States Department of Agriculture and the National Science Foundation.
I thank A. Ah Fong for helpful comments.
Footnotes
Published ahead of print 24 August 2012
REFERENCES
- 1. Ah-Fong A, Xiang Q, Judelson HS. 2007. Architecture of the sporulation-specific Cdc14 promoter from the oomycete Phytophthora infestans. Eukaryot. Cell 6:2222–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ah-Fong AM, Bormann-Chung CA, Judelson HS. 2008. Optimization of transgene-mediated silencing in Phytophthora infestans and its association with small-interfering RNAs. Fungal Genet. Biol. 45:1197–1205 [DOI] [PubMed] [Google Scholar]
- 3. Andrews JH. 1975. Distribution of label from 3H-glucose and 3H-leucine in lettuce cotyledons during the early stages of infection with Bremia lactucae. Can. J. Bot. 53:1103–1115 [Google Scholar]
- 4. As-sadi F, et al. 2011. Transcriptomic analysis of the interaction between Helianthus annuus and its obligate parasite Plasmopara halstedii shows single nucleotide polymorphisms in CRN sequences. BMC Genomics 12:498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baurain D, et al. 2010. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol. Biol. Evol. 27:1698–1709 [DOI] [PubMed] [Google Scholar]
- 6. Baxter L, et al. 2010. Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science 330:1549–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beakes GW, Glockling SL, Sekimoto S. 2012. The evolutionary phylogeny of the oomycete “fungi.” Protoplasma 249:3–19 [DOI] [PubMed] [Google Scholar]
- 8. Beakes GW, Sekimoto S. 2009. The evolutionary phylogeny of oomycetes—insights gained from studies of holocarpic parasites of algae and vertebrates, p 1–24 In Lamour K, Kamoun S. (ed), Oomycete genetics and genomics: diversity, interactions, and research tools. John Wiley and Sons, Hoboken, NJ [Google Scholar]
- 9. Bowler C, et al. 2008. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456:239–244 [DOI] [PubMed] [Google Scholar]
- 10. Bozkurt TO, Schornack S, Banfield MJ, Kamoun S. 2012. Oomycetes, effectors, and all that jazz. Curr. Opin. Plant Biol. 15:483–492 [DOI] [PubMed] [Google Scholar]
- 11. Chamnanpunt J, Shan WX, Tyler Brett M. 2001. High frequency mitotic gene conversion in genetic hybrids of the oomycete Phytophthora sojae. Proc. Natl. Acad. Sci. U. S. A. 98:14530–14535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chan CX, Reyes-Prieto A, Bhattacharya D. 2011. Red and green algal origin of diatom membrane transporters: insights into environmental adaptation and cell evolution. PLoS One 6:e29138 doi:10.1371/journal.pone.0029138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen C, Gentles AJ, Jurka J, Karlin S. 2002. Genes, pseudogenes, and Alu sequence organization across human chromosomes 21 and 22. Proc. Natl. Acad. Sci. U. S. A. 99:2930–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cvitanich C, Salcido M, Judelson HS. 2006. Concerted evolution of a tandemly arrayed family of mating-specific genes in Phytophthora analyzed through inter- and intraspecific comparisons. Mol. Genet. Genomics 275:169–184 [DOI] [PubMed] [Google Scholar]
- 15. Damasceno CM, et al. 2008. Structure of the glucanase inhibitor protein (GIP) family from Phytophthora species suggests coevolution with plant endo-beta-1,3-glucanases. Mol. Plant Microbe Interact. 21:820–830 [DOI] [PubMed] [Google Scholar]
- 16. Donaldson ME, Saville BJ. 2012. Natural antisense transcripts in fungi. Mol. Microbiol. 85:405–417 [DOI] [PubMed] [Google Scholar]
- 17. Dong S, et al. 2012. The NLP toxin family in Phytophthora sojae includes rapidly evolving groups that lack necrosis-inducing activity. Mol. Plant Microbe Interact. 25:896–909 [DOI] [PubMed] [Google Scholar]
- 18. Dong S, et al. 2011. Sequence variants of the Phytophthora sojae RXLR effector Avr3a/5 are differentially recognized by Rps3a and Rps5 in soybean. PLoS One 6:e20172 doi:10.1371/journal.pone.0020172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Erwin DC, Ribeiro OK. 1996. Phytophthora diseases worldwide. APS Press, St. Paul, MN [Google Scholar]
- 20. Feschotte C, Pritham EJ. 2007. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 41:331–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fontanillas P, Hartl DL, Reuter M. 2007. Genome organization and gene expression shape the transposable element distribution in the Drosophila melanogaster euchromatin. PLoS Genet. 3:e210 doi:10.1371/journal.pgen.0030210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fry WE. 2008. Phytophthora infestans: the plant (and R gene) destroyer. Mol. Plant Pathol. 9:385–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fukuoka Y, Inaoka H, Kohane IS. 2004. Inter-species differences of co-expression of neighboring genes in eukaryotic genomes. BMC Genomics 5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gardner MJ, et al. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gaulin E, et al. 2008. Transcriptome of Aphanomyces euteiches: new oomycete putative pathogenicity factors and metabolic pathways. PLoS One 3:e1723 doi:10.1371/journal.pone.0001723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gijzen M, Nurnberger T. 2006. Nep1-like proteins from plant pathogens: recruitment and diversification of the NPP1 domain across taxa. Phytochemistry 67:1800–1807 [DOI] [PubMed] [Google Scholar]
- 27. Gilroy EM, et al. 2011. Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2-like in Phytophthora infestans determine virulence on R2 plants. New Phytol. 191:763–776 [DOI] [PubMed] [Google Scholar]
- 28. Grenville-Briggs L, et al. 2011. A molecular insight into algal-oomycete warfare: cDNA analysis of Ectocarpus siliculosus infected with the basal oomycete Eurychasma dicksonii. PLoS One 6:e24500 doi:10.1371/journal.pone.0024500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grunwald NJ, Garbelotto M, Goss EM, Heungens K, Prospero S. 2012. Emergence of the sudden oak death pathogen Phytophthora ramorum. Trends Microbiol. 20:131–138 [DOI] [PubMed] [Google Scholar]
- 30. Gu W, et al. 1993. Measurement of nuclear DNA contents of Mexican isolates of Phytophthora infestans. Mycol. Res. 97:857–860 [Google Scholar]
- 31. Haas BJ, et al. 2009. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461:393–398 [DOI] [PubMed] [Google Scholar]
- 32. Horner NR, Grenville-Briggs LJ, van West P. 2012. The oomycete Pythium oligandrum expresses putative effectors during mycoparasitism of Phytophthora infestans and is amenable to transformation. Fungal Biol. 116:24–41 [DOI] [PubMed] [Google Scholar]
- 33. Jones JD, Dangl JL. 2006. The plant immune system. Nature 444:323–329 [DOI] [PubMed] [Google Scholar]
- 34. Judelson HS, Ah-Fong AM. 2010. The kinome of Phytophthora infestans reveals oomycete-specific innovations and links to other taxonomic groups. BMC Genomics 11:700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Judelson HS, et al. 2008. Gene expression profiling during asexual development of the late blight pathogen Phytophthora infestans reveals a highly dynamic transcriptome. Mol. Plant Microbe Interact. 21:433–447 [DOI] [PubMed] [Google Scholar]
- 36. Judelson HS, Ah-Fong AMV. 2009. Progress and challenges in oomycete transformation, p 435–454 In Lamour K, Kamoun S. (ed), Oomycete genetics and genomics: diversity, interactions and research tools. John Wiley and Sons, Hoboken, NJ [Google Scholar]
- 37. Judelson HS, Blanco FA. 2005. The spores of Phytophthora: weapons of the plant destroyer. Nat. Rev. Microbiol. 3:47–58 [DOI] [PubMed] [Google Scholar]
- 38. Judelson HS, Narayan RD, Tani S. 2009. Metabolic adaptation of Phytophthora infestans during growth on leaves, tubers, and artificial media. Mol. Plant Pathol. 10:843–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Judelson HS, Tani S. 2007. Transgene-induced silencing of the zoosporogenesis-specific PiNIFC gene cluster of Phytophthora infestans involves chromatin alterations. Eukaryot. Cell 6:1200–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kalmykova AI, Nurminsky DI, Ryzhov DV, Shevelyov YY. 2005. Regulated chromatin domain comprising cluster of co-expressed genes in Drosophila melanogaster. Nucleic Acids Res. 33:1435–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kamoun S. 2006. A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44:41–60 [DOI] [PubMed] [Google Scholar]
- 42. Kasuga T, et al. 2012. Phenotypic diversification is associated with host-induced transposon derepression in the sudden oak death pathogen Phytophthora ramorum. PLoS One 7:e34728 doi:10.1371/journal.pone.0034728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kay J, Meijer HJ, ten Have A, van Kan JA. 2011. The aspartic proteinase family of three Phytophthora species. BMC Genomics 12:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Keeling PJ. 2009. Chromalveolates and the evolution of plastids by secondary endosymbiosis. J. Eukaryot. Microbiol. 56:1–8 [DOI] [PubMed] [Google Scholar]
- 45. Kemen E, et al. 2011. Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLoS Biol. 9:e1001094 doi:10.1371/journal.pbio.1001094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kojima KK, Jurka J. 2011. Crypton transposons: identification of new diverse families and ancient domestication events. Mob. DNA 2:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kristiansson E, Thorsen M, Tamas MJ, Nerman O. 2009. Evolutionary forces act on promoter length: identification of enriched cis-regulatory elements. Mol. Biol. Evol. 26:1299–1307 [DOI] [PubMed] [Google Scholar]
- 48. Kroon LP, Brouwer H, de Cock AW, Govers F. 2012. The genus Phytophthora anno 2012. Phytopathology 102:348–364 [DOI] [PubMed] [Google Scholar]
- 49. Kunjeti SG, et al. 2012. RNA-Seq reveals infection-related global gene changes in Phytophthora phaseoli, the causal agent of lima bean downy mildew. Mol. Plant Pathol. 13:454–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lamour K, et al. 20 June 2012, posting date Genome sequencing and mapping reveal loss of heterozygosity as a mechanism for rapid adaptation in the vegetable pathogen Phytophthora capsici. Mol. Plant Microbe Interact. http://dx.doi.org/10.1094/MPMI-02-12-0028-R [DOI] [PMC free article] [PubMed]
- 51. Levesque CA, et al. 2010. Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol. 11:R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lisch D, Bennetzen JL. 2011. Transposable element origins of epigenetic gene regulation. Curr. Opin. Plant Biol. 14:156–161 [DOI] [PubMed] [Google Scholar]
- 53. Madoui MA, Bertrand-Michel J, Gaulin E, Dumas B. 2009. Sterol metabolism in the oomycete Aphanomyces euteiches, a legume root pathogen. New Phytol. 183:291–300 [DOI] [PubMed] [Google Scholar]
- 54. Martens C, Vandepoele K, Van de Peer Y. 2008. Whole-genome analysis reveals molecular innovations and evolutionary transitions in chromalveolate species. Proc. Natl. Acad. Sci. U. S. A. 105:3427–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Martin F. 1995. Meiotic instability of Pythium sylvaticum as demonstrated by inheritance of nuclear markers and karyotype analysis. Genetics 139:1233–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Martin FM. 1995. Electrophoretic karyotype polymorphisms in the genus Pythium. Mycologia 87:333–353 [Google Scholar]
- 57. Maruyama S, Matsuzaki M, Misawa K, Nozaki H. 2009. Cyanobacterial contribution to the genomes of the plastid-lacking protists. BMC Evol. Biol. 9:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Meijer HJ, Govers F. 2006. Genomewide analysis of phospholipid signaling genes in Phytophthora spp.: novelties and a missing link. Mol. Plant Microbe Interact. 19:1337–1347 [DOI] [PubMed] [Google Scholar]
- 59. Morris PF, et al. 2009. Multiple horizontal gene transfer events and domain fusions have created novel regulatory and metabolic networks in the oomycete genome. PLoS One 4:e6133 doi:10.1371/journal.pone.0006133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Morrison HG, et al. 2007. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317:1921–1926 [DOI] [PubMed] [Google Scholar]
- 61. Moustafa A, et al. 2009. Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science 324:1724–1726 [DOI] [PubMed] [Google Scholar]
- 62. Parfrey LW, et al. 2010. Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst. Biol. 59:518–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Park J, et al. 2008. FTFD: an informatics pipeline supporting phylogenomic analysis of fungal transcription factors. Bioinformatics 24:1024–1025 [DOI] [PubMed] [Google Scholar]
- 64. Ploch S, Thines M. 2011. Obligate biotrophic pathogens of the genus Albugo are widespread as asymptomatic endophytes in natural populations of Brassicaceae. Mol. Ecol. 20:3692–3699 [DOI] [PubMed] [Google Scholar]
- 65. Raffaele S, et al. 2010. Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science 330:1540–1543 [DOI] [PubMed] [Google Scholar]
- 66. Raffaele S, Win J, Cano LM, Kamoun S. 2010. Analyses of genome architecture and gene expression reveal novel candidate virulence factors in the secretome of Phytophthora infestans. BMC Genomics 11:637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Richards TA, et al. 2011. Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc. Natl. Acad. Sci. U. S. A. 108:15258–15263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Robold AV, Hardham AR. 2005. During attachment Phytophthora spores secrete proteins containing thrombospondin type 1 repeats. Curr. Genet. 47:307–315 [DOI] [PubMed] [Google Scholar]
- 69. Roy S. 2011. A multidisciplinary approach for identifying stage-specific transcription factor binding sites in the Irish potato famine pathogen, Phytophthora infestans. Ph.D. thesis University of California, Riverside, Riverside, CA [Google Scholar]
- 70. Runge F, et al. 2011. The inclusion of downy mildews in a multi-locus-dataset and its reanalysis reveals a high degree of paraphyly in Phytophthora. IMA Fungus 2:163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Savory EA, et al. 2012. mRNA-Seq analysis of the Pseudoperonospora cubensis transcriptome during cucumber (Cucumis sativus L.) infection. PLoS One 7:e35796 doi:10.1371/journal.pone.0035796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Savory EA, et al. 2012. Alternative splicing of a multi-drug transporter from Pseudoperonospora cubensis generates an RXLR effector protein that elicits a rapid cell death. PLoS One 7:e34701 doi:10.1371/journal.pone.0034701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Schornack S, et al. 2010. Ancient class of translocated oomycete effectors targets the host nucleus. Proc. Natl. Acad. Sci. U. S. A. 107:17421–17426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Seidl MF, Van den Ackerveken G, Govers F, Snel B. 2011. A domain-centric analysis of oomycete plant pathogen genomes reveals unique protein organization. Plant Physiol. 155:628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Seidl MF, Van den Ackerveken G, Govers F, Snel B. 2012. Reconstruction of oomycete genome evolution identifies differences in evolutionary trajectories leading to present-day large gene families. Genome Biol. Evol. 4:199–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Shan W, Cao M, Leung D, Tyler BM. 2004. The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol. Plant Microbe Interact. 17:394–403 [DOI] [PubMed] [Google Scholar]
- 77. Shen D, Ye W, Dong S, Wang Y, Dou D. 2011. Characterization of intronic structures and alternative splicing in Phytophthora sojae by comparative analysis of expressed sequence tags and genomic sequences. Can. J. Microbiol. 57:84–90 [DOI] [PubMed] [Google Scholar]
- 78. Sohn J, Voegele RT, Mendgen K, Hahn M. 2000. High level activation of vitamin B1 biosynthesis genes in haustoria of the rust fungus Uromyces fabae. Mol. Plant Microbe Interact. 13:629–636 [DOI] [PubMed] [Google Scholar]
- 79. Stassen J, Van den Ackerveken G. 2011. How do oomycete effectors interfere with plant life? Curr. Opin. Plant Biol. 14:407–414 [DOI] [PubMed] [Google Scholar]
- 80. Stiller JW, Huang J, Ding Q, Tian J, Goodwillie C. 2009. Are algal genes in nonphotosynthetic protists evidence of historical plastid endosymbioses? BMC Genomics 10:484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Tani S, Judelson HS. 2006. Activation of zoosporogenesis-specific genes in Phytophthora infestans involves a 7-nucleotide promoter motif and cold-induced membrane rigidity. Eukaryot. Cell 5:745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tian M, et al. 2011. 454 genome sequencing of Pseudoperonospora cubensis reveals effector proteins with a QXLR translocation motif. Mol. Plant Microbe Interact. 24:543–553 [DOI] [PubMed] [Google Scholar]
- 83. Torto TA, Rauser L, Kamoun S. 2002. The pipg1 gene of the oomycete Phytophthora infestans encodes a fungal-like endopolygalacturonase. Curr. Genet. 40:385–390 [DOI] [PubMed] [Google Scholar]
- 84. Torto-Alalibo T, et al. 2005. Expressed sequence tags from the oomycete fish pathogen Saprolegnia parasitica reveal putative virulence factors. BMC Microbiol. 5:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tsai HK, Huang PY, Kao CY, Wang D. 2009. Co-expression of neighboring genes in the zebrafish (Danio rerio) genome. Int. J. Mol. Sci. 10:3658–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tyler BM, et al. 2006. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313:1261–1266 [DOI] [PubMed] [Google Scholar]
- 87. Vadnagara K. 2010. A tale of three phytopathogens: impact of transposable elements on genome evolution. M.S. thesis University of Texas, Arlington, Arlington, TX [Google Scholar]
- 88. van West P. 2006. Saprolegnia parasitica, an oomycete pathogen with a fishy appetite: new challenges for an old problem. Mycologist 20:99–104 [Google Scholar]
- 89. van West P, et al. 2008. Internuclear gene silencing in Phytophthora infestans is established through chromatin remodelling. Microbiology 154:1482–1490 [DOI] [PubMed] [Google Scholar]
- 90. Vetukuri RR, et al. 2011. Silencing of the PiAvr3a effector-encoding gene from Phytophthora infestans by transcriptional fusion to a short interspersed element. Fungal Biol. 115:1225–1233 [DOI] [PubMed] [Google Scholar]
- 91. Whitaker JW, McConkey GA, Westhead DR. 2009. The transferome of metabolic genes explored: analysis of the horizontal transfer of enzyme encoding genes in unicellular eukaryotes. Genome Biol. 10:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Xiang Q, Judelson HS. 2010. Myb transcription factors in the oomycete Phytophthora with novel diversified DNA-binding domains and developmental stage-specific expression. Gene 453:1–8 [DOI] [PubMed] [Google Scholar]
- 93. Xiang Q, Roy S, Kim KS, Judelson HS. 2009. A motif within a complex promoter from the oomycete Phytophthora infestans determines transcription during an intermediate stage of sporulation. Fungal Genet. Biol. 46:400–409 [DOI] [PubMed] [Google Scholar]
- 94. Zhan S, Horrocks J, Lukens LN. 2006. Islands of co-expressed neighboring genes in Arabidopsis thaliana suggest higher-order chromosome domains. Plant J. 45:3347–3357 [DOI] [PubMed] [Google Scholar]