Abstract
Streptococcus agalactiae (group B Streptococcus or GBS) is a common colonizer of the gastrointestinal and genital tracts and an important cause of invasive infections in newborn infants and in adults with predisposing chronic conditions or advanced age. Attachment to epithelial surfaces at mucosal sites is a critical step in the successful colonization of a human host, and regulation of this process is likely to play an important role in both commensalism and dissemination to cause invasive disease. We found that inactivation of the CsrRS (or CovRS) two-component system increased GBS adherence to epithelial cells derived from human vaginal, cervical, and respiratory epithelium, as well as increasing adherence to extracellular matrix proteins and increasing biofilm formation on polystyrene. Neutral (as opposed to acidic) pH enhanced GBS binding to vaginal epithelial cells and to fibrinogen and fibronectin, effects that were partially dependent on CsrRS. The regulatory effects of CsrRS and environmental pH on bacterial adherence correlated with their effects on the expression of multiple surface adhesins, as assessed by quantitative reverse transcription-PCR. We conclude that GBS adherence to epithelial and abiotic surfaces is regulated by the CsrRS two-component system and by environmental pH through their regulatory effects on the expression of bacterial surface adhesins. Dynamic regulation of GBS adherence enhances the organism's adaptability to survival in multiple niches in the human host.
INTRODUCTION
Streptococcus agalactiae (group B Streptococcus or GBS) is a commensal bacterium that colonizes the gastrointestinal and/or genital tracts of 15 to 30% of normal adults (7). GBS is also an important cause of invasive infection in neonates, pregnant women, and elderly or immunocompromised persons. Neonates acquire GBS from a colonized mother shortly before or during birth by aspiration of infected amniotic fluid or vaginal secretions or by bacterial contamination of the skin or mucosal surfaces (7, 41). The development of GBS disease reflects the penetration of epithelial barriers, resistance to immune clearance allowing survival in the bloodstream, and, in cases of meningitis, the ability to breach the endothelial blood-brain barrier.
Adherence to host tissue surfaces is an essential step in the initiation of colonization. GBS is adapted to survive in the human gastrointestinal and genital tracts, where attachment to mucosal surfaces is likely to enhance persistence. Furthermore, bacterial growth on a solid support such as an epithelial surface can be associated with elaboration of proteins, polysaccharides, and nucleic acids that form a biofilm, which, in turn, contributes to resistance to physical removal of entrapped bacteria, to resistance to clearance by immune effector mechanisms, and to antibiotic resistance. Several GBS proteins have been identified as adhesins that participate in bacterial attachment to host cells and/or extracellular matrix (ECM) (Table 1). These include proteins that bind to host cells, such as serine-rich repeat family proteins (32, 50) and BibA (36), as well as proteins that bind specifically to components of host ECM, such as fibrinogen, fibronectin, and laminin (4, 10, 31, 37, 38, 43, 48). Some enzymes and transport binding proteins also have been suggested to be involved in GBS adherence through various mechanisms (16, 22, 27, 29, 47). GBS pili also have been shown to contribute to adherence, although recent work suggests that adherence and biofilm formation may be exclusively functions of PI-2a pili and not of PI-1 or PI-2b pili (30).
Table 1.
Adherence factors included in this study
| Protein category | Locusa | Protein | Function/substrate | Reference(s) |
|---|---|---|---|---|
| AgI/II family | sag1283 | Ssp-5 | Agglutinin receptor | 27, 49 |
| ALP family | sag0433 | Rib | Cell surface protein | 44, 45 |
| Enzymes | sag0823 | GapN | GAPDH,b NADH dependent | 16 |
| sag1197 | HylB | Hyaluronate lyase, degrades ECM that is abundant in placental tissues | 21, 29 | |
| sag1768 | GAPDH | Plasminogen-binding protein | 27, 40 | |
| sag1790 | DltA | Incorporation of d-Ala into LTAc | 26, 28 | |
| Fibrinogen-binding protein | sag0832 | FbsB | Fibrinogen binding | 10, 31 |
| sag1052 | FbsA | Fibrinogen binding | 31, 37, 48 | |
| sag1127 | Fib | Fibrinogen-binding, streptokinase-like protein | 24 | |
| Fibrinogen-binding protein (cleavage) | sag2053 | CspA | Serine protease that generates a fibrin-like product by cleavage of fibrinogen | 11 |
| Fibronectin-binding protein | sag1190 | PavA | Fibronectin binding | 25 |
| sag1236 | ScpB | Fibronectin binding | 4 | |
| Laminin-binding protein | sag1233 | Lmb | Laminin binding | 43 |
| Host cell-binding protein | sag1462 | Srr1 | Serine-rich repeat family, binding to human keratin 4 and epithelial cells. | 27, 32, 50 |
| sag2063 | BibA | Binding to human C4-binding protein and epithelial cells | 36 | |
| Regulatory function | sag1463 (sal1543) | Rga | Regulation of binding to keratin 4, fibrinogen, and epithelial cells | 33 |
| sag1490 | RovS | Regulation of binding to fibrinogen and epithelial cells | 34 | |
| sag1792 | DltR | Regulation of d-Ala LTA biosynthesis | 8, 28 | |
| sag1957, sag1958, sag1959 | RgfA, RgfB, RgfC | rgfBDAC regulatory system, regulation of fbsA and fbsB and binding to fibrinogen | 1, 42 | |
| Transport and binding proteins | sag1007 | Iron compound-binding protein, ABC transporter | 16 | |
| sag1466 | GlnP | Glutamine ABC transporter, glutamine-binding protein/permease protein | 16 | |
| sag1467 | GlnQ | Glutamine ABC transporter, ATP-binding protein | 47 | |
| Other adhesins | sag0765 | Penicillin-binding protein 2b | 16 | |
| sag1331 | Sar5 | R5 protein, surface antigen | 49 | |
| sag1350 | Unidentified surface antigen | 16 | ||
| sag0971 | Lipoprotein of unknown function | 16 |
Locus numbers refer to the genome sequence of GBS strain 2603 (49).
GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
LTA, lipoteichoic acid.
The highly diverse display of surface proteins enables multiple interactions of GBS with different host components and likely increases the organism's adaptability in occupying different anatomic sites of the host. Transcriptional regulators such as RovS, a member of the Rgg protein family (34); Rga, a RofA-like protein (33); and the RgfA/C two-component system (1, 42) have been found to modulate GBS adherence to host cells or ECM, but the molecular basis of the coordinated expression of GBS adherence factors remains incompletely understood.
The CsrRS (or CovRS) two-component regulatory system controls the expression of multiple virulence factors in GBS (14, 15, 19, 20). Inactivation of CsrRS in GBS strain NEM316 was associated with increased adherence to epithelial cells (19), although the specific adhesins responsible for this phenotype have not been defined. In the course of studying the relationship of pilus function to GBS adherence, we found that CsrR negatively regulates the adherence of GBS strain 2603V/R (referred to here as 2603) to human epithelial cells (13). The chromosome of GBS 2603 harbors two pilus operons, PI-1 and PI-2a. However, only PI-1 pili are expressed; PI-2a pili are not produced at a detectable level because of inactivating mutations in the transcriptional activators RogB and/or Rga, which control PI-2a gene expression (6, 9, 33). In an investigation of the function and regulation of the PI-1 pilus, we found that its expression was significantly increased in csrR mutant strain 2603ΔcsrR. However, increased expression of PI-1 pili was not responsible for the hyperadherence phenotype of 2603ΔcsrR. It appeared, rather, that increased adherence associated with inactivation of csrR is mediated by effects on the expression or exposure of other GBS adhesins.
In this investigation, we studied the regulatory mechanisms and environmental factors that control GBS adherence to human epithelial cells and immobilized ECM proteins. We demonstrate that CsrRS plays a major role in regulating GBS adherence to cells, ECM, and abiotic surfaces and that adherence correlates with the CsrRS-regulated expression of multiple adherence factors. We also report evidence that the environmental pH modulates the expression of adherence factors and affects GBS attachment to host cells and ECM in a both CsrRS-dependent and CsrRS-independent manner.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The GBS strains used in this study were type Ia strain 515; type V strain 2603 (2603V/R); their derivative mutants 515ΔcsrR, 2603ΔPI-1, 2603ΔcsrR, 2603ΔcsrR/ΔPI-1, and 2603ΔcsrS; and a repaired ΔcsrR mutant strain, 2603rep (13, 14). Unless otherwise specified, GBS bacteria were grown in Todd-Hewitt broth (THB; Difco) or on Trypticase soy agar (TSA) supplemented with 5% defibrinated sheep blood (PML Microbiologicals). For experiments testing the effects of pH, GBS was cultured at 37°C in complex medium (10 g/liter proteose peptone, 5 g/liter Trypticase peptone, 5 g/liter yeast extract, 2.5 g/liter KCl, 1 mM urea, 1 mM arginine). For assays testing the effect of pH on gene expression or bacterial adherence, GBS was first grown at pH 7.4 to an A600 of 0.35. The GBS cells, divided into two aliquots, were incubated in fresh medium at either pH 7.4 or pH 5.0. After 30 min of treatment, GBS cells were collected and mRNA was isolated for quantitative reverse transcription-PCR (qRT-PCR). For adherence assays, after 60 min of treatment, GBS cells were collected, resuspended at pH 7.4 or pH 5.0 in the appropriate medium, and used in adherence assays as described below.
Cell culture.
VK2, a human vaginal epithelial cell line; ME180, a human cervical carcinoma cell line; and A549, a human alveolar basal epithelial carcinoma cell line, were cultured as described previously (13).
GBS adherence to human epithelial cells.
Adherence assays were performed as described previously (13). Briefly, cells were seeded into 24-well tissue culture plates at densities of 5 × 105 to 8 × 105 per well and cultured at 37°C in 5% CO2 for 5 days to establish a monolayer. GBS cells were grown to an A600 of 0.35 in THB and then washed once with phosphate-buffered saline (PBS). The washed GBS cells were resuspended in RPMI with 2% heat-inactivated fetal bovine serum (FBS) for assays with ME180 or A549 cells or in keratinocyte-SF medium (Invitrogen) with 0.4 mM CaCl2 for assays with VK2 cells. The cell monolayers were infected with 5 × 106 to 8 × 106 GBS cells per well (multiplicity of infection of 10) for 1 h at 37°C under 5% CO2. The monolayer was washed five times with PBS, detached with 0.2 ml of 0.25% trypsin–EDTA for 10 min, and then lysed with 0.8 ml of 0.025% Triton X-100. The lysate was vigorously pipetted to liberate cell-associated bacteria. The serially diluted lysate was cultured on blood agar for enumeration of viable bacteria. Assays were repeated at least three times in triplicate. The percentage of adherent GBS cells was calculated as follows: (number of CFU of adherent GBS/number of CFU in initial inoculum) × 100.
GBS adherence to human ECM proteins.
To investigate the adhesion of GBS to immobilized ECM components, adherence assays were performed with 24-well polystyrene plates coated with individual ECM proteins. Plates coated with fibronectin or laminin were purchased from BD Biosciences. To prepare fibrinogen-coated plates, uncoated plates (BD Biosciences) were incubated with 10 μg/ml fibrinogen in PBS, pH 7.4, overnight at 4°C. The ECM component-coated plates were washed three times with PBS to remove unbound protein. GBS cells were grown to an A600 of 0.35, washed once with PBS, and then resuspended in RPMI supplemented with 2% FBS. Each well of the plate was inoculated with 5 × 106 to 8 × 106 GBS cells for 1 h at 37°C under 5% CO2. The plate was washed five times with PBS to remove unattached GBS, and then GBS cells were detached from the wells with 0.3 ml of 0.25% trypsin–EDTA for 15 min. The serially diluted bacterial suspension was cultured on blood agar to enumerate viable bacteria. Assays were repeated at least three times in triplicate. The percentage of adherent GBS was calculated as follows: (number of CFU of adherent GBS/number of CFU in initial inoculum) × 100.
Biofilm formation assay.
Biofilm assays were performed with 96-well polystyrene flat-bottom microtiter plates (Costar) essentially as described previously (18, 30). GBS cultures grown overnight were diluted 1:20 in fresh LB medium containing 1% glucose. Each well was seeded with 100 μl of GBS suspension after being rinsed three times with PBS. Wells filled with growth medium but without GBS were included as negative controls. Plates were incubated without shaking at 37°C for 24 h aerobically in 5% CO2. Before biofilm quantification, the growth of wild-type and mutant strains was assessed by measuring the A600 of the cultures in the wells with a spectrophotometer. The contents of the wells were then decanted, and the wells were rinsed with PBS. Wells were air dried, and adherent bacteria were stained for 10 min with a 0.5% (wt/vol) solution of crystal violet. The wells were rinsed with water, and then bound dye was released from stained cells using 30% glacial acetic acid. Biofilm formation was quantified by measuring the A540 of the solution with a microplate reader. Each assay was performed in triplicate and repeated at least three times.
qRT-PCR.
GBS strains grown overnight on TSA blood agar plates were inoculated in 10 ml of THB. The GBS cells were collected in the mid-exponential growth phase (A650 of 0.35) by centrifugation (3,200 × g, 5 min). RNA isolation and qRT-PCR were performed as described previously (13).
Statistical analysis.
Data are reported as means ± standard deviations (SD) unless otherwise stated. Statistical analysis was performed using Prism 5.0 (GraphPad Software Inc.). Differences between groups were analyzed using a two-tailed t test. Differences with P values of <0.05 were considered statistically significant. Asterisks in the figures represent ranges of P values for differences between groups (NS [not significant], P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
RESULTS
GBS adherence to human epithelial cells is controlled by the CsrR regulator.
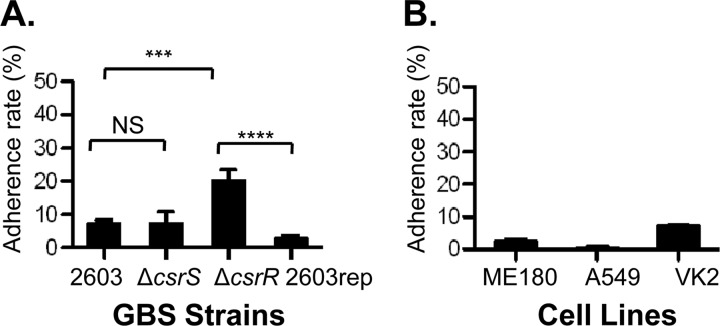
CsrRS (or CovRS) is a global two-component regulatory system that regulates the expression of approximately 7% of GBS genes (14, 15, 19, 20). A hyperadherence phenotype has been observed in GBS mutant strains that lack a functional CsrR regulator (13, 19). To further explore the effect of CsrRS on GBS adherence, we compared the association of wild-type GBS 2603 with VK2, an immortalized human vaginal epithelial cell line, with that of 2603ΔcsrR, an isogenic csrR deletion mutant. After 1 h of exposure to GBS, cell monolayers were washed to remove the unbound bacteria, detached from wells, and lysed and quantitative cultures of the lysates were performed to enumerate the cell-bound bacteria. Although we did not distinguish internalized bacteria from those attached to the cell surface, previous studies have shown that GBS cells internalized by epithelial cells represent 1% or less of the cell-associated population (4, 10). We observed significantly greater adherence of 2603ΔcsrR than of wild-type strain 2603 (Fig. 1A). To confirm that the observed increase in bacterial attachment was due to the deletion of csrR and not to other, unknown, mutations, we performed the same assay using a repaired strain, 2603rep, in which the mutant csrR allele was replaced with a copy of the wild-type gene (14). As shown in Fig. 1A, in the repaired strain, adherence was restored to a level similar to that of the original wild type. This result provided evidence that the observed increase in adherence was a result of csrR inactivation. Finally, we investigated whether CsrS, the sensor component of the CsrRS two-component regulatory system, is also involved in the regulation of GBS adherence. In contrast to the result obtained with the csrR mutant, the adherence of a csrS deletion mutant, 2603ΔcsrS, was not significantly different from that of the wild type (Fig. 1A). Together, these data indicate that CsrR represses the adherence of GBS 2603 to human vaginal epithelial cells while involvement of CsrS, under these experimental conditions, was not evident.
Fig 1.
Adherence of GBS strains to human epithelial cells. (A) Adherence to VK2 cells was compared among GBS strains 2603, 2603ΔcsrR (a ΔcsrR mutant), 2603ΔcsrS (a ΔcsrS mutant), and 2603rep (in which the csrR deletion mutation was repaired). (B) Adherence of strain 2603 to three types of epithelial cells, ME180, A549, and VK2. Adherence is shown as a percentage of the initial inoculum (mean ± SD). Each assay was performed at least three times in triplicate. NS, P > 0.05; ****, P < 0.0001; ***, P < 0.001.
Because GBS may bind to mucosal surfaces at various anatomic sites during colonization or infection, it was of interest to study GBS attachment to different types of human epithelial cells. We compared the adherence of strain 2603 to epithelial cells from three different body sites, ME180 (cervical carcinoma cell line), A549 (lung adenocarcinoma basal alveolar epithelial cell line), and VK2 (vaginal epithelial cells). As shown in Fig. 1B, we observed the highest rate of GBS adherence to VK2 cells (11.7%), an intermediate rate of adherence to ME180 cells (2.9%), and the lowest rate of adherence to A549 (0.7%). These results are compatible with the known predilection of GBS to colonize the female lower genital tract, while infection of the respiratory tract may reflect primarily the effects of aspiration of infected fluid rather than preferential attachment to respiratory epithelial cells. Inactivation of CsrR also increased GBS adherence to ME180 and A549 cells (13), which is in agreement with the results obtained with VK2 cells. Together, these data suggest that the CsrR regulator plays an important role in controlling GBS adherence to human epithelial cells.
GBS adherence to human epithelial cells is modulated by the environmental pH.
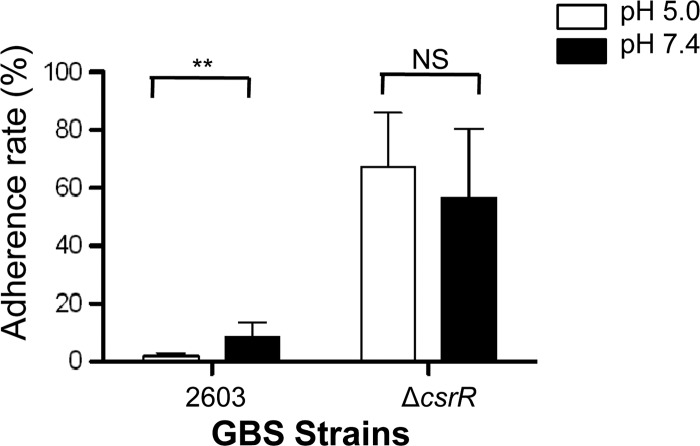
In the course of colonization and infection of a human host, GBS encounters a wide range of environmental conditions, from the acidic milieu of the vagina to the near neutral pH in the neonatal lung or bloodstream. To test whether GBS adherence changes in response to the environmental pH, we assessed the relative binding of strain 2603 to human epithelial cells at acidic versus neutral pHs. In pilot studies, we found that exposure to an acidic pH had cytotoxic effects on ME180 and A549 cells, even in the absence of GBS, whereas VK2 cells tolerated acidic medium without exhibiting overt cytotoxicity. Therefore, we used the VK2 cell line as a model to study pH effects on GBS adherence. The adherence of wild-type strain 2603 was approximately 4-fold higher at pH 7.4 than at pH 5.0 (Fig. 2). On the other hand, pH did not affect the binding of the csrR deletion mutant 2603ΔcsrR (Fig. 2), a result that suggested that pH regulation of GBS adherence is, at least in part, CsrRS dependent.
Fig 2.
Effect of pH on GBS adherence to vaginal epithelial cells. The influence of pH on GBS adherence to VK2 cells was compared between strains 2603 and 2603ΔcsrR (a ΔcsrR mutant) after exposure to pH 5.0 or 7.4. Adherence is shown as a percentage of the initial inoculum (mean ± SD). Each assay was performed at least three times in triplicate. **, P < 0.01; NS, P > 0.05.
Factors influencing GBS adhesion to human ECM.
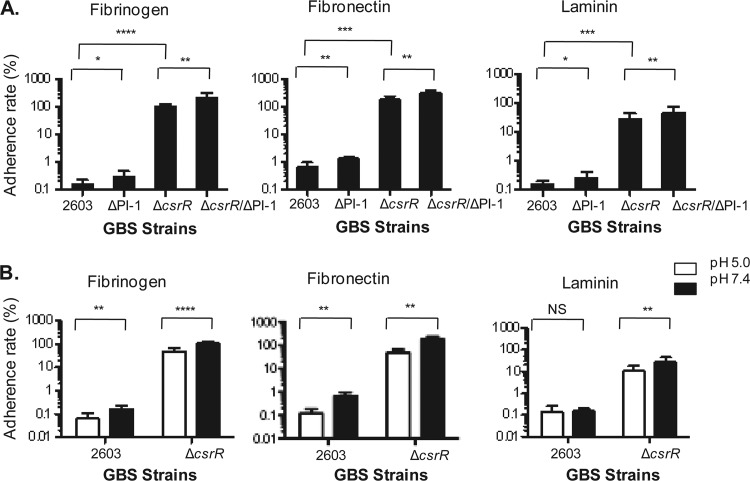
GBS-host tissue interactions involve the attachment of the bacterium to human ECM molecules such as fibronectin, fibrinogen, and laminin, which, in turn, bind host cell surface integrins (12). To investigate GBS adherence to ECM, we used 24-well plates coated with one of three ECM proteins, fibronectin, fibrinogen, or laminin. Coated wells were each inoculated with approximately 6 × 106 CFU of GBS, and the adherence rate was calculated as the ratio of the number of CFU of GBS bound to the number of CFU in the initial inoculum. We found markedly greater adherence of the csrR deletion mutant 2603ΔcsrR than of wild-type GBS 2603 to all three types of immobilized ECM components (Fig. 3A), a result that suggests that CsrR represses the expression of GBS factors that mediate adherence to human ECM proteins.
Fig 3.
Adherence of GBS strains to immobilized human ECM proteins. (A) Relative adherence to immobilized human fibrinogen, fibronectin, or laminin by GBS strains 2603, 2603ΔPI-1 (a ΔPI-1 mutant), 2603ΔcsrR (a ΔcsrR mutant), and 2603ΔcsrR/ΔPI-1 (a ΔcsrR/ΔPI-1 double mutant). (B) Influence of pH on GBS adherence to immobilized human fibrinogen, fibronectin, or laminin by strain 2603 or 2603ΔcsrR after exposure to pH 5.0 or 7.4. Adherence is shown as a percentage of the initial inoculum (mean ± SD). Each assay was performed at least three times in triplicate. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; NS, P > 0.05.
Pili have been shown to contribute to the adherence of multiple bacterial species, including GBS (6, 22, 23, 39). However, our previous studies of the function of the PI-1 pilus showed that it does not mediate the adhesion of GBS 2603 to human epithelial cells (13). To evaluate the role of the PI-1 pilus in GBS attachment to human ECM, we compared the relative association of wild-type 2603 and strain 2603ΔPI-1 to fibronectin, fibrinogen, or laminin. We found that pilus loss did not result in reduced GBS adhesion to any of the immobilized ECM components tested (Fig. 3A). As the expression of the PI-1 pilus is repressed by CsrR in strain 2603 (13), we also assessed the importance of PI-1 pili in adherence to ECM in the background of the 2603ΔcsrR mutant, in which PI-1 pilus production is derepressed, comparing its adherence to that of the double mutant 2603ΔcsrRΔPI-1. As shown in Fig. 3A, inactivation of the PI-1 pilus in 2603ΔcsrR did not reduce GBS adherence to the immobilized ECM proteins. Rather, our data revealed that GBS strains lacking pili (ΔPI-1 and ΔPI-1ΔcsrR) consistently exhibited binding to ECM proteins that was stronger than that of the respective parent strains that express PI-1 pili (wild-type 2603 and ΔcsrR) (13). It is possible that loss of pili enhances the exposure of adhesins on the bacterial cell surface, thereby promoting GBS adherence. Taken together, these data indicated that PI-1 pili do not contribute to GBS adherence to ECM components such as fibrinogen, fibronectin, and laminin. CsrR may regulate GBS adherence to ECM by modulating the expression or surface exposure of other adherence factors.
To study the effect of the environmental pH on GBS binding to ECM, we evaluated the adherence of strain 2603 to ECM proteins at acidic and neutral pHs. GBS binding to immobilized fibrinogen and fibronectin was greater at a neutral pH than at an acidic pH (Fig. 3B). In contrast, binding to laminin was similar at pH 7.4 and pH 5.0 (Fig. 3B). To investigate whether the effect of pH on GBS binding requires CsrRS, we further tested the adherence of the ΔcsrR mutant to ECM proteins at different pHs. We found that neutral pH also enhanced the adherence of 2603ΔcsrR to fibrinogen and fibronectin but to a lesser extent than the effect on wild-type strain 2603. The results suggest that pH regulation of GBS adherence to ECM is partially dependent on CsrRS.
In summary, GBS adherence to human ECM is upregulated in 2603ΔcsrR and at neutral pH. Neutral pH enhances the adhesion of wild-type 2603 to fibrinogen and fibronectin but not to laminin. PI-1 pili do not mediate GBS attachment to ECM, which is similar to results showing no role for PI-1 pili in binding to human epithelial cells. It is likely that CsrR regulates ECM binding of GBS through effects on the expression or exposure of other (nonpilus) adherence factors.
Factors influencing biofilm formation in GBS.
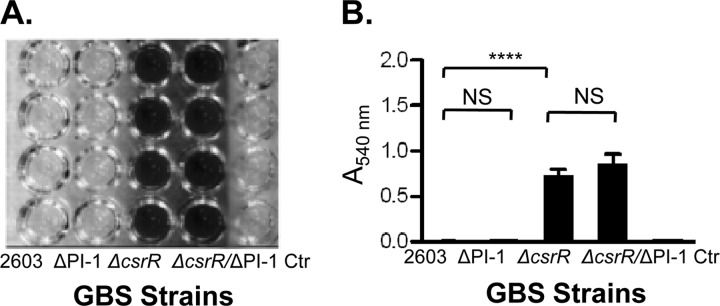
Biofilm is an aggregate of microorganisms in which cells adhere to each other on a surface, typically embedded within a self-produced matrix of exopolysaccharide, proteins, and nucleic acids. GBS bacteria have been isolated from the population of biofilm-forming bacteria on intrauterine devices (5) and have been shown to form biofilms on abiotic and cell surfaces in vitro (17, 18, 30). Biofilm formation by GBS appears to be associated with the production of PI-2a pili but not with the production of PI-1 or PI-2b (18, 30). Since PI-1 pilus expression is tightly repressed by CsrR (13), the poor formation of biofilm might result from the low-level production of the PI-1 pilus in GBS strains investigated before. In this study, we investigated whether 2603, a GBS strain that expresses only PI-1 pili, has the ability to form biofilm, as assessed by the deposition of bacterial biomass adherent to polystyrene microtiter wells. GBS strains were grown in LB medium supplemented with 1% glucose in polystyrene plates, and biofilm formation was detected by crystal violet staining, followed by dye solubilization with acetic acid and measurement of A540. As shown in Fig. 4, strain 2603 produced minimal biofilm, consistent with a previous report (30). Similar results were obtained with pilus mutant strain 2603ΔPI-1. In contrast, biofilm formation by the 2603ΔcsrR mutant was greatly enhanced (Fig. 4). To test whether the increased biofilm production of 2603ΔcsrR is attributable to increased pilus expression, we compared it with a double mutant, 2603ΔcsrR/ΔPI-1. However, the two strains showed no significant difference in biofilm formation, a result indicating that the increased biofilm formation of 2603ΔcsrR is not due to the better expression of the PI-1 pilus. In summary, these data indicate that the PI-1 pilus does not contribute to biofilm formation by GBS 2603 even when its expression is derepressed in a csrR mutant. CsrR negatively regulates biofilm formation, probably through the modulation of expression of nonpilus adhesins.
Fig 4.
Biofilm formation by GBS strains. Biofilm formation was compared among GBS strains 2603, 2603ΔPI-1 (a ΔPI-1 mutant), 2603ΔcsrR (a ΔcsrR mutant), and 2603ΔcsrR/ΔPI-1 (a ΔcsrR/ΔPI-1 double mutant). GBS strains were grown in liquid medium in 96-well polystyrene plates. Wells containing growth medium but no GBS were included as a negative control (Ctr). (A) Photograph of a microtiter plate after adherent bacteria were stained with crystal violet. (B) Quantification of biofilm by measurement of A540 after the release of biofilm and bound crystal violet from each well using glacial acetic acid. ****, P < 0.0001; NS, P > 0.05.
The CsrRS system controls the expression of multiple adherence factors in GBS.
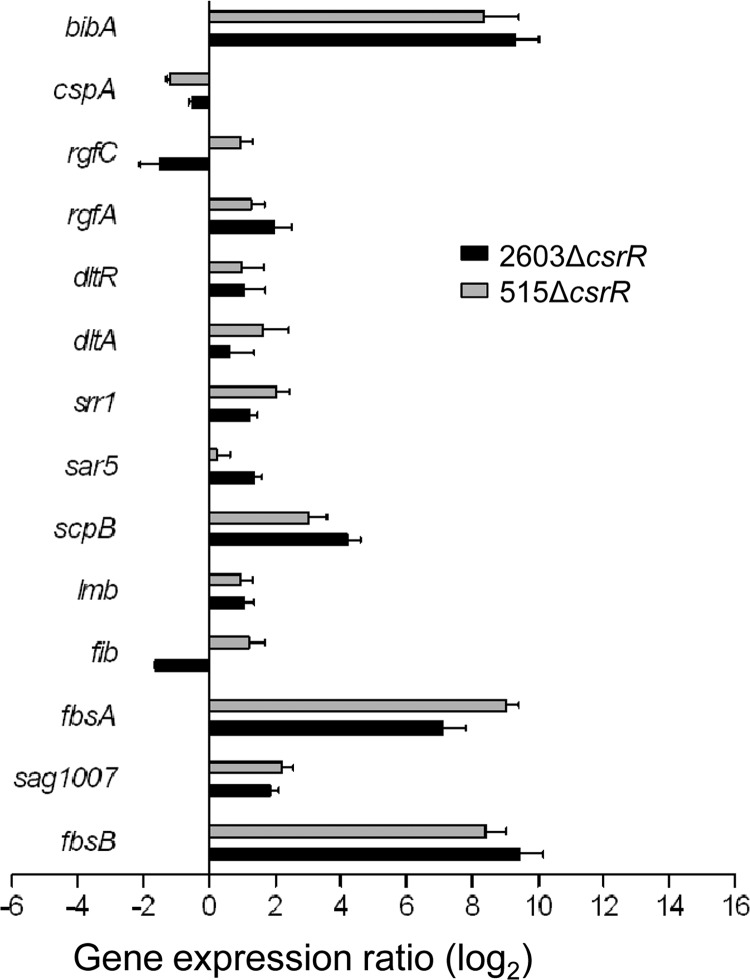
As demonstrated above, GBS adherence to biotic and abiotic surfaces is greatly enhanced in mutant strain 2603ΔcsrR but the increase in adherence is not attributable to increased expression of PI-1 pili. Rather, it seems likely that inactivation of csrR results in derepressed expression of CsrRS-regulated nonpilus adhesins that promote bacterial attachment. To test that hypothesis, we used qRT-PCR to compare the expression of 28 candidate genes that encode known and putative GBS adherence proteins and related regulators in wild-type 2603 and 2603ΔcsrR (Table 1). The specific oligonucleotide primers used for qRT-PCR are listed in Table 2. Using a threshold of a 2-fold change in gene expression between the mutant and the wild type, we found evidence that CsrR regulates the expression of 12 of the 28 genes tested in strain 2603 (Fig. 5 and Table 3). The same comparison was performed in an independent GBS strain background, type Ia strain 515, with similar results; 13 genes were found to be regulated by CsrR in strain 515, including 9 genes that were similarly regulated in strain 2603 (Fig. 5 and Table 3). Furthermore, among the adhesin-encoding genes regulated by CsrR, most were found to be upregulated in 2603ΔcsrR compared to wild-type 2603, consistent with the enhanced adherence of the ΔcsrR mutant (Fig. 1 to 4).
Table 2.
Oligonucleotide primers used in PCRs
| Locus | Protein | Primer | Directiona | Sequence (5′-3′) |
|---|---|---|---|---|
| sag1283 | Ssp-5 | 1330 | F | CAATACCGAGGGATTAAAGCAG |
| 1329 | R | TAGCTGCTTCATCTGGCAATAA | ||
| sag0433 | Rib | 1321 | F | TCCACGTACAGATGCCGATA |
| 1322 | R | TGGAGTTGCATTCTCACCTG | ||
| sag0823 | GapN | 1309 | F | GGAATGCGTCCAATTATGCT |
| 1310 | R | GCGTTGTCCGGAGTAATCAT | ||
| sag1197 | HylB | 1351 | F | GGAACGACGTGACTATTGGTAA |
| 1352 | R | TCCAGTTTGATAGCTTCGATGT | ||
| sag1768 | GAPDH | 1334 | F | ACCACGATATCCTTGATGGAAC |
| 1333 | R | CACCAGTGTATGCGTGGATAGT | ||
| sag1790 | DltA | 1318 | F | TGACAGTTGCTCAACCAAGC |
| 1317 | R | AACATTTGCGGTCTTTCAGG | ||
| sag0832 | FbsB | 1363 | F | CGGCGTATAAATCAACAGTCAA |
| 1364 | R | TTCAATAGCTTGGTAAGCACGA | ||
| sag1052 | FbsA | 1313 | F | ATTTTCATTGCGTCTCAAACC |
| 1314 | R | ATTACCTAAAACAGGTGATGATCAAA | ||
| sag1127 | Fib | 1327 | F | AACGTTCAATACAGCAACATCG |
| 1328 | R | ATTTGTTTGGACCGTCTTTAGG | ||
| sag2053 | CspA | 1335 | F | ATACGATGAGGAACGAGCAAAT |
| 1336 | R | CAACGAGTAAGGCTGCTTTCTT | ||
| sag1190 | PavA | 1331 | F | TCCTTCCTGGTTCGACTTACAT |
| 1332 | R | AGGGCTTAGATCATTGGTTTGA | ||
| sag1236 | ScpB | 955 | F | CTGAAACTGCTACGGTCA |
| 845 | R | ACGTTCAATAAGGGCAATC | ||
| sag2063 | BibA | 985 | F | AGAGCATCACAGGATACA |
| 984 | R | ACGTCTTGACTGCTTACC | ||
| sag1233 | Lmb | 1324 | F | TATGACGCGGATTTGTTTGT |
| 1323 | R | CAATGCCTTGTGTGACTTCC | ||
| sag1463 (sal1543) | RgA | 1368 | F | CTTTACAGGCAATGGTCGTG |
| 1367 | R | TGCAGCTGGACCAATAATGT | ||
| sag1490 | RovS | 1326 | F | TTAGCAGATGAGCACCTATCCA |
| 1325 | R | AGTGTGCGCCTTAGAATGAATC | ||
| sag1792 | DltR | 1320 | F | TGCGGCTTGGAAATATAAGG |
| 1319 | R | CTGCTTTGCATTTCGCATAA | ||
| sag1957 | RgfA | 1344 | F | GCGTTACAGCCAGGTACAATTA |
| 1343 | R | AAGTGGCTGTGAATTCCATTCT | ||
| sag1958 | RgfB | 1341 | F | TGAAGAGCTGGTGTACCTTCAA |
| 1342 | R | TAGCAGAGCATATCTTCCGTGA | ||
| sag1959 | RgfC | 1346 | F | AGTGTCACTCCAGCACGTTTTA |
| 1345 | R | GTGTCACCCAAACAAAGTTGAA | ||
| sag1462 | Srr1 | 1316 | F | CAATGTCAGCCAGCACAAGT |
| 1315 | R | CTTGCTGATGTCGATGCACT | ||
| sag1007 | 1312 | F | AAAGGAGTTGCTGAGCGTGT | |
| 1311 | R | ACCAAAACGACCAGAAGGTG | ||
| sag1466 | GlnP | 1353 | F | TTGCTGCTACTATCGCTCTTTC |
| 1354 | R | CTACGACTTGCTTCCATTTGAC | ||
| sag1467 | GlnQ | 1389 | F | GCATGGGATGGAATTACTTG |
| 1390 | R | CGCTAAGCTTCTTGCGATAG | ||
| sag0765 | 1307 | F | CCGAAGGGATGTATGCAGTT | |
| 1308 | R | GTCGCACCTTTCACAACAGA | ||
| sag1331 | Sar5 | 1340 | F | GCAACTTTAGCAGCAACATCAC |
| 1339 | R | CGAACACTGTCAGCATATGGAT | ||
| sag1350 | 1304 | F | CTCAACCACAAGCCCGTATT | |
| 1303 | R | CTTGCCCTCCTTGTTCAGTC | ||
| sag0971 | 1305 | F | TCTTGTTTCCGTTGCTGAGA | |
| 1306 | R | TCGTTAACTGCATCGTTTGG |
F, forward; R, reverse.
Fig 5.
Regulation by CsrR of the expression of adhesin genes in GBS. Pairs of bars represent the ratios of transcript abundance in strains 2603ΔcsrR (black bars) and 515ΔcsrR (gray bars) to that in wild-type strains 2603 and 515, respectively. Each qRT-PCR experiment was performed at least three times in duplicate.
Table 3.
Adherence factors regulated by CsrR in group B Streptococcus
| Locusa | Gene | Relative expression (log2)b |
Function | |
|---|---|---|---|---|
| 2603ΔcsrR | 515ΔcsrR | |||
| sag0832 | fbsB | 9.4 | 8.4 | Fibrinogen binding |
| sag1007 | sag1007 | 1.9 | 2.2 | Iron compound binding |
| sag1052 | fbsA | 7.1 | 9.0 | Fibrinogen binding |
| sag1127 | fib | −1.6 | 1.2 | Fibrinogen binding |
| sag1233 | lmb | 1.1 | 1.0 | Laminin binding |
| sag1236 | scpB | 4.2 | 3.0 | Fibronectin binding |
| sag1331 | sar5 | 1.4 | 0.2 | R5 protein surface antigen |
| sag1462 | srr-1 | 1.2 | 2.0 | Serine-rich repeat family, binding to human keratin 4 and epithelial cells |
| sag1790 | dltA | 0.6 | 1.6 | Biosynthesis of d-Ala lipoteichoic acid |
| sag1792 | dltR | 1.1 | 1.0 | Regulation of d-Ala lipoteichoic acid biosynthesis |
| sag1957 | rfgA | 2.0 | 1.3 | Regulator of rgfAC system |
| sag1959 | rfgC | −1.5 | 1.0 | Sensor of rgfAC system |
| sag2053 | cspA | −0.5 | −1.2 | Fibrinogen binding (cleavage) |
| sag2063 | bibA | 9.3 | 8.4 | Binding to human C4-binding protein and epithelial cells |
Locus names refer to the genome sequence of GBS strain 2603 (49).
Values represent log2 n-fold differences in transcript abundance in 2603ΔcsrR or 515ΔcsrR relative to that in 2603 or 515, respectively.
Several highly regulated genes encode ECM-binding proteins (Fig. 5 and Table 3). Among these, fbsA encodes a protein that interacts with fibrinogen via 16-amino-acid repeats, the number of which varies among GBS strains (31, 38). In strain 2603, the fbsA locus (sag1052), however, encodes a truncated protein that lacks these repeats. The truncated protein consists of only 47 amino acids, which are identical to the C terminus of FbsA of strain NEM316. In strain 515, the fbsA locus (sal1159) encodes a complete protein of 281 amino acids containing 10 repeat units. Transcription of fbsA is markedly upregulated in csrR deletion mutants in both strain backgrounds: the expression of the truncated and intact forms of fbsA was 137- and 512-fold higher in 2603ΔcsrR and 515ΔcsrR than in the respective parent strains (Table 3). Similarly, fbsB, which encodes an independent fibrinogen-binding protein that binds to human fibrinogen by its N-terminal 388 amino acids (31), exhibits strikingly higher expression in ΔcsrR mutants, 676-fold higher in 2603ΔcsrR and 338-fold higher in 515ΔcsrR than in the respective wild-type strains. The C5a peptidase encoded by scpB has been shown to function not only as a protease for the chemoattractant complement protein C5a but also as a surface-associated fibronectin-binding protein (2, 4, 24). qRT-PCR results showed that the expression of scpB is 18- and 8-fold higher in the 2603ΔcsrR and 515ΔcsrR mutants than in the respective parent strains. Finally, a moderate (about 2-fold) increase in the expression of lmb, which encodes a laminin-binding protein, was observed in both ΔcsrR mutants.
We also observed in both GBS ΔcsrR mutants a marked increase in the expression of bibA, which encodes a protein that binds to human C4-binding protein and human epithelial cells (36). Expression of the serine-rich repeat protein Srr-1, which binds human keratin 4 and promotes adherence to epithelial Hep-2 cells and brain microvascular endothelial cells (32), is also upregulated in both csrR mutants. Interestingly, the RgfA/C two-component system, which regulates fbsA and fbsB and modulates GBS binding to fibrinogen (1, 42), is under the negative control of CsrR (Table 3), indicating a complex regulatory cascade for GBS adherence. Other adherence proteins regulated by CsrR include Sar5 (R5 protein), Sag1007 (iron compound binding), Fib (fibrinogen binding), and CspA (fibrinogen binding, cleavage) (Fig. 5 and Table 3). Together, these results demonstrate that CsrRS regulates multiple GBS adhesins and support a central role for this two-component system in the control of GBS adherence during colonization and infection.
Environmental pH modulates the expression of adherence factors in a both CsrRS-dependent and -independent manner.
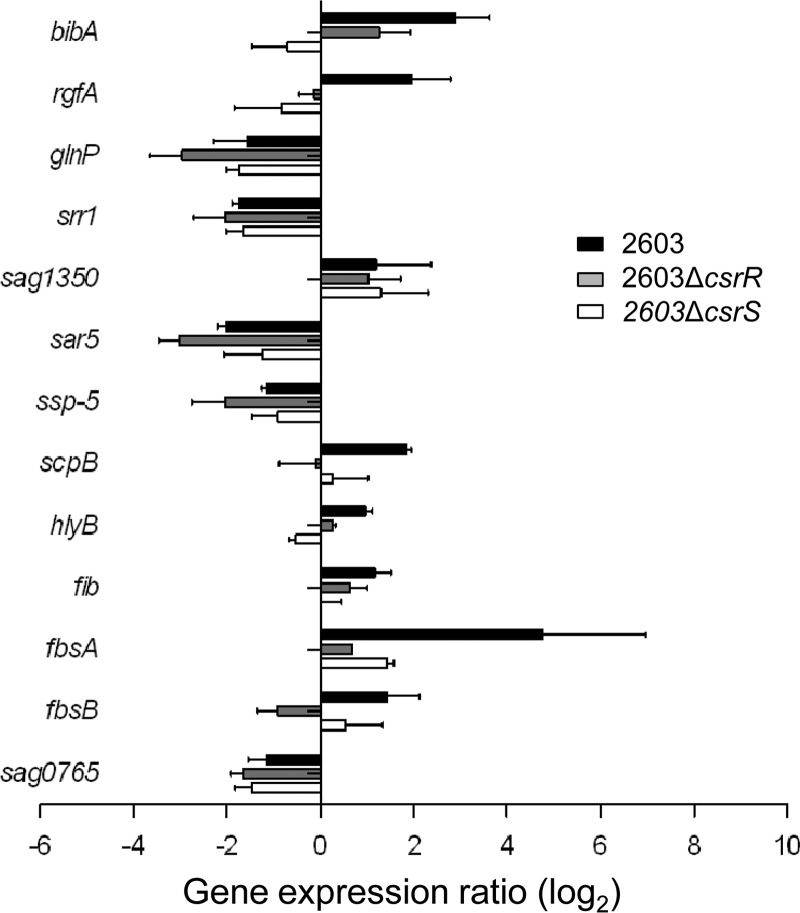
As shown earlier, GBS adherence to host cells and ECM is modulated by the environmental pH (Fig. 2 and 3B). To gain insight into the underlying molecular mechanism, we compared the expression of 28 adhesin-encoding genes (Table 1) at acidic and neutral pHs. Again, using a 2-fold change as the threshold, we found that 13 of the 28 genes in GBS 2603 were differentially expressed at neutral and acidic pHs: 8 genes were upregulated and 5 genes were downregulated at pH 7.4 compared to their expression at pH 5.0 (Fig. 6 and Table 4). Among the genes upregulated at pH 7.4, most are ECM binding proteins, such as fibrinogen- or fibronectin-binding proteins FbsB/A, Fib, and ScpB. In addition, the expression of BibA, RgfA, HylB (hyaluronate lyase), and Sag1350 (an unidentified surface protein) is also promoted by a neutral pH. The regulation of the serine-rich family protein Srr-1 and four other proteins showed the opposite pattern, i.e., downregulation at a neutral pH. Laminin-binding protein (Lmb) was unaffected by pH in strain 2603, which is consistent with the observation noted above that pH did not alter the laminin-binding ability of GBS 2603 (Fig. 3B). In summary, while the effect of pH on the expression of individual adhesins is mixed, on balance, a neutral pH promotes increased adherence to host epithelial cells and ECM proteins (Fig. 2 and 3B).
Fig 6.
Modulation by pH of the expression of adhesin genes in GBS. Differential regulation of gene expression in GBS strain 2603 (black bars) versus that in isogenic mutant strains 2603ΔcsrR (gray bars) and 2603ΔcsrS (white bars) after exposure to pH 7.4 or 5.0. Values represent the ratio of transcript abundance at pH 7.4 to that at pH 5.0. Each qRT-PCR experiment was performed at least three times in duplicate.
Table 4.
Adherence factors regulated by environmental pH in GBS strain 2603
| Locusa | Gene | Expression ratio (log2), pH 7.4/5.0b |
Function | ||
|---|---|---|---|---|---|
| 2603 | 2603ΔcsrR | 2603ΔcsrS | |||
| CsrR dependent | |||||
| sag0832 | fbsB | 1.4 | −0.9 | 0.5 | Fibrinogen binding |
| sag1052 | fbsA | 4.8 | 0.7 | 1.4 | Fibrinogen binding |
| sag1127 | fib | 1.2 | 0.6 | 0.5 | Fibrinogen binding |
| sag1197 | hylB | 1.0 | 0.3 | −0.5 | Hyaluronate lyase |
| sag1236 | scpB | 1.8 | −0.1 | 0.3 | Fibronectin binding |
| sag1957 | rgfA | 2.0 | −0.1 | −0.8 | Response regulator |
| sag2063 | bibA | 2.9 | 1.3 | −0.7 | Binding to human C4-binding protein and epithelial cells |
| CsrR independent | |||||
| sag0765 | −1.1 | −1.7 | −1.5 | Penicillin-binding protein 2b | |
| sag1283 | ssaP-5 | −1.1 | −2.0 | −0.9 | Agglutinin receptor |
| sag1331 | sar5 | −2.0 | −3.0 | −1.2 | R5 protein, surface antigen |
| sag1350 | 1.2 | 1.0 | 1.3 | Unidentified surface antigen | |
| sag1462 | srr-1 | −1.7 | −2.0 | −1.7 | Serine-rich repeat family, binding to human keratin 4 and epithelial cells |
| sag1466 | glnP | −1.6 | −3.0 | −1.7 | Glutamine ABC transporter |
Locus names refer to the genome sequence of GBS strain 2603 (49).
Values represent log2 n-fold change in transcript abundance at pH 7.4 relative to that at pH 5.0.
To investigate whether pH modulation is mediated through CsrRS, we compared the expression of adherence factors in 2603ΔcsrR and 2603ΔcsrS at neutral and acidic pHs (Fig. 6 and Table 4). Exposure of strain 2603ΔcsrR to different pHs was associated with less extensive changes in gene transcription than that of the wild type. Of the 13 pH-regulated adhesin genes of strain 2603, 7 showed differential expression in 2603ΔcsrR while 6 did not (Table 4), indicating that environmental pH regulates adherence factor expression in a both CsrR-dependent and CsrR-independent manner. This result is consistent with our earlier observation that pH alteration exerted no effect on the adherence of 2603ΔcsrR to host cells and less of an effect on binding to ECM components (fibrinogen and fibronectin) in 2603ΔcsrR than in the wild type (Fig. 2 and 3B). Furthermore, our data revealed a similar trend of pH regulation in the sensor mutant, ΔcsrS, relative to that in the ΔcsrR mutant (Table 4), a result that indicates that the CsrRS system is involved but is not the exclusive pathway for pH regulation of adherence in GBS 2603.
DISCUSSION
The present investigation provides clear evidence that CsrRS plays an important role in controlling GBS adherence and biofilm formation. We found that deletion of CsrR resulted in a significant increase in the adherence of GBS strain 2603 to human epithelial cells. Adherence was reduced to a level similar to that of the wild type by repair of the csrR mutation, confirming the involvement of CsrR in the regulation of GBS adherence. These results are consistent with a previous report that showed increased adherence in a CsrRS mutant of GBS strain NEM316 (19). GBS also binds to host ECM, a macromolecular structure underlying epithelia and surrounding tissue cells, which can mediate secondary attachment to host cells. We found that mutation of CsrR led to a dramatic increase in GBS binding to the immobilized ECM components fibrinogen, fibronectin, and laminin. In addition, CsrR controls adherence to abiotic surfaces in GBS 2603, as deletion of CsrR markedly increased biofilm formation on polystyrene plates. GBS encounters various pH environments during transmission from the vagina of a colonized mother to the lungs or blood of an infected neonate. Our results revealed that such changes in environmental pH exert a significant effect on the expression of multiple adhesins and on the attachment of GBS to both epithelial cells and ECM components.
Other transcriptional regulators have been implicated in the modulation of GBS adherence, in addition to CsrR. One of the Rgg family proteins, RovS, negatively regulates the expression of fbsA in type III strain 6313. Deletion of rovS resulted in increased GBS attachment to immobilized fibrinogen and to A549 cells (34). In addition, Rga regulates adherence factors of strain NEM316 in at least two ways; i.e., it activates the expression of Srr-1 and PilA, a pilus protein, and represses expression of FbsA. Inactivation of Rga reduced GBS binding to epithelial Hep-2 cells and to immobilized human keratin 4 but increased binding to immobilized fibrinogen and A549 cells (33). The RgfA/C two-component system regulates FbsA and FbsB differentially; it inhibits the expression of FbsA but activates that of FbsB. Deletion of RgfA/C resulted in fibrinogen-binding ability lower (52 to 68%) than that of wild-type strains (1). Expression of RgfA/C is regulated by CsrRS, so some regulatory effects of CsrRS on GBS adherence may be indirect.
Several previous studies have implicated pili in GBS adherence to human epithelial cells and abiotic surfaces. We recently reported that the PI-1 pilus is overexpressed in the hyperadherent 2603ΔcsrR mutant (13). While this observation initially suggested that the PI-1 pilus mediates GBS adherence, we found that deletion of the PI-1 pilus operon, either from wild-type strain 2603 or from its isogenic ΔcsrR mutant, did not reduce GBS adherence to host cells, to ECM proteins, or to polystyrene plates (13). On the contrary, loss of PI-1 pili slightly enhanced the adhesion of GBS. It is possible that removal of pili enhances the exposure of other adherence factors on the bacterial surface to increase adherence of pilus mutants. In the present study, we investigated the role of CsrRS in the regulation of the expression of 28 known or putative adherence factors, as well as regulators implicated in their expression. By comparison of gene expression in the wild type and ΔcsrR mutants using qRT-PCR, we found that CsrR regulates the expression of multiple alternative adherence factors in strain 2603. The most highly regulated genes include those that encode ECM-binding proteins such as FbsA, FbsB, and ScpB and host cell-binding proteins such as BibA and Srr-1. The expression of these genes was increased in 2603ΔcsrR, from 18-fold for scpB to 676-fold for fbsB. These results support the hypothesis that CsrR plays a central role in regulating GBS adherence by controlling the expression of multiple adhesins. To test the generality of these findings, we also analyzed the expression of adherence factors in strain 515 and its isogenic ΔcsrR mutant. We observed a similar effect of CsrR on adherence factor expression in the 515 background (Fig. 5). Microarray analysis had suggested the increased expression of some of these factors in csrR or csrRS mutants in earlier studies (15, 19, 20); however, qRT-PCR analysis for specific adhesin genes in the present investigation provided a more comprehensive and accurate assessment of the regulation of these genes by CsrRS.
Earlier studies reported conflicting results on the effects of pH on GBS adherence. In 1979, Botta et al. found that the adherence of GBS strains (types Ia, Ib, II, and III) to human vaginal cells was greater at neutral pH than at acidic pH (3). In the same year, another study showed that the adherence of a type III GBS strain to human vaginal cells was maximal at pH 5.5 to 6.5 and declined significantly at pH values below and above this range (51). Both studies used vaginal cells directly collected from volunteers and performed adherence assays using suspensions of cells and bacteria. Using a different approach, Rubens' group studied the pH effect on GBS adherence to a cell monolayer, which was fixed with glutaraldehyde to maintain stability at acidic pH. Their results showed that the adherence of type III GBS strain COH1 to A549 and ME180 cells was greater at pH 4.0 than at neutral pH (46). These apparently contradictory results may reflect differences in the GBS strains, epithelial cells, and assay methods used in these experiments. In the present study, we pretreated GBS strains with either neutral or acidic pH for 1 h before adherence assays in order to model pH effects on the expression of adherence factors during bacterial adaptation, as well as that on bacterium-host cell interaction. Under these conditions, neutral pH promoted GBS adherence to both epithelial cells and ECM proteins. Using qRT-PCR to analyze gene transcription at pHs 5.0 and 7.4, we found that pH regulated the expression of 13 adhesin-encoding genes in strain 2603. The involvement of CsrRS in pH modulation was also studied in this investigation. We found that CsrRS is required for the regulation of some adhesin-encoding genes but not for others (Table 4), results consistent with a model in which the environmental pH regulates GBS adherence in a both CsrRS-dependent and CsrRS-independent manner. This formulation is consistent with conclusions of an earlier study on the effects of pH on global gene expression in GBS (35). That work identified some, but not all, of the pH-regulated genes described in the present investigation, a difference that may be attributable to the greater sensitivity and accuracy of the targeted qRT-PCR method used in this study than transcriptional profiling by microarray hybridization, as well as technical differences in the experimental approaches used.
Taken together, results of the present study identify the CsrRS two-component system and the environmental pH as key factors that regulate GBS adherence to cells, to ECM proteins, and to abiotic surfaces. GBS adherence is modulated by the effects of these two factors on the expression of multiple GBS surface proteins that participate in bacterial attachment. The finding that adherence is increased during growth at pH 7.4 suggests that transit of GBS from the female genital tract to potential sites of invasive infection such as amniotic fluid or the fetal lung is associated with an enhanced capacity for bacterial attachment to host tissues in these neutral-pH environments. Thus, pH regulation of adherence may promote invasive infection. For strains such as 2603, pili appear not to participate directly in bacterial attachment, but rather, they may block adhesive interactions of cell wall adhesins with their cognate binding partners on cells or host ECM components. The multiplicity of adhesin molecules produced by GBS and the complexity of adherence regulation underscore the importance of bacterial attachment in GBS interactions with the human host.
ACKNOWLEDGMENT
This work was supported in part by Public Health Service grant R01 AI59502 (M.R.W.) from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print 4 September 2012
REFERENCES
- 1. Al Safadi R, et al. 2011. Two-component system RgfA/C activates the fbsB gene encoding major fibrinogen-binding protein in highly virulent CC17 clone group B Streptococcus. PLoS One 6:e14658 doi:10.1371/journal.pone.0014658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beckmann C, Waggoner JD, Harris TO, Tamura GS, Rubens CE. 2002. Identification of novel adhesins from Group B streptococci by use of phage display reveals that C5a peptidase mediates fibronectin binding. Infect. Immun. 70:2869–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Botta GA. 1979. Hormonal and type-dependent adhesion of group B streptococci to human vaginal cells. Infect. Immun. 25:1084–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cheng Q, Stafslien D, Purushothaman SS, Cleary P. 2002. The group B streptococcal C5a peptidase is both a specific protease and an invasin. Infect. Immun. 70:2408–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dramsi S, et al. 2006. Assembly and role of pili in group B streptococci. Mol. Microbiol. 60:1401–1413 [DOI] [PubMed] [Google Scholar]
- 7. Edwards MS, Baker CJ. 2001. Group B streptococcal infections, p 1091–1156 In Remington JS, Klein JO. (ed), Infectious diseases of the fetus and newborn, 5th ed W. B. Saunders, Philadelphia, PA [Google Scholar]
- 8. Goldschmidt JCJ, Panos C. 1984. Teichoic acids of Streptococcus agalactiae: chemistry, cytotoxicity, and effect on bacterial adherence to human cells in tissue culture. Infect. Immun. 43:670–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gutekunst H, Eikmanns BJ, Reinscheid DJ. 2003. Analysis of RogB-controlled virulence mechanisms and gene repression in Streptococcus agalactiae. Infect. Immun. 71:5056–5064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gutekunst H, Eikmanns BJ, Reinscheid DJ. 2004. The novel fibrinogen-binding protein FbsB promotes Streptococcus agalactiae invasion into epithelial cells. Infect. Immun. 72:3495–3504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harris TO, Shelver DW, Bohnsack JF, Rubens CE. 2003. A novel streptococcal surface protease promotes virulence, resistance to opsonophagocytosis, and cleavage of human fibrinogen. J. Clin. Invest. 111:61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hauck CR, Ohlsen K. 2006. Sticky connections: extracellular matrix protein recognition and integrin-mediated cellular invasion by Staphylococcus aureus. Curr. Opin. Microbiol. 9:5–11 [DOI] [PubMed] [Google Scholar]
- 13. Jiang S, Park SE, Yadav P, Paoletti LC, Wessels MR. 2012. Regulation and function of pilus island 1 in group B streptococcus. J. Bacteriol. 194:2479–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jiang SM, Cieslewicz MJ, Kasper DL, Wessels MR. 2005. Regulation of virulence by a two-component system in group B Streptococcus. J. Bacteriol. 187:1105–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang SM, et al. 2008. Variation in the group B Streptococcus CsrRS regulon and effects on pathogenicity. J. Bacteriol. 190:1956–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johri AK, et al. 2007. Transcriptional and proteomic profiles of group B Streptococcus type V reveal potential adherence proteins associated with high-level invasion. Infect. Immun. 75:1473–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaur H, Kumar P, Ray P, Kaur J, Chakraborti A. 2009. Biofilm formation in clinical isolates of group B streptococci from north India. Microb. Pathog. 46:321–327 [DOI] [PubMed] [Google Scholar]
- 18. Konto-Ghiorghi Y, et al. 2009. Dual role for pilus in adherence to epithelial cells and biofilm formation in Streptococcus agalactiae. PLoS Pathog. 5:e1000422 doi:10.1371/journal.ppat.1000422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lamy MC, et al. 2004. CovS/CovR of group B streptococcus: a two-component global regulatory system involved in virulence. Mol. Microbiol. 54:1250–1268 [DOI] [PubMed] [Google Scholar]
- 20. Lembo A, et al. 2010. Regulation of CovR expression in group B Streptococcus impacts blood-brain barrier penetration. Mol. Microbiol. 77:431–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin WS, Cunneen T, Lee CY. 1994. Sequence analysis and molecular characterization of genes required for the biosynthesis of type 1 capsular polysaccharide in Staphylococcus aureus. J. Bacteriol. 176:7005–7016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maisey HC, Hensler M, Nizet V, Doran KS. 2007. Group B streptococcal pilus proteins contribute to adherence to and invasion of brain microvascular endothelial cells. J. Bacteriol. 189:1464–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mandlik A, Swierczynski A, Das A, Ton-That H. 2008. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 16:33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Margarit I, et al. 2009. Preventing bacterial infections with pilus-based vaccines: the group B streptococcus paradigm. J. Infect. Dis. 199:108–115 [DOI] [PubMed] [Google Scholar]
- 25. Mitchell TJ. 2003. The pathogenesis of streptococcal infections: from tooth decay to meningitis. Nat. Rev. Microbiol. 1:219–230 [DOI] [PubMed] [Google Scholar]
- 26. Nealon TJ, Mattingly SJ. 1984. Role of cellular lipoteichoic acids in mediating adherence of serotype III strains of group B streptococci to human embryonic, fetal, and adult epithelial cells. Infect. Immun. 43:523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nobbs AH, Lamont RJ, Jenkinson HF. 2009. Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 73:407–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Poyart C, Lamy MC, Boumaila C, Fiedler F, Trieu-Cuot P. 2001. Regulation of d-alanyl-lipoteichoic acid biosynthesis in Streptococcus agalactiae involves a novel two-component regulatory system. J. Bacteriol. 183:6324–6334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pritchard DG, Lin B, Willingham TR, Baker JR. 1994. Characterization of the group B streptococcal hyaluronate lyase. Arch. Biochem. Biophys. 315:431–437 [DOI] [PubMed] [Google Scholar]
- 30. Rinaudo CD, et al. 2010. Specific involvement of pilus type 2a in biofilm formation in group B Streptococcus. PLoS One 5:e9216 doi:10.1371/journal.pone.0009216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rosenau A, et al. 2007. Evaluation of the ability of Streptococcus agalactiae strains isolated from genital and neonatal specimens to bind to human fibrinogen and correlation with characteristics of the fbsA and fbsB genes. Infect. Immun. 75:1310–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Samen U, Eikmanns BJ, Reinscheid DJ, Borges F. 2007. The surface protein Srr-1 of Streptococcus agalactiae binds human keratin 4 and promotes adherence to epithelial HEp-2 cells. Infect. Immun. 75:5405–5414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Samen U, et al. 2011. Rga is a regulator of adherence and pilus formation in Streptococcus agalactiae. Microbiology 157(Pt 8):2319–2327 [DOI] [PubMed] [Google Scholar]
- 34. Samen UM, Eikmanns BJ, Reinscheid DJ. 2006. The transcriptional regulator RovS controls the attachment of Streptococcus agalactiae to human epithelial cells and the expression of virulence genes. Infect. Immun. 74:5625–5635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Santi I, et al. 2009. CsrRS regulates group B Streptococcus virulence gene expression in response to environmental pH: a new perspective on vaccine development. J. Bacteriol. 191:5387–5397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Santi I, et al. 2007. BibA: a novel immunogenic bacterial adhesin contributing to group B Streptococcus survival in human blood. Mol. Microbiol. 63:754–767 [DOI] [PubMed] [Google Scholar]
- 37. Schubert A, et al. 2004. The fibrinogen receptor FbsA promotes adherence of Streptococcus agalactiae to human epithelial cells. Infect. Immun. 72:6197–6205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schubert A, et al. 2002. A fibrinogen receptor from group B Streptococcus interacts with fibrinogen by repetitive units with novel ligand binding sites. Mol. Microbiol. 46:557–569 [DOI] [PubMed] [Google Scholar]
- 39. Scott JR, Zahner D. 2006. Pili with strong attachments: Gram-positive bacteria do it differently. Mol. Microbiol. 62:320–330 [DOI] [PubMed] [Google Scholar]
- 40. Seifert KN, McArthur WP, Bleiweis AS, Brady LJ. 2003. Characterization of group B streptococcal glyceraldehyde-3-phosphate dehydrogenase: surface localization, enzymatic activity, and protein-protein interactions. Can. J. Microbiol. 49:350–356 [DOI] [PubMed] [Google Scholar]
- 41. Spellerberg B. 2000. Pathogenesis of neonatal Streptococcus agalactiae infections. Microbes Infect. 2:1733–1742 [DOI] [PubMed] [Google Scholar]
- 42. Spellerberg B, Rozdzinski E, Martin S, Weber-Heynemann J, Lutticken R. 2002. rgf encodes a novel two-component signal transduction system of Streptococcus agalactiae. Infect. Immun. 70:2434–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Spellerberg B, et al. 1999. Lmb, a protein with similarities to the LraI adhesin family, mediates attachment of Streptococcus agalactiae to human laminin. Infect. Immun. 67:871–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stålhammar-Carlemalm M, Areschoug T, Larsson C, Lindahl G. 1999. The R28 protein of Streptococcus pyogenes is related to several group B streptococcal surface proteins, confers protective immunity and promotes binding to human epithelial cells. Mol. Microbiol. 33:208–219 [DOI] [PubMed] [Google Scholar]
- 45. Stålhammer-Carlemalm M, Stenberg L, Lindahl G. 1993. Protein Rib: a novel group B streptococcal cell surface protein that confers protective immunity and is expressed by most strains causing invasive infections. J. Exp. Med. 177:1593–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tamura GS, Kuypers JM, Smith S, Raff H, Rubens CE. 1994. Adherence of group B streptococci to cultured epithelial cells: roles of environmental factors and bacterial surface components. Infect. Immun. 62:2450–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tamura GS, Nittayajarn A, Schoentag DL. 2002. A glutamine transport gene, glnQ, is required for fibronectin adherence and virulence of group B streptococci. Infect. Immun. 70:2877–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tenenbaum T, Bloier C, Adam R, Reinscheid DJ, Schroten H. 2005. Adherence to and invasion of human brain microvascular endothelial cells are promoted by fibrinogen-binding protein FbsA of Streptococcus agalactiae. Infect. Immun. 73:4404–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tettelin H, et al. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. U. S. A. 99:12391–12396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van Sorge NM, et al. 2009. The group B streptococcal serine-rich repeat 1 glycoprotein mediates penetration of the blood-brain barrier. J. Infect. Dis. 199:1479–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zawaneh SM, Ayoub EM, Baer H, Cruz AC, Spellacy WN. 1979. Factors influencing adherence of group B streptococci to human vaginal epithelial cells. Infect. Immun. 26:441–447 [DOI] [PMC free article] [PubMed] [Google Scholar]