Abstract
The morphological transformation of Trichomonas vaginalis from an ellipsoid form in batch culture to an adherent amoeboid form results from the contact of parasites with vaginal epithelial cells and with immobilized fibronectin (FN), a basement membrane component. This suggests host signaling of the parasite. We applied integrated transcriptomic and proteomic approaches to investigate the molecular responses of T. vaginalis upon binding to FN. A transcriptome analysis was performed by using large-scale expressed-sequence-tag (EST) sequencing. A total of 20,704 ESTs generated from batch culture (trophozoite-EST) versus FN-amoeboid trichomonad (FN-EST) cDNA libraries were analyzed. The FN-EST library revealed decreased amounts of transcripts that were of lower abundance in the trophozoite-EST library. There was a shift by FN-bound organisms to the expression of transcripts encoding essential proteins, possibly indicating the expression of genes for adaptation to the morphological changes needed for the FN-adhesive processes. In addition, we identified 43 differentially expressed proteins in the proteomes of FN-bound and unbound trichomonads. Among these proteins, cysteine peptidase, glyceraldehyde-3-phosphate dehydrogenase (an FN-binding protein), and stress-related proteins were upregulated in the FN-adherent cells. Stress-related genes and proteins were highly expressed in both the transcriptome and proteome of FN-bound organisms, implying that these genes and proteins may play critical roles in the response to adherence. This is the first report of a comparative proteomic and transcriptomic analysis after the binding of T. vaginalis to FN. This approach may lead to the discovery of novel virulence genes and affirm the role of genes involved in disease pathogenesis. This knowledge will permit a greater understanding of the complex host-parasite interplay.
INTRODUCTION
Trichomonosis, caused by Trichomonas vaginalis, is the most common nonviral sexually transmitted infection (STI), and more than 170 million women are infected annually worldwide (63). This infection is associated with poor health outcomes for women, including vaginitis, preterm delivery, low-birth-weight infants, infertility, susceptibility to human papillomavirus (HPV) and herpesvirus infection, and cervical cancer (36). As there is no immunity to T. vaginalis, a common trait of trichomonosis is persistence (29). In males, T. vaginalis infection is usually asymptomatic, although in some cases, urethritis and chronic prostatitis are observed (7, 28). More recently, seropositivity for T. vaginalis was found to be associated with late-stage and lethal prostate cancer (64, 65). This STI is important because of the high prevalence and increased risk of human immunodeficiency virus transmission (51, 67).
Most mucosal pathogens bind to host tissues and possess mechanisms to evade host immune surveillance to initiate infection. Adherence to vaginal epithelial cells (VECs) and interactions with extracellular matrix (ECM) glycoproteins are essential for T. vaginalis to establish and maintain infection (46). T. vaginalis transforms from an ellipsoid form to an amoeboid form after contact with both VECs and fibronectin (FN) (2, 6), suggesting a signaling of parasites upon interactions with the host. Also, the ability of trichomonads to coat their surface with host proteins, including FN, may be important for both immune evasion and host colonization (59–61). Trichomonad surface proteins play crucial roles in adherence to mucosal surfaces. The binding of T. vaginalis to VECs is mediated by surface adhesin proteins (5). In addition, T. vaginalis adherence to host cervical epithelial cells appears to be partially mediated by surface lipoglycan and galectin-1 (57). Cysteine proteinase (CP) activity, which was previously hypothesized to unmask adhesins for function, is also essential for adherence to epithelial cells (4, 26, 34). A recent report showed that glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a receptor for FN (45), revealing yet another glycolytic enzyme as a surface-associated protein with an alternative function. T. vaginalis adherence mediates differential gene expression in human VECs, reflecting the host responses triggered by the parasites (41), and there is also correspondingly upregulated expression of adhesins and other trichomonad proteins upon binding to VECs (46).
While the roles of adhesion-associated proteins in the pathogenesis of and host responses to T. vaginalis have been elucidated, the complex regulatory network of the parasite in response to adherence is as yet unknown. In this study, we used an integrated comparative transcriptomic and proteomic approach in an attempt to decipher for the first time the putative network regulating the transformation of batch-grown trophozoites into amoeboid organisms adherent on immobilized FN (16). We hypothesized that this approach would identify genes involved in the regulation of transformation and/or of functional proteins necessary for ECM associations. In this way, we may learn about the reprogramming of gene transcription using high-throughput technologies, such as expressed sequence tags (ESTs), to provide insights into the expression patterns and the physiological responses of the FN-bound organisms compared to those of batch culture trophozoites.
Proteomic tools now permit us to observe global cellular events by directly visualizing the proteins being expressed. Neither genome nor transcriptome analysis alone permits the analysis of such complex parasite responses. The T. vaginalis proteome reference map has been established (37), and the surface proteome was recently examined (17). Despite the importance of the transformation into amoeboid organisms upon binding to VECs and FN, there are no comparative data identifying and/or characterizing the genes and proteins of amoeboid and FN-bound versus free-swimming parasites. This study provides for the first time insights into molecular components expressed by adherent FN-bound organisms. We hope that the data will help us to elucidate the roles of these proteins and genes in pathogenesis and permit a better understanding of the complex interplay between T. vaginalis and host cells, tissues, and basement membrane components like FN.
MATERIALS AND METHODS
T. vaginalis culture conditions.
T. vaginalis cultures were maintained in YIS medium (pH 5.8) containing 10% heat-inactivated fetal calf serum at 37°C (18). The number of viable cells was determined based on trypan blue exclusion hemocytometer counts. Cells grown to a late logarithmic phase with more than 90% viable cells were harvested for the construction of the unsynchronized trophozoite-EST library. To induce transformation from the trophozoite to the amoeboid form, 1 × 10 8 cells concentrated in 10 ml medium were added to FN-coated T-75 flasks at 37°C for 3 h, a time sufficient for all adherent organisms to become amoeboid, as evidenced by phase microscopy inspection of the flasks. After washing twice with phosphate-buffered saline (PBS) to remove unbound organisms, the FN-adherent amoeboid trichomonads were harvested for the construction of the amoeboid-form FN-EST library for comparison with the trophozoite-EST library of batch-grown organisms.
cDNA construction and EST sequencing.
We constructed two cDNA libraries from mRNA isolated from the trophozoite and amoeboid forms. Total RNA was extracted from harvested cells by using TRIzol reagent (GE Healthcare), and the residual genomic DNA was digested with DNase I. Poly(A)+ RNA was isolated by using the poly(A)+ tract mRNA isolation kit (Promega). cDNA primed with oligo(dT) was synthesized by using a Zap-cDNA synthesis kit and directionally cloned into the EcoRI and XhoI sites of Uni-Zap XR (Stratagene). Single and well-separated plaques were cored out from agar plates and transferred into 96-well microtiter plates containing SM buffer (100 mM NaCl, 8 mM MgSO4 · 7H2O, 50 mM Tris-Cl [pH 7.5], and 0.002% gelatin). The phage stocks were used as templates for cDNA insert amplification with 1 nM (each) the T3 and T7 primers. Amplified products were separated in 1.5% agarose gels, and clones that yielded single PCR-amplified bands were collected for sequencing. Single-pass sequencing from the 5′ end of the cDNA insert was carried out with the T3 primer by using the ABI Prism BigDye Terminator cycle sequencing kit (Applied Biosystems). The sequencing products were resolved and analyzed by using either an ABI Prism 377 (Applied Biosystems) or a Megabace (GE Healthcare) DNA sequencer. The nucleotide sequences obtained were processed with the Phred/Phrap/Consed package (23, 24, 31). Assembled sequence contigs were compared against known Trichomonas mRNAs, putative open reading frames (ORFs) of the T. vaginalis G3 genome (11), as well as NCBI's nonredundant (nr) nucleotide (E value of ≦10−15) and Swiss-Prot (E value of ≦10−10) databases using BLAST tools. Interpro and Gene Ontology analyses were used to classify genes into functional categories.
Protein extraction for isoelectric focusing and two-dimensional gel electrophoresis (2-DE).
Mid-logarithmic-phase trophozoites and FN-induced amoeboid cells were harvested by centrifugation at 3,000 rpm for 15 min and washed with saline three times. One milliliter of lysis buffer {8 M urea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS)} containing protease inhibitors (Roche Diagnostics) was added to the pellet containing 1 × 108 trichomonads. Cells were disrupted by sonication in an ice bath for eight cycles. After centrifugation at 13,000 rpm for 15 min at 4°C, the supernatant was kept at −70°C until use. The 2D-Clean Up kit (GE Healthcare) was used to remove impurities from the protein samples. The protein concentration was determined by using the Bio-Rad protein assay kit.
Isoelectric focusing (IEF) of protein samples in the first dimension as well as separation by SDS-PAGE in the second dimension were performed as previously described (37). Briefly, 250 μg of protein was diluted to a final volume of 250 μl in rehydration buffer (8 M urea, 2% CHAPS) containing a trace amount of bromophenol blue. Samples were applied onto 13-cm immobilized pH gradient (IPG) gel strips (GE Healthcare) with a linear separation range of pH 4 to 7. The focused IPG strips were equilibrated for 15 min in buffer (50 mM Tris-HCl [pH 8.8], 6 M urea, 30% glycerol, 2% SDS, and a trace amount of bromophenol blue) containing 1% (wt/vol) dithiothreitol (DTT), followed by 15 min of equilibration in buffer containing 2.5% (wt/vol) iodoacetamide. Equilibrated IPG strips were separated across 15% SDS-PAGE gels. Gels were run at 35 mA/gel and at 4°C until the tracking dye migrated to the bottom of the gel.
Image analysis.
The silver-stained 2-DE gels were scanned and analyzed by using Phoretix 2D analysis software. The automatic analysis protocol for the images of the trophozoite-stage and FN-induced amoeboid-stage 2-DE gels included spot detection, background subtraction, matching, and reference gel creation. Spot volumes were normalized against the total volumes of all the spots in the gel. Differential expression analysis was performed by comparing the normalized volumes of matched spots in each pair of samples.
Protein identification by mass spectrometry (MS).
Differentially expressed protein spots were selected by Phoretix 2D analysis software and excised from the trophozoite- and amoeboid-stage 2-DE gels. The gel pieces were first destained in a solution containing 30 mM potassium ferricyanide and 100 mM sodium thiosulfate, followed by washing and shrinking steps using 50 mM ammonium bicarbonate and acetonitrile (ACN). The gel pieces were dehydrated completely by a vacuum centrifuge and rehydrated in 3 μl of trypsin (Promega) solution (20 μg/ml in 25 mM ammonium bicarbonate). In-gel trypsin digestion was performed at 37°C for 16 h. Peptide extraction was performed twice for 15 min by sonication with 2 μl of extraction buffer (100% ACN with 1% trifluoroacetic acid [TFA]). Peptides were eluted and then cocrystallized with a saturated solution on a matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) sample plate using 0.5 μl of sample and 0.5 μl of matrix. The peptides were analyzed by using an Ultraflex MALDI-TOF mass spectrometer (Bruker Daltonic). An automated database search was performed as previously described (37). A global MASCOT score greater than 62 was considered significant (P < 0.05). The theoretical molecular weight (Mr) and pI of each spot in the 2-DE gels were also obtained by using the MASCOT search engine.
Quantitative PCR.
Total RNA was isolated from batch culture trophozoites and FN-bound amoeboid organisms. Reverse transcription (RT) was carried out with a reaction mixture containing 5 μg total RNA, 50 nM RT primer, 0.25 mM deoxynucleoside triphosphates (dNTPs), 0.75 U/μl ThermoScript III reverse transcriptase, 0.2 U/μl RNase Out, and 0.05 M DTT (ThermoScript III RT-PCR system; Invitrogen). The RT reaction mixture was incubated at 50°C for 30 min, and the reaction was then stopped at 70°C for 15 min. Quantitative PCR (qPCR) was performed by using the Ampliqon III RealQ-PCR master mix kit on an MX3000 system (Stratagene). The 20-μl PCR mixtures contained 1 μg reverse transcription product, master mix, and 0.5 μM real-time forward and reverse primers. Primer pairs used in this study are listed in Table S2 in the supplemental material. The reactions were performed as previously described (48). 60S rRNA was used as an internal control for normalization in all experimental groups.
Nucleotide sequence accession numbers.
The trophozoite-EST data were deposited in the NCBI dBEST database under accession numbers BQ621379 to BQ621732, BQ640771 to BQ640943, CO576867 to CO578496, and JK965400 to JK972143, and the FN-EST data were deposited under accession numbers JG708501 to JG718466, which can be retrieved from TrichDB (http://trichdb.org/trichdb/).
RESULTS AND DISCUSSION
Transcriptional profiling of FN-bound amoeboid versus batch-grown trophozoite forms.
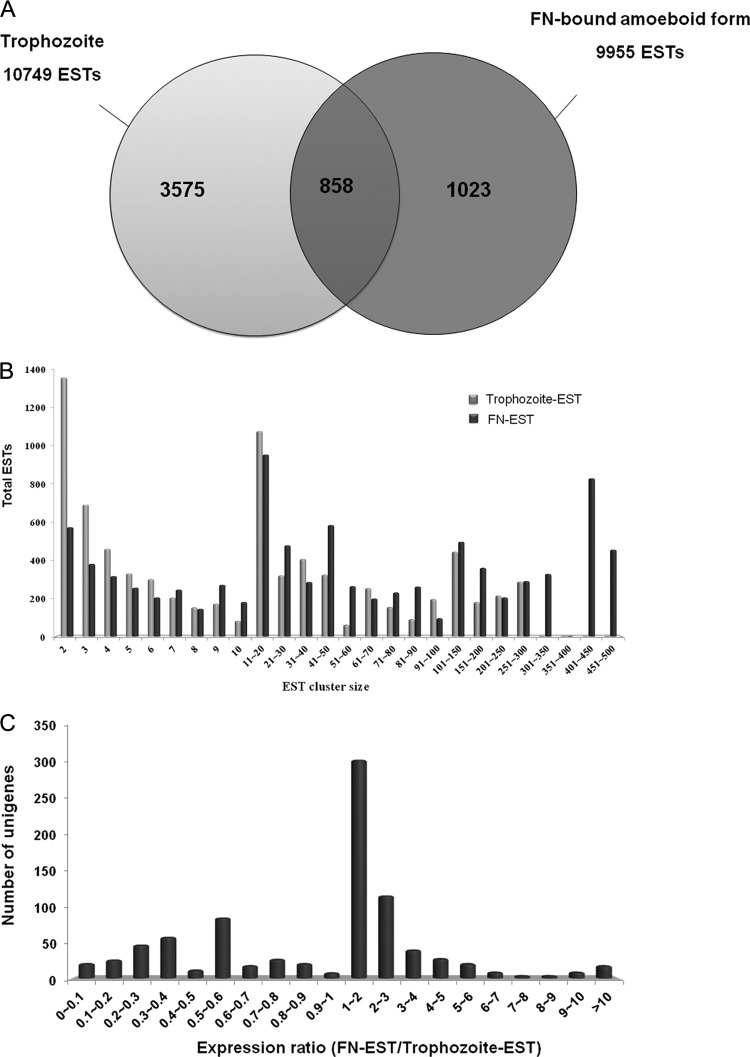
We initiated an EST project to gain insight into the gene expression profiles following parasite binding to FN. cDNA libraries of unsynchronized, batch-grown T. vaginalis trichomonads (trophozoite-EST library) and FN-adherent amoeboid cells (FN-EST library) were constructed, generating a total of 20,704 ESTs (Table 1 and Fig. 1A). The ESTs were clustered and assembled into 4,433 and 1,881 unique gene sequences (unigenes) from the trophozoite-EST and FN-EST cDNA libraries, respectively. Unigenes containing only 1 EST are defined as singletons, whereas unigenes containing at least 2 ESTs are defined as tentative clusters (TCs). Trophozoite-ESTs expressed 3,575 unigenes, whereas FN-ESTs expressed 1,023 unigenes. The expression of unigenes was represented as a number of ESTs per 10,000 clones. Among the transcripts specifically expressed in the FN-EST transcriptome, the most abundant transcripts were heat shock protein 20 (TVAG_381290), tryparedoxin peroxidase (TXNPx) (TVAG_350540), and iron superoxide dismutase (SOD) (TVAG_161300). The expression ratio of trophozoite cells/FN-adherent cells of 858 unigenes expressed at both stages was analyzed (Fig. 1C). Among them, TXNPx (TVAG_038090) was the most highly expressed transcript in the FN-EST transcriptome compared to the trophozoite-EST transcriptome. Global analysis of the distribution of ESTs showed that 7,657 trophozoite-ESTs (71.23%) and 8,884 FN-ESTs (89.24%) were in TCs, leaving 3,092 trophozoite-EST (28.77%) and 1,071 FN-EST (10.76%) singletons. This suggests that a large number of low-abundance genes of trophozoites were globally shut down in the FN-amoeboid organisms. The cluster size represents the number of ESTs assembled in the cluster. In a nonnormalized cDNA library, the cluster size is equal to the abundance of the transcript in the transcriptome. An analysis of EST cluster sizes will shed light on the gene expression patterns among different cDNA libraries and provide information on the kinetics of transcription that are globally activated or shut down under different conditions. The distribution of the EST cluster sizes from both EST libraries is shown in Fig. 1B. There were 4 TCs containing more than 300 ESTs, which constituted 16.2% of the total ESTs in the FN-EST cDNA library (Fig. 1B; see also Table S1 in the supplemental material). In contrast, the largest TC in the trophozoite-EST cDNA library contains only 287 ESTs. Table S1 in the supplemental material shows the 100 most sequenced ESTs from the FN-EST library. The predominant abundant transcripts in the FN-bound, amoeboid stage were GAPDH (TVAG_146910), TXNPx (TVAG_114310), a hypothetical protein, and actin (TVAG_200190). The top 20 highly expressed transcripts in the FN-EST library contain 3,635 ESTs, almost 36.4% of all identified ESTs. In contrast, the top 20 highly expressed transcripts in the trophozoite-EST library contain 1,832 ESTs (18.3%), almost half the number obtained for FN-ESTs. Combined with the analysis described above, the results suggest strongly that the FN-adherent, amoeboid trichomonads had decreased transcript levels of trophozoite-ESTs already present in small amounts concomitant with a shift to the expression of certain genes that may have essential functions in response to the specific association with FN. The fact that GAPDH, a surface-associated, FN-binding protein of T. vaginalis (48), was readily detected in the FN-EST library reinforces the idea that the expressions of genes/proteins essential for FN anchoring are upregulated.
Table 1.
Summary of EST sequencing data from the T. vaginalis trophozoite and FN-bound amoeboid cDNA libraries
| Parameter | Value for cDNA library |
|
|---|---|---|
| TvEb | TvFc | |
| No. of ESTs | 10,749 | 9,955 |
| No. of TCsa | 1,341 | 810 |
| No. of ESTs in TCs | 7,657 | 8,884 |
| % ESTs in TCs | 71.23 | 89.24 |
| No. of singletons | 3,092 | 1,071 |
| % ESTs in singletons | 28.77 | 10.76 |
| No. of unigenes | 4,433 | 1,881 |
| % TCs | 30.25 | 43.06 |
| % singletons | 69.75 | 56.94 |
Tentative consensus sequences (TCs) refer to collapsed ESTs in virtual transcripts, which may contain full or partial cDNA sequences; unigenes are unique genes.
TvE, cDNA library constructed from trophozoites.
TvF, cDNA library constructed from FN-induced amoeboid cells.
Fig 1.
Transcriptional profiling of the FN-binding cDNA library compared with the trophozoite cDNA library. (A) Number of total ESTs and unigenes clustered in the FN-EST and trophozoite-EST transcriptomes of T. vaginalis. (B) Cluster size (the number of ESTs present in this cluster) distribution of the annotated ESTs in the FN-EST and trophozoite-EST data sets of T. vaginalis. (C) Unigenes expressed in both trophozoite-EST and FN-EST transcriptomes were compared (FN-EST/trophozoite-EST ratio).
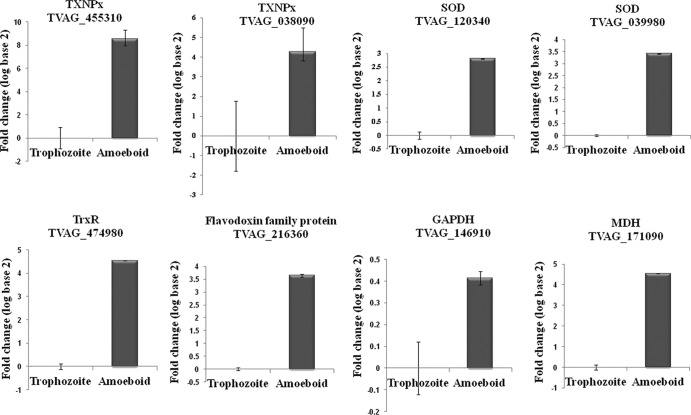
To validate the transcriptomic data, we utilized qPCR analysis to estimate the gene expression levels of the TXNPx (TVAG_038090), SOD (TVAG_039980, TVAG_120340), thioredoxin reductase (TrxR) (TVAG_474980), and malate dehydrogenase (MDH) (TVAG_171090) transcripts (see Fig. 6). Consistent with data from our EST transcriptomic analysis, these transcripts were highly expressed in the FN-bound, amoeboid parasites. Based on the trophozoite-EST and FN-EST data sets, we clustered the genes that had expression levels 10-fold (32 genes) higher in the FN-EST transcriptome than in the trophozoite-EST transcriptome according to their biological function (Fig. 2). The results showed that the largest category of the differentially expressed genes in the FN-adherent, amoeboid organisms encodes stress-related proteins (39%), suggesting that the stress response is one of the most important physiological adaptations of T. vaginalis during the morphological transformation and host colonization processes.
Fig 6.
qPCR analysis validating the comparative transcriptome and proteome data. mRNA expression levels of eight genes with differential expression in our FN-binding transcriptome and proteome results were confirmed by qPCR analysis compared with the trophozoite stage. Data are expressed as log fold changes (log base 2) ± standard deviations of the means from three independent experiments. Transcripts selected for qPCR validation were tryparedoxin peroxidase (TXNPx) (TVAG_455310 and TVAG_038090), iron superoxide dismutase (SOD) (TVAG_039980 and TVAG_120340), thioredoxin reductase (TrxR) (TVAG_474980), a flavodoxin family protein (TVAG_216360), GAPDH (TVAG_146910), and malate dehydrogenase (MDH) (TVAG_171090).
Fig 2.
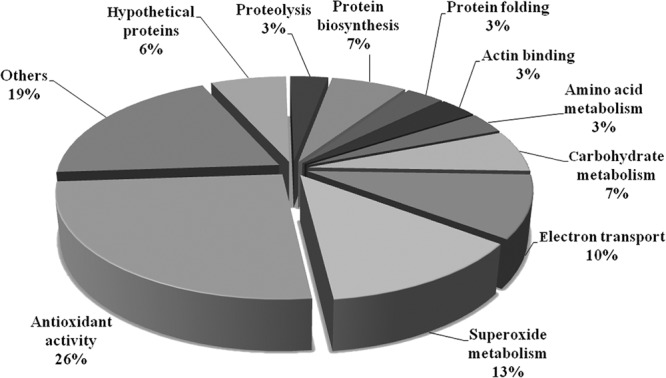
Functional categorization of upregulated genes in FN-bound amoeboid cells compared with trophozoites of T. vaginalis. Differentially expressed genes which showed 10-fold-higher expression levels in the FN-EST transcriptome than in the trophozoite-EST transcriptome were clustered and classified according to the Gene Ontology index (http://www.geneontology.org/).
Functional classification of transcripts in T. vaginalis after FN contact.
Upon an examination of the FN-EST data generated from amoeboid T. vaginalis cells bound to FN, we assigned FN-EST classifications to 10 categories. The categories with a listing of representative EST genes include the categories of adherence, cytoskeleton, membrane-associated proteins, metabolism, proteinases/protease inhibitors, oxygen radicals, protein trafficking/signaling, growth factors, transcription/translation, and unknown. The fact that the trichomonad adhesins and the receptors for host basement membrane glycoproteins are surface metabolic enzymes (5, 45, 52) illustrates the overlap of categories for EST genes.
FN-binding-related ESTs.
We previously reported that the cytoadherence of T. vaginalis to immortalized MS-74 VECs induces the expression of trichomonad genes that encode proteins with specific functions (42). As expected, ESTs for the established metabolic enzymes that function as adhesins or receptors for FN were readily detected in the FN-EST transcriptome of FN-bound organisms (20, 27, 42). Of interest is that particular members of these multigene families were expressed under different environmental conditions, and this may indicate that subsets of these family members are regulated based upon different environmental signals. This finding suggests that these proteins are unique from typical housekeeping proteins. Not unexpectedly, other genes that were upregulated were those for the transcription and translation of the genes encoding the adhesins and other essential proteins. Predictably evident was the increased expression level of the α-actinin protein, which is required for cytoskeletal rearrangements for morphological changes following adherence to VECs (6) and binding to FN (12, 13, 45). That the expression level of an α-tubulin-1 gene is increased after FN binding is another example of expression among subsets of cytoskeletal genes based on host tissue interactions. These FN-ESTs are not surprising given that the α-β-tubulin heterodimer is the structural subunit of microtubules, which are cytoskeletal elements essential for intracellular transport, the maintenance of cell shape, and cell division (50, 55). Finally, the calponin protein is an extensively studied actin-binding protein that has been isolated from Caenorhabditis elegans (30) and the animal parasite Onchocerca volvulus (39). The calponin protein has been suggested to function as a scaffolding protein for cytoskeletal structures, and an alternative function is an involvement in signal transduction, properties which are both important for associations of parasites with cells and basement membrane proteins.
Interestingly, there is a multidrug resistance protein induced in the FN-EST transcriptome (40). This may be important for any future development of therapeutics. This same FN-EST identified the phenotypically variable P270 protein immunogen of T. vaginalis that alternates between surface and cytoplasmic expression in the type II, double-stranded RNA (dsRNA) virus-infected trichomonads (1). The alternating expressions of immunogens and adhesins for type II parasites are regulated by iron, which upregulates the expression and surface placement of adhesins in a coordinated fashion (1). In this case, however, FN binding is mediated by surface GAPDH, the level of which, as mentioned above, is increased in this FN-EST transcriptome. It is not inconceivable that the C-type transmembrane lectin may be analogous to similar molecules in mammalian cells that facilitate the efficient export of secretory glycoproteins from the endoplasmic reticulum (54). Finally, the large family of BspA-like proteins with an as-yet-undefined function is found in the transcriptome of FN-bound organisms (35). There have been 656 BspA-like hypothetical proteins identified in the T. vaginalis genome, sharing the Treponema pallidum leucine-rich repeat (TpLRR) domain of the BspA surface proteins (11). TpLRR-containing proteins in several mucosal bacteria have been demonstrated to modulate binding to host cells and ECM proteins (38). Recently, BspA-like proteins were identified in the surface proteome of T. vaginalis (17), suggesting the potential role of these proteins in mediating adherence and, thus, pathogenicity.
Growth factors.
The FN-EST transcriptome indicates that the parasite genes share identities with mammalian homologues, including insulin-like growth factor (GF). Epidermal growth factor (EGF)-like motifs in Plasmodium vivax merozoite surface protein 1 play a role in erythrocyte invasion (33), and the domains are critical for the binding between the parasite ligand and erythrocyte receptor. Insulin-like GF was suggested previously to regulate cell proliferation, differentiation, and metabolism in mammalian cells (8), but whether such a factor exerts similar properties in trichomonads remains to be experimentally determined. Alternatively, it is conceivable that, if secreted by the parasite, as has been found for numerous other proteins (43), this insulin-like GF may influence the host cell. Of interest in the future will be whether host hormones signal for trichomonal growth and multiplication.
Oxygen radicals and stress.
The presence of T. vaginalis thioredoxin peroxidase to detoxify potentially damaging oxidants and iron-containing superoxide dismutase (SOD), which allow the parasites to survive in the host through superoxide detoxification, has been established (68). The FN-EST transcriptome makes evident the many enzymes that are involved in countering the oxidative stress possibly encountered by trichomonads during infection. The heretofore unreported glutathione peroxidase and alkyl hydroperoxide reductase were also found in the FN-EST library. Importantly, alkyl hydroperoxide reductase was shown previously to protect Helicobacter pylori from iron-promoted DNA damage (70), and this enzyme may play a similar role in trichomonads, given the importance of iron for optimal growth and multiplication through the regulation of gene expression. These findings illustrate the extensive antioxidant defenses that permit parasite survival, and these data may indicate that the parasite experiences a more aggressive host immune response and a more severe hostile environment than previously appreciated.
Proteinases and degrading enzymes.
T. vaginalis organisms possess many immunogenic cysteine proteinases (CPs) expressed under various environmental conditions (9, 53). These CPs have been found to be secreted during infection and have promiscuous substrate specificities that altogether confer immune evasion capabilities upon trichomonads (15, 62). Equally noteworthy is the role that these proteinases play in unmasking adhesins to promote efficient VEC binding (3, 26). Not surprisingly, this large family of CPs was found to be synthesized under various environmental conditions. This notwithstanding, there were numerous CPs that were uniquely expressed in the FN-EST transcriptome. Metalloproteinases that are known to play important roles in the virulence and pathogenesis of other parasites (10, 25, 49) were equally evident in this FN-EST library, although most were not ordinarily expressed. This fact illustrates the need for a better understanding of the unique environments that the parasite experiences during infection. Indeed, there are so many peptidases of different substrate specificities and that are expressed at very different times under various environmental conditions that it is difficult to establish patterns. In Leishmania major, leishmanolysin, a member of the M8 surface metalloproteinases, is highly expressed on Leishmania promastigotes. During tissue invasion, leishmanolysin degrades extracellular matrix proteins and prevents the lysis of promastigotes by complement (72). Trypanosoma brucei leishmanolysin was reported previously to be involved in virulence by removing surface glycoprotein, resulting in antigenic variation (44). Recently, it was shown that Entamoeba histolytica metallosurface protease 1 regulates amoebic adherence, motility, and phagocytosis (66), reaffirming the role of metalloproteinases in the pathogenesis of the protozoan parasites.
Protein trafficking/signaling.
It is now well established that iron and contact with VECs regulate the expressions of numerous T. vaginalis genes (14, 41, 42). Calcium is required for the regulation of binding to FN (13). Signaling following adherence to VECs results in a dramatic transformation in morphology (6) as well as the synthesis and surface placement of adhesins. Iron modulates the compartmentalization of adhesins outside the hydrogenosome organelle, where the proteins function as enzymes for the oxidative decarboxylation of pyruvate (27). Not surprisingly, therefore, numerous FN-ESTs encoding chaperones, kinases, and phosphatases were detected in the transcriptome of FN-amoeboid parasites. Likewise, the FN-EST transcriptomes had a large number of transcripts for the GTPase superfamily. ADP-ribosylation factor (ARF)-like proteins are members of the subfamily of small GTPases that play various roles within cells through protein modifications. One such function is to regulate membrane traffic and structure (19), which is accomplished through the modification of membrane lipids and the recruitment of proteins, including coat proteins and actin, to the surface of membranes. Interestingly, soluble inorganic phosphatase was uniquely expressed by parasites bound to FN. The large number of trafficking and signaling pathway enzymes clearly shows that this organism is capable of responding to the highly complex, constantly changing environment of the vagina, as was hypothesized previously (47).
Metabolism.
Metabolic enzymes were the most frequently identified ESTs in both the trophozoite-EST and FN-EST transcriptomes, and cytosolic as well as hydrogenosomal metabolic enzyme ESTs were well represented. Beyond the obvious importance of energy-generating metabolic pathways regardless of their cellular location, these enzymes have alternative functions related to infection, such as GAPDH receptor binding of FN (45) and α-enolase receptor binding of plasminogen (52), which are important properties during infection. It may not be surprising that the α-enolase expression level is increased upon FN binding by trichomonads, as this enzyme also binds other basement membrane glycoproteins in a specific receptor-ligand fashion (63).
Another enzyme of interest is carboxymuconolactone decarboxylase, which is known to be important for the degradation of aromatic compounds (22). Important for glycolytic flux in cells is phosphoglycerate mutase (21), which may play a similar role within trichomonads. Finally, there are numerous carbohydrate-degrading and/or -modifying enzymes, some of which are highly expressed in the FN-EST transcriptome, and these enzymes may play an important role in aborting any lectin-type interactions between the parasite and the host cell.
2-DE profiling of the FN-binding amoeboid stage.
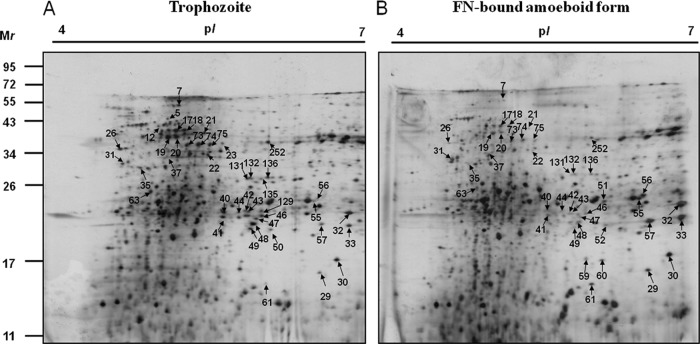
We previously established proteome reference maps of T. vaginalis and identified a total of 247 protein spots representing 164 nonredundant proteins (37). To further assess the differentially expressed proteins between the adherent amoeboid forms and free-swimming trophozoites, a comparative proteomic analysis of the FN-bound parasites and the free-swimming trophozoites was performed by using 2-DE combined with MALDI-TOF MS. The majority of the protein spots are distributed in the pH range of 4 to 7 on the T. vaginalis proteome reference map (37). Hence, proteins extracted from the FN-associated amoeboid forms versus batch culture trophozoites were separated at pH 4 to pH 7. Representative 2-DE gels are shown in Fig. 3. Approximately 500 protein spots were detected on these 2-DE gels between these two types of trichomonads. A total of 63 differentially expressed protein spots for the FN-amoeboid organisms were excised and analyzed by MALDI-TOF peptide mass fingerprinting (PMF) for protein identification. We successfully identified 43 differentially expressed proteins in the proteome of FN-amoeboid organisms (Fig. 3). Among them, 31 proteins were downregulated, whereas 12 proteins were upregulated. The locus, annotation, protein scores, theoretical pI values and molecular masses, and sequence coverage of all identified proteins are provided in Tables 2 and 3.
Fig 3.
2-DE maps of trophozoites and FN-bound amoeboid cells of T. vaginalis. Proteins extracted from trophozoites (A) and FN-induced amoeboid cells (B) were separated. IEF in the first dimension was performed with 250 μg of proteins using 13-cm pH 4 to 7 strips, which was followed by separation in 15% polyacrylamide gel for the second dimension. Proteins were visualized by silver staining. Differentially expressed protein spots in the amoeboid stage compared with the trophozoite stage are marked by arrows and protein spot numbers.
Table 2.
List of downregulated proteins in FN-bound amoeboid cells identified by MALDI-TOF MS
| Spot | Protein | Locus tag | MASCOT score | Coverage (%) | Theoretical molecular mass (kDa)/pI |
|---|---|---|---|---|---|
| 17 | Actin | TVAG_160060 | 108 | 38 | 42.2/5.05 |
| 18 | Actin | TVAG_172680 | 86 | 34 | 42.1/5.05 |
| 19 | Actin | TVAG_160060 | 155 | 56 | 42.2/5.05 |
| 20 | Actin | TVAG_054030 | 90 | 17 | 42.1/5.12 |
| 73 | Actin | TVAG_160060 | 189 | 53 | 42.2/5.05 |
| 74 | Actin | TVAG_160060 | 274 | 67 | 42.2/5.05 |
| 75 | Actin | TVAG_054030 | 149 | 53 | 42.1/5.12 |
| 26 | Actinin | TVAG_156680 | 87 | 25 | 69.6/4.89 |
| 22 | Alpha-tubulin 1 | TVAG_196270 | 120 | 32 | 50.7/4.9 |
| 23 | Alpha-tubulin 1 | TVAG_196270 | 92 | 33 | 50.7/4.9 |
| 5 | ATP synthase beta subunit, putative | TVAG_420260 | 78 | 17 | 68.3/5.14 |
| 12 | ATP synthase beta subunit, putative | TVAG_420260 | 109 | 29 | 68.3/5.14 |
| 63 | Beta-tubulin | TVAG_148390 | 85 | 41 | 28.5/5.21 |
| 135 | Carbamate kinase | TVAG_261970 | 64 | 38 | 40.5/5.48 |
| 35 | Chaperone, Hsp70, endoplasmic reticulum | TVAG_092490 | 76 | 20 | 71.6/5.06 |
| 40 | Fructose-1,6-bisphosphate aldolase | TVAG_360700 | 66 | 34 | 36.4/5.79 |
| 41 | Fructose-1,6-bisphosphate aldolase | TVAG_043070 | 87 | 41 | 36.4/5.79 |
| 42 | Fructose-1,6-bisphosphate aldolase | TVAG_043070 | 50 | 29 | 36.4/5.79 |
| 43 | Fructose-1,6-bisphosphate aldolase | TVAG_300000 | 88 | 48 | 36.5/6.02 |
| 44 | Fructose-1,6-bisphosphate aldolase | TVAG_043170 | 54 | 38 | 36.5/5.84 |
| 46 | Fructose-1,6-bisphosphate aldolase | TVAG_043170 | 124 | 49 | 36.5/5.84 |
| 49 | Fructose-1,6-bisphosphate aldolase | TVAG_300000 | 110 | 48 | 36.5/6.02 |
| 50 | Fructose-1,6-bisphosphate aldolase | TVAG_043170 | 78 | 47 | 36.5/5.84 |
| 131 | Fructose-1,6-bisphosphate aldolase | TVAG_043070 | 64 | 23 | 36.4/5.79 |
| 132 | Fructose-1,6-bisphosphate aldolase | TVAG_360700 | 67 | 28 | 36.4/5.88 |
| 136 | Fructose-1,6-bisphosphate aldolase | TVAG_043070 | 189 | 50 | 36.4/5.79 |
| 252 | Ketopantoate reductase | TVAG_538040 | 81 | 38 | 37.8/5.69 |
| 47 | Malate dehydrogenase | TVAG_171090 | 111 | 45 | 37.3/5.77 |
| Malate dehydrogenase | TVAG_171100 | 111 | 45 | 37.3/5.96 | |
| 7 | Peptidase M20/M25/M40-containing protein | TVAG_224980 | 87 | 29 | 51.9/5.03 |
| 21 | Peptidase M20/M25/M40-containing protein | TVAG_224980 | 96 | 37 | 51.9/5.03 |
| 48 | Phosphoglycerate kinase | TVAG_268050 | 77 | 25 | 45.5/6.58 |
| Phosphoglycerate kinase | TVAG_383940 | 77 | 25 | 45.5/6.82 |
Table 3.
List of upregulated proteins in FN-bound amoeboid cells identified by MALDI-TOF MS
| Spot | Protein | Locus tag | MASCOT score | Coverage (%) | Theoretical molecular mass (kDa)/pI |
|---|---|---|---|---|---|
| 61 | Flavodoxin family protein | TVAG_216360 | 103 | 69 | 14.9/6.32 |
| 51 | Glyceraldehyde-3-phosphate dehydrogenase | TVAG_347410 | 88 | 39 | 39.7/7.03 |
| 55 | Glyceraldehyde-3-phosphate dehydrogenase | TVAG_146910 | 137 | 42 | 39.7/7.98 |
| Glyceraldehyde-3-phosphate dehydrogenase | TVAG_366380 | 137 | 42 | 39.7/7.03 | |
| 56 | Glyceraldehyde-3-phosphate dehydrogenase | TVAG_347410 | 78 | 36 | 39.7/7.03 |
| Glyceraldehyde-3-phosphate dehydrogenase | TVAG_146910 | 78 | 36 | 39.7/7.98 | |
| Glyceraldehyde-3-phosphate dehydrogenase | TVAG_366380 | 78 | 36 | 39.7/7.98 | |
| 31 | Heat shock protein 70 | TVAG_092490 | 100 | 23 | 71.6/5.06 |
| 32 | PDase, cathepsin L-like cysteine peptidase | TVAG_057000 | 70 | 28 | 35.2/8.4 |
| 29 | Thiol peroxidase | TVAG_165690 | 74 | 46 | 18.1/6.05 |
| 30 | Thiol peroxidase | TVAG_165690 | 195 | 83 | 18.1/6.05 |
| 60 | Thiol peroxidase | TVAG_165690 | 124 | 58 | 18.1/6.05 |
| Thiol peroxidase | TVAG_165610 | 124 | 58 | 18.1/6.05 | |
| 33 | Thioredoxin peroxidase | TVAG_484570 | 104 | 54 | 22.1/6.31 |
| 59 | Thioredoxin reductase | TVAG_474980 | 135 | 55 | 32.8/6.02 |
| 57 | Tryparedoxin peroxidase | TVAG_455310 | 132 | 69 | 22.1/6.05 |
Identification of differentially expressed proteins after parasite binding to FN.
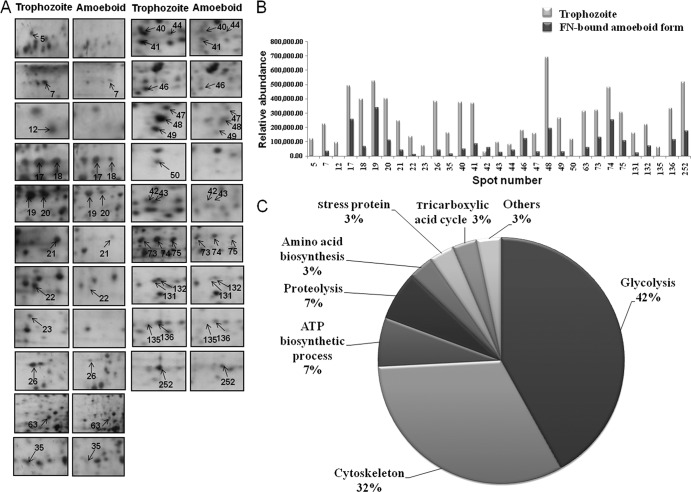
We previously demonstrated that carbohydrate metabolism represented the most abundant category in the T. vaginalis reference proteome (37). In addition, ∼10% of the parasite reference proteome was composed of cytoskeletal proteins. These proteins, involved in energy production and cell motility, are continuously expressed in batch culture trichomonads. However, among the 33 downregulated protein spots following trichomonad binding to FN, 13 and 10 spots were classified as glycolysis and cytoskeletal proteins, respectively (Fig. 4). These data suggest that T. vaginalis may shut down some physiological functions performed in free-living organisms to adapt to the morphological changes and colonization events evident in VEC adherence and binding to basement membrane components.
Fig 4.
Downregulated proteins in FN-bound amoeboid cells compared with trophozoites. (A) Magnified regions of the 2-DE maps showing the 31 downregulated proteins in the FN-binding proteome. Protein spots with a lower abundance in the amoeboid stage are numbered and indicated by arrows. (B) Each bar represents the relative abundance expressed as the normalized spot volume calculated by using Phoretix 2D analysis software. Details of their information are shown in Table 2. (C) Functional categories of the downregulated proteins identified by MALDI-TOF PMF classified according to the Gene Ontology index (http://www.geneontology.org/).
CPs are the major proteolytic enzymes expressed by T. vaginalis, and some of them are involved in cytotoxicity, hemolysis (15), immune response evasion (62), and cytoadherence (4). CPs are considered to be essential for the efficient adhesion of T. vaginalis to target cells and are thought to play significant roles in the pathogenesis of trichomonosis. In our proteomic results, we observed that CP (spot 32) was of greater abundance after adherence to FN than in free-swimming trophozoites (Fig. 5).
Fig 5.
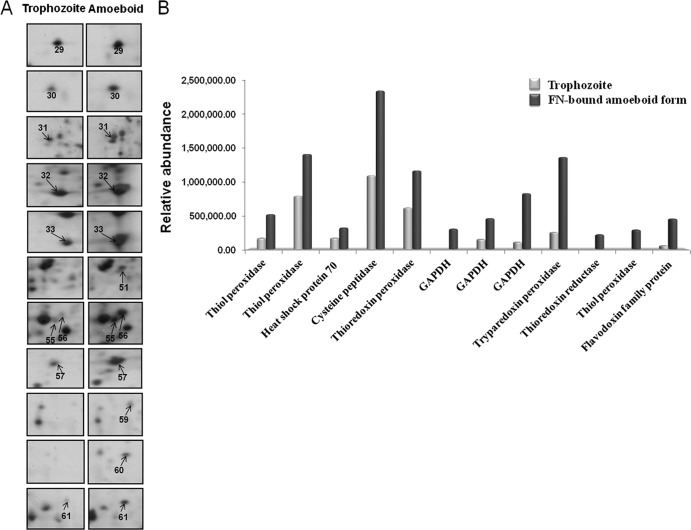
Upregulated proteins in FN-bound amoeboid cells compared with trophozoites. (A) Magnified regions of the 2-DE maps showing the 12 upregulated proteins in the FN-binding amoeboid stage of T. vaginalis. Protein spots with a higher abundance in the amoeboid stage are numbered and indicated by arrows. (B) Upregulated proteins identified by MALDI-TOF PMF are shown, and each bar represents the relative abundance expressed as the normalized spot volume calculated by using Phoretix 2D analysis software. Details of their information are listed in Table 3.
It was demonstrated previously that metabolic enzymes in some cases were expressed on the surfaces of bacteria, yeast, and parasites (32, 45, 71), and these enzymes have alternative functions as novel virulence factors. Previous studies showed that some pathogens possess GAPDH on their cell surfaces, and they have been identified as adhesion proteins. For instance, group A streptococcus surface GAPDH shows multiple binding activities, including binding with plasminogen, lysozyme, myosin, actin, and FN (16, 58, 71). In addition, the surface-associated GAPDH of Candida albicans was found previously to bind to laminin and FN (32). We recently showed that GAPDH was a surface-associated, FN-binding protein of T. vaginalis (45). In this study, and consistent with the FN-EST transcriptome data, we observed that GAPDH is upregulated in the FN-binding proteome (Fig. 5), a finding consistent with a role of this glycolytic enzyme in basement membrane-FN associations.
Moreover, we observed that 8 stress-related proteins were upregulated after parasite binding to FN (Fig. 5 and Table 3). For instance, the level of a protein identified as TXNPx (spot 57) (TVAG_455310) increased ∼5.5-fold after trichomonads bound FN compared with levels in free-swimming trophozoites. In Leishmania donovani, not only is TXNPx crucial for survival during oxidative stress, it also enhances infectivity (69). For Trypanosoma cruzi, there is increasing evidence showing that antioxidant enzymes are critical for immune evasion by protecting the parasite from macrophage-derived reactive oxygen species (ROS) (56). In addition, the expression level of thioredoxin peroxidase (spot 33) (TVAG_484570) increased ∼1.9-fold. TrxR (spot 59) (TVAG_474980), which functions together with thioredoxin and thioredoxin peroxidase to detoxify potentially damaging oxidants, was expressed only in the proteome of FN-bound parasites. TrxR is a strong antioxidant that protects T. vaginalis from oxidative attack by the activated host phagocyte and monocyte during tissue adherence and invasion (8, 47). Other proteins with antioxidant functions showing increased expression levels included thiol peroxidase (spots 29, 30, and 60) (TVAG_165610 and TVAG_165690) and a flavodoxin family protein (spot 61) (TVAG_216360). These data are consistent with our transcriptome results in that the proteome of FN-associated organisms shows increases in the levels of antioxidant proteins.
Functional classification of differentially expressed proteins.
Putative functional annotations for the 43 identified proteins were classified into the following categories: glycolysis, protein binding, proteolysis, stress proteins, amino acid biosynthesis, tricarboxylic acid cycle, and hypothetical proteins. Of the 31 proteins downregulated in the proteome of FN-bound parasites, glycolysis (42%) represented the most abundant category, followed by cytoskeletal proteins (32%), ATP biosynthetic process (7%), proteolysis (7%), amino acid biosynthesis (3%), stress proteins (3%), tricarboxylic acid cycle (3%), and others (3%) (Fig. 4 and Table 2). On the contrary, of the 12 proteins upregulated after FN binding, the largest category (67%) comprised proteins involved in stress responses (Fig. 5 and Table 3).
Analysis of mRNA expression of differentially expressed proteins.
Finally, we felt that it was important to test whether the differentially expressed proteins in the proteome of FN-amoeboid trichomonads are regulated at the transcription level. The amounts of mRNA of GAPDH (TVAG_146910), TXNPx (TVAG_455310), TrxR (TVAG_474980), and a flavodoxin family protein (TVAG_216360) were determined by qPCR analysis (Fig. 6). Our qPCR data show that results for enzymes with antioxidant activities correlate well with our proteome results. It seems clear that these antioxidant enzymes were upregulated at both the gene and protein expression levels after parasite binding to FN, as evidenced collectively by our 2-DE, EST, and qPCR results. Stress responses following FN associations may suggest that such antioxidant proteins have critical roles in host-pathogen interactions.
In summary, our work provided insights into the genes and proteins that are upregulated upon binding to FN by T. vaginalis. We identified novel genes that adapt or respond to the dramatic transformation in morphology that takes place upon the adherence of these parasites to immobilized FN, suggesting a parasite response preparatory to host colonization. The continued characterization of these proteins will elucidate the roles of these proteins as virulence factors involved in mechanisms of pathogenesis of trichomonosis.
Supplementary Material
ACKNOWLEDGMENTS
Funding for this project was provided by the Chang Gung Memorial Hospital (CMRPD170481-3) to P.T.
We thank Yi-Ywan Margaret Chen for her critical review of and comments on the manuscript.
Footnotes
Published ahead of print 27 August 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Alderete JF. 1999. Iron modulates phenotypic variation and phosphorylation of P270 in double-stranded RNA virus-infected Trichomonas vaginalis. Infect. Immun. 67:4298–4302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alderete JF, Benchimol M, Lehker MW, Crouch ML. 2002. The complex fibronectin-Trichomonas vaginalis interactions and trichomonosis. Parasitol. Int. 51:285–292 [DOI] [PubMed] [Google Scholar]
- 3. Arroyo R, Alderete JF. 1989. Trichomonas vaginalis surface proteinase activity is necessary for parasite adherence to epithelial cells. Infect. Immun. 57:2991–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arroyo R, Alderete JF. 1995. Two Trichomonas vaginalis surface proteinases bind to host epithelial cells and are related to levels of cytoadherence and cytotoxicity. Arch. Med. Res. 26:279–285 [PubMed] [Google Scholar]
- 5. Arroyo R, Engbring J, Alderete JF. 1992. Molecular basis of host epithelial cell recognition by Trichomonas vaginalis. Mol. Microbiol. 6:853–862 [DOI] [PubMed] [Google Scholar]
- 6. Arroyo R, Gonzalez-Robles A, Martinez-Palomo A, Alderete JF. 1993. Signalling of Trichomonas vaginalis for amoeboid transformation and adhesion synthesis follows cytoadherence. Mol. Microbiol. 7:299–309 [DOI] [PubMed] [Google Scholar]
- 7. Bennett JR, Barnes WG, Coffman S. 1989. The emergency department diagnosis of Trichomonas vaginitis. Ann. Emerg. Med. 18:564–566 [DOI] [PubMed] [Google Scholar]
- 8. Blüher S, Kratzsch J, Kiess W. 2005. Insulin-like growth factor I, growth hormone and insulin in white adipose tissue. Best Pract. Res. Clin. Endocrinol. Metab. 19:577–587 [DOI] [PubMed] [Google Scholar]
- 9. Bozner P, Gombosova A, Valent M, Demes P, Alderete JF. 1992. Proteinases of Trichomonas vaginalis: antibody response in patients with urogenital trichomoniasis. Parasitology 105(Pt 3):387–391 [DOI] [PubMed] [Google Scholar]
- 10. Branquinha MH, Vermelho AB, Goldenberg S, Bonaldo MC. 1996. Ubiquity of cysteine- and metalloproteinase activities in a wide range of trypanosomatids. J. Eukaryot. Microbiol. 43:131–135 [DOI] [PubMed] [Google Scholar]
- 11. Carlton JM, et al. 2007. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315:207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Crouch ML, Alderete JF. 1999. Trichomonas vaginalis interactions with fibronectin and laminin. Microbiology 145(Pt 10):2835–2843 [DOI] [PubMed] [Google Scholar]
- 13. Crouch ML, Benchimol M, Alderete JF. 2001. Binding of fibronectin by Trichomonas vaginalis is influenced by iron and calcium. Microb. Pathog. 31:131–144 [DOI] [PubMed] [Google Scholar]
- 14. Crouch MV, Alderete JF. 2001. Trichomonas vaginalis has two fibronectin-like iron-regulated genes. Arch. Med. Res. 32:102–107 [DOI] [PubMed] [Google Scholar]
- 15. Dailey DC, Chang TH, Alderete JF. 1990. Characterization of Trichomonas vaginalis haemolysis. Parasitology 101(Pt 2):171–175 [DOI] [PubMed] [Google Scholar]
- 16. D'Costa SS, Boyle MD. 2000. Interaction of group A streptococci with human plasmin(ogen) under physiological conditions. Methods 21:165–177 [DOI] [PubMed] [Google Scholar]
- 17. de Miguel N, et al. 2010. Proteome analysis of the surface of Trichomonas vaginalis reveals novel proteins and strain-dependent differential expression. Mol. Cell. Proteomics 9:1554–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Diamond LS, Clark CG, Cunnick CC. 1995. YI-S, a casein-free medium for axenic cultivation of Entamoeba histolytica, related Entamoeba, Giardia intestinalis and Trichomonas vaginalis. J. Eukaryot. Microbiol. 42:277–278 [DOI] [PubMed] [Google Scholar]
- 19. Donaldson JG. 2005. Arfs, phosphoinositides and membrane traffic. Biochem. Soc. Trans. 33:1276–1278 [DOI] [PubMed] [Google Scholar]
- 20. Engbring JA, Alderete JF. 1998. Three genes encode distinct AP33 proteins involved in Trichomonas vaginalis cytoadherence. Mol. Microbiol. 28:305–313 [DOI] [PubMed] [Google Scholar]
- 21. Engel M, Mazurek S, Eigenbrodt E, Welter C. 2004. Phosphoglycerate mutase-derived polypeptide inhibits glycolytic flux and induces cell growth arrest in tumor cell lines. J. Biol. Chem. 279:35803–35812 [DOI] [PubMed] [Google Scholar]
- 22. Eulberg D, Lakner S, Golovleva LA, Schlömann M. 1998. Characterization of a protocatechuate catabolic gene cluster from Rhodococcus opacus 1CP: evidence for a merged enzyme with 4-carboxymuconolactone-decarboxylating and 3-oxoadipate enol-lactone-hydrolyzing activity. J. Bacteriol. 180:1072–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ewing B, Green P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186–194 [PubMed] [Google Scholar]
- 24. Ewing B, Hillier L, Wendl MC, Green P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175–185 [DOI] [PubMed] [Google Scholar]
- 25. Florent I, et al. 1998. A Plasmodium falciparum aminopeptidase gene belonging to the M1 family of zinc-metallopeptidases is expressed in erythrocytic stages. Mol. Biochem. Parasitol. 97:149–160 [DOI] [PubMed] [Google Scholar]
- 26. Garcia AF, Benchimol M, Alderete JF. 2005. Trichomonas vaginalis polyamine metabolism is linked to host cell adherence and cytotoxicity. Infect. Immun. 73:2602–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Garcia AF, et al. 2003. Iron and contact with host cells induce expression of adhesins on surface of Trichomonas vaginalis. Mol. Microbiol. 47:1207–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gardner WA, Jr, Culberson DE, Bennett BD. 1986. Trichomonas vaginalis in the prostate gland. Arch. Pathol. Lab. Med. 110:430–432 [PubMed] [Google Scholar]
- 29. Gatski M, Kissinger P. 2010. Observation of probable persistent, undetected Trichomonas vaginalis infection among HIV-positive women. Clin. Infect. Dis. 51:114–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goetinck S, Waterston RH. 1994. The Caenorhabditis elegans muscle-affecting gene unc-87 encodes a novel thin filament-associated protein. J. Cell Biol. 127:79–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gordon D, Abajian C, Green P. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195–202 [DOI] [PubMed] [Google Scholar]
- 32. Gozalbo D, et al. 1998. The cell wall-associated glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is also a fibronectin and laminin binding protein. Infect. Immun. 66:2052–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Han HJ, et al. 2004. Epidermal growth factor-like motifs 1 and 2 of Plasmodium vivax merozoite surface protein 1 are critical domains in erythrocyte invasion. Biochem. Biophys. Res. Commun. 320:563–570 [DOI] [PubMed] [Google Scholar]
- 34. Hernandez H, et al. 2004. Monoclonal antibodies against a 62 kDa proteinase of Trichomonas vaginalis decrease parasite cytoadherence to epithelial cells and confer protection in mice. Parasite Immunol. 26:119–125 [DOI] [PubMed] [Google Scholar]
- 35. Hirt RP, Harriman N, Kajava AV, Embley TM. 2002. A novel potential surface protein in Trichomonas vaginalis contains a leucine-rich repeat shared by microorganisms from all three domains of life. Mol. Biochem. Parasitol. 125:195–199 [DOI] [PubMed] [Google Scholar]
- 36. Hobbs MM, Sena AC, Swygard H, Schwebke JR. 2008. Trichomonas vaginalis and trichomoniasis, p 771–793 In Holmes KK, et al. (ed), Sexually transmitted diseases. McGraw-Hill Medical, New York, NY [Google Scholar]
- 37. Huang KY, et al. 2009. A proteome reference map of Trichomonas vaginalis. Parasitol. Res. 104:927–933 [DOI] [PubMed] [Google Scholar]
- 38. Ikegami A, Honma K, Sharma A, Kuramitsu HK. 2004. Multiple functions of the leucine-rich repeat protein LrrA of Treponema denticola. Infect. Immun. 72:4619–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Irvine M, Huima T, Prince AM, Lustigman S. 1994. Identification and characterization of an Onchocerca volvulus cDNA clone encoding a highly immunogenic calponin-like protein. Mol. Biochem. Parasitol. 65:135–146 [DOI] [PubMed] [Google Scholar]
- 40. Jones PM, George AM. 2005. Multidrug resistance in parasites: ABC transporters, P-glycoproteins and molecular modelling. Int. J. Parasitol. 35:555–566 [DOI] [PubMed] [Google Scholar]
- 41. Kucknoor A, Mundodi V, Alderete JF. 2005. Trichomonas vaginalis adherence mediates differential gene expression in human vaginal epithelial cells. Cell. Microbiol. 7:887–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kucknoor AS, Mundodi V, Alderete JF. 2005. Adherence to human vaginal epithelial cells signals for increased expression of Trichomonas vaginalis genes. Infect. Immun. 73:6472–6478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kucknoor AS, Mundodi V, Alderete JF. 2007. The proteins secreted by Trichomonas vaginalis and vaginal epithelial cell response to secreted and episomally expressed AP65. Cell. Microbiol. 9:2586–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. LaCount DJ, Gruszynski AE, Grandgenett PM, Bangs JD, Donelson JE. 2003. Expression and function of the Trypanosoma brucei major surface protease (GP63) genes. J. Biol. Chem. 278:24658–24664 [DOI] [PubMed] [Google Scholar]
- 45. Lama A, Kucknoor A, Mundodi V, Alderete JF. 2009. Glyceraldehyde-3-phosphate dehydrogenase is a surface-associated, fibronectin-binding protein of Trichomonas vaginalis. Infect. Immun. 77:2703–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lehker MW, Alderete JF. 2000. Biology of trichomonosis. Curr. Opin. Infect. Dis. 13:37–45 [DOI] [PubMed] [Google Scholar]
- 47. Lehker MW, Alderete JF. 1992. Iron regulates growth of Trichomonas vaginalis and the expression of immunogenic trichomonad proteins. Mol. Microbiol. 6:123–132 [DOI] [PubMed] [Google Scholar]
- 48. Lin WC, et al. 2009. Malate dehydrogenase is negatively regulated by miR-1 in Trichomonas vaginalis. Parasitol. Res. 105:1683–1689 [DOI] [PubMed] [Google Scholar]
- 49. Maizels RM, Gomez-Escobar N, Gregory WF, Murray J, Zang X. 2001. Immune evasion genes from filarial nematodes. Int. J. Parasitol. 31:889–898 [DOI] [PubMed] [Google Scholar]
- 50. Mandelkow E, Mandelkow EM. 1995. Microtubules and microtubule-associated proteins. Curr. Opin. Cell Biol. 7:72–81 [DOI] [PubMed] [Google Scholar]
- 51. McClelland RS, et al. 2007. Infection with Trichomonas vaginalis increases the risk of HIV-1 acquisition. J. Infect. Dis. 195:698–702 [DOI] [PubMed] [Google Scholar]
- 52. Mundodi V, Kucknoor AS, Alderete JF. 2008. Immunogenic and plasminogen-binding surface-associated alpha-enolase of Trichomonas vaginalis. Infect. Immun. 76:523–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Neale KA, Alderete JF. 1990. Analysis of the proteinases of representative Trichomonas vaginalis isolates. Infect. Immun. 58:157–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Neve EP, Lahtinen U, Pettersson RF. 2005. Oligomerization and interacellular localization of the glycoprotein receptor ERGIC-53 is independent of disulfide bonds. J. Mol. Biol. 354:556–568 [DOI] [PubMed] [Google Scholar]
- 55. Nogales E, Wolf SG, Downing KH. 1998. Structure of the alpha beta tubulin dimmer by electron crystallography. Nature 391:199–203 [DOI] [PubMed] [Google Scholar]
- 56. Nogueira FB, Ruiz JC, Robello C, Romanha AJ, Murta SM. 2009. Molecular characterization of cytosolic and mitochondrial tryparedoxin peroxidase in Trypanosoma cruzi populations susceptible and resistant to benznidazole. Parasitol. Res. 104:835–844 [DOI] [PubMed] [Google Scholar]
- 57. Okumura CY, Baum LG, Johnson PJ. 2008. Galectin-1 on cervical epithelial cells is a receptor for the sexually transmitted human parasite Trichomonas vaginalis. Cell. Microbiol. 10:2078–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pancholi V, Fischetti VA. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176:415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Peterson KM, Alderete JF. 1982. Host plasma proteins on the surface of pathogenic Trichomonas vaginalis. Infect. Immun. 37:755–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Peterson KM, Alderete JF. 1984. Iron uptake and increased intracellular enzyme activity follow host lactoferrin binding by Trichomonas vaginalis receptors. J. Exp. Med. 160:398–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Peterson KM, Alderete JF. 1984. Trichomonas vaginalis is dependent on uptake and degradation of human low density lipoproteins. J. Exp. Med. 160:1261–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Provenzano D, Alderete JF. 1995. Analysis of human immunoglobulin-degrading cysteine proteinases of Trichomonas vaginalis. Infect. Immun. 63:3388–3395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schwebke JR, Burgess D. 2004. Trichomoniasis. Clin. Microbiol. Rev. 17:794–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Stark JR, et al. 2009. Prospective study of Trichomonas vaginalis infection and prostate cancer incidence and mortality: Physicians' Health Study. J. Natl. Cancer Inst. 101:1406–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sutcliffe S, et al. 2009. Trichomonosis and subsequent risk of prostate cancer in the Prostate Cancer Prevention Trial. Int. J. Cancer 124:2082–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Teixeira JE, Sateriale A, Bessoff KE, Huston CD. 2012. Control of Entamoeba histolytica adherence involves metallosurface protease 1, an M8 family surface metalloprotease with homology to leishmanolysin. Infect. Immun. 80:2165–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Van Der Pol B, et al. 2008. Trichomonas vaginalis infection and human immunodeficiency virus acquisition in African women. J. Infect. Dis. 197:548–554 [DOI] [PubMed] [Google Scholar]
- 68. Viscogliosi E, et al. 1998. Cloning and expression of an iron-containing superoxide dismutase in the parasitic protist, Trichomonas vaginalis. FEMS Microbiol. Lett. 161:115–123 [DOI] [PubMed] [Google Scholar]
- 69. Walker J, et al. 2006. Comparative protein profiling identifies elongation factor-1beta and tryparedoxin peroxidase as factors associated with metastasis in Leishmania guyanensis. Mol. Biochem. Parasitol. 145:254–264 [DOI] [PubMed] [Google Scholar]
- 70. Wang G, et al. 2005. Oxidative stress defense mechanisms to counter iron-promoted DNA damage in Helicobacter pylori. Free Radic. Res. 39:1183–1191 [DOI] [PubMed] [Google Scholar]
- 71. Winram SB, Lottenberg R. 1996. The plasmin-binding protein Plr of group A streptococci is identified as glyceraldehyde-3-phosphate dehydrogenase. Microbiology 142:2311–2320 [DOI] [PubMed] [Google Scholar]
- 72. Yao C, Donelson JE, Wilson ME. 2003. The major surface protease (MSP or GP63) of Leishmania sp. Biosynthesis, regulation of expression, and function. Mol. Biochem. Parasitol. 132:1–16 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.