Abstract
Salmonella enterica serovar Typhimurium is able to resist antimicrobial peptide killing by induction of the PhoP-PhoQ and PmrA-PmrB two-component systems and the lipopolysaccharide (LPS) modifications they mediate. Murine cathelin-related antimicrobial peptide (CRAMP) has been reported to inhibit S. Typhimurium growth in vitro and in vivo. We hypothesize that infection of human monocyte-derived macrophages (MDMs) with Salmonella enterica serovar Typhi and S. Typhimurium will induce human cathelicidin antimicrobial peptide (CAMP) production, and exposure to LL-37 (processed, active form of CAMP/hCAP18) will lead to upregulation of PmrAB-mediated LPS modifications and increased survival in vivo. Unlike in mouse macrophages, in which CRAMP is upregulated during infection, camp gene expression was not induced in human MDMs infected with S. Typhi or S. Typhimurium. Upon infection, intracellular levels of ΔphoPQ, ΔpmrAB, and PhoPc S. Typhi decreased over time but were not further inhibited by the vitamin D3-induced increase in camp expression. MDMs infected with wild-type (WT) S. Typhi or S. Typhimurium released similar levels of proinflammatory cytokines; however, the LPS modification mutant strains dramatically differed in MDM-elicited cytokine levels. Overall, these findings indicate that camp is not induced during Salmonella infection of MDMs nor is key to Salmonella intracellular clearance. However, the cytokine responses from MDMs infected with WT or LPS modification mutant strains differ significantly, indicating a role for LPS modifications in altering the host inflammatory response. Our findings also suggest that S. Typhi and S. Typhimurium elicit different proinflammatory responses from MDMs, despite being capable of adding similar modifications to their LPS structures.
INTRODUCTION
Cationic antimicrobial peptides are an evolutionarily conserved component of the innate immune system that aid the host in defense against invading bacteria, viruses, and fungi through their ability to directly kill invading pathogens and modulate the host innate and adaptive immune responses. The antimicrobial activity of these peptides comes from the ability of these molecules to insert into the microbial membrane, resulting in membrane destabilization and microbial lysis (3, 15, 18). Antimicrobial peptides are small amphipathic molecules that are classified based on their secondary structure, and they can be separated into categories such as α- or β-defensins and cathelicidins. Cathelicidin antimicrobial peptide (CAMP) is the only member of the cathelicidin family expressed in humans. camp encodes the precursor protein, hCAP18, which is cleaved to release LL-37, a cationic 37-amino-acid antimicrobial peptide (3). In addition to having potent killing activity toward many different pathogens, LL-37 is also able to inhibit immunostimulatory effects of various bacterial components, including lipopolysaccharide (LPS) and lipoteichoic acid (3, 18). LL-37 has been shown to be expressed by epithelial cells, specific lymphocyte populations, neutrophils, monocytes, and macrophages.
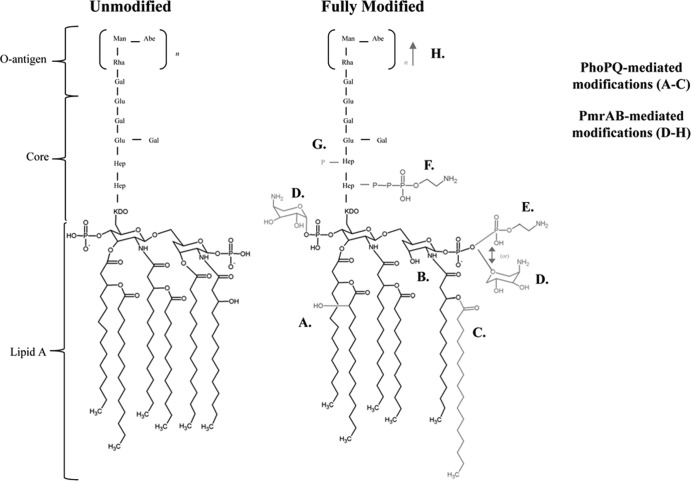
The innate immune system recognizes and responds to pathogens through the detection of conserved microbially associated molecular patterns such as LPS. LPS is a major component of the outer membrane of Gram-negative bacteria. LPS consists of phospholipids and polysaccharides, and its structure is divided into three different regions: lipid A, core, and O antigen (Fig. 1). The conserved hexa-acylated lipid A of enteric bacteria is responsible for anchoring the LPS molecule to the outer membrane. The lipid A portion of LPS is recognized by Toll-like receptor 4 (TLR4) in complex with MD2 present on multiple cell types, including epithelial cells and macrophages. The binding of lipid A by TLR4/MD2 triggers a cascade of events that leads to the production of proinflammatory cytokines and antimicrobial peptides (7, 17). Modification of the lipid A portion of LPS alters signaling through TLR4/MD2. Removal of fatty acid chains or altering the charges of the phosphate groups present on lipid A has been shown to alter the inflammatory activity of LPS (11, 12, 17).
Fig 1.
Unmodified and fully modified Salmonella lipopolysaccharide. LPS can be modified by PhoP-PhoQ-regulated mechanisms (modifications A to C are shown in gray) or by PmrA-PmrB-regulated mechanisms (modifications D to H are shown in gray). PhoPQ-regulated genes include lpxO, whose product results in the addition of 2-hydroxymyristate to the 3′ position of lipid A (A); pagL, whose product results in deacylation at the 3′ position of lipid A (B); and pagP, whose product results in the addition of a palmitate chain to the 2′ position of lipid A (C). PmrAB-regulated genes include pmrHFIJKL, whose product results in the addition of aminoarabinose to the 1 and 4′ phosphates of lipid A (D); pmrC, whose product results in the addition of phosphoethanolamine to the 1 and 4′ phosphates of lipid A (E); cptA, whose product results in the addition of phosphoethanolamine to the LPS core (F); pmrG, whose product results in the addition of phosphate to heptose present in the LPS core (G); and cld, whose product results in an O-antigen chain length determinant (H). Collectively, these LPS modifications are generally thought to aid Salmonella survival by providing resistance to killing by host antimicrobial peptides.
Salmonella enterica serovar Typhimurium is capable of infecting a variety of hosts. While S. Typhimurium causes salmonellosis in humans, it causes a typhoid-like disease in susceptible mice, making this a common animal model for studying typhoid fever (4, 6, 20). Salmonella enterica serovar Typhi is a human-specific pathogen that causes enteric or typhoid fever (16). Both S. Typhimurium and S. Typhi invade through the M cells present in intestinal Peyer's patches. S. Typhimurium and S. Typhi interact with antimicrobial peptides during infection of the intestinal mucosa and macrophages. S. Typhimurium and S. Typhi are able to resist killing by antimicrobial peptides, primarily through the induction of the PhoP-PhoQ (PhoPQ) and PmrA-PmrB (PmrAB) two-component regulatory systems (TCRS) and through the LPS modifications mediated by their regulated genes (Fig. 1) (8; S. Richards et al., submitted for publication). Much of our current understanding of Salmonella LPS modifications and resistance to antimicrobial peptides comes from work performed with S. Typhimurium using in vitro models.
Prior research has demonstrated a role for the murine cathelin-related antimicrobial peptide (CRAMP) in the innate immune response of mouse macrophages to S. Typhimurium infection, showing that CRAMP inhibits S. Typhimurium growth both in vitro and in vivo (19). Our lab has recently shown that S. Typhi is also able to use the PhoPQ and PmrAB-regulated gene products to modify its LPS in a manner similar to that of S. Typhimurium (Fig. 1 and data not shown) (Richards et al., submitted). We hypothesized that similar to the response of murine macrophages to S. Typhimurium infection, infection of human monocyte-derived macrophages (MDMs) with Salmonella would induce camp expression, leading to LL-37 production, that exposure to low levels of LL-37 would lead to upregulation of PmrAB-mediated LPS modifications in Salmonella, and that exposure to high levels of LL-37 would cause bacterial killing. However, our results indicate that unlike mouse macrophages, in which CRAMP has been reported to be upregulated during S. Typhimurium infection, camp was not induced in human MDMs infected with wild-type (WT) or LPS modification mutant strains of S. Typhi or S. Typhimurium, nor did it significantly affect bacterial survival. Exposure to LPS purified from these strains also did not induce camp expression. Our findings indicate that CAMP/LL-37 is not induced or required for bacterial clearance during Salmonella infection of human MDMs. However, the cytokine responses from MDMs infected with S. Typhi and S. Typhimurium LPS modification mutants are significantly different, indicating a role for LPS modifications in altering the host inflammatory response.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The Salmonella strains used are listed in Table 1. Salmonella strains were maintained using Luria-Bertani medium (LB) plus antibiotics, and when appropriate, antibiotics were used at the following concentrations: chloramphenicol, 25 μg/ml; kanamycin, 45 μg/ml; and tetracycline, 15 μg/ml. Prior to infection of MDMs or monocytes, bacteria were grown overnight (O/N) at 37°C with aeration in LB containing low levels of Mg2+ (25) or were cultured in N-minimal medium (NMM) (pH 5.5) plus 10 μM MgCl2 to induce PhoPQ- and PmrAB-mediated LPS modifications (the presence of LPS modifications was confirmed by matrix-assisted laser desorption ionization–time of flight mass spectrometry [MALDI-TOF MS] [data not shown]). Bacteria were pelleted and washed with RPMI 1640 and resuspended in RPMI 1640 plus 1% autologous serum for infection experiments.
Table 1.
Salmonella enterica serovar Typhimurium and Typhi strains used in this study
| Strain | Relevant characteristics | Source or reference |
|---|---|---|
| JSG210 | WT S. Typhimurium: ATCC 14028s (CDC) | ATCC |
| JSG421 | ΔpmrA S. Typhimurium: ATCC 14028s + pmrA:: Tn10d-tet (made by transducing Tn10d-tet pool into pagB::MudJ strain) | 8b |
| JSG1049 | S. Typhimurium ATCC 14028s; pmrF::Tn10d (Tet) | 8a |
| JSG206 | ΔphoP S. Typhimurium; ATCC 14028s + phoP::Tn10d-cam (CS015) (Cam) | Richards et al., submitted |
| JSG208 | PhoPc S. Typhimurium | 19 |
| JSG698 | WT S. Typhi: Ty2 (EX542) | ATCCa |
| JSG3028 | ΔpmrAB S. Typhi (JSG698) | Richards et al., submitted |
| JSG3070 | ΔpmrF S. Typhi (JSG698) | Richards et al., submitted |
| JSG3079 | ΔphoPQ S. Typhi (JSG698) | Richards et al., submitted |
| JSG700 | PhoPc S. Typhi | 1 |
| JSG1213 | tviB::Kan, Kan cassette inserted into Vi Ag gene of S. Typhi (Ty2) | 23ab |
Gift from R. Morona (University of Adelaide, Australia).
Gift from M. Popoff (Pasteur Institute, France).
Isolation and culture of human monocytes and macrophages.
Blood was obtained from healthy human volunteers using a protocol approved by The Ohio State University Institutional Review Board and was processed as previously described (10). Briefly, peripheral blood mononuclear cells (PBMCs) were isolated from heparinized blood over a Ficoll-Paque PLUS (Amersham Biosciences/GE Healthcare, Pittsburgh, PA) gradient and were cultured in Teflon wells (Savillex, Minneapolis, MN) for 1 to 5 days. Approximately 2 × 106 PBMCs/ml were seeded into each well in the presence of RPMI 1640 (Gibco, Grand Island, NY) containing 20% autologous serum and were incubated at 37°C with 5% CO2 (2). Cells were considered to be monocytes during days 1 to 4 and had differentiated into monocyte-derived macrophages by day 5. On day 5, MDMs were removed from the Teflon wells, and 4 × 106 PBMCs/ml were seeded into 12-well tissue culture plates and incubated for 3 h in RPMI 1640 containing 10% human AB serum (Lonza BioWhittaker, Basel, Switzerland) at 37°C with 5% CO2. After 3 h, cells were washed with warm RPMI 1640 to remove any nonadherent cells, and remaining adherent cells were incubated for up to an additional 7 days in RPMI 1640 containing 20% human AB serum. The cells were then used for experiments (referred to as day 12 MDMs). None of the volunteers had received the Ty21a or ViCPS vaccine or had a history of typhoid fever.
Purification of S. Typhi lipopolysaccharide.
LPS was isolated from WT Ty2, ΔpmrAB, ΔpmrF, and ΔphoPQ S. Typhi grown O/N in either LB or NMM (pH 5.5) plus 10 μM MgCl2. LPS was purified using a TRIzol-based protocol adapted from the method of Yi and Hackett (26). Contaminating lipids were removed from the LPS samples by Folch extraction. LPS samples were analyzed by SDS-PAGE, followed by silver staining, and endotoxic activities of the LPS samples were confirmed by an endotoxin assay kit (GenScript, Piscataway, NJ) (data not shown).
Monocyte and MDM infection and LPS stimulation studies.
Day 12 MDMs were used for all macrophage infections and LPS stimulations. For infection studies involving monocytes, cells were obtained on day 1 of maturation in Teflon wells. MDMs or monocytes were incubated at 37°C with 5% CO2 in RHH medium (RPMI 1640 plus 10 mM HEPES [Invitrogen, Grand Island, NY] plus 0.4% human serum albumin [CSL Behring LLC, King of Prussia, PA]) or RPMI 1640 plus 1% autologous serum during infection or LPS stimulation. For infection studies, MDMs or monocytes were incubated with Salmonella (multiplicity of infection [MOI], 10:1) in triplicate. Tissue culture wells were centrifuged briefly (180 × g for 1 min) following the addition of bacteria to synchronize infection. MDMs or monocytes were incubated with bacteria for 2 h, and medium was then removed from each well and replaced with medium containing either 50 μg/ml or 10 μg/ml gentamicin (Gibco) to kill any salmonellae that had not been internalized with the MDMs or monocytes. Wells that received 50 μg/ml gentamicin were incubated for an additional 30 min, after which the MDMs or monocytes were processed for RNA isolation, protein isolation, or enumeration of intracellular salmonellae (the samples collected after a total of 2.5 h of Salmonella exposure represent the initial infection time point). Wells that received 10 μg/ml gentamicin were incubated for additional lengths of time. These wells were processed for RNA isolation, protein isolation, or enumeration of intracellular salmonellae after 10 and 24 total hours of bacterial exposure. Total crude protein isolated from MDMs that had been infected with S. Typhi or exposed to LPS purified from WT S. Typhi was used for Western blot detection of LL-37 using a rabbit anti-LL-37 antibody (Phoenix Pharmaceuticals, Burlingame, CA) and a goat anti-rabbit secondary antibody (Bio-Rad, Hercules, CA). Purified LL-37 (AnaSpec, Fremont, CA) was used as a positive control (see Fig. S1 in the supplemental material). The proinflammatory cytokines tumor necrosis factor alpha (TNF-α) and interleukin 6 (IL-6) were detected in cell culture supernatant by enzyme-linked immunosorbent assay (ELISA) (Biolegend, San Diego, CA, and R&D Systems, Minneapolis, MN, respectively). For LPS stimulation experiments, MDMs were incubated in medium containing 100 ng/ml purified S. Typhi LPS for 2.5, 10, or 24 h. RNA was isolated from MDMs. As a positive control for the induction of LL-37 gene expression, monocytes or MDMs were stimulated with 100 nM 1α,25-dihydroxyvitamin D3 (1,25D3) (Enzo Life Sciences, Farmingdale, NY) for 24 h in either RHH or RPMI 1640 plus 1% autologous serum (14, 24).
Gene expression studies by qRT-PCR.
Gene expression levels of camp and gapdh were monitored during Salmonella infection and LPS stimulation of MDMs and monocytes. To isolate RNA, monocytes and MDMs were lysed in TRIzol, and then total RNA was isolated using the Qiagen RNeasy column method. The quantity of RNA was determined using the NanoDrop spectrophotometer (NanoDrop Products, Wilmington, DE). RNA (500 ng) was reverse transcribed to cDNA by reverse transcriptase enzyme (Superscript III, Invitrogen). camp gene expression was determined by quantitative real-time PCR (qRT-PCR) using SYBR green PCR master mixture in the Bio-Rad CFX (Bio-Rad, Hercules, CA). camp amplification was normalized to gapdh expression (threshold cycle [ΔCT]). Relative copy number (RCN) was calculated as described by Gavrilin et al. (5). PCR was performed by using the following oligonucleotide primers: camp, 5′-TGCCCAGGTCCTCAGCTAC-3′ and 5′-GTGACTGCTGTGTCGTCCT-3′, and gapdh, 5′-AAGGTGAAGGTCGGAGTCAAC-3′ and 5′-GGGGTCATTGATGGCAACAATA-3′.
Infection studies with 1,25D3-pretreated and untreated MDMs.
MDMs were obtained as previously described and allowed to mature until day 11. On day 11, RPMI containing 20% AB serum was removed and replaced with RPMI 1640 containing 1% autologous serum plus either 100 nM 1,25D3 or an equivalent amount of vehicle (1,25D3 is dissolved in ethanol). MDMs were incubated at 37°C for 24 h in the presence of RPMI 1640 plus 1,25D3 or RPMI 1640 plus vehicle. After 24 h of incubation, MDMs were gently washed three times with warm RPMI 1640 and were then infected with WT (grown in LB [low Mg2+] or in NMM with low pH and low Mg2+ prior to infection to induce LPS modifications), ΔpmrAB, ΔpmrF, ΔphoPQ, or PhoPc S. Typhi (in RPMI 1640 plus 1% autologous serum) for 2 h, and then medium was removed from each well and replaced with RPMI 1640 plus 1% autologous serum and either 50 μg/ml or 10 μg/ml gentamicin (as described above) to kill any S. Typhi organisms that had not been internalized with the MDMs or monocytes. MDMs were processed for enumeration of intracellular S. Typhi organisms 2.5 and 10 h postinfection (hpi).
RESULTS
Exposure to purified LPS from S. Typhi does not induce camp gene expression in human MDMs.
The effect of S. Typhimurium LPS modifications on mouse antimicrobial peptide expression has been well studied, while the effects of S. Typhi and S. Typhimurium LPS modifications on human antimicrobial peptide expression remain unknown. The studies presented here address the effects of S. Typhi and S. Typhimurium LPS modifications on the expression of camp. To investigate the role of LPS modifications, LPS was isolated and purified from WT Ty2, ΔpmrAB, ΔpmrF, and ΔphoPQ S. Typhi using a TRIzol-based technique (26) and then was repurified by Folch extraction. These strains were grown in LB (which contained low levels of Mg2+) to induce LPS modifications (25). Primary human MDMs were allowed to mature for 12 days prior to stimulation with 100 ng of purified LPS. MDMs were exposed to LPS for 2.5, 10, and 24 h. Expression of camp was determined by quantitative real-time PCR (qRT-PCR) at each time point. Our studies revealed that exposure to LPS purified from WT Ty2, ΔpmrAB, ΔpmrF, and ΔphoPQ S. Typhi did not induce camp gene expression (Fig. 2). MDMs were exposed to 100 nM 1,25D3 for 24 h as a positive control for camp gene expression (14, 24). Western blot analysis on crude protein collected from S. Typhi LPS-stimulated MDMs also did not show an increase in LL-37 production, while exposure to 1,25D3 clearly induced the precursor protein, hCAP18 (see Fig. S1 in the supplemental material). A limulus assay was performed to verify that the purified LPS samples had retained endotoxic activity (data not shown). To confirm that the MDMs were responsive to LPS exposure, the proinflammatory cytokines IL-6 and TNF-α were measured from the cell culture supernatants of the LPS-stimulated MDMs. Exposure to 100 ng LPS did induce the secretion of both IL-6 and TNF-α (two different donors [data not shown]). Despite variations in the levels of proinflammatory cytokines triggered by LPS exposure, MDMs from both donors responded with a robust increase in cytokine production, suggesting that although the MDMs are capable of initiating a proinflammatory response to S. Typhi LPS (with and without various LPS modifications), an increase in camp gene expression does not appear to be a part of this innate immune response.
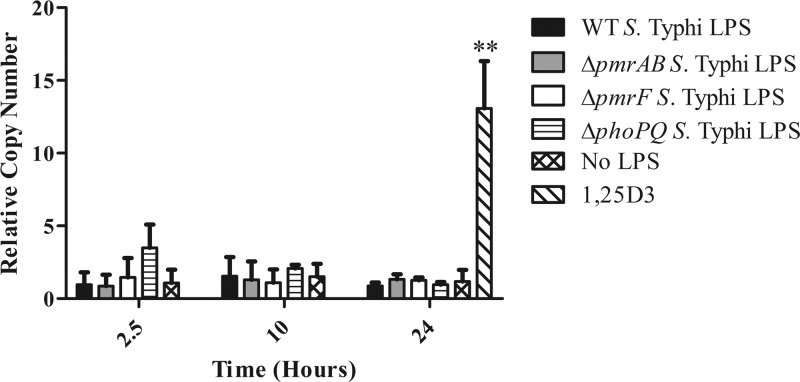
Fig 2.
Levels of camp gene expression induced by exposure to LPS purified from S. Typhi. Primary human MDMs were allowed to mature for 12 days. On day 12, MDMs were exposed to 100 ng LPS purified from WT Ty2, ΔpmrAB, ΔpmrF, and ΔphoPQ S. Typhi for 2.5, 6, or 10 h. MDMs were also incubated with 100 nM 1,25D3 (hormonal form of vitamin D) to serve as a positive control for camp expression. The graph represents the pooled results from two donors; however, these results have been confirmed using MDMs from additional donors stimulated with selected LPS species (n = 3). Statistical significance was determined by Student's t test comparing RCNs from a 24-h no-LPS sample to a 24-h 1,25D3-stimulated sample. **, P < 0.0001.
Infection with S. Typhi does not induce camp expression in human MDMs.
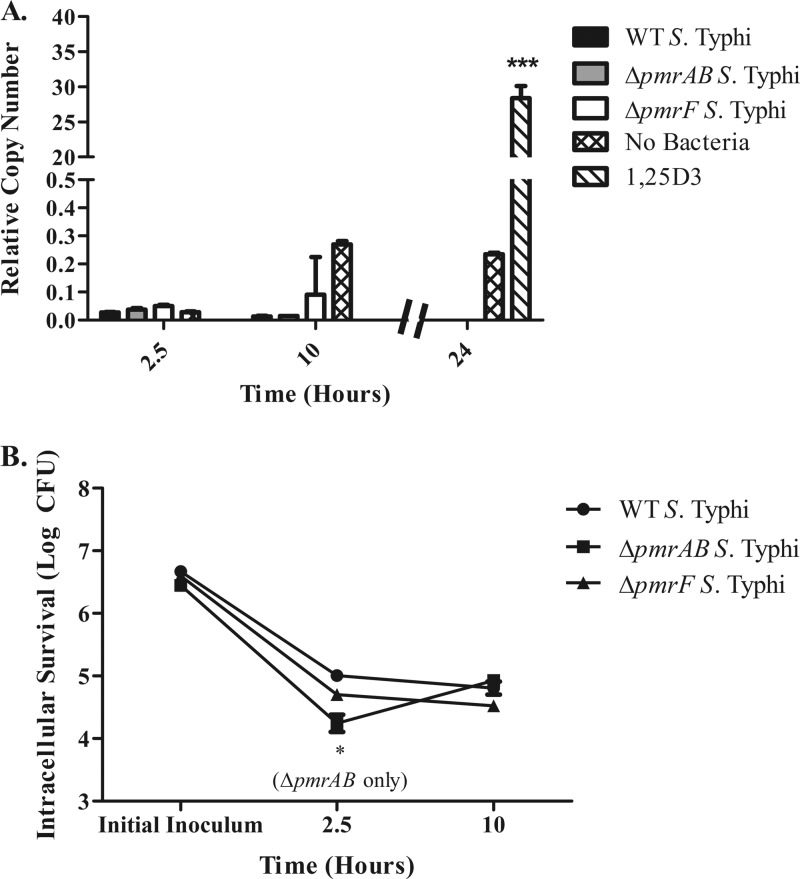
Although stimulation with purified LPS did not increase camp gene expression, it is possible that exposure to LPS free in tissue culture medium does not represent the normal context in which MDMs would recognize and respond to S. Typhi LPS during infection. Prior research had indicated a role for cathelicidin during S. Typhimurium infection of mouse macrophages (19). To address the potential effect of S. Typhi LPS modifications during infection, day 12 MDMs were infected with WT Ty2, ΔpmrAB, ΔpmrF, ΔphoPQ, or PhoPc S. Typhi at an MOI of 1:10 (MDM/bacteria) for 2.5, 10, or 24 h. Infection with WT or LPS modification mutants did not induce an increase in camp expression, while exposure to 1,25D3 caused a significant increase in camp expression (Fig. 3A). Western blots were used to detect LL-37 from crude protein samples collected from infected MDMs. The Western blots did not show increases in the precursor protein, hCAP18, or in the presence of processed LL-37 during infection with any S. Typhi strain (see Fig. S1 in the supplemental material). Differences between the findings presented here and the above-described published work demonstrating the role of CRAMP during S. Typhimurium infection of mouse macrophages may be due to differences between S. Typhi and S. Typhimurium. The presence of the Vi antigen on the surface of S. Typhi may alter the interactions between the macrophage and the bacterium and may be capable of suppressing camp expression. To address this possibility, day 12 MDMs were infected with a ΔtviB S. Typhi mutant lacking the Vi antigen. MDMs infected with ΔtviB S. Typhi also did not respond by increasing camp expression (Fig. 3A), suggesting that the presence of the Vi antigen does not mask the ability of LPS to induce camp.
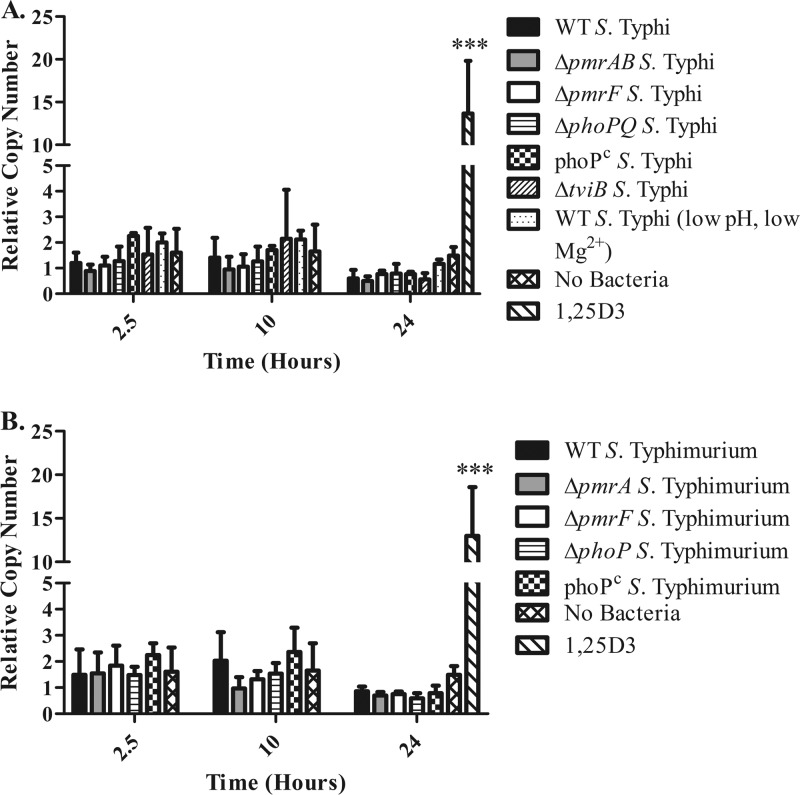
Fig 3.
Levels of camp gene expression induced by infection with S. Typhi and S. Typhimurium. Day 12 MDMs were infected in duplicate with S. Typhi (A) or S. Typhimurium (B) at an MOI of 1:10 (MDM/bacteria) for 2.5, 10, or 24 h prior to collection of eukaryotic RNA. MDMs were also treated with 100 nM 1,25D3 for 24 h as a positive control for the induction of camp gene expression. The graphs represent the pooled results from two donors; however, these results have been confirmed using MDMs from additional donors infected with select Salmonella strains (n = 3). Statistical significance was determined by Student's t test comparing RCNs from a 24-h uninfected sample to a 24-h 1,25D3-stimulated sample. ***, P < 0.00005.
Infection with S. Typhimurium does not induce camp expression in human MDMs.
Although the presence of the Vi antigen does not appear to dampen camp expression, it is possible that other differences between S. Typhi and S. Typhimurium may account for the induction of CRAMP in response to S. Typhimurium but not camp in response to S. Typhi. Day 12 MDMs were infected with WT, ΔpmrA, ΔpmrF, ΔphoP, or PhoPc S. Typhimurium at an MOI of 1:10 (MDM/bacteria) for 2.5, 10, or 24 h. Infection with WT or LPS modification mutants of S. Typhimurium also did not induce camp expression in human MDMs (Fig. 3B).
Intramacrophage survival of S. Typhi and S. Typhiurium in human MDMs.
In addition to investigating changes in camp expression during infection with S. Typhi and S. Typhimurium, levels of intramacrophage survival were also monitored. All tested strains appeared to be phagocytosed by the MDMs to similar extents. The levels of WT, ΔpmrF, and ΔtviB S. Typhi recovered at each time point were similar and remained constant from 2.5 to 24 h of infection. As expected, intracellular levels of ΔphoPQ and PhoPc S. Typhi decreased over time compared to levels of WT S. Typhi (Fig. 4A). Intracellular survival of the ΔpmrAB mutant was significantly lower than that of WT S. Typhi after 10 h postinfection (hpi); however, this growth defect was only noticed at the 10-h time point, and levels of ΔpmrAB S. Typhi were similar to those of the WT by 24 hpi (Fig. 4A).
Fig 4.
Intramacrophage survival S. Typhi and S. Typhimurium. Day 12 MDMs were infected with either S. Typhi (A) or S. Typhimurium (B) at an MOI of 1:10 for 2.5, 10, or 24 h prior to bacterial enumeration. MDMs were infected in duplicate with S. Typhi and S. Typhimurium (WT and LPS modification mutants). The graphs represent the pooled results from two donors; however, these results have been confirmed using MDMs from additional donors infected with selected Salmonella strains (n = 3). Statistical significance was determined by Student's t test comparing intramacrophage survival levels between LPS modification mutants and the WT strain enumerated at that time point. *, P < 0.05.
Interestingly, most of the tested strains of S. Typhimurium appeared to increase in number over time, unlike the S. Typhi strains tested, which remained constant over time and did not appear to be replicating (Fig. 4B). This finding was unexpected, as S. Typhi is thought to survive inside the human macrophage and progress to systemic infection, while S. Typhimurium causes an infection that is limited to the gastrointestinal mucosa and is controlled at the level of the reticuloendothelial system (1a). Only the PhoPc strain of S. Typhimurium did not increase in CFU over time; however, its survival defect was not as severe as the defect seen for PhoPc S. Typhi (Fig. 4). Surprisingly, ΔphoP S. Typhimurium did not show a survival defect in MDMs. To address this concern, a differently constructed ΔphoP S. Typhimurium strain was used to infect MDMs; it showed similar results (data not shown).
Proinflammatory cytokines released from S. Typhi- and S. Typhimurium-infected MDMs are different.
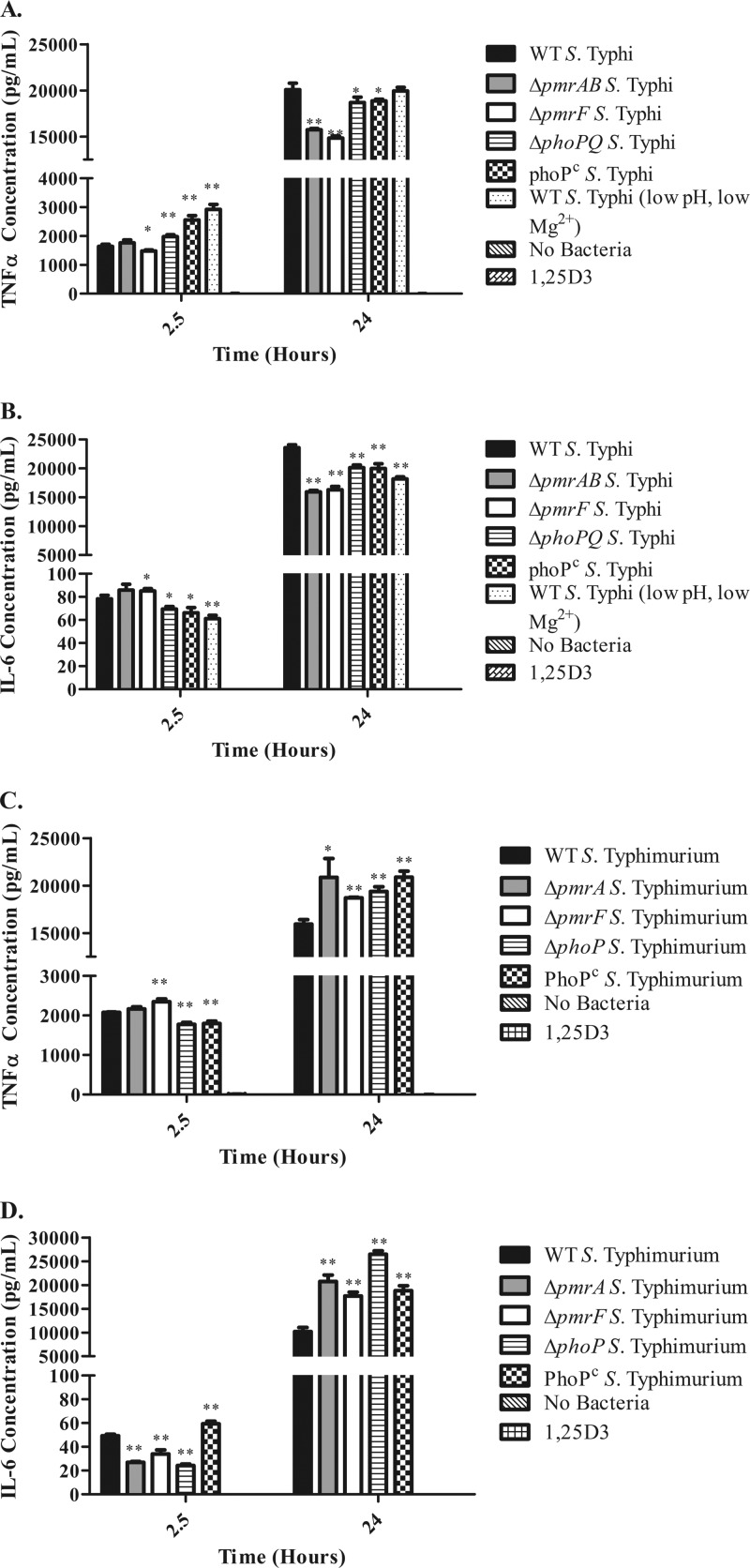
Although camp expression does not appear to be increased during the macrophage response to intracellular Salmonella, it is possible that LPS modifications alter TLR4 activation, leading to altered proinflammatory cytokine production during infection. To address whether the presence of LPS modifications alter the production of proinflammatory cytokines, TNF-α and IL-6 were measured from the cell culture supernatants of infected MDMs. Infection with all strains tested triggered the release of TNF-α and IL-6 from MDMs, while uninfected or 1,25D3-treated MDMs did not release significant levels of TNF-α or IL-6 (Fig. 5). After 2.5 h of infection, WT and ΔpmrAB S. Typhi-infected MDMs released similar levels of TNF-α, while ΔpmrF S. Typhi-infected MDMs released significantly lower levels of TNF-α (Fig. 5A). Interestingly, ΔpmrF S. Typhi-infected MDMs released significantly lower levels of IL-6 than did WT-infected MDMs after 2.5 h (Fig. 5B). The ΔphoPQ and PhoPc S. Typhi-infected MDMs released significantly greater amounts of TNF-α but significantly lower levels of IL-6 than did WT S. Typhi-infected MDMs after 2.5 h of infection (Fig. 5A and B). MDMs infected with the WT S. Typhi grown at low pH and low Mg2+ released the most TNF-α and the least IL-6 at the 2.5-h time point (Fig. 5A and B). After 24 h of infection, WT-infected MDMs released significantly greater amounts of TNF-α and IL-6 than did MDMs infected with ΔpmrAB, ΔpmrF, ΔphoPQ, or PhoPc S. Typhi, with TNF-α and IL-6 levels released from ΔpmrAB and ΔpmrF S. Typhi-infected MDMs being the lowest of all (Fig. 5A and B). MDMs infected with WT S. Typhi grown under low-pH and low-Mg2+ conditions showed significantly lower levels of IL-6 but not TNF-α (Fig. 5A and B). Interestingly, the trends for TNF-α and IL-6 release from MDMs infected with WT and mutant S. Typhimurium differed dramatically from those in cells infected with S. Typhi. By 24 h postinfection, MDMs infected with LPS modification mutant strains of S. Typhimurium released significantly greater levels of TNF-α and IL-6 than did WT-infected MDMs (Fig. 5C and D). These findings suggest that although LPS modifications can affect the proinflammatory response to Salmonella, additional factors must influence the MDM response to intracellular Salmonella, as the production of TNF-α and IL-6 differed dramatically between S. Typhi- and S. Typhimurium-infected MDMs (despite these strains being capable of modifying their LPS in similar manners).
Fig 5.
Levels of proinflammatory cytokines released from S. Typhi- or S. Typhimurium-infected MDMs. Shown are levels of TNF-α (A) and IL-6 (B) released from MDMs after 2.5 and 24 h of S. Typhi infection and TNF-α (C) and IL-6 (D) released from MDMs after 2.5 and 24 h of S. Typhimurium infection. Statistical significance was determined by Student's t test comparing cytokine levels between LPS modification mutant-infected samples and the WT-infected sample collected at that time point. *, P < 0.05; **, P < 0.005.
Infection with S. Typhi does not induce camp expression in human monocytes.
LL-37 is produced and stored as the inactive propeptide hCAP18. Proteinase-3 is an enzyme known to cleave hCAP18 to its active form and has been identified in neutrophils and monocytes (21). Although proteinase-3 has not been identified in MDMs, it is still likely that this cell type possesses other enzymes that are capable of cleaving hCAP18 into the active antimicrobial peptide, LL-37. Because human monocytes are known to possess proteinase-3, S. Typhi infection experiments were also performed using monocytes. Day 1 monocytes were infected with WT Ty2, ΔpmrAB, or ΔpmrF S. Typhi at an MOI of 1:10 (monocyte/bacteria) for 2.5 or 10 h. Infection with WT or LPS modification mutants did not induce an increase in camp expression, while exposure to 1,25D3 for 24 h caused a significant increase in camp (Fig. 6A). S. Typhi uptake and intracellular survival in monocytes had trends similar to those in infected MDMs; however, the overall amounts of intracellular S. Typhi detected at each time point were slightly lower than those seen in MDMs (Fig. 6B and Fig. 4A).
Fig 6.
camp gene expression and intracellular survival of S. Typhi in infected monocytes. Monocytes were isolated from 2 different donors and were infected with WT S. Typhi and S. Typhi LPS modification mutants in duplicate. All experiments showed similar results. The graphs represent the results from one donor. Student's t test compared results from a 24-h uninfected sample and a 24-h 1,25D3-stimulated sample (A) and intracellular survival levels between LPS modification mutant and the WT strain enumerated at that time point (B). *, P < 0.05; ***, P < 0.00001.
Pretreatment with 1,25D3 does not inhibit S. Typhi intracellular survival.
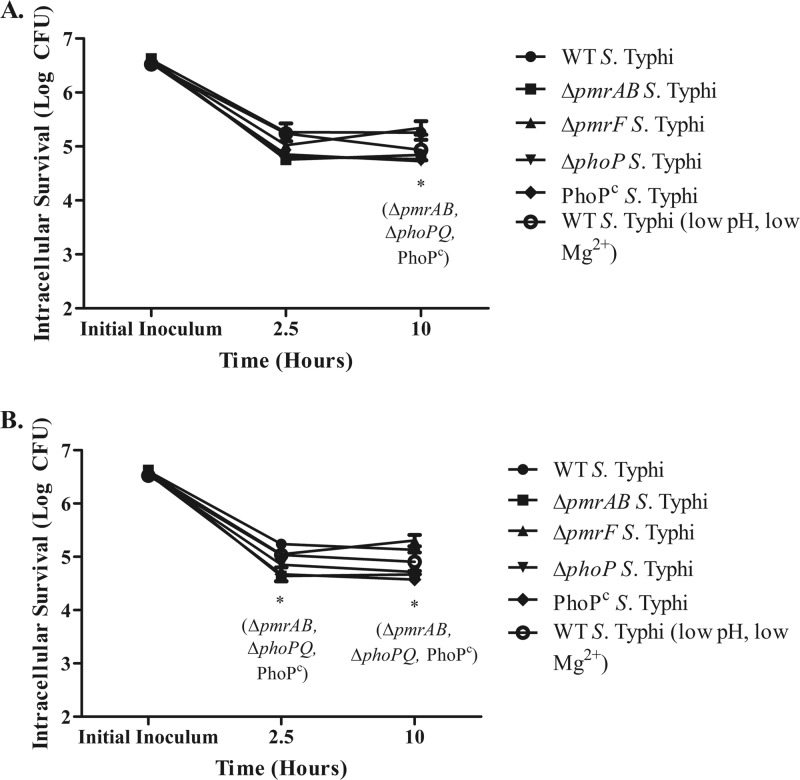
The ability of the hormonal form of vitamin D to induce camp expression has been demonstrated by numerous research groups and has been confirmed in our current studies (14, 24). To investigate the effects of elevated camp expression on WT S. Typhi and LPS modification mutants, MDMs were stimulated to upregulate camp expression by exposure to 1,25D3 for 24 h prior to infection with S. Typhi. Day 11 MDMs were incubated in RPMI 1640 plus 1% autologous serum containing 100 mM 1,25D3 for 24 h. After this treatment, MDMs were washed and then infected at an MOI of 1:10 for 2.5 and 10 h with WT, ΔpmrAB, ΔpmrF, or ΔphoPQ S. Typhi. Levels of intracellular survival for all strains were similar regardless of pretreatment with 1,25D3 (Fig. 7). As shown above, MDMs infected with ΔpmrAB, ΔphoPQ, and PhoPc S. Typhi showed impaired intracellular survival compared to those infected with WT S. Typhi. Overall, our studies suggest that camp expression is not induced or required for bacterial clearance during S. enterica infection of human MDMs.
Fig 7.
Intramacrophage survival of S. Typhi in MDMs previously stimulated with 1,25D3 (A) versus MDMs without 1,25D3 stimulation prior to infection (B). Day 11 MDMs were treated with 100 nM 1,25D3 for 24 h to induce camp gene expression or with medium alone. On day 12 (24 h after incubation with and without 1,25D3), MDMs were infected with WT S. Typhi and S. Typhi LPS modification mutants at an MOI of 1:10 for 2.5, 10, or 24 h prior to bacterial enumeration. MDMs were isolated from 2 different donors and were infected with S. Typhi in duplicate. All experiments showed similar results. The graphs represent the results from one donor. Student's t test compared intracellular survival levels between the LPS modification mutant and the WT strain enumerated at that time point. *, P < 0.05.
DISCUSSION
CRAMP, the murine cathelicidin and human CAMP ortholog, has previously been implicated in S. Typhimurium killing in mouse macrophages. Additionally, these studies demonstrated that a PhoP-null strain survived better in CRAMP KO macrophages than WT macrophages (19). These data suggest that CRAMP interacts with Salmonella within macrophages and suggest that LPS modifications play a role in providing bacterial resistance to antimicrobial peptides. Based on the published role of CRAMP in the murine macrophage response to Salmonella infection, we predicted that CAMP/LL-37 would play a significant role during Salmonella infection of human macrophages. Surprisingly, camp was not found to be induced following exposure to purified LPS or during S. Typhi or S. Typhimurium infection of human macrophages or monocytes, regardless of the presence or absence of different LPS modifications.
Mouse macrophages infected with S. Typhimurium rapidly induce CRAMP expression (19), while the work presented here showed that S. Typhi does not induce camp in human macrophages. It is possible that differences between S. Typhi and S. Typhimurium account for the differences between these two macrophage models. We addressed this concern by expanding our MDM infection studies to include WT and LPS modification mutant strains of S. Typhimurium. Infection with S. Typhimurium did not result in induction of camp expression, suggesting that the different results obtained from mouse versus human macrophages are most likely due to differences in the host immune response and not to differences between S. Typhi and S. Typhimurium. Furthermore, the Vi antigen of S. Typhi, which has previously been shown to elicit anti-inflammatory properties (9), did not impact the induction of camp expression.
Survival within macrophages is thought to be a critical step in the establishment of typhoid fever. Unexpectedly, S. Typhimurium survived better in human MDMs than did S. Typhi. Although both S. Typhi and S. Typhimurium were able to survive being internalized by MDMs, S. Typhimurium organisms appeared to increase in number over time, while S. Typhi was only able to persist. As expected, the persistence of ΔphoPQ and PhoPc strains of S. Typhi was significantly inhibited by MDMs compared to that of the WT. The ΔpmrAB strain of S. Typhi, but not S. Typhimurium, was significantly inhibited in MDMs compared to the WT. Only the PhoPc S. Typhimurium strain displayed a growth defect in MDMs. The ΔphoP S. Typhimurium strain was predicted to show a growth defect as well; however, while multiple ΔphoP strains were tested, none showed a survival defect. Further investigation is needed to understand why this defect was not observed.
Overall, these findings suggest that the loss of either TCRS or overexpression of LPS modifications encoded by PhoPQ leaves S. Typhi at a significant disadvantage during human macrophage infection. It is surprising to find that several of the intracellular survival patterns seen with S. Typhi are dramatically different when the infecting strain is S. Typhimurium (even when the strains lack the same LPS modifications). These findings may also suggest that our current views on Salmonella pathogenesis may need to be altered to allow for a model in which S. Typhimurium is also capable of surviving the harsh and restrictive internal environment of the macrophage, even in the absence of PhoPQ-mediated LPS modifications. It is also possible that the S. Typhimurium intracellular survival trends are an artifact that arises from studying macrophage infection in vitro. Infections occurring in vivo would be much more complex, and it is likely that numerous factors play a role in the ultimate elimination of Salmonella in the host environment.
Not only does LL-37 not appear to be induced during Salmonella infection but also the forced induction of camp expression by exposure to the hormonal form of vitamin D did not improve bacterial clearance by MDMs, demonstrating that LL-37 is not required for clearance of intracellular S. Typhi. The results presented here not only highlight differences between the mouse and human responses to human pathogens but also highlight the importance of studying the role of Salmonella LPS modifications in vivo. Although the ability to modify LPS increases resistance to LL-37 in vitro, this finding may not be relevant during infection of macrophages. It is still possible, however, that LPS modifications alter antimicrobial peptide expression during other stages of Salmonella infection.
Although LPS modifications did not appear to alter camp expression, infection with WT and LPS modification mutant strains triggered the release of proinflammatory cytokines, but to differing extents. LPS modifications can alter TLR4 activation and could contribute to the altered levels of proinflammatory cytokines released from infected MDMs. For example, PhoPQ-mediated modifications such as those mediated by PagL (which catalyzes deacylation at the 3 position of lipid A) alter recognition by TLR4 (11–13, 22, 23). Based on structural studies of LPS interactions with TLR4/MD2, the LPS modifications have been suggested to affect key points of contact (17). The PmrAB-mediated modifications would be predicted to affect LPS contact with TLR4 due to the addition of aminoarabinose or phosphoethanolamine to the 1′ and 4′ phosphate groups on lipid A. Consistent with this prediction, the MDMs infected with WT S. Typhimurium released significantly lower levels of TNF-α than did those infected with mutant strains unable to modify the phosphate groups present on lipid A. However, the MDMs infected with WT and LPS modification mutant strains of S. Typhi did not show the same trends. Infection with the WT triggered a greater release of IL-6 and TNF-α than did infection with ΔpmrAB and ΔpmrF S. Typhi; thus, the LPS modifications do play a role in altering the host innate immune response, but it is likely that other factors, in addition to LPS modification, play significant roles in the initiation of the proinflammatory response during S. Typhi and S. Typhimurium infection.
Although much insight and understanding have been and can be obtained by using mouse models to study S. Typhimurium infection, it is possible that some of what is known about the mouse immune response to S. Typhimurium cannot be directly applied to the human response to S. Typhi or S. Typhimurium. Overall, the findings presented here highlight the importance of studying Salmonella using human cells. Although LL-37 was not found to be involved in the human MDM response to S. Typhi or S. Typhimurium, it is possible that this important antimicrobial peptide plays a role at other tissue sites and during other stages of salmonellosis. It is also likely that other antimicrobial peptides play important roles in the innate immune response to Salmonella. Additional experiments investigating Salmonella infection in MDMs overexpressing LL-37 and in CAMP-deficient MDMs are necessary to better understand the role of LL-37 during infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank the current and former members of the Gunn lab and the Department of Microbial Infection and Immunity/Center for Microbial Interface Biology at The Ohio State University for their guidance and support. Special thanks are due to Larry Schlesinger and lab members for providing assistance with MDM isolation, Robert Ernst for performing MALDI-TOF analysis of lipid A samples, and Mark Wewers for guidance and support.
Funding for this work was provided by the National Institutes of Health (NIH) through an RO1 grant awarded to J.S.G. (AI043521), The Ohio State University Graduate School through the Dean's Distinguished University Fellowship to S.M.R., and T32 Lung Inflammation Training Grant (HL007946) awarded to K.L.S.
We do not have any conflicts of interest to disclose.
Footnotes
Published ahead of print 27 August 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Baker SJ, Daniels C, Morona R. 1997. PhoP/Q regulated genes in Salmonella typhi identification of melittin sensitive mutants. Microb. Pathog. 22:165–179 [DOI] [PubMed] [Google Scholar]
- 1a. Barrow PA, Huggins MB, Lovell MA. 1994. Host specificity of Salmonella infection in chickens and mice is expressed in vivo primarily at the level of the reticuloendothelial system. Infect. Immun. 62:4602–4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beharka AA, et al. 2002. Pulmonary surfactant protein A up-regulates activity of the mannose receptor, a pattern recognition receptor expressed on human macrophages. J. Immunol. 169:3565–3573 [DOI] [PubMed] [Google Scholar]
- 3. Bucki R, Leszczynska K, Namiot A, Sokolowski W. 2010. Cathelicidin LL-37: a multitask antimicrobial peptide. Arch. Immunol. Ther. Exp. (Warsz.) 58:15–25 [DOI] [PubMed] [Google Scholar]
- 4. Carter PB, Collins FM. 1974. The route of enteric infection in normal mice. J. Exp. Med. 139:1189–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gavrilin MA, et al. 2006. Internalization and phagosome escape required for Francisella to induce human monocyte IL-1beta processing and release. Proc. Natl. Acad. Sci. U. S. A. 103:141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gibbons HS, Kalb SR, Cotter RJ, Raetz CR. 2005. Role of Mg2+ and pH in the modification of Salmonella lipid A after endocytosis by macrophage tumour cells. Mol. Microbiol. 55:425–440 [DOI] [PubMed] [Google Scholar]
- 7. Gioannini TL, Weiss JP. 2007. Regulation of interactions of Gram-negative bacterial endotoxins with mammalian cells. Immunol. Res. 39:249–260 [DOI] [PubMed] [Google Scholar]
- 8. Gunn JS. 2008. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 16:284–290 [DOI] [PubMed] [Google Scholar]
- 8a. Gunn JS, et al. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27:1171–1182 [DOI] [PubMed] [Google Scholar]
- 8b. Gunn JS, Miller SI. 1996. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178:6857–6864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Haneda T, et al. 2009. The capsule-encoding viaB locus reduces intestinal inflammation by a Salmonella pathogenicity island 1-independent mechanism. Infect. Immun. 77:2932–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Horwitz MA, Silverstein SC. 1980. Legionnaires' disease bacterium (Legionella pneumophila) multiplies intracellularly in human monocytes. J. Clin. Invest. 66:441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kawasaki K, Ernst RK, Miller SI. 2004. 3-O-deacylation of lipid A by PagL, a PhoP/PhoQ-regulated deacylase of Salmonella typhimurium, modulates signaling through Toll-like receptor 4. J. Biol. Chem. 279:20044–20048 [DOI] [PubMed] [Google Scholar]
- 12. Kawasaki K, Ernst RK, Miller SI. 2004. Deacylation and palmitoylation of lipid A by Salmonellae outer membrane enzymes modulate host signaling through Toll-like receptor 4. J. Endotoxin Res. 10:439–444 [DOI] [PubMed] [Google Scholar]
- 13. Kawasaki K, Ernst RK, Miller SI. 2005. Inhibition of Salmonella enterica serovar Typhimurium lipopolysaccharide deacylation by aminoarabinose membrane modification. J. Bacteriol. 187:2448–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu N, et al. 2009. Vitamin D induces innate antibacterial responses in human trophoblasts via an intracrine pathway. Biol. Reprod. 80:398–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matsuzaki K, et al. 1997. Membrane permeabilization mechanisms of a cyclic antimicrobial peptide, tachyplesin I, and its linear analog. Biochemistry 36:9799–9806 [DOI] [PubMed] [Google Scholar]
- 16. Ohl ME, Miller SI. 2001. Salmonella: a model for bacterial pathogenesis. Annu. Rev. Med. 52:259–274 [DOI] [PubMed] [Google Scholar]
- 17. Park BS, et al. 2009. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458:1191–1195 [DOI] [PubMed] [Google Scholar]
- 18. Radek K, Gallo R. 2007. Antimicrobial peptides: natural effectors of the innate immune system. Semin. Immunopathol. 29:27–43 [DOI] [PubMed] [Google Scholar]
- 19. Rosenberger CM, Gallo RL, Finlay BB. 2004. Interplay between antibacterial effectors: a macrophage antimicrobial peptide impairs intracellular Salmonella replication. Proc. Natl. Acad. Sci. U. S. A. 101:2422–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Scherer CA, Miller SI. 2001. Molecular pathogenesis of salmonellae, p 266–316 In Groisman EA. (ed), Principles of bacterial pathogenesis. Academic Press, San Diego, CA [Google Scholar]
- 21. Sørensen OE, et al. 2001. Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood 97:3951–3959 [DOI] [PubMed] [Google Scholar]
- 22. Tran AX, et al. 2005. Resistance to the antimicrobial peptide polymyxin requires myristoylation of Escherichia coli and Salmonella typhimurium lipid A. J. Biol. Chem. 280:28186–28194 [DOI] [PubMed] [Google Scholar]
- 23. Trent MS, Pabich W, Raetz CR, Miller SI. 2001. A PhoP/PhoQ-induced Lipase (PagL) that catalyzes 3-O-deacylation of lipid A precursors in membranes of Salmonella typhimurium. J. Biol. Chem. 276:9083–9092 [DOI] [PubMed] [Google Scholar]
- 23a. Virlogeux I, Waxin H, Ecobichon C, Popoff MY. 1995. Role of the viaB locus in synthesis, transport and expression of Salmonella typhi Vi antigen. Microbiology 141(Pt 12):3039–3047 [DOI] [PubMed] [Google Scholar]
- 24. Wang TT, et al. 2004. Cutting edge: 1,25-dihydroxyvitamin D3 is a direct inducer of antimicrobial peptide gene expression. J. Immunol. 173:2909–2912 [DOI] [PubMed] [Google Scholar]
- 25. Wee S, Wilkinson BJ. 1988. Increased outer membrane ornithine-containing lipid and lysozyme penetrability of Paracoccus denitrificans grown in a complex medium deficient in divalent cations. J. Bacteriol. 170:3283–3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yi EC, Hackett M. 2000. Rapid isolation method for lipopolysaccharide and lipid A from gram-negative bacteria. Analyst 125:651–656 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.