Abstract
Eukaryotic microbes are highly diverse, and many lineages remain poorly studied. One such lineage, the diplomonads, a group of binucleate heterotrophic flagellates, has been studied mainly due to the impact of Giardia intestinalis, an intestinal, diarrhea-causing parasite in humans and animals. Here we describe the development of a stable transfection system for use in Spironucleus salmonicida, a diplomonad that causes systemic spironucleosis in salmonid fish. We designed vectors in cassette format carrying epitope tags for localization (3×HA [where HA is hemagglutinin], 2× Escherichia coli OmpF linker and mouse langerin fusion sequence [2×OLLAS], 3×MYC) and purification of proteins (2× Strep-Tag II–FLAG tandem-affinity purification tag or streptavidin binding peptide–glutathione S-transferase [SBP-GST]) under the control of native or constitutive promoters. Three selectable gene markers, puromycin acetyltransferase (pac), blasticidin S-deaminase (bsr), and neomycin phosphotransferase (nptII), were successfully applied for the generation of stable transfectants. Site-specific integration on the S. salmonicida chromosome was shown to be possible using the bsr resistance gene. We epitope tagged six proteins and confirmed their expression by Western blotting. Next, we demonstrated the utility of these vectors by recording the subcellular localizations of the six proteins by laser scanning confocal microscopy. Finally, we described the creation of an S. salmonicida double transfectant suitable for colocalization studies. The transfection system described herein and the imminent completion of the S. salmonicida genome will make it possible to use comparative genomics as an investigative tool to explore specific, as well as general, diplomonad traits, benefiting research on both Giardia and Spironucleus.
INTRODUCTION
Eukaryotic microorganisms make up the majority of eukaryotic diversity and have attained a magnificent array of complexity during the billion years since their emergence (4, 29). Sequencing the genomes of diverse microbial eukaryotes illuminates the stunning complexity and dynamic nature of the eukaryotic gene repertoire. Even though there is an expanse of diversity within eukaryotes, very little is known about how this diversity shapes the cell biology of individual taxa. For example, at present there are only around 10 genome sequences available for excavate organisms, a diverse group of single-celled flagellated heterotrophs that includes significant human pathogens, such as Leishmania, Trypanosomes, Trichomonas, and Giardia. Moreover, the vast majority of eukaryotic microbial lineages remain unexplored at the molecular level, and important unexpected discoveries have been made by studying such organisms, e.g., the discoveries of telomerase (11) and RNA editing (3).
The excavate order Diplomonadida belongs to an undersampled part of eukaryotic diversity, and studies performed so far have indicated substantial diversity within the order, including alternative genetic codes (21), differences in the number of nuclei, and large genomic differences between morphologically similar organisms (2, 7, 15, 30).
Model organisms, such as yeast, have been instrumental in determining the inner workings of many basal eukaryotic features. However, generalizations of cellular process are difficult to make because of the distant ancestry of many model organisms to excavate parasites and gene loss among many eukaryotic lineages upon niche adaptation (8). Genomic streamlining and adaptive reduction seem to be important factors driving the evolution within diplomonads (24), along with an influx of genes from within microbial communities by lateral gene transfer (1).
The availability of genome sequence information has profoundly changed the way in which research on eukaryotic microbes is performed. The manipulation of genomes and the expression of transgenes are now possible in an ever-increasing number of organisms. However, to date, there are only two diplomonads for which transfection systems have been constructed. Giardia intestinalis was the first transfected diplomonad (32), and it is by far the most-studied organism, benefiting from the wealth of genomic information that is available (7, 15, 24). Spironucleus vortens, the likely cause of the “hole-in-the-head” disease in tropical fish species (26), has been successfully transfected and has many favorable characteristics as a model organism, such as wide temperature tolerance and extremely rapid growth in vitro to high cell densities (6, 23, 31). However, the S. vortens genome project reported severe problems with assembly, and it has been put on hold. Spironucleus salmonicida, a causative organism of severe systemic infection in farmed Atlantic salmon (Salmo salar), farmed Chinook salmon (Oncorhynchus tshawytscha), and farmed Arctic charr (Salvelinus alpinus) (19), has favorable characteristics, similar to those of S. vortens, as a model organism, and has been reported to have a small genome with limited divergence between its two nuclei (30). We have participated in the sequencing of the genome of S. salmonicida and are aiming to provide reagents for the creation of a second diplomonad model system to support research on diplomonad parasites. Here we describe the development of methodologies for the stable transfection of S. salmonicida, employing three different selection markers (pac, bsr, and nptII), and we demonstrate the utility of this transfection system by tagging, expressing, and localizing six proteins in S. salmonicida. Further, we describe the creation of site-specific integration on the S. salmonicida chromosome using the bsr selectable marker. Finally, we describe the creation of S. salmonicida double transfectants by enabling experiments involving colocalization studies. By creating tools and methods for transfection in the diplomonad S. salmonicida, we hope to accelerate research on this group of curious and fascinating organisms.
MATERIALS AND METHODS
Reagents and cell culture.
Unless otherwise indicated, the reagents were obtained from Sigma Chemical Co. S. salmonicida (ATCC 50377, previously ATCC 50380), isolated from a muscle abscess in an Atlantic salmon from Vesteraalen, northern Norway (previously known as Spironucleus barkhanus), was obtained from the American Type Culture Collection (ATCC) and grown in axenic culture in a modified liver digest, yeast extract, and iron (LYI) culture medium.
Spironucleus salmonicida was cultivated in slanted polystyrene screw-cap tubes (Nunc) in 10 ml of LYI medium at 16°C with tightly capped tubes. Two hundred fifty milliliters basal medium (0.25 g K2HPO4, 0.15 g KH2PO4, 0.25 g NaCl, 6.25 g yeast extract [product no. 70161; Fluka], 1.25 g liver digest neutralized [Oxoid], 0.25 g l-cysteine-HCl, 2.5 g glucose, 0.05 g ascorbic acid, and 2.5 ml 2.28-mg/ml ferric ammonium citrate solution) was added. The pH was set to 6.8 using 5 M NaOH, and the medium was filter sterilized using 0.45-μm cellulose acetate (CA) filter units (Corning). The basal medium was kept at 4°C for immediate use or stored frozen at −20°C. The medium was completed by supplementing it with 10% bovine serum and 2.5% Diamond Vitamin Tween 80 40× solution (catalog no. 58980C; SAFC Biosciences). The completed medium was used within 72 h.
The cells were inspected by inverted light microscopy and passaged upon reaching confluence (∼106 cells/ml). Prior to passage, the cells were placed on ice for 15 min, and the tube was inverted several times both before and after the passage. Cells were passaged once per week by diluting the cultures to ∼104 cells/ml in complete LYI medium.
Optimization of the concentration for selection drugs.
Susceptibilities to different eukaryotic selection agents were determined by testing various concentrations of puromycin (10, 25, 50, and 100 μg/ml), G418 (50, 150, 300, and 500 μg/ml), paromomycin (100, 250, 300, and 350 μg/ml), hygromycin B (50, 100, 200, and 300 μg/ml), and blasticidin S (5, 10, 15, and 20 μg/ml). The drugs were added to tubes with 105 cells/ml in growth medium, and the cells were counted manually in a hemocytometer on days 3, 5, and 7 of culture. The cells were fixed with 1% formaldehyde to halt cell motility in the chamber. The results were used for calculating the percentage of growth inhibition compared to that of control samples grown in the absence of drugs. The equation used to determine the percent growth inhibition was [(U − T)/U] × 100, where U stands for the number of untreated cells at a given time point, and T stands for the number of treated cells for the same indicated time. The experiments were performed in duplicate.
Construction of vectors and transfection of S. salmonicida using electroporation.
Detailed descriptions of vector construction are available in section S3 in the supplemental material. The transfection of S. salmonicida largely followed the methods used successfully to transfect G. intestinalis (32) and S. vortens (6). Late exponentially growing cells (∼106 cells/ml) were placed in ice slush for 15 min, followed by repeated tapping of the tube to dislodge the cells. The cells were pelleted at 500 × g for 5 min at 4°C, the culture medium was removed, and the pellet was resuspended in fresh ice-cold LYI medium to a cell density of 107 cells/ml. For each transfection, 300 μl of S. salmonicida cell suspension was placed in a prechilled 4-mm electroporation cuvette (Bio-Rad) on ice. Episomal (20 to 40 μg) or linearized (10 to 20 μg) plasmids in a volume not larger than 30 μl were added and gently mixed with the cells. The optimal condition used for electroporation using a Gene Pulser instrument (Bio-Rad) was 320 V, 800 Ω, and 960 μF, which delivered an electric pulse for 80 to 98 ms. However, the transfectants could also be obtained at the other voltage settings (350 V, 400 V, 420 V) at a lower frequency.
The electroporated cells were incubated on ice for 10 min, followed by transfer to culture tubes with 10 ml LYI medium precooled to 16°C. The cells were grown at 16°C for 24 h before the appropriate selection drug was added (50 μg/ml puromycin, 15 μg/ml blasticidin S, or 150 μg/ml G418).
The transfected cells were given fresh media and selective drug each week until confluence was reached, allowing them to be passaged as the wild-type cells. To avoid contamination during the transfection, 100 μg/ml gentamicin (Gibco) was added to the culture tubes until confluence was reached. The transfected S. salmonicida strains were cryopreserved in LYI medium containing 10% dimethyl sulfoxide (DMSO) and stored first at −80°C, and then they were promptly moved to liquid nitrogen for long-term stabilization.
Preparation of linearized plasmid DNA for integration on the chromosome.
Plasmid DNA prepared using a scaled-up miniprep protocol (see section S3 in the supplemental material) was linearized for integration on the chromosome. The pSpiro-BSR-BiP-2×OLLAS-C (PstI), pSpiro-BSR-Sec61α-2×OLLAS-C (PstI), and pSpiro-BSR-PDI2-2×OLLAS-C (PsyI) vectors were linearized at the indicated unique restriction site. Forty micrograms of plasmid DNA was digested 4 h to overnight at 37°C using 5 FastDigest units (FDU) of enzyme in 1× FastDigest buffer. The digested plasmid was purified and concentrated by ethyl alcohol (EtOH) precipitation by adding 2.5 volumes of −20°C absolute EtOH and 1/10 the aqueous volume of 5 M sodium acetate (pH 5.5), followed by 1 h of incubation at −80°C. The precipitated material was pelleted by centrifugation at 13,000 rpm for 30 min at 4°C. The pellet was washed by ice-cold 70% EtOH, briefly dried, and resuspended in 20 μl double-distilled water (ddH2O). The digested plasmid was analyzed by agarose gel electrophoresis to confirm successful linearization. The concentration of plasmid was determined by NanoDrop measurement.
Methanol-acetone fixation of cells.
Droplets of 10 to 20 μl of cells washed in phosphate-buffered saline (PBS) were placed in the wells of poly-l-lysine-coated slides (Thermo Scientific), allowed 5 min to attach, and fixed in a −20°C methanol-acetone (1:1) mixture for 10 min. The slide was dried at room temperature, and the cells were rehydrated by adding a 15-μl drop of PBS, followed by incubation for 10 min. The cells were blocked with 1% bovine serum albumin (BSA) (albumin bovine fraction V; BDH) in PBS for 1 h at room temperature or, alternatively, overnight at 4°C in a humidity pan.
Paraformaldehyde fixation of cells.
Transfected cells were centrifuged at 500 × g for 5 min, and the cells were washed three times in PBS. Droplets of 15 to 20 μl of resuspended cells were placed on poly-l-lysine-coated slides (Thermo Scientific) and left to attach for 5 min at 16°C. The cells were fixed in 2% paraformaldehyde (PFA) in PBS for 30 min at 16°C. Cell fixative was quenched in 0.1 M glycine in PBS followed by two 5-min washes with PBS. The cells were permeabilized by adding PBS containing 0.2% Triton X-100, incubated for 30 min, and washed with PBS three times for 5 min each time. The cells were blocked in 2% BSA in PBS overnight at 4°C in a humidity pan.
Western blot analysis.
Western blotting was performed essentially as described in reference 16. Polyvinylidene difluoride (PVDF) membranes were blocked in either 1% BSA in PBS-0.1% Tween 20 (PBS-T) for detection of the hemagglutinin (HA) tag or 5% milk in PBS-T for the Escherichia coli OmpF linker and mouse langerin fusion sequence (OLLAS) tag or anti-centrin 20H5 antibodies. The HA tag was detected by mouse monoclonal anti-HA (1:10,000) (catalog no. H9658; Sigma-Aldrich) in 1% BSA in PBS-T. The OLLAS tag was detected by incubating the blocked membranes with rabbit anti-OLLAS (1:1,000) (catalog no. A01658; GenScript) in 5% milk in PBS-T. The mouse monoclonal anti-centrin 20H5 antibody (1:100) (catalog no. 04-1624; Millipore) was diluted in 5% milk PBS-T. The anti-HA and anti-centrin 20H5 antibodies were detected by anti-mouse horseradish peroxidase (HRP) (1:10,000) (product code P0161; Dako) and the OLLAS tag by anti-rabbit HRP (1:7,500) (product code P0217; Dako) diluted in the blocking buffer used for the primary antibody. The Western blots were developed using either ECL Plus (GE Healthcare) or Western Lightning ECL Pro (Perkin-Elmer). The images were recorded on a ChemiDoc MP+ imaging system (Bio-Rad).
Immunofluorescence.
The blocking solution was removed from the slides, and 15 μl of primary antibody was added. Proteins that were tagged using the three-HA epitope (3×HA) tag were detected by the anti-HA monoclonal antibody (1:500) (product no. H 9658; Sigma-Aldrich) or rabbit monoclonal HA tag (C29F4) antibody (1:1,600) (catalog no. 3724; Cell Signaling Technology) in blocking buffer (PBS, 0.1% Triton X-100, 3% bovine serum). Proteins that were tagged with 2×OLLAS were probed using the OLLAS epitope tag antibody (1:100) (catalog no. NBP1-06713; Novus Biologicals) or rabbit anti-OLLAS (1:2,000) (catalog no. A01658; GenScript) in blocking buffer. Tubulin was labeled using the mouse monoclonal anti-tubulin TAT1 antibody (35) diluted 1:150 in blocking buffer. After incubation for 1 h at room temperature (RT), the wells were washed six times with 15-μl droplets of wash buffer (PBS plus 0.1% Triton X-100), followed by addition of the secondary antibody. The mouse anti-HA or anti-tubulin TAT1 primary antibody was detected by polyclonal rabbit anti-mouse conjugated to fluorescein isothiocyanate (1:50) (code F0232; Dako) goat anti-mouse Alexa Fluor 488 (1:200) (catalog no. A-11029; Invitrogen) in blocking buffer. The OLLAS epitope antibody was detected by highly cross-adsorbed donkey anti-rat IgG (H plus L) conjugated to CF594 (1:200) (catalog no. 20159; Biotium) in blocking buffer. The rabbit anti-OLLAS antibody or the rabbit anti-HA antibody was detected by goat anti-rabbit Alexa Fluor 594 (1:250) (catalog no. A-11037; Invitrogen). The slide was incubated for 1 h at RT, washed eight times with 15 μl wash buffer, and mounted with 3 μl of VectaShield medium containing 4′,6-diamidino-2-phenylindole (DAPI) (catalog no. H-1200; Vector Laboratories). A coverslip was placed over the wells and sealed with nail varnish. The slides were viewed using a Zeiss Axioplan 2 fluorescence microscope or a Zeiss 510 laser scanning confocal microscope. The images were processed using the software AxioVision LE release 4.8.2.0 or Zen 2011 v7.0.0.285 (Carl Zeiss GmbH). Colocalization analyses and three-dimensional reconstruction were performed with BioImageXD v1.0 RC3 software (20).
Cloning of genes into vectors for tagging.
The coding sequences and 200 to 500 bp of the putative promoters of the caltractin, fibrillarin, IFT46, protein disulfide isomerase 2 (PDI2), Sec61α, and binding immunoglobulin protein (BiP) genes were PCR amplified from genomic DNA of S. salmonicida (see sections S1 and S3 in the supplemental material). The PCR products were analyzed on agarose gels to confirm that the sizes were correct. The caltractin, fibrillarin, and IFT46 PCR products were doubly digested with NotI and MluI, followed by ligation into pSpiro-PAC-3×HA-C. The BiP, PDI2, and Sec61α PCR products were digested using PacI (New England BioLabs) and NotI, followed by ligation into pSpiro-BSR-2×OLLAS-C.
Nucleotide sequence accession numbers.
The nucleotide sequences of the pSpiro-PAC-3×HA-C, pSpiro-BSR-2×OLLAS-C, pSpiro-NptII-3×MYC-C, pSpiro-PAC-alphaTub-3×HA-N, pSpiro-PAC-IFT46-3×HA-C, pSpiro-PAC-fibrillarin-3×HA-C, pSpiro-PAC-caltractin-3×HA-C, pSpiro-PAC-ADI-3×HA-C, pSpiro-BSR-BiP-2×OLLAS-C, pSpiro-NptII-ESP-C-SF-TAP, pSpiro-BSR-PDI2-2×OLLAS-C, and pSpiro-BSR-Sec61α-2×OLLAS-C vectors have been submitted to GenBank under the accession numbers JQ768097 through JQ768108.
RESULTS
Construction of episomal vectors for stable transfection of S. salmonicida.
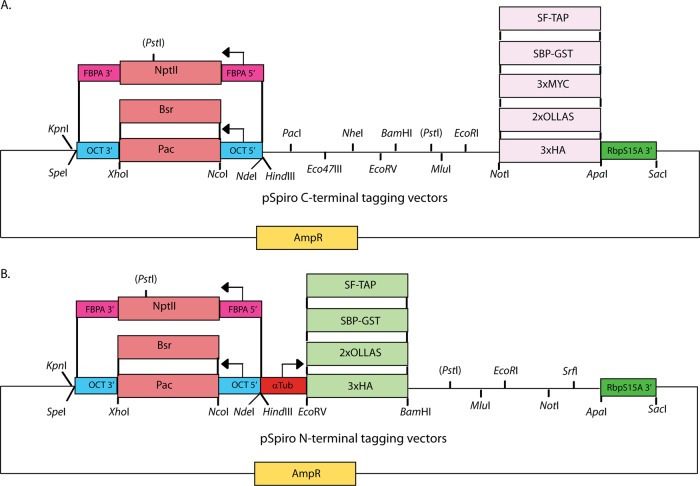
We constructed a suite of plasmid vectors for the expression of the recombinant proteins in S. salmonicida (Fig. 1). The vectors were based on an already-existing vector system in Giardia (16) that was designed in cassette format, enabling exchanges of selection markers, epitope tags, and inserted genes. The starting Giardia vector contained the selectable resistance markers puromycin N-acetyltransferase (pac) for selection in eukaryotic cells and β-lactamase for selection in E. coli. To allow efficient expression, the pac gene was flanked by the 5′ untranslated region (UTR) and the 3′ UTR of S. salmonicida ornithine carbamoyltransferase (OCT), a metabolic gene in S. salmonicida. The pac gene can be replaced by inserting the gene for blasticidin S-deaminase (bsr), since the resistance genes are flanked by the same restriction sites (NcoI and XhoI). The neomycin-phosphotransferase (nptII) resistance gene was fused seamlessly to the up- and downstream regions of the fructose bisphosphate aldolase gene from S. salmonicida by overlap extension PCR in order to keep the native C terminus of the resistance protein (28).
Fig 1.
Schematic pSpiro vector maps showing constructed cassettes and relevant restriction sites. The episomal vectors contain the ampicillin resistance gene (AmpR) for selection in E. coli and the pac, bsr, and nptII genes for selection in S. salmonicida. The pac and bsr genes are flanked by the 5′ UTR and 3′ UTR of the ornithine carbamoyltransferase (OCT) gene, which are cloned between the HindIII/NcoI and XhoI/KpnI sites, respectively. The nptII gene is inserted between the 5′ UTR and 3′ UTR of the fructose bisphosphate aldolase (FBPA) gene, and the entire cassette is cloned between SpeI and NdeI. The three eukaryotic selection markers can be exchanged using combinations of KpnI/SpeI and NdeI/HindIII. The 3′UTR from the ribosomal protein S15A (RbpS15A) gene was cloned between the ApaI and SacI sites. (A) The C-terminal cassettes (3×HA, 2×OLLAS, 3×MYC, SBP-GST, and SF-TAP) are cloned between the NotI and ApaI sites. Ten unique restriction sites are available for cloning in the MCS region of the pSpiro C-terminal tagging vectors. (B) The N-terminal vectors carry the α-tubulin (αTub) 5′ UTR cloned between the HindIII and EcoRV sites. The N-terminal tagging cassettes (3×HA, 2×OLLAS, SBP-GST, and SF-TAP) are cloned at the EcoRV and BamHI sites. Six restriction sites are available for cloning in the MCS region of the pSpiro N-terminal tagging vectors.
The 3′ UTR, including the stop codon, from the ribosomal protein S15A gene was inserted downstream of the multiple-cloning site (MCS) between ApaI and SacI to allow C-terminal tagging and expression using either a constitutive or a native promoter. We constructed a range of C-terminal cassettes (between NotI and ApaI) aimed at providing tools for molecular biology studies of S. salmonicida (see section S2 in the supplemental material). The 3×HA and 2×OLLAS tags have been utilized in immunofluorescence and Western blotting.
For the purposes of protein purification, the Strep-Tag II–FLAG tandem-affinity purification (SF-TAP) (2×Strep-Tag II plus 1×FLAG) and streptavidin binding peptide-glutathione S-transferase (SBP-GST) tags can be utilized in either an N-terminal or a C-terminal configuration (NotI and ApaI for C-terminal constructs and EcoRV and BamHI for N-terminal constructs). The SBP-GST fusion is cleavable by the site-specific PreScission protease. The multiple cloning sites contain 6 unique restriction sites that can be used to mobilize gene fragments to any of the constructed vectors (Fig. 1; see also section S2 in the supplemental material). The promoter (HindIII and EcoRV) and 3′ UTR cassettes (ApaI and SacI) of the expression cassette can also be exchanged between vectors due to the presence of unique restriction sites.
Growth inhibition effects for puromycin, blasticidin S, and G418 on S. salmonicida cells.
The drugs puromycin, blasticidin S, G418, hygromycin, and paromomycin were tested for their effects on the growth of S. salmonicida trophozoites. Four different concentrations of each drug were added to wild-type cells, and the drug-treated cells were counted after 3, 5, and 7 days of incubation. The percentage of growth inhibition was used to estimate the half-maximal inhibitory concentration (IC50) of the drugs after 5 days (Table 1). The determined IC50 values were used to estimate the required concentration of each drug that would yield a growth inhibition of over 90% after 5 days of culture. The suggested levels of drug for the generation of transgenic parasites were 50 μg/ml puromycin, 15 μg/ml blasticidin S, and 150 μg/ml G418 (Table 1). Transgenic puromycin-resistant cells were less sensitive to blasticidin S inhibition. This interaction could be alleviated by increasing the selection concentration of blasticidin S to 70 μg/ml, thereby allowing the generation of double transfectants.
Table 1.
Estimation of IC50s for three selection drugs tested on S. salmonicida cellsa
| Selection drug | IC50 (μg/ml) | Suggested concn for use in selection (μg/ml) | % growth inhibition at selection concn |
|---|---|---|---|
| Puromycin | 18 | 50 | 96 |
| Blasticidin S | 7 | 15 | 92 |
| G418 | 49 | 150 | 94 |
| Hygromycin B | 120 | 300 | 88 |
| Paromomycin | 180 | 350 | 84 |
The cells were treated with the indicated drugs and counted manually at given time points. This table shows the results from the fifth day of drug treatment. The data were used for plotting the percent growth inhibition against the concentrations tested, from which the IC50s were estimated.
Electroporation and generation of transgenic S. salmonicida cells.
Electroporation settings were evaluated to find the optimal conditions for the generation of S. salmonicida transfectants based on known conditions in G. intestinalis and S. vortens (6, 32). The most favorable conditions identified were transfection at 320 V, 800 Ω, and 960 μF using 20 to 40 μg high-quality plasmid vector DNA at a concentration of at least 1 μg/μl. However, transfection was also possible with less material and different voltage settings with lower frequency. Drug-resistant proliferating parasites were apparent 7 to 10 days after transfection, with most strains reaching confluence within 3 to 4 weeks after transfection. The transgenic parasites were subcultured weekly under continuous selection and were stable for long periods of time (at least 10 months).
Genomic DNA prepared from the transfected cells could be used to transform E. coli, whereas genomic DNA of mock-transfected parasites failed to yield E. coli colonies. The recovered plasmids prepared from transformed E. coli were indistinguishable from the starting vector, as assessed by restriction digestion and their capability to retransfect S. salmonicida. These observations are consistent with plasmid replication as episomes in transfected S. salmonicida, as has been demonstrated in G. intestinalis and S. vortens (6, 32).
Localization of epitope-tagged proteins in S. salmonicida.
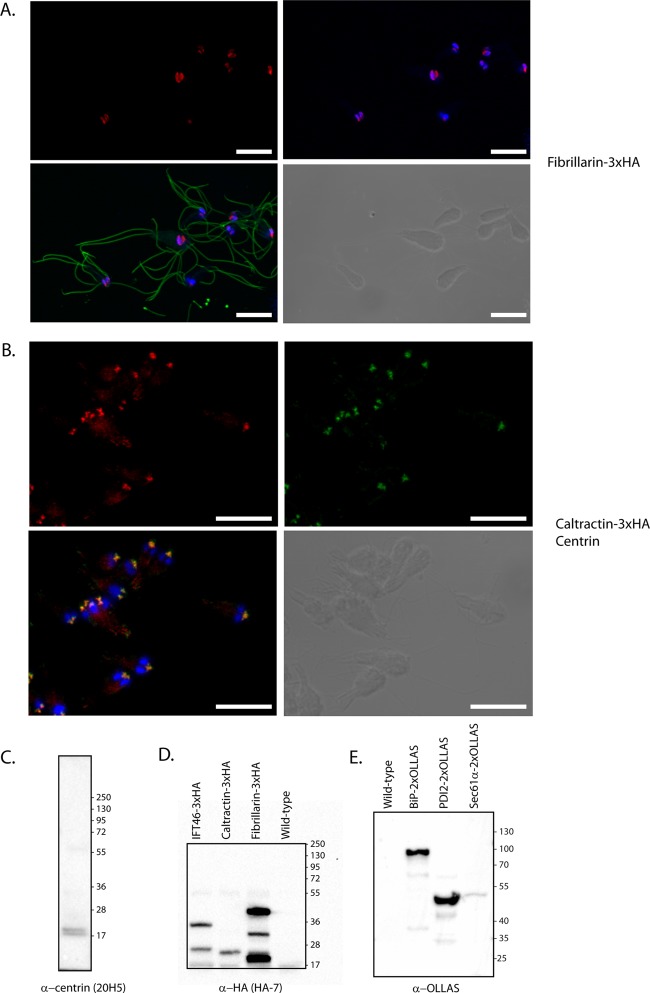
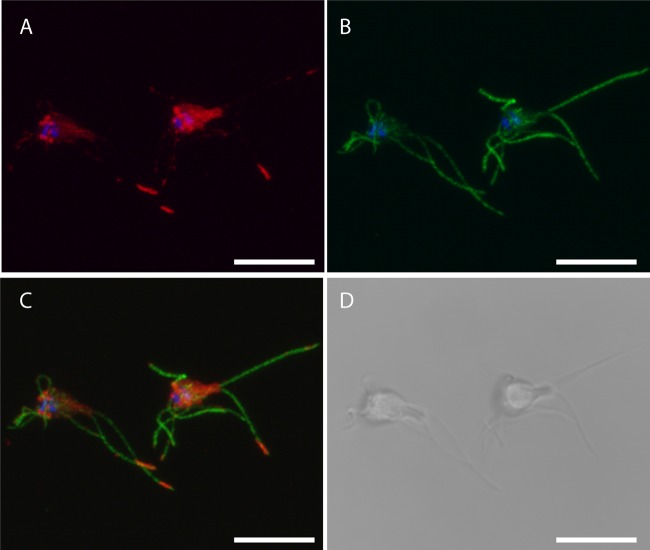
Three proteins were chosen for C-terminal 3×HA epitope tagging (caltractin, IFT46, fibrillarin). The genes were identified using reciprocal tBLASTn searches in a draft assembly of the S. salmonicida genome using the homologs present in the G. intestinalis genomes (7, 15, 24). These proteins are implicated in diverse cellular processes and localize to different cellular structures in other organisms. The predicted full-length protein coding regions, along with 200 to 500 bp of upstream untranslated regions containing putative promoters, were cloned into the pSpiro-PAC-3×HA-C vector. The constructs were transfected into S. salmonicida, and transgenic parasites were generated by puromycin selection. The expression of proteins in line with their expected sizes (caltractin-3×HA, 23.4 kDa; fibrillarin-3×HA, 39.1 kDa; IFT46-3×HA, 30.4 kDa) were detected by Western blotting toward the HA tag in the three transgenic strains (Fig. 2D). Additional lower-molecular-weight protein species were detected for IFT46-3×HA and fibrillarin-3×HA (Fig. 2D). Next, we performed confocal immunofluorescence microscopy to determine the subcellular localization of the epitope-tagged constructs. Caltractin localized to two pyramidal foci above the nuclei (Fig. 2B). The caltractin label colocalized with that of the monoclonal anti-centrin 20H5 antibody (5) (Fig. 2B). The anti-centrin 20H5 monoclonal antibody recognizes two proteins of around 20 kDa in S. salmonicida by Western blotting (Fig. 2C), which is the expected size range for centrin homologs. These two results suggest strongly that these structures represent the basal bodies of S. salmonicida. We localized tagged fibrillarin in S. salmonicida to the nuclei showing partial colocalization to the genomic DNA stain of DAPI (Fig. 2A; see also Fig. S2 in the supplemental material). The tagged homolog of the intraflagellar transport (IFT) complex B protein IFT46 localized to the flagella (Fig. 3) and two C-shaped sheets that run the length of the cell up to the position of the nuclei (see Fig. S1 in the supplemental material). Here the sheets transition into two central cylindrical structures with accompanying pairs of additional cylindrical structures between and in close proximity to the nuclei (Fig. 3; see also Fig. S1 in the supplemental material). The signal in the flagella was present in local foci with enrichment at the flagellar tips in some cells, potentially demonstrating the presence of IFT particles tracking along the microtubules (Fig. 3). Diffuse staining of the cell body was also commonly observed (Fig. 3).
Fig 2.
Expression and localization of epitope-tagged proteins in S. salmonicida. Stably transfected S. salmonicida carrying the pSpiro-PAC-fibrillarin-3×HA and pSpiro-PAC-caltractin-3×HA episomal plasmids were fixed using 2% PFA and methanol and acetone, respectively. (A) Fixed cells were blocked with 2% BSA; they were stained using rabbit anti-HA (1:1,600) and anti-tubulin TAT1 (1:150) and detected using anti-rabbit Alexa Fluor 594 (A594) (1:250) and anti-mouse Alexa Fluor 488 (A488) (1:200), respectively. (B) Cells were stained with mouse monoclonal anti-centrin 20H5 (1:100) and rabbit anti-HA (1:1,600) and detected by anti-mouse A488 and anti-rabbit A594, respectively. The cells were mounted in VectaShield medium containing DAPI and viewed using a Zeiss 510 laser scanning confocal microscope. (A and B, upper left) Maximum intensity projection (MIP) of the A594 signal (red). (A, upper right) MIP of the A594 (red) and DAPI (blue) signals. (B, upper right) MIP of A488 (green). (A and B, lower left) MIP of A488 (green), A594 (red), and DAPI (blue). (A and B, lower right) Bright-field image from the center of the confocal Z-stacks. Fibrillarin-3×HA displays nuclear localization showing partial colocalization with DAPI but enrichment in the regions devoid of DAPI stain. Caltractin-3×HA shows colocalization to the basal body marker centrin in the triangular foci anterior to the nuclei. Scale bars, 5 μm. (C) Western blot of S. salmonicida cells using the mouse monoclonal anti-centrin (20H5) antibody. A doublet of bands is observed at ∼20 kDa. (D) Western blot of S. salmonicida transfectants expressing HA-tagged proteins using the mouse monoclonal anti-HA antibody. From left to right, shown are IFT46-3×HA transfectants (theoretical molecular mass, 30.4 kDa), caltractin-3×HA transfectants (23.4 kDa), fibrillarin-3×HA transfectants (39.1 kDa), and S. salmonicida wild-type cells. (E) Western blot of S. salmonicida transfectants expressing OLLAS-tagged proteins using the rabbit anti-OLLAS antibody. From left to right, shown are S. salmonicida wild-type cells, BiP-2×OLLAS transfectants (75.1 kDa), PDI2-2×OLLAS transfectants (44.5 kDa), and Sec61α-2×OLLAS transfectants (55.6 kDa).
Fig 3.
Localization of 3×HA epitope-tagged IFT46 S. salmonicida trophozoites. Transfectants carrying the IFT46-3×HA construct were fixed in 2% PFA and blocked with 2% BSA; they were stained using rabbit anti-HA (1:1,600) and anti-tubulin TAT1 (1:150) and detected using anti-rabbit Alexa Fluor 594 (A594) (1:250) and anti-mouse Alexa Fluor 488 (A488) (1:200), respectively. The cells were mounted in VectaShield medium containing DAPI and viewed using a Zeiss 510 laser scanning confocal microscope. (A) Maximum intensity projection (MIP) of A594 (red) and DAPI (blue) signals; (B) MIP of A488 (green) and DAPI signals; (C) MIP of A488, A594, and DAPI signals; (D) bright-field image from the center of the confocal Z-stack. IFT46-3×HA displays localization to paired cylindrical foci above the nuclei, to sheets that pass through the cell body, and to foci along the flagella sometimes being enriched at the tips. Diffuse staining of the cell body is commonly observed. Scale bars, 10 μm.
Integration on the chromosome in S. salmonicida.
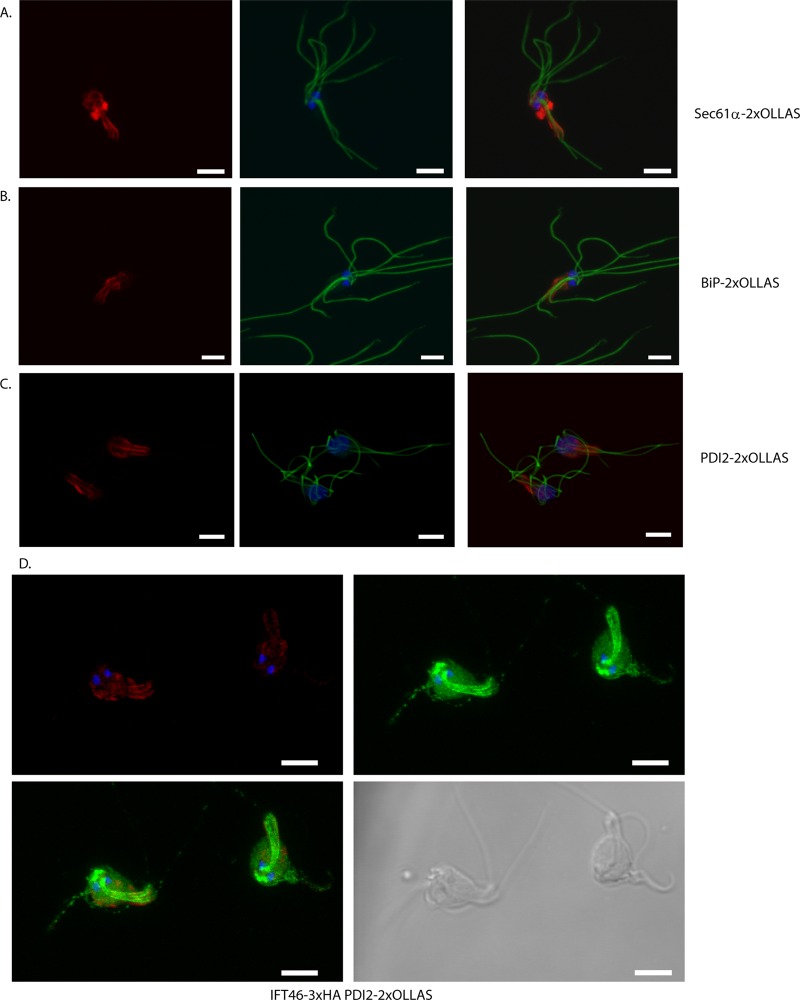
The integration of a selection marker on the chromosome in S. salmonicida was attempted using an approach recently described to be successful in G. intestinalis (10). Three genes potentially associated with the endoplasmic reticulum (ER) (BiP, Sec61α, and protein disulfide isomerase 2 [PDI2]) were cloned into the 2×OLLAS C-terminal tagging vectors, keeping 200 to 500 bp of the endogenous promoters. The plasmid constructs were linearized utilizing a unique restriction site within the cloned region and purified, and 10 to 20 μg of plasmid was used in a regular transfection of S. salmonicida. Stable transfectants were generated by blasticidin S selection, and Western blotting confirmed the expression of the epitope-tagged proteins of expected sizes (BiP-2×OLLAS, 75.1 kDa; Sec61α-2×OLLAS, 55.6 kDa; PDI2-2×OLLAS, 44.5 kDa) in the transgenic cell lines (Fig. 2E). By performing PCR using primers designed to amplify only upon successful integration, we could confirm integration for the BiP-2×OLLAS construct (see Fig. S3 in the supplemental material), whereas PDI2-2×OLLAS and Sec61α-2×OLLAS were not found to be integrated into their respective loci (data not shown). Immunolocalization by confocal microscopy of the three epitope-tagged proteins revealed similar cellular distributions, with signals around the nuclei and around the recurrent flagella throughout the length of the cell (Fig. 4A to C). Some cells displayed an intensely stained region below the nuclei (Fig. 4A to C). These labeling patterns are consistent with the distributions of ER previously reported in electron microscopy studies of S. salmonicida (19).
Fig 4.
Localization of Sec61α-2×OLLAS, BiP-2×OLLAS, PDI2-2×OLLAS, 2×OLLAS, IFT46-3×HA, and PDI2-2×OLLAS in S. salmonicida trophozoites. Cells carrying constructs of Sec61α-2×OLLAS (A), BiP-2×OLLAS (B), PDI2-2×OLLAS (C), and IFT46-3×HA, PDI2-2×OLLAS (D) were fixed using 2% PFA and blocked using 2% BSA in PBS. (A to C) Cells were reacted using the anti-OLLAS rabbit (1:2,000) and anti-tubulin TAT1 (1:150) antibodies and detected using anti-rabbit antibody conjugated to Alexa Fluor 594 (A594) (1:250) and anti-mouse antibody conjugated to Alexa Fluor 488 (A488) (1:200), respectively. (D) Cells were incubated with anti-OLLAS rabbit (1:2,000) and mouse monoclonal anti-HA (1:500) and detected with anti-rabbit antibody conjugated to A594 (1:250) and anti-mouse antibody conjugated to A488 (1:200), respectively. The cells were mounted in VectaShield medium with DAPI and viewed using a Zeiss 510 laser scanning confocal microscope. (A to C, left panel) Maximum intensity projection (MIP) of A594 (red); (A to C, middle) MIP of DAPI (blue) and A488 (green) signals; (A to C, right) overlay of the left and middle panels. The three proteins in panels A to C localize to similar structures enveloping the nuclei and are distributed along the length of the recurrent flagella. (D, upper left) MIP of DAPI (blue) and A594 (red) signals; (D, upper right) MIP of DAPI (blue) and A488 (red) signals. The lower-left panel shows an overlay of the upper panels. The lower-right panel shows the cells in bright-field view. (D) The cells display labeling of foci on the flagella, paired cylindrical signals above the nuclei, two sheets that run the length of the cell to posterior end, and cytoplasmatic staining in the green channel and labeling around the nuclei and along the recurrent flagella in the red channel. Scale bar, 5 μm.
Generation of double transfectants of S. salmonicida.
We explored the option of generating S. salmonicida transfectants carrying two different selectable markers for the purpose of performing colocalization experiments. The transfectants carrying the pSpiro-PAC-IFT46-3×HA-C construct were transfected with the linearized pSpiro-BSR-PDI2-2×OLLAS-C vector, and transfectants were generated by selection with 70 μg/ml blasticidin S, while puromycin selection was maintained. Confocal microscopy of immunostained double transfectants demonstrated the expression of both epitope-tagged proteins in the majority of cells. The localization of the respective proteins was identical to that seen in the two respective single transfectants (Fig. 4D).
DISCUSSION
Very little is known about the genomes and cell biology of diplomonads other than G. intestinalis, the most studied representative of this group of eukaryotes. The purpose of this work was to create a new model system for studying pathogenic diplomonads by constructing vectors and developing methods for the transfection of the fish parasite Spironucleus salmonicida.
We designed a suite of vectors in a cassette format where selectable markers and tags can be exchanged using unique restriction sites (Fig. 1; see also section S2 in the supplemental material). The mislocalization or instability of tagged proteins is a common concern with any tagging strategy (33); thus, we provided the ability to tag both termini of a protein as well as to utilize different epitope tags depending on the task at hand. The 3×HA, 2×OLLAS (25), SBP-GST (16), and SF-TAP (9) tags are available in N-terminal and C-terminal configurations; in addition, there is a C-terminal 3×MYC tag cassette (Fig. 1).
Next, we applied the constructed vectors in order to explore transfection conditions for S. salmonicida cells. Transfection using electroporation with circular DNA proved robust, reproducible, and comparatively fast, with plasmids likely being replicated as episomes. Stable transfection was possible using three different selection markers (pac, nptII, and bsr), and transfected strains appeared stable during extended cultivation under continuous selective pressure.
The performance of the constructed vectors was investigated by establishing stably transfected S. salmonicida strains, using the pac and bsr resistance markers, expressing epitope-tagged proteins as detected by Western blotting for the HA- and OLLAS-tagged constructs (Fig. 2D and E). The subcellular localizations of the epitope-tagged proteins were determined by confocal laser scanning microscopy. Caltractin was shown by colocalization to centrin to reside in the basal bodies (Fig. 2B), as has been observed in Giardia (22). In this species, involvement in the migration and orientation of the basal bodies during mitosis has been suggested as a possible function of caltractin (22). Whether caltractin in S. salmonicida performs a similar role in cell division is unclear.
Fibrillarin exhibited a subnuclear localization distinct from that of genomic DNA in S. salmonicida (Fig. 2A; see also Fig S2 in the supplemental material). Epitope-tagged fibrillarin in Giardia localizes to subcompartments in the nuclei (17), and the present results might suggest the presence of a similar subnuclear organization also in this diplomonad.
IFT46 localized to the flagella and C-shaped sheets running the length of the cell (Fig. 3; see also Fig. S1 in the supplemental material), which terminated in cylindrical structures positioned close to the nuclei. The C-shaped sheet could be interpreted as the striated lamina that partially envelopes the recurrent flagella (18). The potential IFT particles were distributed along the flagella, with enrichment at the tip in some cells (Fig. 3), as has been seen for homologues of IFT complexes A and B in Giardia (12). However, unlike homologues of IFT complex A and B proteins in Giardia that localize to flagellar pores, and to cytoplasmic and membrane-bound regions of the flagellar axonemes (12), IFT46 in S. salmonicida localized prominently to the region above the nuclei where the basal bodies are located. This localization of IFT complex proteins is similar to that observed in other flagellates, such as Chlamydomonas (13, 27). The unregulated maintenance of episomes might lead to elevated and uneven expression levels, as noticed previously in G. intestinalis transfectants (32). We utilized single-crossover homologous recombination, previously shown to be effective in G. intestinalis (10, 14, 34), for the construction of a chromosomally integrated BiP-2×OLLAS strain (see Fig. S3 in the supplemental material). Efforts to integrate the PDI2-2×OLLAS and Sec61α-2×OLLAS constructs site specifically on the chromosome were not successful for reasons unknown to us. Despite this, all three OLLAS-tagged constructs were expressed, and we observed recombinant proteins of the expected size by Western blotting (Fig. 2E). Confocal microscopy of immunostained transfectants expressing the three proteins, PDI2, BiP, and Sec61α, showed similar subcellular localizations (Fig. 4A to C). This pattern is reminiscent of ER distributions inferred from electron microscopy studies in S. salmonicida (19).
Finally, we demonstrated the construction of an S. salmonicida double transfectant using the pac and bsr resistance genes. Even though pac and bsr showed an interaction, this did not preclude the generation of double transfectants by increasing the blasticidin S concentration. Confocal microscopy of the IFT46-3×HA PDI2-2×OLLAS double transfectants revealed protein localizations that matched exactly those determined for single-marker transfectants (Fig. 4D). The option of utilizing several markers for colocalization studies is especially attractive when establishing a new model system for which there is a paucity of reagents and poor performance of heterologous antibodies.
S. salmonicida has been proven amenable to the transfection and generation of stable transfectants, which makes this organism capable of being used as an alternative model system to Giardia in the study of diplomonad biology and their particular solution to being eukaryotic cells.
Supplementary Material
ACKNOWLEDGMENTS
S.G.S., E.E., and J.J.-H. were supported by grants from the Swedish Medical Research Council and FORMAS.
Jan O. Andersson and FeiFei Xu, Department of Cell and Molecular Biology, Uppsala University, are acknowledged as partners in the S. salmonicida genome project and for providing access to a draft assembly of the S. salmonicida genome. Stéphane Gourguechon, Department of Molecular and Cell Biology, UC Berkeley, Berkeley, CA, and Fredrik Söderbom, Department of Molecular Biology, Swedish University of Agricultural Sciences, Uppsala, Sweden, are acknowledged for providing reagents. BioVis, Science for Life Laboratory, Uppsala University, are acknowledged for their assistance with confocal microscopy.
Footnotes
Published ahead of print 14 September 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Andersson JO, Sjogren AM, Davis LA, Embley TM, Roger AJ. 2003. Phylogenetic analyses of diplomonad genes reveal frequent lateral gene transfers affecting eukaryotes. Curr. Biol. 13:94–104 [DOI] [PubMed] [Google Scholar]
- 2. Andersson JO, et al. 2007. A genomic survey of the fish parasite Spironucleus salmonicida indicates genomic plasticity among diplomonads and significant lateral gene transfer in eukaryote genome evolution. BMC Genomics 8:51 doi:10.1186/1471-2164-8-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Benne R. 1994. RNA editing in trypanosomes. Eur. J. Biochem. 221:9–23 [DOI] [PubMed] [Google Scholar]
- 4. Brinkmann H, Philippe H. 2007. The diversity of eukaryotes and the root of the eukaryotic tree. Adv. Exp. Med. Biol. 607:20–37 [DOI] [PubMed] [Google Scholar]
- 5. Correa G, Morgado-Diaz JA, Benchimol M. 2004. Centrin in Giardia lamblia—ultrastructural localization. FEMS Microbiol. Lett. 233:91–96 [DOI] [PubMed] [Google Scholar]
- 6. Dawson SC, et al. 2008. Stable transformation of an episomal protein-tagging shuttle vector in the piscine diplomonad Spironucleus vortens. BMC Microbiol. 8:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Franzen O, et al. 2009. Draft genome sequencing of Giardia intestinalis assemblage B isolate GS: is human giardiasis caused by two different species? PLoS Pathog. 5:e1000560 doi:10.1371/journal.ppat.1000560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fritz-Laylin LK, et al. 2010. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140:631–642 [DOI] [PubMed] [Google Scholar]
- 9. Gloeckner CJ, Boldt K, Schumacher A, Roepman R, Ueffing M. 2007. A novel tandem affinity purification strategy for the efficient isolation and characterisation of native protein complexes. Proteomics 7:4228–4234 [DOI] [PubMed] [Google Scholar]
- 10. Gourguechon S, Cande WZ. 2011. Rapid tagging and integration of genes in Giardia intestinalis. Eukaryot. Cell 10:142–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Greider CW, Blackburn EH. 1987. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell 51:887–898 [DOI] [PubMed] [Google Scholar]
- 12. Hoeng JC, et al. 2008. High-resolution crystal structure and in vivo function of a kinesin-2 homologue in Giardia intestinalis. Mol. Biol. Cell 19:3124–3137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hou Y, et al. 2007. Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J. Cell Biol. 176:653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. House SA, Richter DJ, Pham JK, Dawson SC. 2011. Giardia flagellar motility is not directly required to maintain attachment to surfaces. PLoS Pathog. 7:e1002167 doi:10.1371/journal.ppat.1002167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jerlstrom-Hultqvist J, et al. 2010. Genome analysis and comparative genomics of a Giardia intestinalis assemblage E isolate. BMC Genomics 11:543 doi:10.1186/1471-2164-11-543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jerlstrom-Hultqvist J, Stadelmann B, Birkestedt S, Hellman U, Svard SG. 2012. Plasmid vectors for proteomic analyses in Giardia: purification of virulence factors and analysis of the proteasome. Eukaryot. Cell 11:864–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jimenez-Garcia LF, et al. 2008. Identification of nucleoli in the early branching protist Giardia duodenalis. Int. J. Parasitol. 38:1297–1304 [DOI] [PubMed] [Google Scholar]
- 18. Jorgensen A, Alfjorden A, Henriksen K, Sterud E. 2007. Phylogenetic analysis of the SSU rRNA gene from the piscine diplomonad Spironucleus torosus (Diplomonadida: Hexamitinae). Folia Parasitol. (Praha) 54:277–282 [DOI] [PubMed] [Google Scholar]
- 19. Jorgensen A, Sterud E. 2006. The marine pathogenic genotype of Spironucleus barkhanus from farmed salmonids redescribed as Spironucleus salmonicida n. sp. J. Eukaryot. Microbiol. 53:531–541 [DOI] [PubMed] [Google Scholar]
- 20. Kankaanpää P, et al. 2012. BioImageXD: an open, general-purpose and high-throughput image-processing platform. Nat. Method 9:683–689 [DOI] [PubMed] [Google Scholar]
- 21. Keeling PJ, Doolittle WF. 1997. Widespread and ancient distribution of a noncanonical genetic code in diplomonads. Mol. Biol. Evol. 14:895–901 [DOI] [PubMed] [Google Scholar]
- 22. Meng TC, et al. 1996. Immunolocalization and sequence of caltractin/centrin from the early branching eukaryote Giardia lamblia. Mol. Biochem. Parasitol. 79:103–108 [DOI] [PubMed] [Google Scholar]
- 23. Millet CO, Lloyd D, Williams C, Cable J. 2011. In vitro culture of the diplomonad fish parasite Spironucleus vortens reveals unusually fast doubling time and atypical biphasic growth. J. Fish Dis. 34:71–73 [DOI] [PubMed] [Google Scholar]
- 24. Morrison HG, et al. 2007. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317:1921–1926 [DOI] [PubMed] [Google Scholar]
- 25. Park SH, et al. 2008. Generation and application of new rat monoclonal antibodies against synthetic FLAG and OLLAS tags for improved immunodetection. J. Immunol. Methods 331:27–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paull GC, Matthews RA. 2001. Spironucleus vortens, a possible cause of hole-in-the-head disease in cichlids. Dis. Aquat. Organ. 45:197–202 [DOI] [PubMed] [Google Scholar]
- 27. Pedersen LB, et al. 2005. Chlamydomonas IFT172 is encoded by FLA11, interacts with CrEB1, and regulates IFT at the flagellar tip. Curr. Biol. 15:262–266 [DOI] [PubMed] [Google Scholar]
- 28. Reiss B, Sprengel R, Schaller H. 1984. Protein fusions with the kanamycin resistance gene from transposon Tn5. EMBO J. 3:3317–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roger AJ, Simpson AG. 2009. Evolution: revisiting the root of the eukaryote tree. Curr. Biol. 19:R165–R167 [DOI] [PubMed] [Google Scholar]
- 30. Roxstrom-Lindquist K, et al. 2010. Large genomic differences between the morphologically indistinguishable diplomonads Spironucleus barkhanus and Spironucleus salmonicida. BMC Genomics 11:258 doi:10.1186/1471-2164-11-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sangmaneedet S, Smith SA. 2000. In vitro studies on optimal requirements for the growth of Spironucleus vortens, an intestinal parasite of the freshwater angelfish. Dis. Aquat. Organ. 39:135–141 [DOI] [PubMed] [Google Scholar]
- 32. Singer SM, Yee J, Nash TE. 1998. Episomal and integrated maintenance of foreign DNA in Giardia lamblia. Mol. Biochem. Parasitol. 92:59–69 [DOI] [PubMed] [Google Scholar]
- 33. Terpe K. 2003. Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 60:523–533 [DOI] [PubMed] [Google Scholar]
- 34. Woessner DJ, Dawson SC. 2012. The Giardia median body protein is a ventral disc protein that is critical for maintaining a domed disc conformation during attachment. Eukaryot. Cell 11:292–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Woods A, et al. 1989. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93:491–500 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.