Abstract
The indigenous microbial community of the gastrointestinal (GI) tract determines susceptibility to Clostridium difficile colonization and disease. Previous studies have demonstrated that antibiotic-treated mice challenged with C. difficile either developed rapidly lethal C. difficile infection or were stably colonized with mild disease. The GI microbial community of animals with mild disease was dominated by members of the bacterial family Lachnospiraceae, while the gut community in moribund animals had a predominance of Escherichia coli. We investigated the roles of murine Lachnospiraceae and E. coli strains in colonization resistance against C. difficile in germfree mice. Murine Lachnospiraceae and E. coli isolates were cultured from wild-type mice. The ability of each of these isolates to interfere with C. difficile colonization was tested by precolonizing germfree mice with these bacteria 4 days prior to experimental C. difficile challenge. Mice precolonized with a murine Lachnospiraceae isolate, but not those colonized with E. coli, had significantly decreased C. difficile colonization, lower intestinal cytotoxin levels and exhibited less severe clinical signs and colonic histopathology. Infection of germfree mice or mice precolonized with E. coli with C. difficile strain VPI 10463 was uniformly fatal by 48 h, but only 20% mortality was seen at 2 days in mice precolonized with the Lachnospiraceae isolate prior to challenge with VPI 10463. These findings confirm that a single component of the GI microbiota, a murine Lachnospiraceae isolate, could partially restore colonization resistance against C. difficile. Further study of the members within the Lachnospiraceae family could lead to a better understanding of mechanisms of colonization resistance against C. difficile and novel therapeutic approaches for the treatment and prevention of C. difficile infection.
INTRODUCTION
Clostridium difficile is a Gram-positive toxin-producing bacterium first described as a commensal organism in the fecal microbiota of healthy newborn infants (18). Currently, C. difficile is the most common cause of health care-associated diarrhea and colitis. C. difficile infection (CDI) is responsible for significant morbidity and mortality and increased economic burden in hospitalized patients (9, 23). Risk for the development of CDI is associated with the use of broad-spectrum antibiotic therapy as well as increasing patient age and hospitalization (2).
The human gastrointestinal (GI) microbiota protects the host against colonization by exogenous pathogenic organisms, a function referred to as colonization resistance (17, 38, 40). Administration of broad-spectrum antibiotics is theorized to destroy this protective function of the indigenous microbiota, allowing C. difficile to proliferate and colonize the GI tract (32, 34, 39). In support of this hypothesis, mice or hamsters challenged with C. difficile are not readily susceptible to C. difficile colonization or disease (CDI) (5, 43, 45), while antibiotic administration will render animals susceptible to infection (3, 5, 12, 20, 30).
In previous studies, it was demonstrated that wild-type mice treated with a cocktail of five antibiotics and clindamycin prior to C. difficile challenge would follow one of two clinical courses. At the appropriate challenge dose, mice either would develop rapidly lethal CDI or were stably colonized, with the development of only mild disease (5, 30). We reported that members of the bacterial family Lachnospiraceae dominated the gut communities of animals with mild disease. Members of the Lachnospiraceae were also the primary component of the GI community in untreated mice from our colony. On the other hand, the GI community of moribund animals had a predominance of Escherichia coli (30). Based on this observation, we hypothesize that members of the Lachnospiraceae family (but not E. coli) were responsible for at least a portion of the natural colonization resistance against C. difficile in the murine GI tract.
In order to directly examine this hypothesis, we isolated Lachnospiraceae and E. coli from the ceca of mice and tested their ability to suppress C. difficile colonization, toxin production, and disease in germfree mice. Our results indicate that Lachnospiraceae can play an important role in limiting C. difficile colonization. Further study could lead to new modes of C. difficile suppression and greater insight into the function of these organisms in health and disease.
MATERIALS AND METHODS
Animals and housing.
Wild-type C57BL/6 mice from a breeding colony established using animals purchased from Jackson Laboratories were housed with autoclaved food, bedding, and water under specific-pathogen-free conditions. Cage changes were performed in a laminar flow hood. Infection studies were performed with 6- to 8-week-old germfree Swiss Webster mice from a breeding colony established at the University of Michigan germfree core facility. Mice were housed in sterile soft-sided plastic isolators with autoclaved food, bedding, and water for the duration of the experiments. Each experimental group was housed in a separate isolator. All animal protocols used during the conduct of these experiments were reviewed and approved by the University Committee on Use and Care of Animals of the University of Michigan, Ann Arbor. The protocol was reviewed following guidelines for the care and use of laboratory animals set by the Office of Laboratory Animal Welfare, U.S. Department of Health and Human Services.
Development of Lachnospiraceae 16S rRNA-encoding gene primers.
Partial 16S rRNA gene sequences for Lachnospiraceae from our previous study (30) were used to generate CLUSTALW multiple-sequence alignments. Regions of conserved homology from the most common Lachnospiraceae operational taxonomic units (OTUs) were identified and used for PCR primer design. These conserved regions were compared against 16S rRNA gene sequences of other, non-Lachnospiraceae bacteria to ensure primer specificity. Primer specificity was confirmed by performing PCR amplification on representative 16S rRNA sequences of non-Lachnospiraceae clones, including E. coli, Pseudomonas, Porphyrmonodaceae, Bacteroides, Verrucomicrobia, Lactobacillus, Clostridiaceae, Staphylococcus, Ruminococcaceae, and Peptococcaceae strains. The final Lachnospiraceae-specific forward primer (LachnoF, 5′-ACC GCA TAA GCG CAC AGC-3′) was used with the broad-range reverse bacterial primer 1492R (5′-GGT TAC CTT GTT ACG ACT T-3′) for PCR with the following cycling conditions; initial denaturation at 95°C for 2 min followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 45 s, and extension at 72°C for 90 s. A final extension at 72°C for 10 min was performed. PCR was performed with 20 pmol of each primer (LachnoF and 1492R), 8 mM deoxynucleoside triphosphate (dNTP) master mix (Promega, Madison, WI), 1 unit GoTaq DNA polymerase (Promega), PCR buffer (Promega), and water in a total of 25 μl per reaction mixture.
Bacterial isolation from specific-pathogen-free mice.
The plate wash PCR technique (35) was adapted for the isolation of murine Lachnospiraceae strains. Ceca from wild-type C57BL/6 mice were removed in a sterile manner and immediately transferred to an anaerobic chamber (Coy Industries, Grass Lake, MI). Anaerobically equilibrated 1× phosphate-buffered saline (PBS) was added to the cecum, and the organ was opened with a sterile scalpel to release the cecal contents. Serial dilutions of this cecal suspension were plated in triplicate on various culture media, including Trypticase peptone (BD, San Jose, CA) with 5% blood (blood agar), chocolate agar (consisting of 5% lysed blood) with Trypticase peptone base, reinforced clostridial agar (BD), modified peptone yeast glucose agar (ATCC medium 1237), routine growth media (8), and brain heart infusion agar (BD) with 0.1% cysteine (Sigma-Aldrich, St. Louis, MO) added (BHIS) in combination with aztreonam (Sigma-Aldrich) and gentamicin (Sigma-Aldrich) to determine which media would provide the greatest enrichment of Lachnospiraceae. The surface of one agar plate was scraped to remove all bacterial colonies, and bacterial DNA was extracted using an automated system (Roche MagNA Pure). Enrichment for Lachnospiraceae was determined by performance of the Lachnospiraceae-specific 16S rRNA gene PCR and the amplification of the expected 1,320-bp band. The greatest enrichment for Lachnospiraceae was obtained using BHIS plates supplemented with 2 mg/liter gentamicin and 1 mg/liter aztreonam (BHIS gen/az). Subsequently, individual colonies from the duplicate BHIS gen/az plates were inoculated into 250 μl of BHIS gen/az broth in a sterile 96-well plate and then incubated anaerobically at 37°C, and growth was monitored. PCR using 1 μl of bacterial culture was used to identify putative Lachnospiraceae. Genomic bacterial DNA was extracted using an automated system (Roche MagNA Pure) from cultures of putative Lachnospiraceae isolates. Sanger sequencing of full-length 16S rRNA gene amplicons was obtained using the following primers: 8F (5′-AGA GTT TGA TCC TGG CTC AG-3′), 515F (5′-GTG CCA GCM GCC GCG GTA-3′), E939R (5′-CTT GTG CGG GCC CCC GTC AAT TC-3′), and 1492R (5′-GGT TAC CTT GTT ACG ACT T-3′). DNA sequencing was performed by the DNA Sequencing Core facilities at the University of Michigan.
Isolation of Escherichia coli was performed by selectively plating murine cecal contents on MacConkey agar anaerobically at 37°C. The identity of putative E. coli isolates was confirmed by sequencing the 16S rRNA-encoding gene amplicon. 16S rRNA gene sequence analysis, and taxonomic classification was performed using the RDP Bayesian classifier implemented in mothur (33).
Clostridium difficile strains and growth conditions.
C. difficile spores were prepared from strains VPI 10463 (ATCC 43255) and 630 (ATCC BAA-1382) as follows. C. difficile was cultured overnight anaerobically at 37°C in brain heart infusion broth with 0.1% cysteine (BHIS). On the following day, 100 μl of these cultures was spread onto BHIS plates (four plates per strain) and the plates incubated for 7 days anaerobically at 37°C. The plates were removed from the anaerobic chamber and exposed to ambient oxygen for 24 h at room temperature to kill vegetative cells. Plates were flooded with 15 ml cold water, and bacteria were removed by scraping with a sterile loop. Bacterial suspensions were centrifuged and washed in cold water at least three times. Spore stocks were stored at 4°C in sterile water. As an additional measure to remove vegetative cells, the C. difficile spore preparation was heat treated for 20 min at 65°C prior to experimental animal infection (37). The presence of spores was confirmed using phase-contrast microscopy, and spores were enumerated by plating for viable CFU on taurocholate-cycloserine-cefoxitin-fructose agar (TCCFA).
Germfree mouse infection studies.
Six- to 8-week-old germfree Swiss Webster mice were divided into groups of 3 to 9 animals. Each treatment group was housed in separate sterile isolators. Initially, mice were challenged via oral gavage with various spore doses of two C. difficile strains, VPI 10463 or strain 630, to determine the appropriate dose of spores to use for the remaining experiments. For C. difficile VPI 10463, groups of three to five mice were challenged with 3.8 × 101, 3 × 102, 3.3 × 103, and 1 × 105 spores, while for C. difficile strain 630, mice were challenged with 1 × 101, 1 × 102, 1 × 103, and 1 × 104 spores, administered in a volume of 100 μl (Fig. 1). Based on these experiments, 100 spores were used as the challenge dose for all experiments.
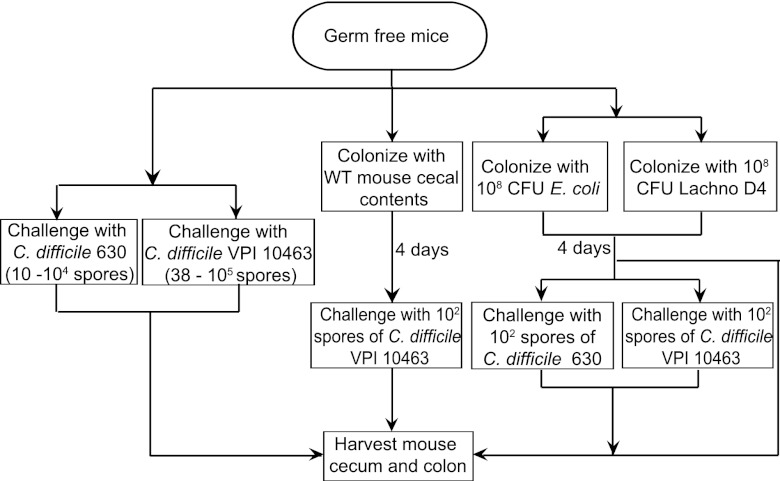
Fig 1.
Schematic for examining the effects of Lachnospiraceae D4 and E. coli on C. difficile colonization in germfree mice. Mice were infected with titrating doses of C. difficile 630 spores (10, 102, 103, and 104; n = 3 per dose) or C. difficile VPI 10463 spores (38, 3 × 102, 3.3 × 103, and 1 × 105; n = 4 per dose) and monitored for colonization. Animals were colonized with cecal contents from a wild-type (WT) mouse for 4 days prior to C. difficile VPI 10463 challenge (n = 5). Additionally, mice were either precolonized with E. coli and then challenged with C. difficile 630 (n = 9) or C. difficile VPI 10463 (n = 7) spores or precolonized with Lachnospiraceae D4 and then challenged with C. difficile 630 (n = 8) or C. difficile VPI 10463 (n = 14) spores. Mice monocolonized with C. difficile 630 (n = 11) or C. difficile VPI 10463 (n = 15) were used as controls. Other groups of mice were colonized with either E. coli (n = 4) or Lachnospiraceae D4 (n = 4) for 4 days and harvested. The cecum and colon from each animal were harvested for bacterial and cytotoxin quantification.
For experiments testing colonization resistance, mice were precolonized via oral gavage with 1 × 108 CFU of either Escherichia coli or Lachnospiraceae for 4 days. As a control, mice were also orally gavaged with a single dose of cecal content homogenate obtained from a wild-type mouse at 4 days prior to C. difficile challenge. The cecal content homogenate was prepared by adding 100 mg of cecal contents to 900 μl of prereduced 1× PBS. One hundred microliters of the homogenate was then orally gavaged into germfree mice. Each treatment group consisted of 5 to 9 animals. Mice were challenged by oral gavage with 100 spores of C. difficile (VPI 10463 or strain 630) and monitored daily for signs of disease such as diarrhea, hunched posture, and weight loss. Control groups consisted of animals challenged with only C. difficile (n = 11 and n = 15 for strains 630 and VPI 10463, respectively), E. coli alone (n = 4), Lachnospiraceae alone (n = 4), cecal content homogenate from wild-type mice (n = 3), or no bacteria (n = 5) (Fig. 1). Bacterial colonization was monitored daily by anaerobic culture of fecal pellets and cecal contents at 37°C for 24 h. E. coli colonization was specifically monitored by culture on MacConkey agar, while the levels of Lachnospiraceae were monitored by culture on BHIS gen/az. The colonization status of C. difficile-challenged animals was monitored by anaerobic culture on TCCFA.
Necropsy and histologic procedures.
Mice were euthanized by CO2 asphyxiation. Cecum and colon tissue were placed intact into histology tissue cassettes, stored in 10% buffered formalin for 24 h, and then transferred to 70% ethyl alcohol. Tissue cassettes were then processed and paraffin embedded, and 5-μm sections were prepared. Hematoxylin-and-eosin (H&E)-stained slides were prepared for histologic examination (McClinchey Histology Lab Inc. Stockbridge, MI.).
Histopathologic examination.
Histologic changes were coded, randomized, and scored in a blinded manner by a board-certified veterinary pathologist. A scoring system was adapted from previously published methods (5, 22). Edema, neutrophilic inflammation, and epithelial damage in colon and cecum were scored from 0 to 4 according to the following defined criteria. Edema scores were graded as follow: 0, no edema; 1, mild edema with minimal (<2× normal width) multifocal submucosal expansion; 2, moderate edema with moderate (2× to 3× normal width) multifocal submucosal expansion; 3, severe edema with severe (>3× normal width) multifocal submucosal expansion; and 4, same as score 3 with diffuse submucosal expansion. Neutrophilic inflammation scores were graded as follows: 0, no inflammation; 1, minimal multifocal neutrophilic inflammation (marginating or perivascular neutrophils in submucosa, minimal intraepithelial and proprial neutrophils); 2, moderate multifocal neutrophilic inflammation (perivascular and interstitial neutrophils in submucosa, mild to moderate intraepithelial and proprial neutrophils); 3, severe multifocal to coalescing neutrophilic inflammation (perivascular and increased interstitial neutrophils in submucosa with or without extension to muscular wall, moderate intraepithelial neutrophils); and 4, same as score 3 with abscesses. Epithelial damage was scored as follows: 0, no epithelial changes; 1, minimal multifocal superficial epithelial damage (vacuolation, apoptotic figures, villus tip attenuation/necrosis); 2, moderate multifocal superficial epithelial damage (vacuolation, apoptotic figures, villus tip attenuation/necrosis); 3, severe multifocal epithelial damage (same as score 2 with or without pseudomembrane [intraluminal neutrophils, sloughed epithelium in a fibrinous matrix]); and 4, same as score 3 with significant pseudomembrane or epithelial ulceration (focal complete loss of epithelium).
C. difficile cytotoxin assay.
Quantification of C. difficile toxin present in luminal contents was performed using a method adapted from that described by Corthier et al. (7). Green African monkey kidney epithelial cells (Vero) (provided by M. Imperiale, University of Michigan) were grown to confluence in Dulbecco modified Eagle medium (DMEM) (Invitrogen, Carlsbad, CA) containing 10% heat-inactivated fetal bovine serum (Invitrogen) and 1% penicillin-streptomycin solution (Invitrogen). The cells were trypsinized using 0.25% trypsin (Invitrogen) and washed with 1 volume of DMEM. Cells were diluted in DMEM, and approximately 1 × 105 cells were distributed per well in a 96-well flat bottom microtiter plate (Corning) and incubated at 37°C for 18 to 24 h. One hundred milligrams of luminal contents was weighed, and 500 μl of 1× PBS was added to make a suspension. Samples were mixed, particulate material was removed by centrifugation at 9,000 × g for 5 min, and then the supernatant was filtered through a 0.2-μm membrane. Each sample was titrated in 10-fold dilutions within the wells to a maximum dilution of 10−12, and each well had a corresponding control to which both neutralizing Clostridium sordelli antitoxin (TechLabs, Blacksburg, VA) and sample were added. After an overnight incubation at 37°C, plates were fixed with 10% buffered formalin for 2 h and then stained with Giemsa stain (50 μl per well) for 15 min, followed by a wash with 1× PBS. Wells with approximately 100% round cells were easily recognized under a magnification of ×200. The cytotoxic titer was defined as the reciprocal of the highest dilution that rounds 100% of Vero cells per gram of sample.
SCFA measurement.
In vivo measurement of short-chain fatty acids (SCFAs) was performed as follows. Groups of four 6- to 8-week-old Swiss Webster germfree mice were colonized with 108 CFU of either E. coli or Lachnospiraceae D4 for 4 days. Additionally, a third group remained germfree as a control. All mice were necropsied, and cecal contents were removed, weighed, flash frozen, and stored at −80°C. Samples were sent to the Michigan Metabolomics and Obesity Center at the University of Michigan for SCFA measurement by gas chromatography-mass spectrometry (GC-MS). GC (Agilent 6890) separation was performed using a ZB-Wax Plus column (0.25 μm by 0.25 mm by 30 m), and a quadruple mass spectrometer (Agilent, Santa Clara, CA) was used to identify and quantitate SCFAs using Agilent Chemstation software.
Statistical analysis.
Statistical analyses were performed using Prism 5 for Mac OS X GraphPad software. The nonparametric Kruskal-Wallis test was used to determine significance for all treatment groups, while Student's t test was used for individual comparisons. Statistical significance was set at a P value of <0.05.
RESULTS
Isolation and characterization of murine Lachnospiraceae and E. coli.
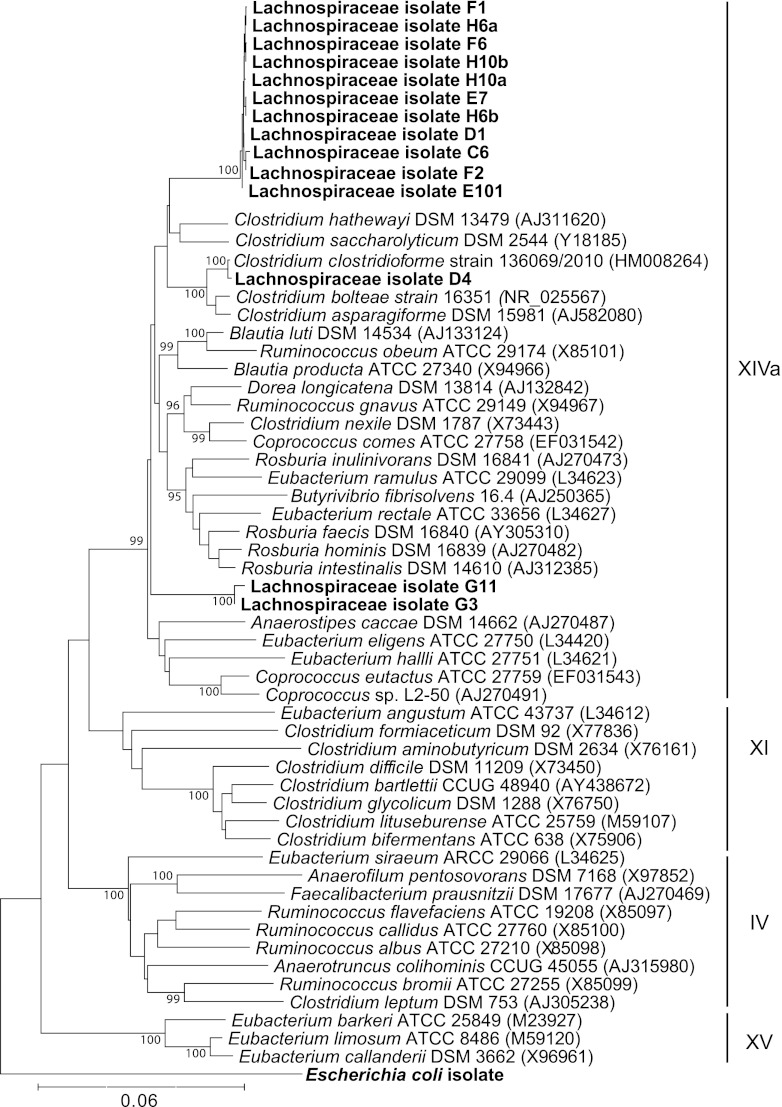
A strain of E. coli was isolated from the murine gut by selectively plating murine cecal contents on MacConkey agar. The identification of potential E. coli cultivars was confirmed by 16S rRNA gene sequence analysis. To isolate Lachnospiraceae, primers targeting 16S-targeted rRNA-encoding genes were designed based on the partial 16S rRNA gene sequences from our previous study (30) to guide cultivation efforts via the plate wash PCR technique (35). The greatest enrichment for Lachnospiraceae was achieved by selectively plating murine cecal contents on brain heart infusion agar supplemented with 0.1% cysteine, 2 mg/liter gentamicin, and 1 mg/liter aztreonam (BHIS gen/az) under anaerobic conditions. A total of 14 Lachnospiraceae isolates were confirmed by 16S rRNA-encoding gene sequence analysis (Fig. 2). We determined the phylogenetic relationship of these isolates to previously characterized members of the low-moles-percent G+C Gram-positive bacteria in the Firmicutes phylum (Fig. 2). All 14 Lachnospiraceae isolates were members of the clostridial cluster XIVa (10).
Fig 2.
Phylogenetic tree showing clostridial clusters of low-moles-percent G+C Gram-positive bacteria based on 16S rRNA sequence. The tree was constructed using the neighbor-joining method, with the murine E. coli isolate used as the outgroup. Newly isolated murine Lachnospiraceae strains are shown in boldface. Accession numbers for sequences are given in parentheses. Bootstrap values greater than 95 (per 500 replicates) are shown at branch points. The scale bar represents genetic distance, and clostridial clusters are indicated by Roman numerals.
These Lachnospiraceae isolates were further characterized based on growth on BHIS gen/az plates and BHIS gen/az broth. Of the 14 Lachnospiraceae isolates, 13 took 3 to 4 days to grow on solid media and grew poorly in broth culture. The remaining isolate (referred to as D4) was most closely related to Clostridium clostridioforme based on 16S rRNA gene sequence (Fig. 2). Lachnospiraceae isolate D4 grew well on solid media and in broth culture, which allowed us to test its ability to inhibit C. difficile growth and toxin production in germfree mice.
Colonization of germfree mice with Lachnospiraceae D4 interferes with subsequent C. difficile colonization.
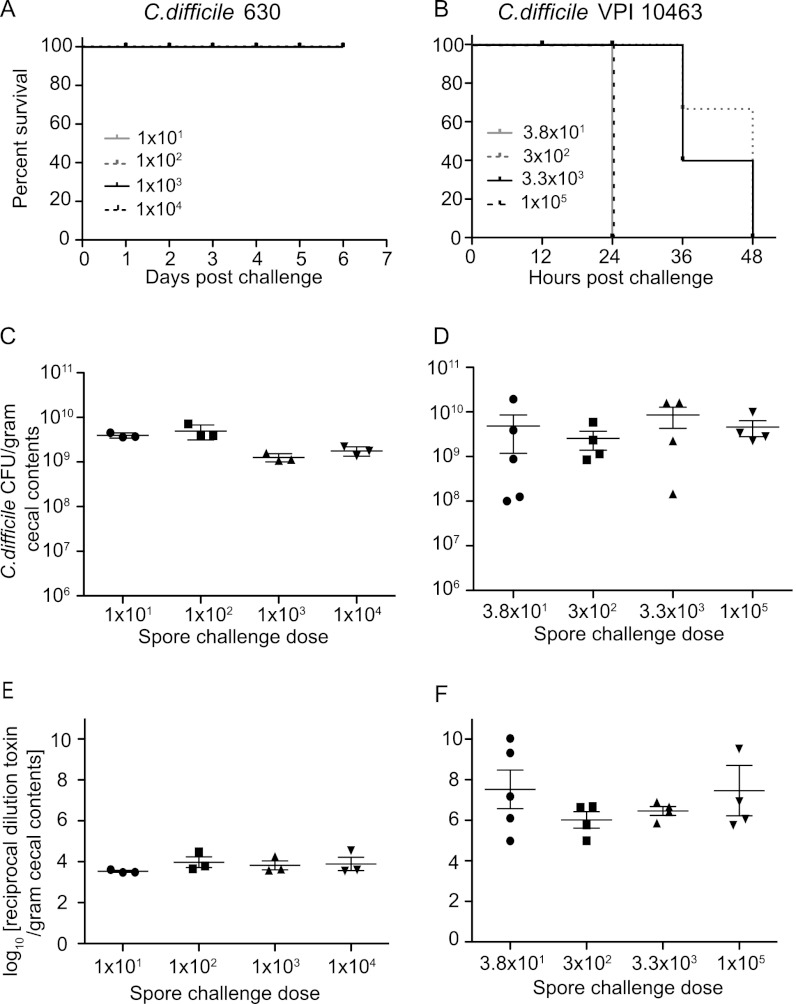
We recently demonstrated that cefoperazone-treated mice were readily colonized with C. difficile strain 630 and developed an acute, histologically apparent colitis but did not exhibit signs of clinically severe CDI such as weight loss, diarrhea, or hunched posture (37). Therefore, infection of cefoperazone-treated mice with C. difficile 630 permits the examination of the effect of Lachnospiraceae D4 and E. coli on C. difficile colonization and cytotoxin production in a nonlethal infection model that mimics many aspects of human infection with the pathogen. We initially challenged germfree mice with various doses of C. difficile 630 spores ranging from 101 to 104 (Fig. 1) to determine if the lack of lethality observed in antibiotic-treated animals would extend to monocolonization with this strain. C. difficile colonization levels were monitored daily by culture of fecal pellets and culture of cecal contents at necropsy. Levels of C. difficile cytotoxin were measured using a Vero cell assay. As with conventional mice, germfree mice challenged with C. difficile 630 did not exhibit clinical signs of CDI and survived the infection (Fig. 3A). Monocolonized animals had high levels of C. difficile colonization (>109 CFU/gram) (Fig. 3C). Similar results were seen regardless of the infectious dose administered. Subsequently, we employed an infectious dose of 100 C. difficile 630 spores for the following experiments.
Fig 3.
C. difficile infection in germfree mice. (A and B) Kaplan-Meier survival plots for mice infected with titrating doses of C. difficile 630 spores (1 × 101, 1 × 102, 1 × 103, and 1 × 104) (A) and C. difficile VPI 10463 spores (3.8 × 101, 3 × 102, 3.3 × 103, and 1 × 105) (B). (C and D) Quantification of C. difficile was determined by culturing cecal contents at the time of necropsy: at day 6 from mice infected with C. difficile 630 for each challenge dose (n = 3) (C) or at days 1 or 2 for mice infected with C. difficile VPI 10463 for each challenge dose (n = 4) (D). (E and F) Vero cell tissue culture was used to determine the log10 reciprocal cytotoxin dilution per gram of cecal contents from mice used for panels C and D infected with C. difficile 630 (E) or C. difficile VPI 10463 (F) for each challenge dose. Points on each graph represent individual animals. Error bars represent standard deviations.
Germfree mice received 108 CFU of either Lachnospiraceae D4 or E. coli via oral gavage. Mice challenged with either bacterium were readily colonized. Animals colonized with E. coli shed 1012 CFU/gram of feces by 4 days postchallenge, while animals that received Lachnospiraceae D4 shed 1010 CFU/gram of feces (data not shown). Four days after precolonization with either Lachnospiraceae D4 or E. coli, animals were challenged with 100 spores of C. difficile strain 630. All mice challenged with C. difficile were successfully colonized, and this colonization did not alter the level of fecal Lachnospiraceae or E. coli shedding. No clinical disease was apparent in any of the experimental groups.
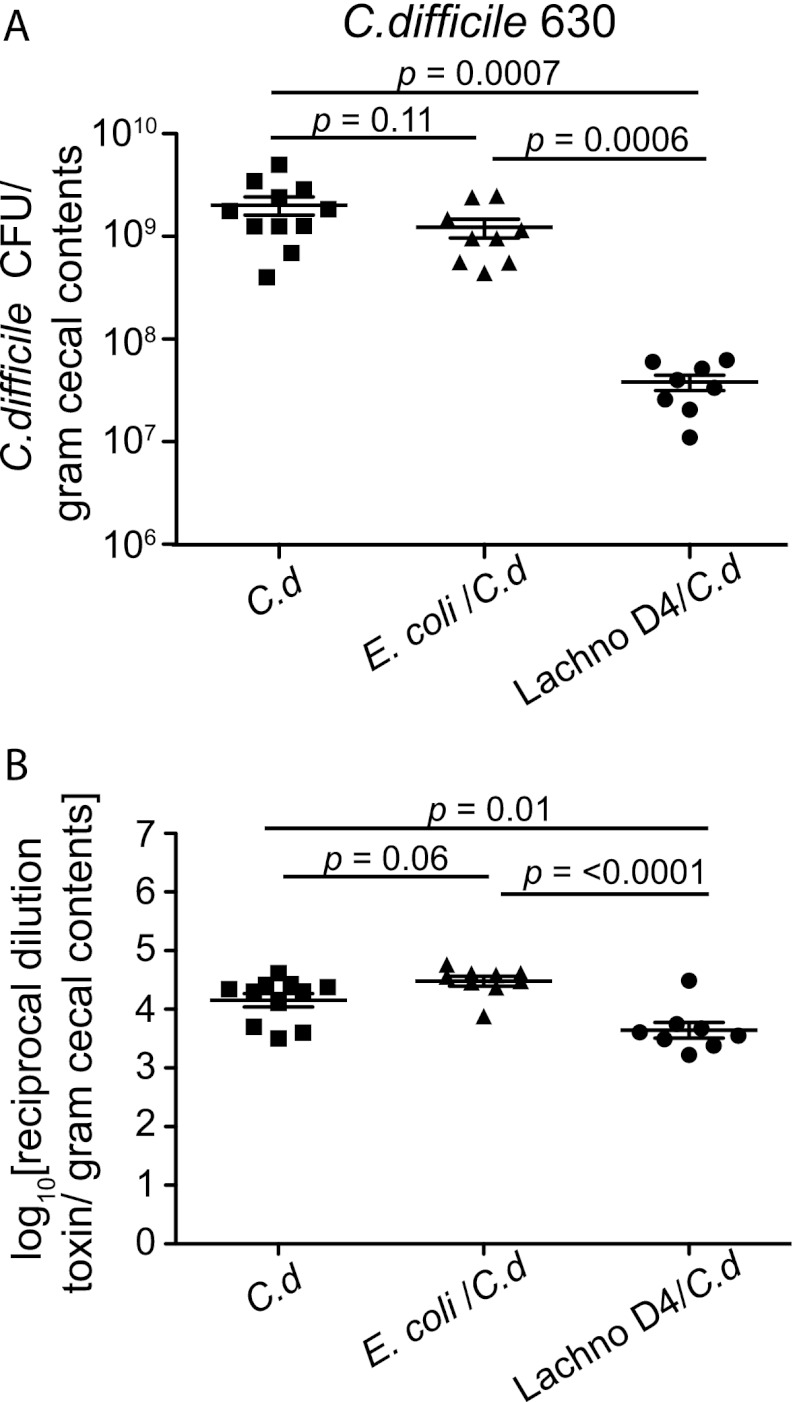
In mice precolonized with E. coli prior to C. difficile challenge, there was no difference in the cecal levels of colonization by C. difficile compared to those in mice monocolonized with C. difficile strain 630 (Fig. 4A). On the other hand, mice precolonized with Lachnospiraceae D4 prior to C. difficile challenge had a significant decrease in the levels of C. difficile colonization (>1.5 log) (Fig. 4A) and a corresponding decrease in the amount of C. difficile cytotoxin in the cecal contents compared to animals monocolonized with C. difficile (Fig. 4B).
Fig 4.
Decreased levels of C. difficile 630 and cytotoxin in Lachnospiraceae D4-precolonized mice. (A) Quantification of C. difficile was determined by culturing cecal contents at the time of necropsy (day 6) from mice infected with C. difficile only (n = 11) or precolonized with either E. coli (n = 9) or Lachnospiraceae D4 (n = 8) and then infected with C. difficile. Each point represents the C. difficile level from an individual animal. Mice precolonized with Lachnospiraceae D4 had significantly decreased levels of C. difficile compared to those in C. difficile-infected controls or E. coli-precolonized mice. Error bars represent standard deviations. Comparisons between groups were performed using the nonparametric Kruskal-Wallis test. (B) Vero cell tissue culture was used to determine the log10 reciprocal cytotoxin dilution per gram of cecal contents from mice used for panel A infected with C. difficile only or precolonized with either E. coli or Lachnospiraceae D4 and then infected with C. difficile. Error bars represent standard deviations. Comparisons between groups were performed using the nonparametric Kruskal-Wallis test. C.d, C. difficile; Lachno D4, Lachnospiraceae D4.
Colonization of germfree mice with Lachnospiraceae isolate D4 decreases subsequent disease severity after challenge with C. difficile strain VPI 10463.
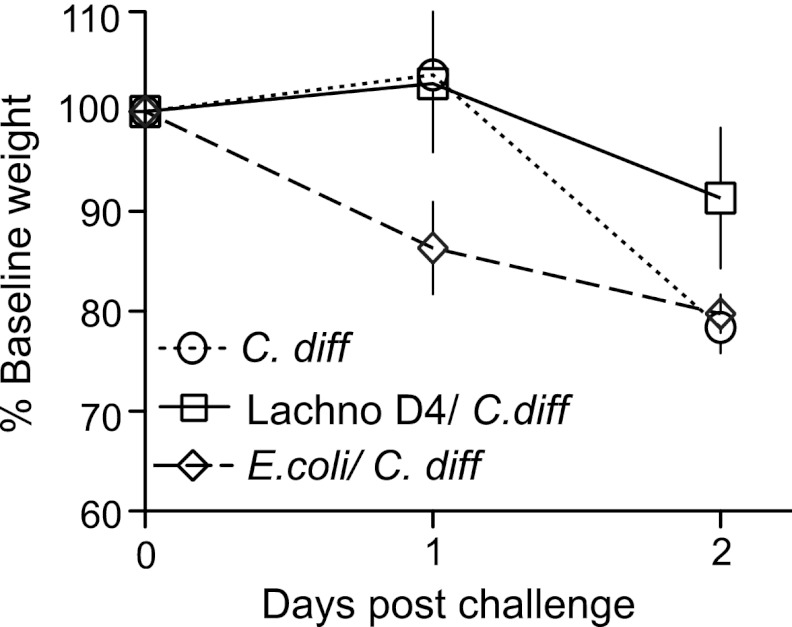
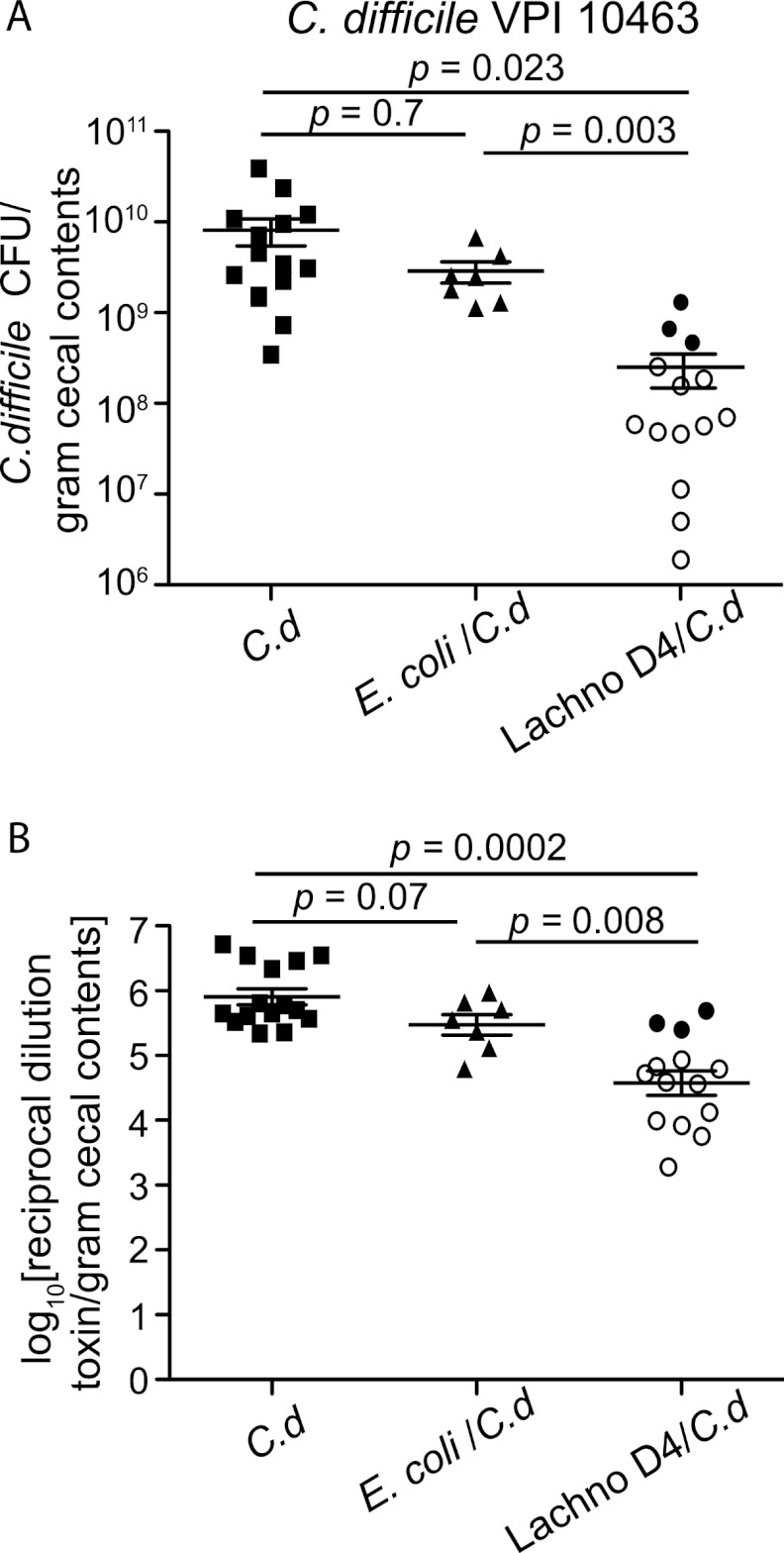
As opposed to C. difficile strain 630, C. difficile strain VPI 10463 causes a hyperacute, severe, and often lethal form of CDI in antibiotic-treated wild-type and germfree mice (21, 30, 37). Germfree mice challenged with various doses of C. difficile VPI 10463 spores (Fig. 3) all developed clinically severe CDI with diarrhea, hunched posture, and significant (>20% from baseline) weight loss. All mice, regardless of challenge dose, were either dead or moribund by 1 to 2 days postchallenge (Fig. 3B). High levels of C. difficile (>109 CFU/gram) (Fig. 3D) and cytotoxin (Fig. 3E and F) were detected in cecal contents at the time of necropsy compared to those in animals monocolonized with C. difficile 630. Similar to the case for animals monocolonized with C. difficile VPI 10463, 100% of mice precolonized with E. coli prior to C. difficile challenge lost >20% of baseline body weight by 2 days postinfection (Fig. 5) and had high levels of C. difficile and cytotoxin production at necropsy (Fig. 6). Conversely, mice precolonized with Lachnospiraceae D4 demonstrated significantly less clinically severe disease following challenge with C. difficile VPI 10463. Of a total of 14 mice, 3 (20%) were moribund and lost significant weight, while the remaining had minimal weight loss and were clinically well at 2 days postinfection (Fig. 5). The 11 mice that survived had lower levels of C. difficile colonization and measureable cytotoxin (Fig. 6) than the moribund mice and mice challenged with C. difficile alone or following prior colonization with E. coli.
Fig 5.
Weight loss in C. difficile-infected mice. Weight loss curves for C. difficile-infected mice (n = 15), mice precolonized with Lachnospiraceae D4 and infected with C. difficile (n = 14), and mice precolonized with E. coli and infected with C. difficile (n = 7) are shown. Lachnospiraceae D4-precolonized mice lost less weight than C. difficile-infected control or E. coli-precolonized mice. Weight loss percentage is based on the starting weight on day 0. Error bars represent the standard deviations of the weights for animals within each group. Lachno D4, Lachnospiraceae D4, C. diff, C. difficile.
Fig 6.
Decreased C. difficile VPI 10643 colonization and cytotoxin levels in Lachnospiraceae D4-precolonized mice. (A) Quantification of C. difficile was determined by culturing cecal contents at the time of necropsy (day 2) from mice infected with C. difficile only (n = 15) or precolonized with E. coli (n = 7) or Lachnospiraceae D4 (n = 14) and then infected with C. difficile. The levels of C. difficile colonization were decreased in Lachnospiraceae D4-precolonized mice compared to those in C. difficile-infected controls or E. coli-precolonized mice. Each point represents the C. difficile level from an individual animal. The open symbols represent animals that had improved CDI signs and did not lose significant weight, while the closed symbols represent animals that were moribund or dead at the time of necropsy. (B) Vero cell tissue culture was used to determine the log10 reciprocal cytotoxin dilution per gram of cecal contents from mice used for panel A infected with C. difficile only or precolonized with E. coli or Lachnospiraceae D4 and then infected with C. difficile. Error bars represent standard deviations. Comparisons between groups were performed using the nonparametric Kruskal-Wallis test. C.d, C. difficile; Lachno D4, Lachnospiraceae D4.
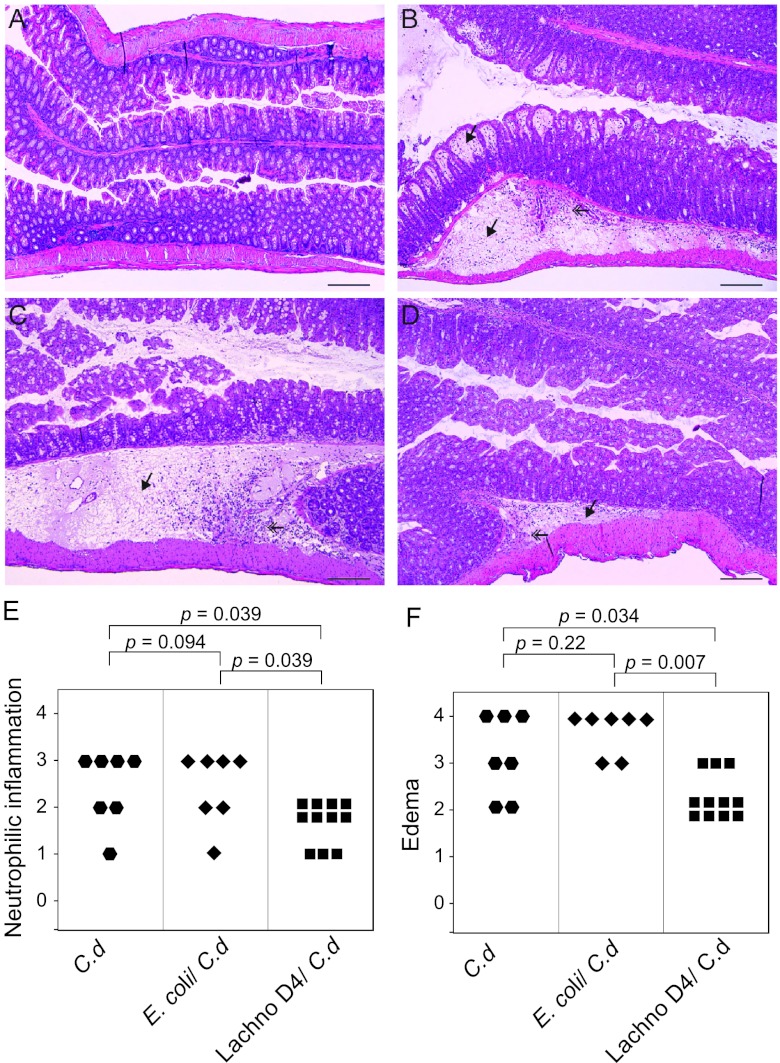
Representative histopathology in the colons of animals challenged with C. difficile VPI 10463 is shown in Fig. 7. Histopathologic changes in animals that developed colitis consisted predominantly of edema within the submucosa and the lamina propria. There was also neutrophilic inflammation perivascularly and interstitially within the submucosa and multifocally within the mucosa. Epithelial damage was not prominent but consisted of vacuolar degeneration and increased loss of apical tip enterocytes. Germfree mice precolonized with Lachnospiraceae D4 prior to challenge with C. difficile VPI 10463 had significantly less colonic inflammation and submucosal edema than either C. difficile-infected controls or animals precolonized with E. coli before C. difficile challenge (Fig. 7E and F). Mice that were maintained germfree and mice monoassociated with either Lachnospiraceae D4 or E. coli had no histologic alterations (data not shown). In addition to the differences in the colon, there was also significantly less mucosal epithelial damage in the ceca of mice precolonized with Lachnospiraceae D4 than in E. coli-precolonized and C. difficile-infected control mice (data not shown).
Fig 7.
Germfree animals precolonized with Lachnospiraceae D4 have decreased colonic histopathology after challenge with C. difficile strain VPI 10463. (A) Colon of an uninfected germfree mouse. (B) Colon of a C. difficile-infected mouse showing severe edema in the submucosa and mucosal lamina propria (arrow) accompanied by neutrophilic inflammation. (C) Colon from a mouse precolonized with E. coli and infected with C. difficile showing submucosal edema (arrow) and neutrophilic inflammation similar to those for a C. difficile-infected mouse. (D) Colon of a mouse precolonized with Lachnospiraceae D4 and infected with C. difficile showing moderate neutrophilic mucosal inflammation and minimal submucosal edema (arrow). All micrographs are of H&E-stained tissue at an original magnification of ×100. Bars represent 100 μm. (E and F) Categorical scores of neutrophilic inflammation (E) and edema (F) in C. difficile-infected controls, C. difficile-infected, E. coli-precolonized mice, and C. difficile-infected, Lachnospiraceae D4-precolonized mice. Comparisons between groups were performed using the nonparametric Kruskal-Wallis test. C.d, C. difficile; Lachno D4, Lachnospiraceae D4.
Prevention of C. difficile colonization in the GI tract of germfree mice inoculated with cecal contents from wild-type mice.
The results described above indicate that Lachnospiraceae strain D4 partially restores colonization resistance against C. difficile in germfree mice. As a control, we tested whether the full complement of cecal microbiota from wild-type mice could completely restore colonization resistance in germfree mice. Germfree mice received cecal contents obtained from a wild-type mouse via oral gavage. Four days later, these mice were challenged with 100 C. difficile VPI 10463 spores (Fig. 1). Unlike C. difficile-monoassociated control mice, mice that were colonized with the cecal content homogenate prior to C. difficile challenge did not exhibit signs of clinical CDI such as weight loss, diarrhea, and hunched posture. No detectable C. difficile was present in feces at 1 day postchallenge or in the cecal contents at 2 days postchallenge or at the time of necropsy (data not shown).
DISCUSSION
The indigenous GI microbiota plays a fundamental role in colonization resistance against C. difficile (41). However, the specific components of the gut microbiota that are important in mediating colonization resistance are not well defined. In this study, we demonstrated that a single component of the murine gut microbiota, a member of the family Lachnospiraceae, is able to partially restore colonization resistance against C. difficile in germfree mice.
Until now, no study has examined the ability of Lachnospiraceae organisms to contribute to colonization resistance against C. difficile or other pathogens. We recently reported that antibiotic-treated mice with clinically severe CDI had a predominance of E. coli in their cecal microbiota, while mice with mild CDI had a predominance of Lachnospiraceae species. Likewise, other studies have associated the microbial gut community of patients with inflammatory bowel disease (IBD) with an increased prevalence of E. coli and decreased prevalence of Lachnospiraceae (15, 24, 25). Furthermore, patients with IBD are at a higher risk for developing CDI (14, 28). These findings prompted us to isolate and investigate the relative roles of Lachnospiraceae and E. coli in mediating C. difficile colonization resistance.
Lachnospiraceae are Gram-positive obligate anaerobes that are mostly non-spore forming (8, 19). Our murine Lachnospiraceae isolates were members of the clostridial cluster XIVa (Fig. 1). Clostridial cluster XIVa represents one of the most abundant phylotypes of the low-moles-percent G+C Gram-positive bacteria (6, 8) and makes up approximately 25% of the total bacterial species found in human fecal specimens (10). Lachnospiraceae organisms found in both humans and mice can have highly similar 16S rRNA gene sequences. In our study, the murine Lachnospiraceae isolate D4 was phylogenetically similar to C. clostridioforme, a Lachnospiraceae organism found in humans (Fig. 1) (13). This organism is present as part of the indigenous gut microbiota in humans and has also been associated with opportunistic infections (13). However, we did not observe any disease or histopathology in our mice monocolonized with Lachnospiraceae D4.
Germfree mice have proven to be a useful tool for studying host-microbe and microbe-microbe interactions within the GI tract (11). Germfree mice have been used to examine how individual bacteria or bacterial communities influence colonization resistance against C. difficile (7, 21, 36, 42, 44). In many cases, the bacteria employed in an attempt to interfere with C. difficile were previously described as “probiotic” organisms or undifferentiated groups of bacteria derived from healthy animals. In the current study, we examined bacteria that were previously observed to be associated with normal or diminished colonization resistance to C. difficile (30). In this way, we used the results of culture-independent study of gut microbial ecology to inform and guide subsequent hypothesis-testing studies utilizing cultured bacterial isolates. We leveraged the plate wash PCR technique to fine-tune our cultivation efforts using 16S rRNA-encoding gene sequence data (35). We feel that this coupling of sequence-based microbial ecology studies with more traditional methods such as bacterial cultivation and experimental animal infection represents a powerful way study bacterial pathogenesis.
Multiple mechanisms that explain how the indigenous microbiota can mediate colonization resistance have been proposed (reviewed in reference 4). It is likely that several factors are involved in mediating colonization resistance, but the production of bacterial products that directly inhibit pathogens has received significant experimental attention. Several investigators have examined the ability of bacterial fermentation products, including short-chain fatty acids (SCFAs), to inhibit C. difficile growth. Some studies have shown that butyrate is capable of inhibiting C. difficile in vitro (26, 31), although contradicting reports exist (36). Lachnospiraceae organisms are notable in that many are capable of fermenting complex carbohydrates to SCFAs, which have an important role in maintaining intestinal homeostasis (1, 8, 29, 46). We investigated whether SCFAs were associated with less C. difficile colonization in Lachnospiraceae-precolonized mice and found that SCFA production was not correlated with lower C. difficile colonization levels (data not shown). Therefore, the decrease in C. difficile colonization levels due to Lachnospiraceae colonization is most likely attributable to the production of other metabolites or the use of other mechanisms.
It has been proposed that rather than specific inhibition by the production of metabolites such as SCFAs or antimicrobial compounds, including bacteriocins, the indigenous microbiota could simply be competing for limiting nutrients, the so-called nutrient niche hypothesis. Stated in brief, this hypothesis maintains that an organism can outcompete another if it utilizes a limiting nutrient more efficiently. It is possible that the diminished levels of C. difficile in the presence of Lachnospiraceae is due to less effective utilization of specific nutrients by the former (16). It should be noted, however, that our experiments do not suggest that there is a simple mass effect with regard to nutrient utilization. When the levels of Lachnospiraceae D4 and E. coli colonization were measured, Lachnospiraceae D4 reached colonization levels 100-fold lower than those of E. coli, suggesting that simply occupying more “space” in the gut, and presumably consuming proportionately more of the available resources, does not necessarily contribute to colonization resistance. Furthermore, competition for adherence to the mucosal epithelium was not apparent, as neither C. difficile, E. coli, nor Lachnospiraceae D4 was found to adhere to the mucosal layer as judged by 16S RNA-based fluorescent in situ hybridization analysis (data not shown).
Corthier and colleagues demonstrated that a neonatal E. coli strain significantly inhibited C. difficile cytotoxin (7). In our studies, an E. coli strain indigenous to wild-type mice had no such effect on C. difficile cytotoxin or colonization levels in germfree mice. Naaber and colleagues examined the effects of over 50 Lactobacillus strains on C. difficile growth inhibition and found only five strains that had antagonistic activity toward C. difficile (27). These studies suggest that although strains may belong to the same genus/species, variation in their individual genetic content results in functional differences. Similarly, it will be important to test the ability of other Lachnospiraceae isolates to inhibit C. difficile in vivo. It is possible that this is an ability that is shared by the Lachnospiraceae as a group or is associated with a function that is more restricted to certain members.
Monocolonization with Lachnospiraceae isolate D4 only partially restored C. difficile colonization resistance, while there was complete restoration following the transfer of cecal contents from a wild-type mouse to germfree mice. This implies that there are likely additive effects of specific microbiota in determining colonization resistance. Each member of the microbiota may partially contribute, but the entire community (or a specific subset) is required for complete colonization resistance. Others have observed only partial restoration of colonization resistance against C. difficile (7, 21). For instance, Itoh and colleagues colonized germfree mice with multiple strains of Bacteroides and lactobacilli and observed little antagonism toward C. difficile. Only when feces containing clostridia were administered to mice was C. difficile eliminated (21).
In summary, our results show that a single component of the gut microbiota, a murine Lachnospiraceae isolate, was able to partially restore colonization resistance against C. difficile and greatly improve clinical CDI outcome. Further investigation of the members within the Lachnospiraceae family potentially in combination with other taxonomically distinct members of the indigenous microbiota could lead to a greater understanding of mechanisms of C. difficile suppression and the role that these organisms play in protection against a variety of other pathogens and disease states.
ACKNOWLEDGMENTS
This work was funded by NIH grant AI090871 (to V.B.Y.) and utilized core services supported in part by NIH grant DK089503.
We thank Sara Poe and Chriss Vowles for technical assistance in experiments performed with germfree mice as well as Judy Opp and Kathy Wozniak for technical assistance. We are grateful to Casey Theriot for helpful discussions. We thank Philip Hanna and Paul Carlson, Jr., for kindly providing C. difficile spores used in this study.
Footnotes
Published ahead of print 13 August 2012
REFERENCES
- 1. Barcenilla A, et al. 2000. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl. Environ. Microbiol. 66:1654–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bartlett JG. 2010. Clostridium difficile: progress and challenges. Ann. N. Y. Acad. Sci. 1213:62–69 [DOI] [PubMed] [Google Scholar]
- 3. Bartlett JG, Onderdonk AB, Cisneros RL, Kasper DL. 1977. Clindamycin-associated colitis due to a toxin-producing species of Clostridium in hamsters. J. Infect. Dis. 136:701–705 [DOI] [PubMed] [Google Scholar]
- 4. Britton RA, Young VB. 2012. Interaction between the intestinal microbiota and host in Clostridium difficile colonization resistance. Trends Microbiol. 20:313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen X, et al. 2008. A mouse model of Clostridium difficile-associated disease. Gastroenterology 135:1984–1992 [DOI] [PubMed] [Google Scholar]
- 6. Collins MD, et al. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812–826 [DOI] [PubMed] [Google Scholar]
- 7. Corthier G, Dubos F, Raibaud P. 1985. Modulation of cytotoxin production by Clostridium difficile in the intestinal tracts of gnotobiotic mice inoculated with various human intestinal bacteria. Appl. Environ. Microbiol. 49:250–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cotta M, Forster R. 2006. The family Lachnospiraceae, including the genera Butyrivibrio, Lachnospira and Rosburia. Prokaryotes 4:1002–1021 [Google Scholar]
- 9. Dubberke E. 2012. Clostridium difficile infection: the scope of the problem. J. Hosp. Med. 7(Suppl. 3):S1–4 [DOI] [PubMed] [Google Scholar]
- 10. Duncan SH, Louis P, Flint HJ. 2007. Cultivable bacterial diversity from the human colon. Lett. Appl. Microbiol. 44:343–350 [DOI] [PubMed] [Google Scholar]
- 11. Falk PG, Hooper LV, Midtvedt T, Gordon JI. 1998. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol. Mol. Biol. Rev. 62:1157–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fekety R, Silva J, Browne RA, Rifkin GD, Ebright JR. 1979. Clindamycin-induced colitis. Am. J. Clin. Nutr. 32:244–250 [DOI] [PubMed] [Google Scholar]
- 13. Finegold SM, et al. 2005. Clostridium clostridioforme: a mixture of three clinically important species. Eur. J. Clin. Microbiol. Infect. Dis. 24:319–324 [DOI] [PubMed] [Google Scholar]
- 14. Frank DN, et al. 2011. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm. Bowel Dis. 17:179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frank DN, et al. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U. S. A. 104:13780–13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Freter R, Ozawa A. 1963. Explanation for limitation of populations of Escherichia coli in broth cultures. J. Bacteriol. 86:904–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gorbach SL, Barza M, Giuliano M, Jacobus NV. 1988. Colonization resistance of the human intestinal microflora: testing the hypothesis in normal volunteers. Eur. J. Clin. Microbiol. Infect. Dis. 7:98–102 [DOI] [PubMed] [Google Scholar]
- 18. Hall I, O'Toole E. 1935. Intestinal flora in newborn infants with the description of a new anaerobic pathogen, Bacillus difficilus. Am. J. Dis. Child. 49:390–402 [Google Scholar]
- 19. Hedberg ME, et al. Lachnoanaerobaculum a new genus in Lachnospiraceae; characterization of Lachnoanaerobaculum umeaense gen. nov., sp. nov., isolated from human small intestine, Lachnoanaerobaculum orale gen. nov., sp. nov., isolated from saliva and reclassification of Eubacterium saburreum (Prevot) Holdeman and Moore 1970 as Lachnoanaerobaculum saburreum comb. nov. Int. J. Syst. Evol. Microbiol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hopkins MJ, Macfarlane GT. 2003. Nondigestible oligosaccharides enhance bacterial colonization resistance against Clostridium difficile in vitro. Appl. Environ. Microbiol. 69:1920–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Itoh K, Lee WK, Kawamura H, Mitsuoka T, Magaribuchi T. 1987. Intestinal bacteria antagonistic to Clostridium difficile in mice. Lab. Anim. 21:20–25 [DOI] [PubMed] [Google Scholar]
- 22. Kelly CP, et al. 1994. Neutrophil recruitment in Clostridium difficile toxin A enteritis in the rabbit. J. Clin. Invest. 93:1257–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Khanna S, Pardi DS. 2010. The growing incidence and severity of Clostridium difficile infection in inpatient and outpatient settings. Expert Rev. Gastroenterol. Hepatol. 4:409–416 [DOI] [PubMed] [Google Scholar]
- 24. Louis P, Flint HJ. 2009. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294:1–8 [DOI] [PubMed] [Google Scholar]
- 25. Manichanh C, Rigottier-Gois L, BE 2006. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 55:205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. May T, Mackie RI, Fahey GC, Jr, Cremin JC, Garleb KA. 1994. Effect of fiber source on short-chain fatty acid production and on the growth and toxin production by Clostridium difficile. Scand. J. Gastroenterol. 29:916–922 [DOI] [PubMed] [Google Scholar]
- 27. Naaber P, et al. 2004. Inhibition of Clostridium difficile strains by intestinal Lactobacillus species. J. Med. Microbiol. 53:551–554 [DOI] [PubMed] [Google Scholar]
- 28. Powell N, Jung SE, Krishnan B. 2008. Clostridium difficile infection and inflammatory bowel disease: a marker for disease extent? Gut 57:1183–1184 (Author reply, 57:1184.) [PubMed] [Google Scholar]
- 29. Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. 2002. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 217:133–139 [DOI] [PubMed] [Google Scholar]
- 30. Reeves AE, et al. 2011. The interplay between microbiome dynamics and pathogen dynamics in a murine model of Clostridium difficile infection. Gut Microbes 2:145–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rolfe RD. 1984. Role of volatile fatty acids in colonization resistance to Clostridium difficile. Infect. Immun. 45:185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rousseau C, et al. 2011. Clostridium difficile colonization in early infancy is accompanied by changes in intestinal microbiota composition. J. Clin. Microbiol. 49:858–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schloss PD, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Spencer RC. 1998. The role of antimicrobial agents in the aetiology of Clostridium difficile-associated disease. J. Antimicrob. Chemother. 41(Suppl. C):21–27 [DOI] [PubMed] [Google Scholar]
- 35. Stevenson BS, Eichorst SA, Wertz JT, Schmidt TM, Breznak JA. 2004. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70:4748–4755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Su WJ, et al. 1987. Role of volatile fatty acids in colonization resistance to Clostridium difficile in gnotobiotic mice. Infect. Immun. 55:1686–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Theriot CM, et al. 2011. Cefoperazone-treated mice as an experimental platform to assess differential virulence of Clostridium difficile strains. Gut Microbes 2:326–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Van der Waaij D, Berghuis JM, Lekkerkerk JE. 1972. Colonization resistance of the digestive tract of mice during systemic antibiotic treatment. J. Hyg. (Lond.) 70:605–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Van der Waaij D, Berghuis JM, Lekkerkerk-van der Wees JEC. 1971. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J. Hyg. (Lond.) 69:405–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vollaard EJ, Clasener HA. 1994. Colonization resistance. Antimicrob. Agents Chemother. 38:409–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wilson KH. 1993. The microecology of Clostridium difficile. Clin. Infect. Dis. 16(Suppl. 4):S214–S218 [DOI] [PubMed] [Google Scholar]
- 42. Wilson KH, Freter R. 1986. Interaction of Clostridium difficile and Escherichia coli with microfloras in continuous-flow cultures and gnotobiotic mice. Infect. Immun. 54:354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wilson KH, Sheagren JN, Freter R. 1985. Population dynamics of ingested Clostridium difficile in the gastrointestinal tract of the Syrian hamster. J. Infect. Dis. 151:355–361 [DOI] [PubMed] [Google Scholar]
- 44. Wilson KH, Sheagren JN, Freter R, Weatherbee L, Lyerly D. 1986. Gnotobiotic models for study of the microbial ecology of Clostridium difficile and Escherichia coli. J. Infect. Dis. 153:547–551 [DOI] [PubMed] [Google Scholar]
- 45. Wilson KH, Silva J, Fekety FR. 1981. Suppression of Clostridium difficile by normal hamster cecal flora and prevention of antibiotic-associated cecitis. Infect. Immun. 34:626–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. 2006. Colonic health: fermentation and short chain fatty acids. J. Clin. Gastroenterol. 40:235–243 [DOI] [PubMed] [Google Scholar]