Abstract
Pathogenic Leptospira spp. are likely to encounter higher concentrations of reactive oxygen species induced by the host innate immune response. In this study, we characterized Leptospira interrogans catalase (KatE), the only annotated catalase found within pathogenic Leptospira species, by assessing its role in resistance to H2O2-induced oxidative stress and during infection in hamsters. Pathogenic L. interrogans bacteria had a 50-fold-higher survival rate under H2O2-induced oxidative stress than did saprophytic L. biflexa bacteria, and this was predominantly catalase dependent. We also characterized KatE, the only annotated catalase found within pathogenic Leptospira species. Catalase assays performed with recombinant KatE confirmed specific catalase activity, while protein fractionation experiments localized KatE to the bacterial periplasmic space. The insertional inactivation of katE in pathogenic Leptospira bacteria drastically diminished leptospiral viability in the presence of extracellular H2O2 and reduced virulence in an acute-infection model. Combined, these results suggest that L. interrogans KatE confers in vivo resistance to reactive oxygen species induced by the host innate immune response.
INTRODUCTION
Leptospira is a diverse genus of spirochete bacteria spanning 20 species (5), including the saprophytic species L. biflexa and the pathogenic species L. interrogans (46). Leptospirosis, caused by pathogenic Leptospira spp., is a widespread zoonotic disease (62) that has emerged as an epidemic in urban slum settings (28). Leptospirosis in susceptible hosts, such as humans and hamsters (44), is manifested by a wide array of clinical signs, including fever and jaundice, with a potential for death from multiple organ failure (29). Rats are asymptomatic carriers and serve as a reservoir for pathogenic Leptospira species (54). In such maintenance hosts, leptospires colonize primarily renal tubules, from where they are shed into the urine and persist in freshwater until they gain access to a new mammalian host (29).
The induction of a host immune response generates a defensive oxidative burst (27). In leptospiral infections, the release of reactive oxygen species (ROS) was demonstrated in vitro with isolated rat Kupffer cells upon exposure to Leptospira (36), and in vivo, it has been shown that cattle diagnosed with leptospirosis display elevated levels of serum oxidative stress biomarkers (16). Together, these studies indicated that Leptospira is exposed to ROS during the infection process. During the host oxidative burst, ROS are generated from free radicals of molecular oxygen (superoxide). Reactive oxygen species can be generated through immune cells, including neutrophils and macrophages (45), which utilize both membrane and cytosolic proteins to generate oxygen radicals from molecular oxygen via the NADPH phagocyte oxidase pathway (1, 61). The generated oxygen radicals can then be used to produce other ROS, including hydrogen peroxide (H2O2), hydroxyl radicals, and hypochlorous acid, all of which mediate bacterial killing.
The molecular mechanism(s) underlying the killing of microorganisms via ROS is incompletely understood (18, 19). For bacteria, the primary cellular target of ROS was thought to be DNA, with extensive ROS-generated DNA damage resulting in a loss of bacterial viability (24). However, recent evidence for Salmonella enterica serovar Typhimurium demonstrated a requirement for the periplasmic superoxide dismutase (SodC), rather than the cytoplasmic enzyme counterparts SodA and SodB, for resistance to extracellular ROS, suggesting a noncytoplasmic target for ROS damage (10). Further evidence comes from the demonstration that membrane lipids are targeted by ROS in Borrelia burgdorferi (4), identifying one potential mechanism for the loss of spirochete viability during infection. Microbial pathogens have evolved various mechanisms to counter the effects of host-generated ROS, including the disruption of ROS delivery to the phagosome (60), the inhibition of ROS production (9), active DNA repair systems (23, 56), and the enzymatic detoxification of ROS (3, 20, 64). Microbial enzymes utilized for ROS detoxification include catalases (6) and peroxidases (50), which degrade hydrogen peroxide, and superoxide dismutases, which detoxify superoxide anions (57).
Catalases of pathogenic bacteria are important for optimal detoxification of H2O2 (15, 22), survival in macrophages (13), resistance to phagocyte-mediated killing (52), and virulence (3, 53, 59, 64). The pathogenic species L. interrogans displays catalase activity, while the saprophytic species L. biflexa displays predominantly peroxidatic activity (7), despite the fact that a katG homolog is present in L. biflexa (65). L. interrogans can degrade H2O2 at concentrations 50-fold higher than those tolerated by L. biflexa (8), consistent with a greater susceptibility of L. biflexa to H2O2-mediated killing (38). This suggests that L. interrogans has evolved an extensive H2O2 detoxification system which is absent from L. biflexa.
Additional evidence suggesting a role for catalase activity in virulence comes from a previous study which showed the upregulation of the catalase KatE by L. interrogans when exposed to “in vivo-like” conditions represented by bacterial growth in medium depleted of iron and containing 10% fetal bovine serum (17). This finding has been supported at the transcriptional level, where katE transcript levels were increased in response to an increase in temperature and the presence of serum (32, 47, 65). These results, together with the potential for the exposure of Leptospira to extracellular ROS during the infection process, prompted us to further characterize the leptospiral response to ROS. In this study, we demonstrate that KatE is a periplasmic catalase that enhances the resistance of L. interrogans to extracellular oxidative stress and is required for virulence in an acute animal model.
MATERIALS AND METHODS
Leptospira strains and culture conditions.
L. interrogans serovar Copenhageni strain Fiocruz L1-130 was isolated from a clinical sample in Salvador, Brazil (28). L. interrogans serovar Pomona L523 was originally isolated from a clinical sample in Australia and was obtained from the World Health Organization Collaborating Centre for Reference and Research on Leptospirosis, Brisbane, Australia. L. interrogans serovar Manilae strain L495 was originally isolated from a clinical sample in the Philippines and was provided by Nobuo Koizumi, National Institute of Infectious Diseases, Tokyo, Japan, and L. biflexa serovar Patoc strain Patoc1 (designated Paris strain) was obtained from a freshwater stream (2) and is stored and maintained at the National Reference Center of Leptospira (Pasteur Institute, Paris, France). All strains were cultured aerobically in Ellinghausen-McCullough-Johnson-Harris (EMJH) medium (26) at 29.5°C.
Catalase assay.
Recombinant KatE (rKatE) was purified by using immobilized metal ion affinity chromatography as described previously (17). This recombinant protein preparation was estimated to have a purity of >95% and appeared as a single band in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analyses. The enzymatic activity was measured by using a catalase assay kit (Cayman Chemical Company, Ann Arbor, MI) at room temperature according to the manufacturer's instructions, with the bovine catalase enclosed in the kit serving as a positive control. Recombinant KatE was added at final concentrations ranging from 0.23 ng/μl to 15 ng/μl. Catalase (KatE) inhibition experiments were conducted by adding 3-amino-1,2,4-triazol (ATZ) (USB Corporation, Cleveland, OH) to the assay buffer (100 mM potassium phosphate, pH 7.0) at a final concentration of 100 mM, prior to conducting the catalase assay. The formaldehyde control assay was performed concurrently according to the manufacturer's protocol. Catalase and formaldehyde standard assays were conducted using duplicate samples. The experiment was performed a total of three times, with comparable results; a representative result from one experiment is presented.
Bacterial oxidative stress conditions and inhibition assays.
Leptospires were enumerated by using a Petroff-Hausser counting chamber (Fisher Scientific, Ottawa, Ontario, Canada) and an Eclipse 50i dark-field microscope (Nikon, Mississauga, Ontario, Canada). Bacteria were grown to >4 × 108 bacteria/ml and diluted to 3 × 108 bacteria/ml in either EMJH medium (without H2O2) or EMJH medium containing H2O2 at a final concentration of 0.1 mM, 1 mM, or 10 mM, with or without 100 mM ATZ. Samples were incubated for 1 h at 37°C. Bacterial oxidative stress assays were also conducted, as described above, without ATZ for 10 min at 37°C. A total of 5 × 109 cells of strain L1-130 from each experimental group (no H2O2 exposure and 0.1, 1, and 10 mM H2O2 exposures) were collected by centrifugation at 2,000 × g, and the bacteria were flash-frozen in liquid nitrogen and stored at −20°C for subsequent immunoblot analyses. Exposures for 30 and 60 min were also tested and yielded similar results.
Bacterial viability assays.
Triplicate samples of Leptospira bacteria exposed to the above-described conditions were dispensed into sterile Falcon 96-well flat-bottom polystyrene microplates (Becton Dickinson, Franklin Lakes, NJ) (250 μl/well). Fifty microliters of an alamarBlue (Biosource, Camarillo, CA) working solution, consisting of 0.4 ml alamarBlue stock solution plus 9.6 ml 0.1 M potassium phosphate buffer (pH 7.4), was added to each well and incubated at 30°C (the optimal temperature for leptospiral growth in vitro) in a humidified Hybaid chamber (ThermoFisher Scientific). In this assay, viable bacteria are able to reduce the nonfluorescent compound resazurin to the fluorescent compound resorufin (43). The chromogenic shift was measured by fluorescence using a 530- ± 25-nm filter for excitation and a 590- ± 20-nm filter for emission in a Synergy HT microplate reader (BioTek) at 0 and 4 h postincubation. Negative controls without bacteria were included. Percent survival was calculated at 4 h by subtracting initial readings (time zero) and the negative control and then by dividing by fluorescence values obtained from leptospires that had not been exposed to H2O2 and were grown in either EMJH medium or EMJH medium with 100 mM ATZ. Leptospiral viability was confirmed via dark-field microscopy, where motile bacteria were deemed viable. Data are representative of a single experiment with three replicates. Experiments were repeated at least two times, with similar results. Statistical analyses were conducted by using the two-tailed Student t test.
Protein fractionation.
Bioinformatic analysis to predict the cellular location of KatE was performed by using pSORTb v3.0 (66). The experimental subcellular localization of KatE in Leptospira was determined by Triton X-114 solubilization of the outer membrane and subsequent fractionation into detergent (DET) and aqueous (AQ) phases, as described previously (21, 48, 68), except that fractionation was performed on approximately 4 × 1010 cells at a concentration of 5 × 109 bacteria/ml.
Immunoblot analyses.
Bacterial cells exposed to conditions of oxidative stress and subcellular protein fractionation samples were resuspended in SDS-PAGE sample loading buffer, subjected to SDS-PAGE, and transferred onto polyvinylidene difluoride (PVDF) membranes, as described previously (17). Membranes were probed at room temperature with rabbit antiserum to either KatE (17), LipL21 (11), FlaA2 (30), or FlaA1 (12) at a 1:3,000 dilution for membranes containing the fractionated protein samples and at a 1:2,000 dilution for membranes containing protein samples from oxidative stress assays, and immunoreactivity was detected with a goat anti-rabbit IgG horseradish peroxidase (HRP)-conjugated antibody (Sigma-Aldrich Chemie GmbH, L'Isle d'Abeau Chesnes, Saint-Quentin Fallavier, France). Blots were developed with a chemiluminescent substrate (SuperSignal West Pico chemiluminescent substrate; Thermo Scientific, Illkirch, France) according to the manufacturer's instructions.
Leptospira insertion mutants.
Transposon mutagenesis was carried out on L. interrogans serovar Manilae strain L495 and L. interrogans serovar Pomona strain L523 with a kanamycin-resistant Himar1 transposon, as described previously (39). Among the transformants, we identified an L. interrogans serovar Manilae mutant (m69) with an insertion into katE (LA1859) 384 bases into the 1,446-bp gene and an L. interrogans serovar Pomona mutant (mutant P3) with a transposon insertion 1,272 bases into the gene.
Genomic DNA was extracted from 20-ml cultures by using the QIAamp DNA blood minikit (Qiagen, Inc., Valencia, CA). Confirmation of the genotype was performed by using PCR with primers 69a (5′-GATACGGAAAGAGATCCGAG-3′) and 69b (5′-ATGATCTGAACACAAAACTTC-3′), which are located in the flanking sequences of the insertion site of the transposon, and Southern blots of EcoRV-digested DNA were probed for hybridization with the kanamycin-resistant cassette.
Quantitative PCR was performed to test the effect of the transposon insertion on the transcription of the gene (lic12031) immediately downstream of katE. Wild-type strains of L. interrogans serovars Pomona and Manilae and the respective katE mutant strains were grown to the same density, and RNA was extracted as previously described (32), with the following modifications. Briefly, Leptospira bacteria were grown to 8 × 108 bacteria/ml, and 20 ml of each culture was centrifuged at 3,200 × g. Each pellet was resuspended in 1 ml TRIzol (Invitrogen, Saint-Aubin, Île-de-France, France), and RNA was purified according to the manufacturer's instructions. DNA digestion was conducted by the addition of 5 μl 10× Turbo DNase buffer (Ambion, Saint-Aubin, Île-de-France, France) and 1 μl Turbo DNase (Ambion) to the sample, followed by a 30-min incubation at 37°C. An additional 3 μl of Turbo DNase was added to the sample, followed by a 30-min incubation at 37°C. Turbo DNase was inactivated by the addition of 11 μl of DNase inactivation reagent (Ambion) to the sample and incubation for 5 min with occasional mixing. The sample was centrifuged at 10,000 × g for 2 min, and the supernatant was transferred into a new microcentrifuge tube.
First-strand cDNA synthesis was performed by using 1 μl RNA extract, 4 μl 5× iScript reaction mix (Bio-Rad, Marnes-la-Coquette, Île-de-France, France), 1 μl iScript reverse transcriptase (Bio-Rad), and 14 μl RNase-free H2O (Bio-Rad). Thermal cycles of 25°C, 42°C, and 85°C for 5, 30, and 5 min, respectively, were performed by using a DNAEngine Peltier thermal cycler (Bio-Rad). A negative-control reaction to control for genomic DNA contamination of RNA samples in downstream quantitative PCR experiments was performed in the absence of reverse transcriptase.
Quantitative PCR analyses were performed with primer pair P1 (5′-TTTCGCATAATCTCCGTTCC) and P2 (5′-CTCCTCCGATCGTCTGAGTC) (amplifies bp 589 to 1837 of the lic12031 open reading frame [ORF]). The flaB1 transcript was used as a normalizing control and was amplified by primer pair fP1 (5′-GAGAGAAACACCGAAGACGG) and fP2 (5′-TGAATAGCAAGAACCCGGAT) (amplifies bp 187 to 287 of the flaB1 [lic11890] ORF). Quantitative PCR was conducted by using 10 μl SsoFast EvaGreen Supermix (Bio-Rad), 5 pmol each primer, and 1 μl of either cDNA or the negative control lacking reverse transcriptase in a total volume of 20 μl. Thermal cycling was performed by using a C1000 Thermal Cycler CFX96 real-time system (Bio-Rad), with a cycling program of 1 cycle at 95°C for 3 min and 39 cycles at 95°C for 10 s, followed by 55°C for 30 s. A melting-curve analysis was performed to assess the amplification of a single amplicon per target by raising the temperature to 95°C for 10 s, followed by a 65°C to 95°C gradient with fluorescence measurements at 0.5°C increments. Triplicate quantitative PCR analyses were performed on each of 2 biological replicates. The Student t test was used to evaluate the differential expression of lic12031 when katE mutant strains of L. interrogans serovars Manilae and Pomona were compared with their respective wild-type strains.
Hamster infections.
L. interrogans strains were tested for virulence in the hamster model of acute leptospirosis. Male hamsters aged 28 days (Janvier) were inoculated intraperitoneally with 106 leptospires (50% infective dose [ID50] of L. interrogans serovar Manilae of <100; n = 8) or 103 leptospires (ID50 of L. interrogans serovar Pomona of <100; n = 10), which reflects the standard in-house operating infectious doses for the two independent laboratories conducting the studies (30). Animals were monitored daily for clinical signs of leptospirosis and euthanized when moribund, in accordance with animal ethics requirements. In the countries involved in this study, the use of clinical disease symptoms as an experimental endpoint is required. Statistical analyses were performed by using Fisher's exact test.
RESULTS
Recombinant KatE displays catalase activity.
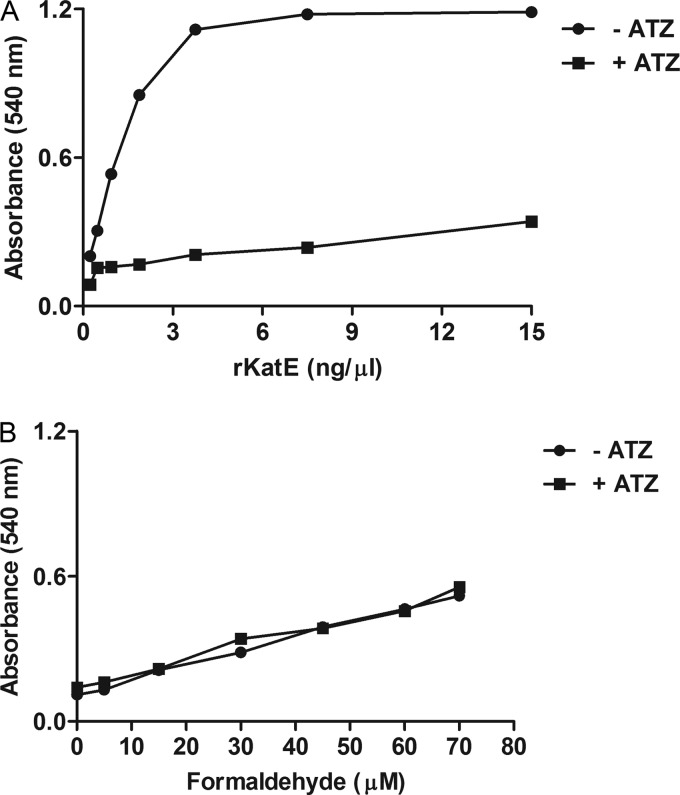
Soluble recombinant KatE (rKatE) was expressed, purified, and utilized in a catalase-specific assay (Fig. 1) that measures the peroxidatic activity of catalase using methanol as an electron donor to decompose H2O2 into formaldehyde and water (25, 63). Other enzymes with peroxidatic activity do not utilize methanol to decompose H2O2 into formaldehyde (25), and therefore, any observed formaldehyde production can be attributed to the presence of catalase. The absorbance increased proportionally to the rKatE concentration, indicating the production of formaldehyde from methanol and H2O2 (Fig. 1A). The inclusion of 100 mM ATZ, a catalase inhibitor (37), diminished the absorbance relative to the rKatE concentration (Fig. 1A) but did not diminish the absorbance due to the formaldehyde concentration (Fig. 1B).
Fig 1.
Recombinant KatE displays catalase activity. Recombinant KatE (rKatE) was analyzed in a catalase assay in the presence or absence of the specific catalase inhibitor ATZ (100 mM). (A) Increased absorbance with increasing rKatE concentrations in the absence of ATZ. No correlation was observed between the absorbance and the rKatE concentration in the presence of ATZ. (B) ATZ did not alter the reaction of formaldehyde standards with chromogen-producing compounds. Data are reported as the means from duplicate absorbance values at 540 nm for a single experiment.
H2O2-induced oxidative stress does not influence KatE expression.
To test whether the expression of KatE was altered during the exposure of Leptospira to H2O2-induced oxidative stress, bacteria were exposed to H2O2 concentrations ranging from 0 to 10 mM. Total cellular protein was separated by SDS-PAGE and subjected to immunoblot analysis using antisera specific for KatE or FlaA1 (flagellar protein A1) (used as a loading control since it is constitutively expressed under various conditions [17, 32, 35]). KatE- and FlaA1-specific antisera recognized bands of molecular masses consistent with the predicted molecular masses of 54.7 kDa and 34.9 kDa for KatE and FlaA1, respectively (Fig. 2). Quantitative fluorescence immunoblot analyses of KatE expression in response to various H2O2 concentrations (normalized to FlaA1 expression) showed that KatE expression levels remained unchanged when cells were exposed to increasing H2O2 concentrations (P > 0.5 for all concentrations).
Fig 2.
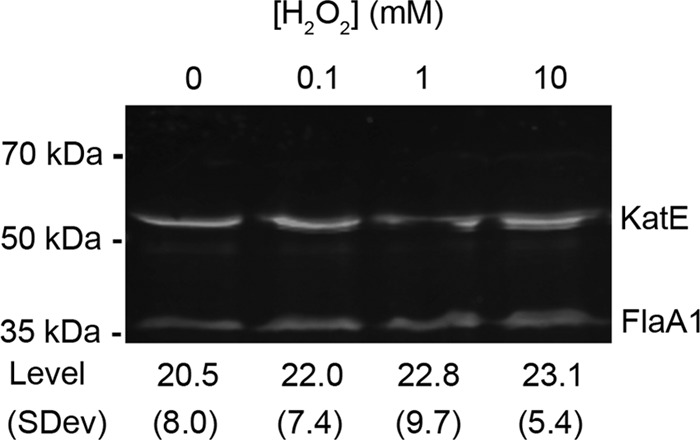
H2O2-induced oxidative stress does not affect KatE expression. Leptospires were exposed to 0, 0.1, 1, or 10 mM H2O2, and immunoblots were performed on total protein. Higher bands represent KatE, and lower bands represent FlaA1, with the latter being used as a loading control. The observed quantitative fluorescence levels are indicated, along with the standard deviations (SDev).
KatE is localized to the periplasmic space.
Bioinformatic analysis using the subcellular localization prediction program PSORTb predicted KatE to be located within the periplasm. To experimentally confirm this prediction, total cellular L. interrogans protein was fractionated to yield a protoplasmic cylinder (PC) containing cytoplasmic and inner membrane proteins, an aqueous (AQ) phase containing periplasmic proteins, and a detergent (DET) phase containing outer membrane proteins (21, 48, 68). Protein fractions were separated by SDS-PAGE (Fig. 3A) and subjected to immunoblot analysis. Antiserum to LipL21 reacted with a protein of approximately 20 kDa in the DET and PC phases (Fig. 3B), in agreement with previous observations showing that LipL21 fractionates with the PC and DET fractions (11). Antiserum to FlaA2 displayed reactivity with a band migrating at approximately 27 kDa in the PC fraction only (Fig. 3B), which is the expected fractionation result for a protein that associates with the flagellum core structure (30). Importantly, KatE-specific antiserum reacted with a band migrating at 52 to 55 kDa in both the AQ and PC phases, but no band was observed in the DET phase (Fig. 3B), consistent with the predicted localization of KatE to the periplasmic space.
Fig 3.
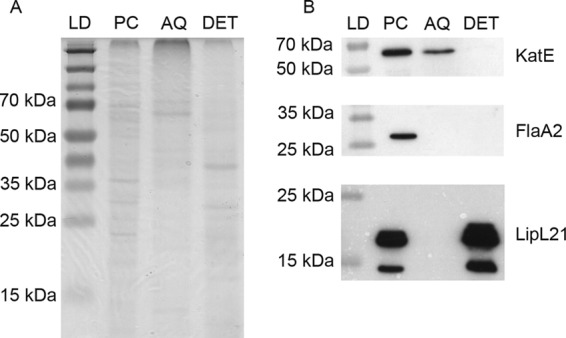
KatE localizes to the periplasmic space. Immunoblot analyses were conducted on L. interrogans fractionated protein samples. (A) SDS-polyacrylamide gel displaying the amount of protein loaded from each fraction. (B) Immunoblots demonstrating the reactivity of antisera with the indicated proteins in each fraction. Antisera to FlaA2 and LipL21 were used as controls to determine fraction purity. LD, protein ladder; PC, protoplasmic cylinder; AQ, aqueous phase; DET, detergent phase. The positions of molecular mass markers are indicated (in kilodaltons).
Construction of katE transposon mutants and confirmation of the lack of transposon-induced downstream polar effects.
L. interrogans transposon mutants disrupted in katE were constructed to assess the viability of these mutants under conditions of oxidative stress and during in vivo infection. Complementation in L. interrogans is extremely difficult and has been documented only twice (49, 67). We compensated for this lack of a genetic “knock-in” capability by constructing independent mutations in two different L. interrogans serovars (L. interrogans serovar Pomona mutant strain P3 and L. interrogans serovar Manilae mutant strain m69) and assessing if similar results were obtained with each of these mutants. To rule out the possibility of downstream polar effects of the katE (lic12032) transposon insertion, we compared the transcript levels of the gene immediately downstream of katE, lic12031, in the two independent mutants and their respective wild-type strains using quantitative PCR. These experiments indicated that lic12031 transcription was not significantly different in the katE mutants compared to their wild-type strains (see Fig. S1 in the supplemental material).
katE enhances L. interrogans viability under conditions of oxidative stress.
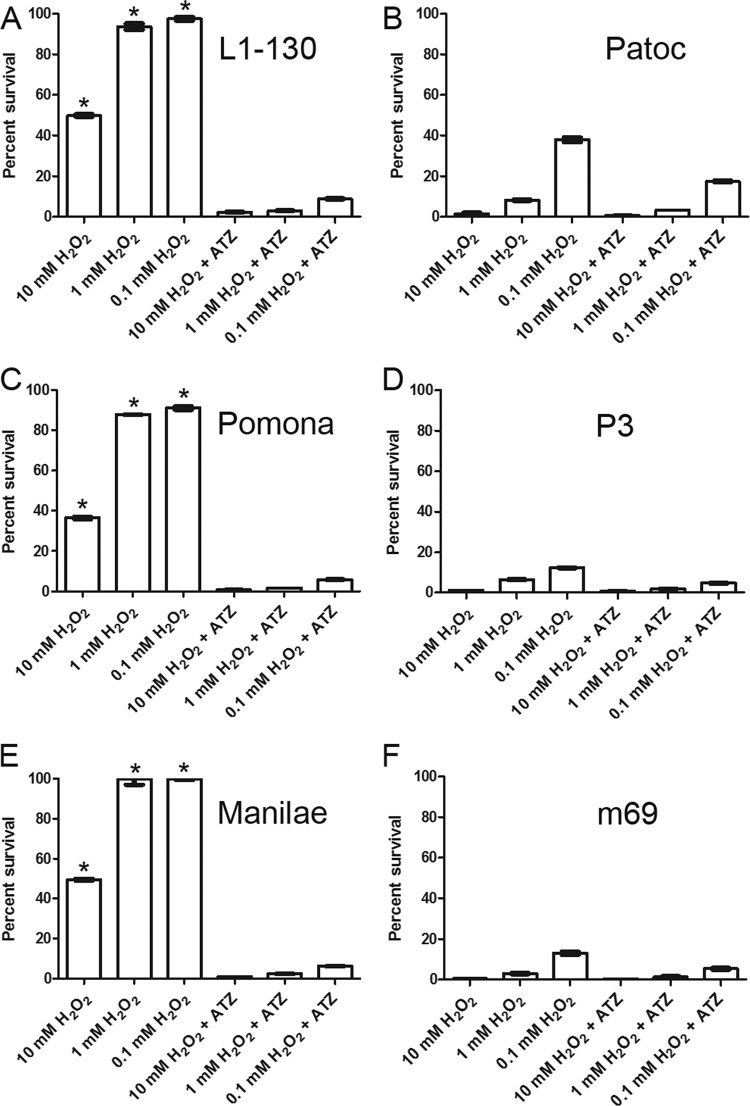
Leptospira strains were exposed to various H2O2 concentrations and tested for viability at 4 h by using an alamarBlue survival assay. Wild-type L. interrogans strains displayed survival rates of 85 to 100% at 0.1 and 1 mM H2O2 and 30 to 50% at 10 mM H2O2 (Fig. 4A, C, and E) compared to Leptospira cells incubated in EMJH medium without exposure to H2O2. L. biflexa displayed survival rates of approximately 40, 10, and 2% (Fig. 4B) at 0.1, 1, and 10 mM H2O2, respectively, while katE mutant strains P3 and m69 showed survival rates of approximately 10, 5, and 2% (Fig. 4D and F) at 0.1, 1, and 10 mM H2O2, respectively, compared to growth in EMJH medium without exposure to H2O2. In the absence of H2O2, the wild-type and mutant strains displayed similar growth curves at 30°C (data not shown).
Fig 4.
KatE enhances resistance to extracellular H2O2. Shown are percent survivals of Leptospira strains exposed to oxidative stress conditions in the presence or absence of the catalase inhibitor ATZ. (A) L. interrogans serovar Copenhageni; (B) L. biflexa serovar Patoc; (C) L. interrogans serovar Pomona; (D) L. interrogans serovar Pomona katE mutant strain P3; (E) L. interrogans serovar Manilae; (F) L. interrogans serovar Manilae katE mutant strain m69. Results are shown as means of triplicate values from a single experiment. Experiments were performed twice, with similar results. Statistical analyses were conducted by comparing the percent survival at a particular H2O2 concentration using the two-tailed Student t test under the following comparative conditions: (i) the presence and absence of ATZ, (ii) L1-130 versus Patoc, and (iii) wild type (L. interrogans serovar Pomona or Manilae) versus mutant (P3 or m69). Standard deviations are indicated, and significant differences between the means (P < 0.0001) are indicated by an asterisk, while the absence of an asterisk indicates no significant difference.
The inhibition of catalase activity with 100 mM ATZ severely reduced the viability of wild-type L. interrogans strains in the presence of H2O2 (Fig. 4A, C, and E) compared to Leptospira cells incubated in EMJH medium without H2O2 but in the presence of 100 mM ATZ. The presence of ATZ had a minimal effect on the survival of L. biflexa bacteria when exposed to H2O2 (Fig. 4B), consistent with the lack of a dependence of this species on catalase production. The survival of L. interrogans mutant strains upon H2O2 exposure in the presence of ATZ was slightly reduced to the background levels observed for the wild-type strains (Fig. 4D and F). No effect on viability was observed for bacteria in the presence of ATZ and in the absence of H2O2 (data not shown).
katE is required for virulence of L. interrogans.
To test the requirement for katE in leptospiral virulence, katE mutant strains of L. interrogans serovar Manilae (m69) and L. interrogans serovar Pomona (P3) were used in infection experiments for comparison to the wild-type strains. Virulence was assessed by using as an experimental endpoint the ability of the leptospiral strains to cause clinical disease symptoms. All hamsters injected with the wild-type strains exhibited signs of disease and were euthanized by day 19 postinfection, whereas all hamsters injected with the m69 or P3 mutant survived to 21 days postinfection and did not exhibit signs of disease (Fig. 5).
Fig 5.
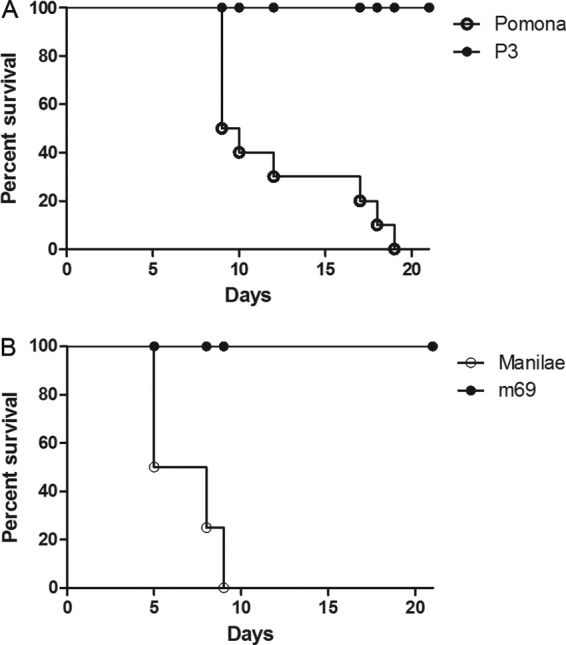
Leptospira katE mutants are attenuated in hamsters. All animals infected with wild-type strains of L. interrogans serovars Pomona (A) and Manilae (B) displayed disease symptoms, and all were euthanized by 19 days postinfection. Animals infected with mutant strains P3 (A) and m69 (B) did not display disease symptoms and exhibited 100% survival at 21 days postinfection (P ≤ 0.0007 by Fisher's exact test).
DISCUSSION
In this study, we investigated the responses of Leptospira species to oxidative stress conditions and, through insertional inactivation mutagenesis, characterized the role of the L. interrogans H2O2-detoxyifying enzyme KatE in protection from oxidative damage and in leptospiral virulence. Here we showed that L. interrogans serovars Manilae and Pomona are more resistant to high H2O2 concentrations than the saprophyte L. biflexa, a finding in agreement with data from a previous report (38). Additionally, we demonstrated that leptospiral resistance to an H2O2-rich environment is mediated by KatE and that the loss of KatE expression leads to an attenuation of virulence for hamsters.
Through insertional inactivation in pathogenic Leptospira strains, seven mutants were previously shown to display attenuated virulence in animal models of infection, including mutations generated in the genes encoding an outer membrane lipoprotein, Loa22 (49); a heme oxygenase, HemO (41); a flagellum motor switch protein, FliY (31); two enzymes involved in lipopolysaccharide (LPS) biosynthesis (40); and the flagellar protein FlaA1 (30). Reduced virulence was achieved upon the inactivation of the gene encoding a putative outer membrane protease ClpB protein (34). Our identification of KatE as an additional virulence factor in Leptospira highlights both the dependence of this pathogen on a diverse repertoire of enzymes for survival and the requirement for a functional ROS resistance mechanism for full leptospiral pathogenesis.
In a previous quantitative proteomic study (17), we detected increased expression levels of KatE in L. interrogans serovar Copenhageni bacteria shifted to medium depleted of iron and supplemented with 10% fetal bovine serum to mimic conditions encountered within the host. Similar independent quantitative proteomic studies demonstrated increased expression levels of KatE in response to a shift in the temperature of L. interrogans from 29.5°C to 37°C (A. Eshghi and C. E. Cameron, unpublished observations), supporting data from a previous report showing that katE was upregulated more than 2-fold upon a shift of L. interrogans serovar Lai cells from environmental to physiological temperatures (32). Interestingly, Lo et al. did not observe an increase in katE expression levels within wild-type Leptospira cells under iron-limiting conditions (33), and similarly, in this study, we did not observe an increase in KatE expression levels upon the exposure of wild-type Leptospira bacteria to hydrogen peroxide-rich conditions. Combined, these studies suggest that in Leptospira temperature may serve as an environmental cue resulting in altered KatE expression, which differs from the requirement for H2O2 for the altered expression of ROS-detoxifying enzymes observed for Salmonella enterica serovar Typhimurium and Escherichia coli (55). An elevated temperature would be an early indicator of a shift to a host environment, resulting in increased leptospiral KatE expression levels and, thus, preexisting protection from a subsequent host-induced oxidative burst response.
Cell fractionation experiments showed that KatE was present in both the aqueous phase and the protoplasmic cylinder, suggesting that this protein localizes, at least partially, to the periplasmic space. Cytoplasmic catalases are utilized for the detoxification of endogenous H2O2 produced from energy generation in the electron transport chain, while periplasmic catalases are utilized for the detoxification of exogenous H2O2 encountered during the oxidative burst response produced by host immune cells (42, 53). A periplasmic location for KatE may thus contribute to leptospiral survival upon infection by protecting periplasmic components against a host-derived oxidative burst response and is consistent with a previously reported observation that establishes the importance of periplasmic, but not cytoplasmic, superoxide dismutase for extracellular ROS resistance (10).
Further evidence supporting KatE-mediated extracellular H2O2 resistance comes from microarray analyses investigating iron-responsive genes. These investigations identified a peroxide stress regulator PerR homolog encoded by the la1857 (lic12034) gene, which lies upstream of katE. The mutation of la1857 resulted in a 4.3-fold increase in the katE expression level and an 8-fold increase in resistance to extracellular H2O2 (33); the perR mutant retained virulence in an animal model (39). To further delineate the critical role of KatE in leptospiral resistance to extracellular oxidative stress conditions, we conducted assays of survival upon exposure to conditions of H2O2-induced stress. The fact that leptospiral survival was significantly reduced when bacteria were exposed to 10 mM H2O2 in the presence of the specific catalase inhibitor ATZ, combined with the fact that the survival of the katE mutants was completely eliminated upon exposure to H2O2-rich conditions, demonstrates that bacterial survival under conditions of oxidative stress is dependent upon catalase activity. The approximately 50% survival rate of L. interrogans cells exposed to 10 mM H2O2 likely indicates that this concentration of H2O2 is on the maximal end of the oxidative stress conditions that L. interrogans can efficiently withstand. Together, these results indicate that KatE expression by Leptospira significantly enhances the ability of the pathogen to survive extracellular H2O2 toxicity.
An early study by Corin et al. (7) demonstrated that L. interrogans lysates can detoxify H2O2 concentrations that are 50-fold higher than those detoxified by L. biflexa lysates. In contrast, our study, which measured bacterial survival, rather than analyzing whole-cell lysates, suggested a range of 2- to 50-fold-higher resistance in L. interrogans than in L. biflexa. Corin et al. concluded that L. interrogans possesses this highly efficient H2O2 detoxification system to detoxify endogenous rather than exogenous H2O2 based on the following two observations. First, L. interrogans, but not L. biflexa, contains cytochrome d, a heme-containing enzyme capable of catalyzing H2O2 to water and oxygen. Second, the extracellular locale of pathogenic Leptospira species would preclude exposure to high concentrations of extracellular H2O2, as would be encountered within a phagosome (7). However, a recent study using a zebrafish embryo system demonstrated that L. interrogans remains in host immune cells for up to 48 h and may use these cells as a means of dissemination to different tissues (14). This observation has been confirmed through an in vitro study demonstrating that Leptospira can survive within macrophages (58). It is therefore plausible that intracellular Leptospira bacteria may be exposed to high concentrations of exogenous H2O2 and other ROS while residing within host immune cells and that KatE expression may confer protection to Leptospira in this environmental context. Consistent with this hypothesis, in vivo studies conducted here with the two independent katE mutant strains L. interrogans serovar Manilae m69 and L. interrogans serovar Pomona P3 showed a strong attenuation of virulence for both mutants in an acute-infection model, demonstrating the essential nature of this enzyme for leptospiral virulence.
Supplementary Material
ACKNOWLEDGMENTS
We thank David A. Haake for the generous gift of L. interrogans serovar Copenhageni strain Fiocruz L1-130 and Tim Witchell for assistance with preparing Leptospira cultures.
This work was funded by a discovery grant from the Natural Sciences and Engineering Research Council of Canada (grant 327186 to C.E.C.), the Institut Pasteur (M.P.), the French Ministry of Research (M.P.; ANR-08-MIE-018), the doctoral program from the Région Ile-de-France (K.L.; DIM Malinf), the Australian Research Council (B.A. and G.L.M.), and the National Health and Medical Research Council (B.A. and G.L.M.). A.E. is a recipient of a University of Victoria fellowship, a graduate scholarship, and a Pacific Century graduate scholarship, and C.E.C. is a Canada research chair in molecular pathogenesis and a Michael Smith Foundation for Health Research scholar.
Footnotes
Published ahead of print 27 August 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Babior BM, Lambeth JD, Nauseef W. 2002. The neutrophil NADPH oxidase. Arch. Biochem. Biophys. 397:342–344 [DOI] [PubMed] [Google Scholar]
- 2. Babudieri B. 1961. Studio serologico del gruppo Semaranga-Patoc di Leptospira biflexa, p 408–414 World Health Organization, Geneva, Switzerland [Google Scholar]
- 3. Bishai WR, Howard NS, Winkelstein JA, Smith HO. 1994. Characterization and virulence analysis of catalase mutants of Haemophilus influenzae. Infect. Immun. 62:4855–4860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boylan JA, Lawrence KA, Downey JS, Gherardini FC. 2008. Borrelia burgdorferi membranes are the primary targets of reactive oxygen species. Mol. Microbiol. 68:786–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cerqueira GM, Picardeau M. 2009. A century of Leptospira strain typing. Infect. Genet. Evol. 9:760–768 [DOI] [PubMed] [Google Scholar]
- 6. Chelikani P, Fita I, Loewen PC. 2004. Diversity of structures and properties among catalases. Cell. Mol. Life Sci. 61:192–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Corin RE, Boggs E, Cox CD. 1978. Enzymatic degradation of H2O2 by Leptospira. Infect. Immun. 22:672–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Corin RE, Cox CD. 1980. Characterization of leptospiral catalase and peroxidase. Can. J. Microbiol. 26:121–129 [DOI] [PubMed] [Google Scholar]
- 9. Cowley SC, Myltseva SV, Nano FE. 1996. Phase variation in Francisella tularensis affecting intracellular growth, lipopolysaccharide antigenicity and nitric oxide production. Mol. Microbiol. 20:867–874 [DOI] [PubMed] [Google Scholar]
- 10. Craig M, Slauch JM. 2009. Phagocytic superoxide specifically damages an extracytoplasmic target to inhibit or kill Salmonella. PLoS One 4:e4975 doi:10.1371/journal.pone.0004975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cullen PA, Haake DA, Bulach DM, Zuerner RL, Adler B. 2003. LipL21 is a novel surface-exposed lipoprotein of pathogenic Leptospira species. Infect. Immun. 71:2414–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cullen PA, et al. 2005. Surfaceome of Leptospira spp. Infect. Immun. 73:4853–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Das D, Bishayi B. 2009. Staphylococcal catalase protects intracellularly survived bacteria by destroying H2O2 produced by the murine peritoneal macrophages. Microb. Pathog. 47:57–67 [DOI] [PubMed] [Google Scholar]
- 14. Davis JM, Haake DA, Ramakrishnan L. 2009. Leptospira interrogans stably infects zebrafish embryos, altering phagocyte behavior and homing to specific tissues. PLoS Negl. Trop. Dis. 3:e463 doi:10.1371/journal.pntd.0000463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elkins JG, Hassett DJ, Stewart PS, Schweizer HP, McDermott TR. 1999. Protective role of catalase in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide. Appl. Environ. Microbiol. 65:4594–4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Erdogan HM, et al. 2008. Serum sialic acid and oxidative stress parameters changes in cattle with leptospirosis. Vet. Res. Commun. 32:333–339 [DOI] [PubMed] [Google Scholar]
- 17. Eshghi A, Cullen PA, Cowen L, Zuerner RL, Cameron CE. 2009. Global proteome analysis of Leptospira interrogans. J. Proteome Res. 8:4564–4578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fang FC. 2011. Antimicrobial actions of reactive oxygen species. mBio 2(5):e00141–11 doi:10.1128/mBio.00141-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fang FC. 2004. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2:820–832 [DOI] [PubMed] [Google Scholar]
- 20. Fang FC, et al. 1999. Virulent Salmonella typhimurium has two periplasmic Cu,Zn-superoxide dismutases. Proc. Natl. Acad. Sci. U. S. A. 96:7502–7507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haake DA, et al. 1991. Changes in the surface of Leptospira interrogans serovar grippotyphosa during in vitro cultivation. Infect. Immun. 59:1131–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Howell ML, et al. 2000. AnkB, a periplasmic ankyrin-like protein in Pseudomonas aeruginosa, is required for optimal catalase B (KatB) activity and resistance to hydrogen peroxide. J. Bacteriol. 182:4545–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Imlay JA, Linn S. 1986. Bimodal pattern of killing of DNA-repair-defective or anoxically grown Escherichia coli by hydrogen peroxide. J. Bacteriol. 166:519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Imlay JA, Linn S. 1988. DNA damage and oxygen radical toxicity. Science 240:1302–1309 [DOI] [PubMed] [Google Scholar]
- 25. Johansson LH, Borg LA. 1988. A spectrophotometric method for determination of catalase activity in small tissue samples. Anal. Biochem. 174:331–336 [DOI] [PubMed] [Google Scholar]
- 26. Johnson RC, Harris VG. 1967. Differentiation of pathogenic and saprophytic leptospires. Growth at low temperatures. J. Bacteriol. 94:27–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kawahara T, et al. 2004. Role of nicotinamide adenine dinucleotide phosphate oxidase 1 in oxidative burst response to Toll-like receptor 5 signaling in large intestinal epithelial cells. J. Immunol. 172:3051–3058 [DOI] [PubMed] [Google Scholar]
- 28. Ko AI, Galvao Reis M, Ribeiro Dourado CM, Johnson WD, Jr, Riley LW. 1999. Urban epidemic of severe leptospirosis in Brazil. Salvador Leptospirosis Study Group. Lancet 354:820–825 [DOI] [PubMed] [Google Scholar]
- 29. Ko AI, Goarant C, Picardeau M. 2009. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat. Rev. Microbiol. 7:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lambert A, et al. 2012. FlaA proteins in Leptospira interrogans are essential for motility and virulence but are not required for formation of flagellum sheath. Infect. Immun. 80:2019–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liao S, et al. 2009. Inactivation of the fliY gene encoding a flagellar motor switch protein attenuates mobility and virulence of Leptospira interrogans strain Lai. BMC Microbiol. 9:253 doi:10.1186/1471-2180-9-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lo M, et al. 2006. Effects of temperature on gene expression patterns in Leptospira interrogans serovar Lai as assessed by whole-genome microarrays. Infect. Immun. 74:5848–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lo M, et al. 2010. Transcriptional response of Leptospira interrogans to iron limitation and characterization of a PerR homolog. Infect. Immun. 78:4850–4859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lourdault K, Cerqueira GM, Wunder EA, Picardeau M. 2011. Inactivation of clpB in the pathogen Leptospira interrogans reduces virulence and resistance to stress conditions. Infect. Immun. 79:3711–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Malmstrom J, et al. 2009. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 460:762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marangoni A, et al. 2006. Production of reactive oxygen species and expression of inducible nitric oxide synthase in rat isolated Kupffer cells stimulated by Leptospira interrogans and Borrelia burgdorferi. World J. Gastroenterol. 12:3077–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Margoliash E, Schejter A. 1962. Kinetics of the irreversible inhibition of catalase by 3-amino-1,2,4-triazole in the presence of hydrogen peroxide and catalase-hydrogen peroxide complex I hydrogen donors. J. Biol. Chem. 237:2359–2363 [PubMed] [Google Scholar]
- 38. Murgia R, Garcia R, Cinco M. 2002. Leptospires are killed in vitro by both oxygen-dependent and -independent reactions. Infect. Immun. 70:7172–7175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Murray GL, et al. 2009. Genome-wide transposon mutagenesis in pathogenic Leptospira species. Infect. Immun. 77:810–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Murray GL, et al. 2010. Mutations affecting Leptospira interrogans lipopolysaccharide attenuate virulence. Mol. Microbiol. 78:701–709 [DOI] [PubMed] [Google Scholar]
- 41. Murray GL, et al. 2009. Leptospira interrogans requires heme oxygenase for disease pathogenesis. Microbes Infect. 11:311–314 [DOI] [PubMed] [Google Scholar]
- 42. Naclerio G, Baccigalupi L, Caruso C, De Felice M, Ricca E. 1995. Bacillus subtilis vegetative catalase is an extracellular enzyme. Appl. Environ. Microbiol. 61:4471–4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nakayama GR, Caton MC, Nova MP, Parandoosh Z. 1997. Assessment of the Alamar Blue assay for cellular growth and viability in vitro. J. Immunol. Methods 204:205–208 [DOI] [PubMed] [Google Scholar]
- 44. Nally JE, Chow E, Fishbein MC, Blanco DR, Lovett MA. 2005. Changes in lipopolysaccharide O antigen distinguish acute versus chronic Leptospira interrogans infections. Infect. Immun. 73:3251–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nathan C, Shiloh MU. 2000. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. U. S. A. 97:8841–8848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Palaniappan RUM, Ramanujam S, Chang Y-F. 2007. Leptospirosis: pathogenesis, immunity, and diagnosis. Curr. Opin. Infect. Dis. 20:284–292 [DOI] [PubMed] [Google Scholar]
- 47. Patarakul K, Lo M, Adler B. 2010. Global transcriptomic response of Leptospira interrogans serovar Copenhageni upon exposure to serum. BMC Microbiol. 10:31 doi:10.1186/1471-2180-10-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pinne M, Haake DA. 2009. A comprehensive approach to identification of surface-exposed, outer membrane-spanning proteins of Leptospira interrogans. PLoS One 4:e6071 doi:10.1371/journal.pone.0006071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ristow P, et al. 2007. The OmpA-like protein Loa22 is essential for leptospiral virulence. PLoS Pathog. 3:e97 doi:10.1371/journal.ppat.0030097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rodriguez-Lopez J, et al. 2001. Mechanism of reaction of hydrogen peroxide with horseradish peroxidase: identification of intermediates in the catalytic cycle. J. Am. Chem. Soc. 123:11838–11847 [DOI] [PubMed] [Google Scholar]
- 51. Reference deleted. [Google Scholar]
- 52. Srinivasa Rao PS, Yamada Y, Leung KY. 2003. A major catalase (KatB) that is required for resistance to H2O2 and phagocyte-mediated killing in Edwardsiella tarda. Microbiology 149:2635–2644 [DOI] [PubMed] [Google Scholar]
- 53. Steele KH, Baumgartner JE, Valderas MW, Roop RM., II 2010. Comparative study of the roles of AhpC and KatE as respiratory antioxidants in Brucella abortus 2308. J. Bacteriol. 192:4912–4922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sterling CR, Thiermann AB. 1981. Urban rats as chronic carriers of leptospirosis: an ultrastructural investigation. Vet. Pathol. 18:628–637 [DOI] [PubMed] [Google Scholar]
- 55. Storz G, Tartaglia LA, Ames BN. 1990. Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science 248:189–194 [DOI] [PubMed] [Google Scholar]
- 56. Suvarnapunya AE, Lagasse HA, Stein MA. 2003. The role of DNA base excision repair in the pathogenesis of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 48:549–559 [DOI] [PubMed] [Google Scholar]
- 57. Tainer JA, Getzoff ED, Richardson JS, Richardson DC. 1983. Structure and mechanism of copper, zinc superoxide dismutase. Nature 306:284–287 [DOI] [PubMed] [Google Scholar]
- 58. Toma C, Okura N, Takayama C, Suzuki T. 2011. Characteristic features of intracellular pathogenic Leptospira in infected murine macrophages. Cell. Microbiol. 13:1783–1792 [DOI] [PubMed] [Google Scholar]
- 59. Tondo ML, Petrocelli S, Ottado J, Orellano EG. 2010. The monofunctional catalase KatE of Xanthomonas axonopodis pv. citri is required for full virulence in citrus plants. PLoS One 5:e10803 doi:10.1371/journal.pone.0010803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vazquez-Torres A, et al. 2000. Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH oxidase. Science 287:1655–1658 [DOI] [PubMed] [Google Scholar]
- 61. Vignais PV. 2002. The superoxide-generating NADPH oxidase: structural aspects and activation mechanism. Cell. Mol. Life Sci. 59:1428–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vinetz JM.2001. Leptospirosis. Curr. Opin. Infect. Dis. 14:527–538 [DOI] [PubMed] [Google Scholar]
- 63. Wheeler CR, Salzman JA, Elsayed NM, Omaye ST, Korte DW., Jr 1990. Automated assays for superoxide dismutase, catalase, glutathione peroxidase, and glutathione reductase activity. Anal. Biochem. 184:193–199 [DOI] [PubMed] [Google Scholar]
- 64. Wilson TM, de Lisle GW, Collins DM. 1995. Effect of inhA and katG on isoniazid resistance and virulence of Mycobacterium bovis. Mol. Microbiol. 15:1009–1015 [DOI] [PubMed] [Google Scholar]
- 65. Xue F, et al. 2010. Transcriptional responses of Leptospira interrogans to host innate immunity: significant changes in metabolism, oxygen tolerance, and outer membrane. PLoS Negl. Trop. Dis. 4:e857 doi:10.1371/journal.pntd.0000857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yu NY, et al. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang L, et al. 2012. The mammalian cell entry (Mce) protein of pathogenic Leptospira species is responsible for RGD motif-dependent infection of cells and animals. Mol. Microbiol. 83:1006–1023 [DOI] [PubMed] [Google Scholar]
- 68. Zuerner RL, Knudtson W, Bolin CA, Trueba G. 1991. Characterization of outer membrane and secreted proteins of Leptospira interrogans serovar Pomona. Microb. Pathog. 10:311–322 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.