Abstract
Clinical and animal studies of coccidioidomycosis have demonstrated that activated CD4+ T lymphocytes are essential for protection against this fungal respiratory disease. We previously reported a vaccine against Coccidioides infection which contained three recombinant CD4+ T cell-reactive proteins and induced a robust, protective immune response in mice. Due to the anticipated high cost of production and clinical assessment of this multivalent vaccine, we generated a single protein which contained immunodominant T cell epitopes of the three polypeptides. Epitopes were initially identified by computational prediction of their ability to bind promiscuously to human major histocompatibility complex class II (MHC II) molecules. Cellular immunoassays confirmed the immunogenicity of the synthesized epitope peptides, while in vitro binding assays revealed a range of peptide affinity for MHC II. A DNA construct was synthesized for bacterial expression of a recombinant protein vaccine which contained five epitopes with the highest affinity for human MHC II, each fused with leader and spacer peptides proposed to optimize epitope processing and presentation to T cell receptors. Recall assays of immune T lymphocytes obtained from human MHC II-expressing HLA-DR4 transgenic mice confirmed that 4 of the 5 epitope peptides were processed. Mice immunized with the epitope-based vaccine admixed with a synthetic oligodeoxynucleotide adjuvant or loaded into yeast glucan particles and then challenged intranasally with Coccidioides showed early lung infiltration of activated T helper-1 (Th1), Th2, and Th17 cells, elevated gamma interferon (IFN-γ) and interleukin (IL)-17 production, significant reduction of fungal burden, and prolongation of survival compared to nonvaccinated mice. This is the first report of an epitope-based vaccine against coccidioidomycosis.
INTRODUCTION
Coccidioides is a desert soil-dwelling mold and causative agent of coccidioidomycosis (also known as San Joaquin Valley fever), which is a potentially life-threatening human respiratory disease (3). Two species of Coccidioides have been identified (Coccidioides immitis and C. posadasii) (14) based mainly on results of multilocus sequence typing and evidence of differential thermotolerances between the species (37). While C. immitis appears to be geographically restricted to central and southern California, C. posadasii is widely distributed in Arizona, Texas, northern Mexico, and parts of Central and South America. In spite of their genetic distinction, no discernible difference between the two species in pathogenicity is recognized. More than 10% of the current U.S. population resides in regions of coccidioidomycosis endemicity in the southwestern United States between west Texas and southern California, and more than 150,000 new coccidioidal infections are estimated to occur annually in the United States alone (4). This respiratory disease typically presents with flu-like symptoms, which in most individuals resolve spontaneously over a few days. However, the pathogen can establish a latent infection that may reactivate months to years later. Solid-organ-transplant patients who reside in the regions of endemicity and undergo immunotherapy to prevent organ rejection face a risk of reactivation of a latent infection or presentation of a new coccidioidal infection acquired from an asymptomatic donor (12). An estimated 5% of healthy, immunocompetent people with a symptomatic response to inhalation of a bolus of Coccidioides spores develop an acute, primary pulmonary infection that can subsequently convert to a life-threatening, disseminated disease. A significantly higher percentage of human immunodeficiency virus type 1 (HIV-1)-infected individuals living in regions where coccidioidomycosis is endemic are at risk of contracting the severe clinical form of this disease (36). No approved human vaccine exists against San Joaquin Valley fever or, for that matter, against any other fungal disease (7). Retrospective evidence from patient studies suggests that people who contract an acute pulmonary or disseminated Coccidioides infection and recover develop lifelong cell-mediated immunity against recurrent coccidioidomycosis. Based on this observation, together with results of protection studies with experimental animals, it has been proposed that generation of a vaccine against this respiratory mycosis is feasible.
Numerous vaccine constructs have been generated and tested in animal models of coccidioidomycosis, including killed or live, attenuated strains of the pathogen, crude immunoreactive cell wall extracts, and purified recombinant antigens (10). Particularly promising results were obtained when the concentrated total protein content of a detergent-extracted parasitic cell wall isolate of Coccidioides was used to vaccinate C57BL/6 mice against a potentially lethal intranasal (i.n.) challenge of the pathogen (47). Since a large body of evidence has indicated that T cell immunity is pivotal for a protective response, we set out in this vaccine study to identify the T cell-reactive proteins present in the protective, antigenic preparation. Our initial strategy included the identification of patient seroreactive polypeptide components of the crude detergent cell wall extract, which were separated by two-dimensional gel electrophoresis and examined by immunoblot analysis. Selected seroreactive gel-stained bands were excised, subjected to trypsin digestion, and sequenced by tandem mass spectrometry. Bioinformatic methods were employed to identify the gene that encoded each of 43 gel-excised proteins by reference to the translated Coccidioides genomic databases (42). The deduced protein sequences then underwent computational analysis using an algorithm (ProPred) (43) which identified putative T cell epitopes predicted to bind promiscuously to human major histocompatibility complex class II (MHC II) molecules. The genes which encoded 3 of the deduced proteins were selected for cloning and bacterial expression based on predictions that each of the gene products contained multiple T cell epitopes, were relatively small in molecular size, and contained signal peptides characteristic of secreted polypeptides (46). Alignment of their amino acid sequences with submitted protein sequences in the National Center for Biotechnology Information (NCBI) database (1) revealed that the three selected antigens were homologs of a previously reported fungal aspartyl protease (Pep1), alpha-mannosidase (Amn1), and phospholipidase B (Plb). T cell reactivity of synthetic peptides (20- to 32-mer) spanning all predicted promiscuous epitopes of each of the three proteins were evaluated by gamma interferon (IFN-γ)-enzyme-linked immunospot (ELISPOT) assays (46, 47). It was evident from the results of these immunoassays that certain epitope peptides induced significantly greater T cell reactivity than others. The full-length recombinant Pep1, Amn1, and Plb proteins were tested singly or as a mixture, and each was combined with a synthetic oligodeoxynucleotide adjuvant (CpG ODN) (31) to vaccinate C57BL/6 mice against coccidioidomycosis (47). The combined-protein vaccine provided significantly better protection than each of the single protein vaccines; approximately 90% of the animals survived beyond 90 days postchallenge, and the majority had cleared the pathogen from their lungs and spleen.
In spite of the positive outcome of these investigations, we realized that a major obstacle would be encountered in the application of this multivalent vaccine to human studies. The cost of preparation of each component protein to achieve necessary standards for good manufacturing practice (GMP) as well as the expense of separate toxicology studies would make the cost of the multivalent recombinant protein vaccine prohibitively exorbitant for clinical trial. We propose here an alternative approach to overcome these limitations in which the most immunogenic T cell epitope peptides of Pep1, Amn1, and Plb are incorporated into a single, epitope-based protein vaccine. In an earlier study, computational prediction of immunodominant T cell epitopes present in a protective 43-kDa glycoprotein (gp43) of Paracoccidioides brasiliensis was introduced as a novel approach to the design of a fungal vaccine (45, 48). CD4+ T cell epitope-driven vaccine constructs against microbial diseases have been shown to be immunologically potent and relatively easy to produce and have been predicted to be safe for human use (41). In this report, we describe a strategy for construction and immunological evaluation of a recombinant epitope-based vaccine (rEBV) against coccidioidomycosis and present initial results of its protective efficacy.
MATERIALS AND METHODS
Fungal strain, growth conditions, and spore preparation.
Coccidioides posadasii (C735) was the clinical isolate used in this study. The saprobic phase was grown on GYE agar (1% glucose, 0.5% yeast extract, 1.5% agar) for 3 to 4 weeks at 30°C to generate a confluent layer of spores on the surface of the culture medium. Spore suspensions used for intranasal challenge of mice as described below were prepared as previously reported (22). All culturing and preparatory procedures which involved live cells of C. posadasii were conducted in a biosafety level 3 (BSL3) laboratory certified by the Centers for Disease Control and Prevention and located at the University of Texas at San Antonio.
Mouse strain.
A breeding colony of HLA-DR4 (DRB1*0401) transgenic mice which express human MHC II (26) was used in this study. The HLA-DR4 mice were genetically engineered from a C57BL/6 background and were backcrossed to MHC II-deficient mice lacking IAβ and IEα alleles to eliminate production of endogenous murine MHC II molecules (26, 34). All mice were housed in a pathogen-free animal facility at the University of Texas at San Antonio and were handled according to the guidelines of the Institutional Animal Care and Use Committee. Mice were relocated to the CDC-certified animal BLS3 (ABSL3) laboratory before challenge with live Coccidioides spores.
Bioinformatics.
Prediction of promiscuous T cell epitope components of the three previously reported immunoreactive, recombinant proteins (Pep1, Amn1, Plb) was based on protein sequence analyses using the ProPred algorithm (http://www.imtech.res.in/raghava/propred/) (43). Examination of epitope conservation between the translated genomes of C. immitis and C. posadasii was made possible by access to the Broad Institute Coccidioides group database (http://www.broadinstitute.org/annotation/genome/coccidioides_group/MultiHome.html) and performed by protein sequence alignment using the BLASTP algorithm (1). Sequence homology between each of the T cell epitopes and human proteins was determined using the nonredundant NCBI protein database (http://www.ncbi.nlm.nih.gov/sites/gquery) as previously reported (46, 47). Search parameters were adjusted for short input sequences. The E-value for each epitope was reported. Identification of single nucleotide polymorphisms (SNP) by alignment of multiple nucleotide sequences of Coccidioides isolates in the Broad Institute database cited above was conducted using “neighborhood quality standard” (NQS) criteria (2) to control for base-calling errors and alignment artifacts as previously reported (37). Nonsynonymous coding SNP were called from sequenced isolates of Coccidioides spp. by alignment of reads with the C. immitis RS strain genome reference sequence using annotation version 3 available from the Broad Institute (37). Mapping with assembly and qualities (Maq) software was employed (30).
Assays employed for selection of T cell epitopes.
Evaluation of T cell reactivity of synthetic peptides in this study was conducted by IFN-γ ELISPOT assays essentially as previously reported (46). In the present study, T lymphocytes were initially isolated from the spleens of HLA-DR4 mice (females, 12 weeks old) which had been immunized with the three combined recombinant, full-length proteins (Pep1, Amn1, Plb) as previously reported (47) and tested for in vitro recall response to synthetic peptides representing the respective epitopes (Table 1) (46, 47). In this assay, the mice were not infected with Coccidioides. Eight of the 25 total epitopes originally identified by ProPred predictive analysis of amino acid sequences of the three immunoreactive proteins were synthesized, evaluated by IFN-γ ELISPOT assays, and shown to have the highest T cell reactivity (47) (Table 1). The second set of IFN-γ ELISPOT assays reported here were employed for further evaluation of the processing of the epitope peptides which were incorporated into the recombinant epitope-based vaccine (rEBV; see Fig. 3 and 7). The assays were conducted as previously reported (46), except that HLA-DR4 mice were immunized with the rEBV protein plus adjuvant and isolated immune T lymphocytes were separately stimulated in vitro with the epitope peptides cited above. An additional 18-mer peptide (GSIACYLHNKYDSSASST), which corresponded to a region of Pep1 not predicted to contain a T cell epitope (46), was synthesized and included in the ELISPOT assays as a negative control. Immune T cells cultured in the presence of concanavalin A (ConA; Sigma, St. Louis, MO) served as a positive control as previously reported (46).
Table 1.
Summary of data used for comparative evaluation of 8 predicted T-cell epitopes of Pep1, Amn1, and Plb antigensa
| Synthetic epitope peptide | Sequence of computationally predicted epitope peptides | ProPred binding prediction (% of alleles) | ELISPOT assay (no. of spots per well) | DELFIA (IC50 [nM])b | % sequence similarity between speciesc | Homology to human proteins (E-value)d | No. of nonsynonymous coding SNPe |
|---|---|---|---|---|---|---|---|
| Pep1-P1 | MRNSILLAATVLLGCTSAKVHL | 100 | 125 | 86.1 | 100 | 10−2 | 0 |
| Pep1-P2 | HVRALGQKYFGSLPSSQQQTV | 85 | 100 | 64.3 | 100 | 10−1 | 0 |
| Amn1-P9 | LFETTIRYLGGMISAYDLLK | 100 | 125 | 1,649.1 | 100 | 10−6 | 0 |
| Amn1-P10 | PAKVDVLLAQSLKLADVLKF | 100 | 100 | 322.8 | 100 | 10−1 | 0 |
| Amn1-P11 | NGLATTGTLVLEWTRLSDIT | 100 | 100 | 166.6 | 100 | 10−2 | 0 |
| Amn1-P13 | YYNLRPEVIESIYYAYRMTK | 92 | 50 | 1,826.7 | 100 | 10−5 | 0 |
| Plb-P2 | AIPLDSNVHIRALPNAPNGY | 90 | 175 | 2,954.4 | 95 | 10−2 | 1 |
| Plb-P6 | TPLVVYIPNYPYTTWSNIST | 90 | 125 | 815.6 | 95 | 10−3 | 1 |
The synthetic epitope peptides listed are those predicted to bind to >80% of 51 human MHC II alleles in the ProPred algorithm. The same peptides were also shown to stimulate IFN-γ production by immune CD4+ T cells (measured by ELISPOT assays) as previously reported (46, 47). The synthetic peptides indicated in bold type were ultimately selected for incorporation into an epitope-based vaccine construct.
Peptide concentrations that showed 50% inhibition of CLIP binding to HLA-DR4 transgenic mouse-derived major histocompatibility class II (MHC II) molecules in the DELFIA.
Results of alignment of peptide sequences of C. immitis and C. posadasii isolates using the Broad Institute Coccidioides Group database.
E-values of the results of an NCBI BLASTP search of peptide sequence similarities (1) with proteins in the Homo sapiens protein database.
Numbers of nonsynonymous coding single-nucleotide polymorphisms (SNP) identified for each peptide sequence (37).
Fig 3.
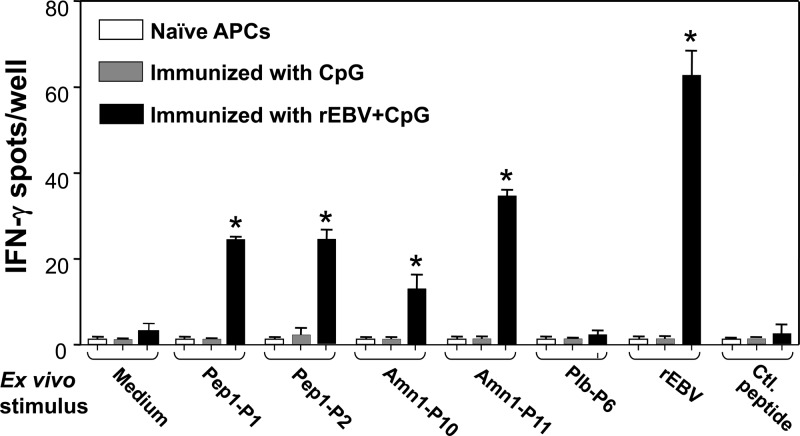
Assessment of IFN-γ production by CD4+ T cells isolated from HLA-DR4 transgenic mice conducted by IFN-γ ELISPOT assays. Mice were either vaccinated with the rEBV protein admixed with CpG or immunized with CpG alone. Asterisks indicate statistically significant differences between recall responses of rEBV-immune T cells coincubated with the indicated synthetic epitope peptide (10 μg/ml) and those of immune T cells incubated in culture medium alone. The control (Ctl.) peptide was derived from a sequence of the aspartyl protease (Pep1) protein which was not predicted to contain a T cell epitope (46). Immune T cells coincubated with the full-length rEBV protein (5 μg/ml) or ConA (not shown) served as positive controls. T cells from mice immunized with the CpG adjuvant alone served as a negative control. Naïve antigen-presenting cells (APCs) coincubated with peptides or rEBV also served as a negative control. The data presented are representative of the results of three independent experiments. Error bars indicate standard deviations.
Fig 7.
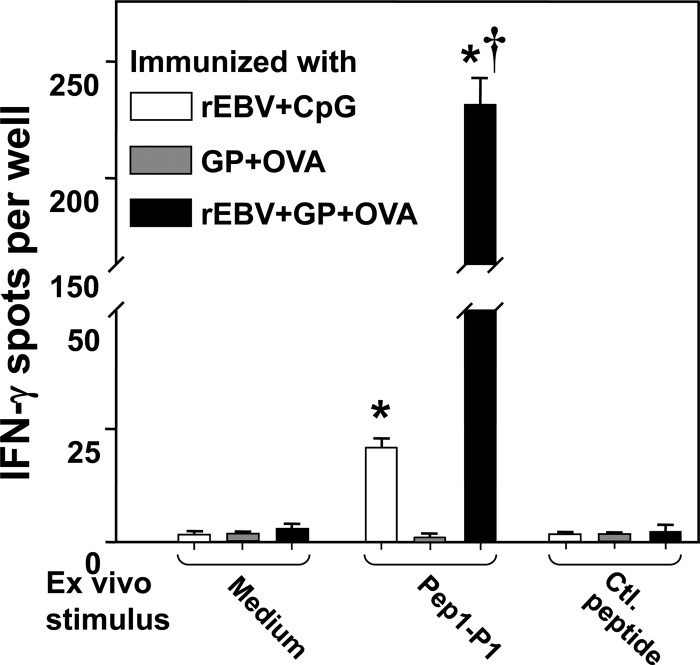
IFN-γ ELISPOT assay of immune CD4+ T cell recall response to a representative epitope peptide component (Pep1-P1; 10 μg/ml) of the recombinant vaccine protein. Transgenic mice were immunized with rEBV that was either admixed with CpG or uploaded into glucan particles. CD4+ T cells derived from mice immunized with GP plus OVA alone served as a negative control. Immune T cells incubated in growth medium alone or in the presence of the same control peptide as described for Fig. 3 also served as negative controls. Immune CD4+ T cells incubated with ConA (not shown) served as a positive control. Asterisks represent significant difference in the stimulation of immune CD4+ T cells with Pep1-P1 compared to the control peptide; the dagger represents significant difference in the stimulation of CD4+ cells derived from rEBV plus GP plus OVA-vaccinated mice versus rEBV plus CpG-vaccinated mice.
An in vitro human MHC II binding assay was also performed with each of the 8 synthetic peptides (Table 1) and served as a pivotal selection criterion for the epitope peptides that were ultimately incorporated into the epitope-based vaccine. The MHC II binding characteristics of the synthetic peptides were determined using a dissociation-enhanced lanthanide fluoroimmunoassay (DELFIA). This competitive inhibition assay was performed essentially as previously reported (15). In brief, purified HLA-DRB1*0401 (MHC II) molecules derived from a cultured HLA-DR4 fibroblast cell line were incubated with a biotinylated reference class II-associated invariant chain peptide (CLIP; residues 97 to 120) at a fixed concentration (250 nM) together with serial dilutions of each biotinylated synthetic peptide (10-fold titration between 1 nM and 100 μM). The purified MHC II molecules, CLIP, and epitope peptides were coincubated for 48 h in phosphate-buffered saline (PBS) containing 5% dimethyl sulfoxide (DMSO) and 0.05% Nonidet P-40 in the presence of a protease inhibitor mixture. The molarity of each synthetic peptide which accounted for 50% inhibition (IC50) of CLIP binding to MHC II molecules was determined by plotting the percent inhibition versus the concentration of the test peptide. The CLIP epitope has an average IC50 of approximately 500 nM. Five of the 8 synthetic peptides identified by the DELFIA to have an IC50 of less than 1,000 nM (2 times the IC50 of CLIP) were considered to bind with highest affinity to the human MHC II molecules.
Design, expression, and purification of the recombinant epitope-based vaccine.
A single, bacterium-expressed, recombinant epitope-based vaccine (rEBV) was designed to contain the 5 promiscuous, immunodominant T cell epitopes derived from Pep1, Amn1, and Plb. The specific epitopes (Pep1-P1, Pep1-P2, Amn1-P10, Amn1-P11, and Plb-P6) were selected for incorporation into the rEBV protein based upon the following criteria: (i) computational prediction of promiscuous binding to the human MHC II receptor, (ii) demonstration of the ability to be processed by antigen-presenting cells (APCs) and presented to CD4+ T cells as revealed by results of ELISPOT assays, and (iii) the ability to bind with high affinity to human MHC II molecules as demonstrated by DELFIA. The N terminus of each epitope was flanked by an Ii-Key fragment of the murine invariant chain protein (LRMKLPKS), which has been reported to enhance MHC II presentation (21), while the C termini of 4 of the 5 epitope peptides were flanked by a GPGPG spacer to avoid processing of junctional epitopes (32). The upstream 20-residue segment of the protein construct is a component of the translated plasmid expression vector (pET28b+; Novagen, Madison, WI) and includes 6 histidine residues used for nickel-affinity chromatographic purification of the rEBV as previously described (11). The nucleotide sequence designed to encode the vaccine protein was codon optimized for translation by Escherichia coli and was synthesized by EZBiolab, Inc. (Carmel, IN). The 507 bp included a 492-bp coding sequence, a stop codon, and two restriction sites (NdeI and XhoI) engineered to permit the gene to be ligated into the E. coli expression vector in the correct translation frame. The plasmid construct was used to transform E. coli strain BL21(DE3) for protein expression as previously reported (25). The recombinant protein (rEBV) was purified under denaturing conditions as previously described (31). Endotoxin was separated from the purified rEBV by passage of the protein through an ActiClean Etox affinity column (Sterogene Bioseparations, Inc., Carlsbad, CA). The amount of residual endotoxin was determined using a Limulus amebocyte lyase pyrochrome kit (Associates of Cape Cod, Inc., East Falmouth, MA). Stock solutions of rEBV contained approximately 0.003 endotoxin units per microgram of purified protein. Confirmation of the correct amino acid sequence of rEBV was performed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) in the Proteomics Center of the University of Texas at San Antonio.
Vaccination protocol, animal challenge, and evaluation of protection.
HLA-DR4 mice were immunized subcutaneously in the abdominal region with the rEBV protein plus adjuvant. For the latter, either a synthetic oligodeoxynucleotide containing unmethylated CpG dinucleotides (CpG ODN) or a preparation of rEBV-loaded yeast cell wall-derived glucan particles (GPs) was used. Details of the adjuvant composition and administration are described below. The mice were immunized with 10 μg of rEBV plus adjuvant followed by two boosts at 2-week intervals using the same amounts of vaccine protein and adjuvant preparation. Mice were challenged intranasally with a suspension of 40 to 50 viable spores of C. posadasii in 30 μl of PBS 4 weeks after completion of the immunization protocol as previously reported (46). Nonvaccinated mice were immunized with adjuvant alone. Mice were sacrificed at 9 days postchallenge for determination of the fungal burden in their lungs and spleen as previously described (22, 23). Survival studies of vaccinated versus nonvaccinated mice were conducted over 3 weeks postchallenge as previously reported (52).
Adjuvant preparations and administration.
Synthesized CpG ODN (Integrated DNA Technologies, Inc., Coralville, IA) (10 μg) was solubilized in 100 μl of incomplete Freund's adjuvant (IFA; Sigma) and admixed with the recombinant vaccine protein (rEBV) for each immunization as previously described (31). The same CpG ODN mouse-specific nucleotide sequence was employed as previously reported (11). Glucan particles derived from cell wall preparations of Saccharomyces cerevisiae (Fleischmann's baker's yeast) were prepared essentially as previously reported (20). One hundred batches of rEBV-GP antigen formulations were prepared and aliquoted in 5-dose batches containing 50 μg rEBV in volumes of 1 mg GPs/0.5 ml and stored frozen. The antigen loading process consisted of a series of hydration and lyophilization steps to absorb the antigen inside the cavity of the GPs. Dry GPs (20 mg) were swollen with 0.5 mg of the chromatographically purified rEBV solubilized in 100 μl of a 6 M urea aqueous solution and incubated for 2 h at 4°C to allow diffusion of the rEBV into the hollow GP cavity. The GPs were then frozen and lyophilized. To ensure that the antigen was predominantly inside the hollow GP cavity, the dry rEBV-GP powder was again swollen in 50 μl water to hydraulically push the antigen into the GP cavity as the particles absorbed the water. After incubation for 1 h at 4°C, the GPs were again frozen and lyophilized. A carrier protein, purified chicken ovalbumin (OVA; Worthington Biochemical Corporation, Lakewood, NJ) was used to protect the antigen and together with yeast tRNA formed a complex that trapped the rEBV inside the GP (tRNA was derived from torula yeast, type VI; Sigma-Aldrich, St. Louis, MO). The tRNA interacted with the proteins to form complexes which were too large to diffuse out through the porous glucan shell. The rEBV-GPs were swollen in 100 μl OVA (25 mg/ml in water) for 1 h at 4°C and then frozen and lyophilized. The dry rEBV-GP-OVA particles were again swollen in 50 μl water to hydraulically push the antigen and OVA into the GP cavity. After incubation for 1 h at 4°C, the GPs were frozen and lyophilized. To trap the rEBV-OVA inside the particles, the GPs were hydrated in the presence of 25 mg/ml yeast tRNA in 0.15 M NaCl and incubated at 50°C for a total of 90 min followed by a series of saline washes to remove urea, untrapped rEBV, and excess OVA and tRNA. The series of steps described above resulted in the loading of 5 μg rEBV/0.2 mg GP dose. To achieve the targeted 10 μg rEBV/0.2 mg GP dose, the rEBV-GP-OVA-tRNA formulation was lyophilized and the entire multistep loading process was repeated. The rEBV-GP-OVA-tRNA suspension (10 μg rEBV/0.2 mg GP dose) was sterilized in 70% ethanol and washed with saline solution. The concentration of GPs was determined using a hemocytometer, and 0.5-ml aliquots (each containing 5 × 108 particles) were diluted and frozen. Confirmation of the successful incorporation of rEBV into the glucan particles was conducted by SDS-PAGE using representative aliquots of treated GPs for detection of the 24-kDa vaccine protein and 43-kDa OVA.
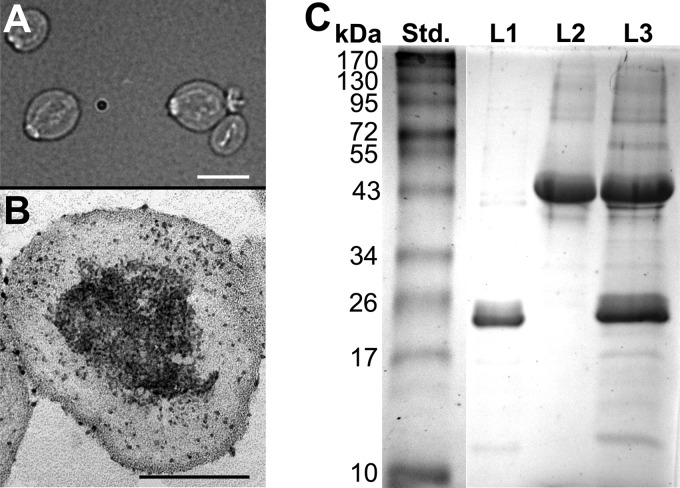
Light and electron microscopy.
Hydrated glucan particles which had been loaded with rEBV plus OVA and tRNA were observed by bright-field light microscopy. Thin sections of resin-embedded aliquots of these same glucan particles were also examined by electron microscopy. In the latter case, GPs were chemically fixed with 6% glutaraldehyde (Sigma) prepared in 0.1 M sodium cacodylate buffer for 2 h at room temperature. After fixation, the particles were stained with 0.5% uranyl acetate prepared in the same buffer for 2 h at 4°C and then stained again with 2% uranyl acetate–70% ethanol for 2 h at 4°C. After further alcohol dehydration, the GPs were embedded in Spurr's low viscosity resin (9) and thin sections of the samples were examined by transmission electron microscopy at the University of Texas Health Sciences Center.
Immunoblot and ELISA.
Immunoblot analyses of Coomassie blue-stained and unstained sister gel electrophoresis separations of the bacterium-expressed, recombinant vaccine protein were conducted using either pooled sera from patients with confirmed Coccidioides infection or sera from an equal number of healthy individuals as previously reported (46). Indirect enzyme-linked immunosorbent assays (ELISAs) of the purified rEBV protein were also conducted as previously described (31). Sera from patients with coccidioidomycosis and from healthy controls were tested for total IgG antibody reactivity with the purified rEBV protein. Patient sera (n = 48) were kindly provided by Craig Rundbaken (Valley Fever Clinic, Phoenix, AZ). Control sera (n = 15) were obtained from Innovative Research, Inc. (Novi, MI). Wells of high-binding enzyme immunoassay (EIA) microtiter plates (Corning Life Sciences, Lowell, MA) were coated overnight with 50 ng of the purified rEBV protein in 100 μl PBS. The wells were blocked with PBS containing 1% bovine serum albumin (BSA) for 2 h, washed with PBS–0.05% Tween 20, and incubated for 1 h at room temperature with human serum samples diluted 1:200 with PBS. Secondary antibody (goat anti-human IgG) conjugated with horseradish peroxidase (Southern Biotech, Birmingham, AL) and diluted 1:4,000 in 100 μl PBS was added to the wells and incubated for 1 h as described above. After a final washing step, bound antibody was detected in the wells by addition of 100 μl of SureBlue TMB peroxidase substrate (KPL, Gaithersburg, MD), and the absorbance was measured spectroscopically at a wavelength of 450 nm. The assays of patient and control sera were conducted in triplicate.
In vitro assays of cytokine secretion by immune splenocytes.
At 7 days postchallenge, spleens of nonvaccinated HLA-DR4 mice immunized with adjuvant alone (CpG or GP plus OVA) or of mice vaccinated with rEBV plus CpG or rEBV plus GP plus OVA (3 animals per group) were separately harvested, pooled, and macerated for isolation of immune splenocytes as previously reported (52). The cells were cultured in 24-well plates (8 × 106 cells/well) in 2 ml RPMI 1640 containing 10% heat-inactivated fetal bovine serum (FBS), 1% gentamicin, and 1% l-glutamine to which 1 μg/ml or 10 μg/ml of either purified OVA or the rEBV protein was added. Examination of in vitro stimulation was conducted by incubation of the immune splenocytes in growth medium alone or with the addition of either OVA or the vaccine protein for 48 h at 37°C in the presence of 5% CO2. Splenocytes incubated in growth medium alone or growth medium plus OVA served as negative controls. After incubation, a cocktail of protease inhibitors (cOmplete, EDTA-free; Roche Diagnostics, Pleasanton, CA) was added to each well and culture supernatants were collected from the centrifuged samples (11,950 × g, 10 min at 4°C). The supernatants were stored at −80°C until they were subjected to cytokine analysis. The concentration of selected cytokines was determined using a Bio-Plex suspension array system (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer's instructions. Three separate splenocyte preparations and cytokine assays were performed for each of the four groups of mice listed above.
FACS analysis and intracellular cytokine staining of pulmonary leukocytes.
Total pulmonary leukocytes were isolated from nonvaccinated HLA-DR4 mice immunized with adjuvant alone (CpG or GP plus OVA) or mice vaccinated with either rEBV plus CpG or rEBV plus GP plus OVA and sacrificed at 9 days postchallenge (4 mice per group) as previously reported (22). Pulmonary leukocytes were also isolated from naïve (untreated) mice. The total number of viable pulmonary leukocytes from each lung sample was visualized by the trypan blue exclusion test and quantified by light microscopy using a hemocytometer. Standard methodology was employed for direct monoclonal antibody (MAb) labeling and enumeration of selected pulmonary immune T cell phenotypes by fluorescence-activated cell sorter (FACS) analysis with a FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ) as previously described (22). In brief, isolated pulmonary leukocytes of each group of mice were separately transferred to RPMI medium containing 10% heat-inactivated FBS for cell sorting analysis. Leukocytes from nonvaccinated, vaccinated, and naïve mice (each in a volume of approximately 0.5 × 106 cells/ml) were aliquoted to a 96-well plate and incubated with appropriate fluorochrome-conjugated MAbs for determination of relative numbers of activated, lung-infiltrated T cells as previously reported (22). The expression level of CD44, an adhesion molecule that binds to hyaluronic acid, is elevated in activated T cells and was used as an activation marker (22). Absolute numbers of each selected T cell subpopulation were determined by multiplying the percentage of each gated population by the total number of viable pulmonary leukocytes derived from hemocytometer counts as described above. Intracellular cytokine staining (ICS) was performed by fluorocytometry to identify subpopulations of specific cytokine-producing T cells as previously reported (22). Permeabilized leukocytes were stained with a cocktail of fluorochrome-conjugated anti-IFN-γ, anti-interleukin-5 (IL-5), or anti-IL-17A. The percentage of specific cytokine-producing CD4+ T cells at 9 days postchallenge relative to total numbers of activated CD4+ T cells which had infiltrated the lungs was determined by analysis of FACS data using the FlowJO software package (Tree Star, Inc., Ashland, OR). Pulmonary leukocytes obtained from age- and gender-matched naïve (untreated) mice were included in these studies for determination of baseline absolute numbers and percentages of selected T cell phenotypes. Three separate pulmonary leukocyte isolations and FACS analyses were performed for each of the five groups of mice listed above.
Histopathology.
Comparative histopathology analysis was conducted with excised lungs from Coccidioides-infected, nonvaccinated HLA-DR4 mice immunized with the GP plus OVA adjuvant alone and from infected mice vaccinated with rEBV plus GP plus OVA. The animals (3 mice per group) were sacrificed at 9 days postchallenge as previously reported (52). The lungs of age-matched naïve mice were included for tissue comparison. Fixation of lung tissue and embedding procedures were performed as described previously (52), and tissue sections (5 μm thick) were stained with hematoxylin and eosin (H&E) by a standard procedure. Paraffin sections were examined using a Leica DMI6000 microscope equipped with an automated TurboScan stage (Objective Imaging, Ltd., Cambridge, United Kingdom), and microscope images of infected lung tissue were acquired and analyzed using Surveyor software (Objective Imaging) as previously reported (23).
Statistical analyses.
The Student t test was used to compare differences with respect to ELISA results, ELISPOT assays, cytokine concentrations, calculations of absolute numbers of lung-infiltrated immune cells, and percentages of specific cytokine-producing T cells as previously reported (22). The Mann-Whitney U test was used to determine differences between the fungal burdens of nonvaccinated and vaccinated mice measured as CFU as previously reported (52). Survival data were examined by the Kaplan-Meier test using log rank analysis to compare survival plots as reported previously (52). A P value of <0.05 was considered statistically significant.
Nucleotide sequence accession numbers.
The genomic and cDNA sequences of the PEP1, Amn1, and Plb genes which encode the previously reported T cell-reactive proteins of C. posadasii were deposited in GenBank (accession no. DQ164306, DQ176863, and DQ188099, respectively). The sequence of the synthesized gene encoding the recombinant, epitope-based vaccine protein (rEBV) that was ligated into the pET28b+ expression vector has also been deposited in GenBank (accession no. JX103829).
RESULTS
Combined prediction and validation permitted selection of candidate T cell epitopes.
The T cell epitopes which were incorporated into our experimental vaccine were initially predicted by computational analyses of amino acid sequences of three previously reported immunoreactive proteins (40) and validated here by a set of selection criteria. This study was an extension of our previously reported investigations of the T cell reactivity and protective efficacy of the three recombinant proteins, Pep1, Amn1, and Plb, predicted on the basis of ProPred analyses to contain a total of 25 human MHC II-binding epitopes (46, 47). Synthetic peptides (20- to 22-mer) which corresponded to 8 of these deduced epitope peptides were selected on the basis of their having been predicted to bind to a minimum of 80% of the MHC II molecules expressed by the 51 representative HLA-DR alleles included in the ProPred algorithm (Table 1). These 8 synthetic peptides were first evaluated by IFN-γ ELISPOT assays to validate in vitro epitope processing and presentation to immune CD4+ T cells. The immune T lymphocytes were derived from HLA-DR4 mice which had been vaccinated with the combined three full-length proteins. The results indicated that each of the 8 computationally selected epitope peptides was recognized by immune T cells but that the peptides elicited different levels of in vitro lymphocyte activation and cytokine response (Table 1). As a second selection criterion, each of the 8 T cell-reactive peptides was tested in a competitive binding assay (DELFIA) to determine their relative affinities for human MHC II molecules. The results revealed that, of the 8 synthetic peptides, 3 (previously designated Amn1-P9, Amn1-P13, and Plb-P2 [46, 47]) had relatively low affinity for MHC II complexes in this competitive inhibition assay compared to the other 5 peptides (Table 1). Bioinformatic methods were applied for further selection of T cell epitopes. Results of amino acid sequence alignment of the 8 peptides with annotated genomes of C. immitis and C. posadasii performed using the BLASTp algorithm revealed conservation of residues between the two species except for Plb-P2 and Plb-P6 (Table 1). The single amino acid substitution at residue 7 in Plb-P2 (N to D) resulted in reduction of the ProPred-predicted MHC II-binding promiscuity of this peptide, while the substitution at residue 15 of Plb-P6 (W to F) did not. The amino acid and nucleotide sequences of the peptides were also examined for homology with human proteins and occurrences of nonsynonymous single nucleotide polymorphisms (SNPs), respectively (Table 1). A BLASTp search of the Homo sapiens protein database using the 8 peptides as probes revealed significantly higher human protein homology with both Amn1-P9 and Amn1-P13 than with the other epitopes. SNP calling from the multiple alignment analysis revealed single nonsynonymous coding polymorphisms in the nucleotide sequences of Plb-P2 and Plb-P6 (Table 1), which in the case of Plb-6 did not change the amino acid sequences of the peptide due to degeneracy of the genetic code. On the basis of these combined assays of the original 8 ProPred-predicted promiscuous epitopes, the 5 (Pep1-P1, Pep1-P2, Amn1-P10, Amn1-P11, and Plb-P6) which best fulfilled our selection criteria for vaccine candidacy were identified.
The epitope-based vaccine construct.
The amino acid sequence of the recombinant epitope-based vaccine (rEBV) was designed to include the 5 selected epitope peptides described above as well as N-terminal leader peptides (fragment of the invariant chain) (16, 28) and glycine/proline spacer sequences (GPGPG) (32) for enhanced processing and presentation to MHC II complexes (Fig. 1A). The 20-residue N-terminal segment of the construct was designed to accommodate the requirements for optimal E. coli expression and purification of the recombinant vaccine protein as described above in Materials and Methods.
Fig 1.
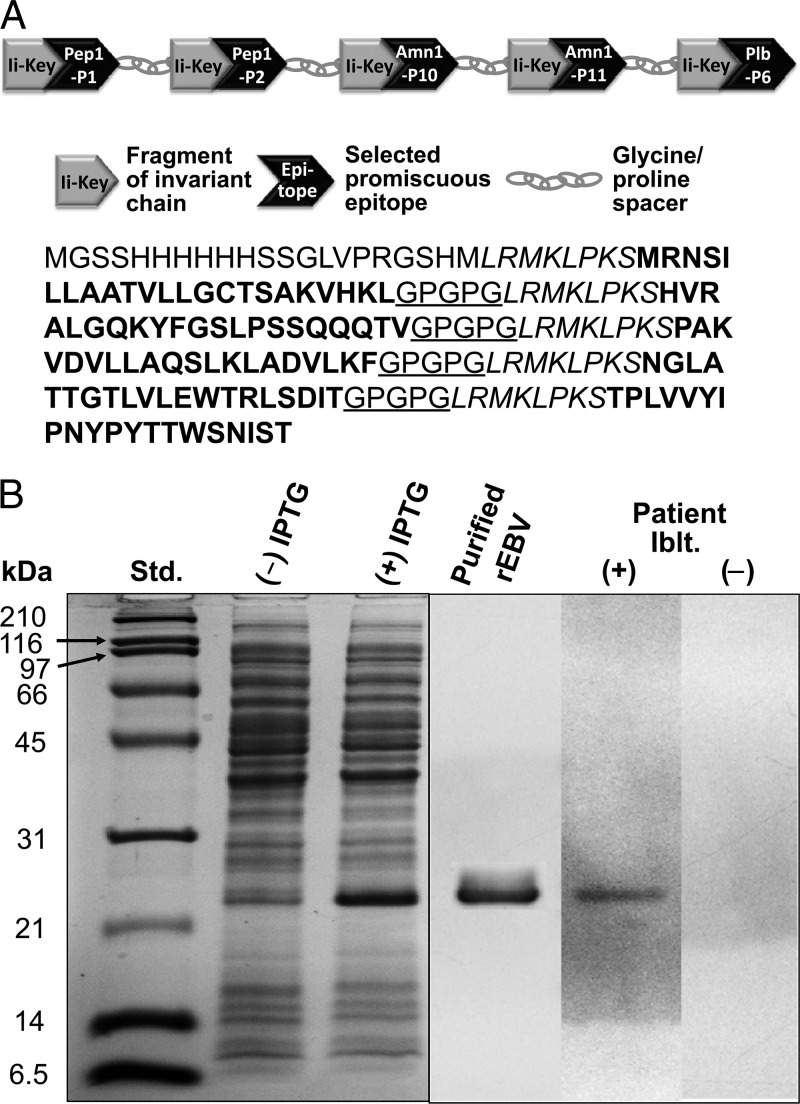
Schematic and amino acid sequence of the epitope-based protein vaccine (A) and its expression, purification, and patient seroreactivity (B). (A) The five selected promiscuous epitope peptides are indicated in bold type and identified according to the convention previously reported (i.e., Pep1-P1, Pep1-P2, etc.) (46, 47). The Ii-Key fragment of the murine invariant chain, indicated in italics, is located at the N terminus of each epitope and includes the core (LRMK) plus linker sequence (LPKS). The underlined glycine/proline spacer sequence (GPGPG) is located at the C terminus of each epitope, except for Plb-P6. The 21-residue N-terminal sequence of the vaccine protein is derived from the translated pET28b+ plasmid expression vector and includes a histidine motif for nickel-affinity purification of the E. coli-expressed recombinant protein. (B) SDS-PAGE separation and immunoblot (Iblt.) of the E. coli-expressed recombinant epitope-based vaccine (rEBV) protein. Standards (Std.) and lysates of E. coli transformed with the pET28b-EBV plasmid vector construct in the presence (+) or absence (−) of isopropyl β-d-thiogalactopyranoside (IPTG) and the nickel affinity-purified rEBV are shown. The immunoblot of rEBV was incubated with either pooled sera from patients with confirmed Coccidioides infection (+) or pooled sera from healthy individuals (−).
The purified recombinant protein vaccine was recognized by patient antibody.
The synthesized 507-bp DNA which encoded the recombinant epitope-based vaccine protein was expressed by E. coli transformed with a pET-28b-EBV plasmid construct. The predicted molecular size of the recombinant protein was 20 kDa but in SDS-PAGE gels appeared as an approximately 24-kDa polypeptide (Fig. 1B). This difference in size was probably due to the high proline content of rEBV, which has been shown to result in anomalous estimates of the molecular size of proteins based on electrophoresis gel separations (40). The chromatographically purified recombinant protein also sometimes appeared as a dimer in electrophoresis gels (not shown in Fig. 1B). Results of amino acid sequence analysis of both the monomeric and dimeric forms of rEBV were in agreement with the predicted sequence of the vaccine protein. Immunoblot assays (Iblt.) of the purified rEBV conducted with pooled sera from either patients with confirmed coccidioidal disease (+) or healthy volunteers (−) showed that only antibody from Coccidioides-infected individuals recognized the recombinant protein. We also performed ELISAs to test the range of reactivity of patient sera with the rEBV protein (Fig. 2). Sera from all 48 individual patients diagnosed with pulmonary Coccidioides infection recognized the recombinant protein, and the median absorbance determined as total bound IgG-specific antibody was 0.20 compared to 0.09 for the control sera (P < 0.001). The patient outliers with high absorbance values were individuals diagnosed with disseminated coccidioidomycosis.
Fig 2.
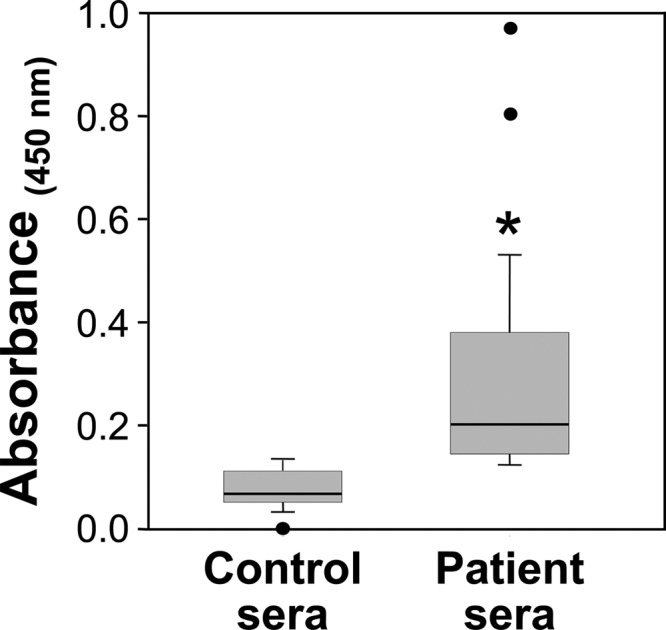
Results of ELISA of human control sera (n = 15) and sera from patients with confirmed coccidioidal infection (n = 48), each reacted with the purified rEBV protein adsorbed to the wells of microtiter plates (50 ng/well). The data are presented as box plots which indicate the 25th and 75th percentiles. Bars above and below the boxes indicate the 90th and 10th percentiles, respectively. The mean absorbance at 450 nm is indicated by a horizontal line within the boxes. Outliers are indicated as solid circles. The asterisk indicates a statistically significant difference between the mean absorbance values of control and patient sera.
Immune CD4+ T cells isolated from mice vaccinated with rEBV plus CpG exhibited in vitro recall response to synthetic peptide epitopes.
Synthetic peptides corresponding to the 5 selected epitopes incorporated into the vaccine construct were used separately to test the in vitro response of immune CD4+ T cells derived from HLA-DR4 mice vaccinated with rEBV plus CpG (Fig. 3). IFN-γ ELISPOT assays of each peptide (10 μg/ml) and full-length rEBV (5 μg/ml) were compared to that of a control peptide (10 μg/ml) not predicted to be T cell reactive. Immune CD4+ T cells showed significant response to in vitro stimulation with Pep1-P1, Pep1-P2, Amn1-P10, Amn1-P11, and the full-length rEBV compared to the control peptide. However, in vitro stimulation of immune T cells with Plb-P6 failed to elicit a response. No statistically significant difference was observed between responses of immune CD4+ T cells cultured in growth medium alone and those of T cells cocultured with the control peptide (P = 0.87). Immune CD4+ T cells cultured in the presence of ConA (0.5 μg/ml) generated a significant immune response (>100 spots/well; not shown).
Splenocytes isolated from infected mice vaccinated with rEBV plus CpG secreted protective cytokines upon in vitro stimulation with the recombinant vaccine protein.
An in vitro recall assay was conducted (Table 2) to determine whether immune splenocytes isolated from mice immunized with rEBV plus CpG at 7 days after challenge could be induced to secrete an array of cytokines that correlated with a protective immune response to Coccidioides infection as previously defined in a study of vaccinated C57BL/6 mice (22). Representative proinflammatory, regulatory, and T helper (Th)-type cytokines were all secreted at significantly higher concentrations by in vitro-stimulated immune cells isolated from vaccinated than by those isolated from nonvaccinated mice. The concentration of IL-6 in supernatants of rEBV-stimulated splenocytes showed an 8-fold increase upon exposure to the vaccine protein at 1 μg/ml, while that of IL-17A revealed a 14-fold increase upon coincubation with 10 μg/ml of rEBV. A modest 3- to 4-fold increase in the concentration of IL-10 was recorded in supernatants of in vitro-stimulated immune splenocytes derived from mice vaccinated with rEBV plus CpG compared to nonvaccinated mice. Cytokines representative of a Th1-type response (IL-2 and IFN-γ) showed concentration increases of up to 43- and 50-fold, respectively, in supernatants of immune cells isolated from vaccinated compared to nonvaccinated mice. The concentrations of secreted Th2-type cytokines (IL-4, IL-5) revealed less-striking increases compared to those of the Th1-type cytokines (up to 9- and 19-fold increases in concentration, respectively) upon stimulation of immune splenocytes isolated from mice vaccinated with rEBV plus CpG.
Table 2.
Concentrations (pg/ml) of cytokines (pg/ml) secreted in vitro by rEBV-stimulated immune splenocytes isolated from nonvaccinated mice versus mice vaccinated with rEBV plus CpG and sacrificed at 7 days postchallengea
| Secreted cytokine |
In vitro stimulation of immune splenocytes from mice in indicated group |
|||||
|---|---|---|---|---|---|---|
| Growth medium alone |
rEBV |
|||||
| 1 μg/ml |
10 μg/ml |
|||||
| Nonvaccinated | Vaccinated | Nonvaccinated | Vaccinated | Nonvaccinated | Vaccinated | |
| Proinflammatory | ||||||
| IL-6 | 5.9 ± 2.9 | 44.2 ± 2.9* | 281.3 ± 7.5 | 2,225.0 ± 130.9* | 814.3 ± 49.2 | 2,042.0 ± 152.7* |
| IL-17A | 4.9 ± 0.4 | 23.8 ± 2.2* | 28.2 ± 0.9 | 273.5 ± 13.6* | 23.2 ± 4.8 | 325.9 ± 13.2* |
| Regulatory | ||||||
| IL-10 | 9.0 ± 0.8 | 34.8 ± 3.6* | 55.4 ± 4.1 | 233.8 ± 14.9* | 221.6 ± 10.9 | 769.9 ± 18.6* |
| Th1 type | ||||||
| IL-2 | 97.0 ± 8.6 | 176.4 ± 10.3* | 185.4 ± 5.6 | 2,101.3 ± 45.6* | 97.2 ± 2.9 | 4,195.6 ± 142.5* |
| IFN-γ | 7.8 ± 2.0 | 8.6 ± 1.6 | 60.5 ± 7.8 | 3,035.0 ± 95.7* | 71.4 ± 6.6 | 2,531.0 ± 188.8* |
| Th2 type | ||||||
| IL-4 | 4.5 ± 0.7 | 13.0 ± 0.2* | 5.4 ± 0.2 | 15.3 ± 0.9* | 3.6 ± 0.1 | 34.0 ± 0.2* |
| IL-5 | 2.1 ± 0.6 | 47.5 ± 0.9* | 10.5 ± 1.4 | 214.5 ± 6.8* | 9.8 ± 0.8 | 166.3 ± 7.5* |
rEBV, recombinant vaccine protein was added to immune cells in growth medium and the mixture was incubated for 48 h. Nonvaccinated mice were immunized with CpG alone. Asterisks indicate a significant difference between cytokine concentrations (P < 0.05) in culture supernatants of in vitro stimulated immune splenocytes derived from nonvaccinated mice versus mice vaccinated with rEBV plus CpG.
Activated T cells infiltrated infected lungs of mice vaccinated with rEBV plus CpG in significantly higher numbers than lungs of nonvaccinated mice.
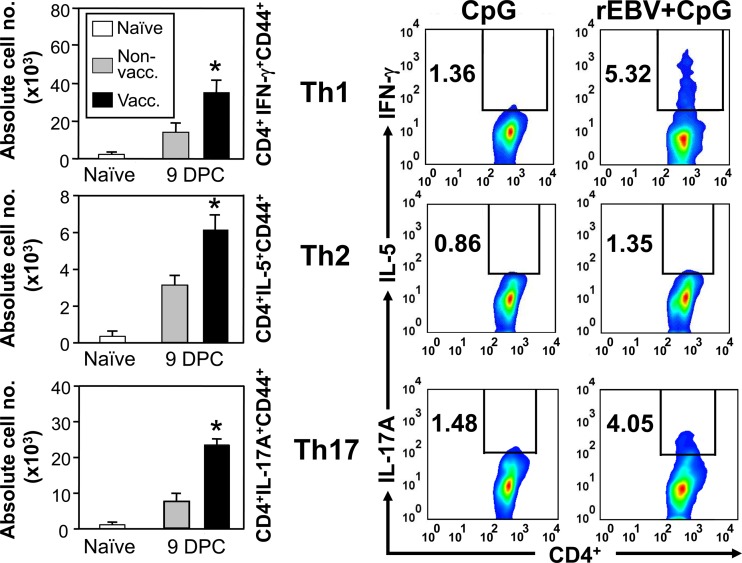
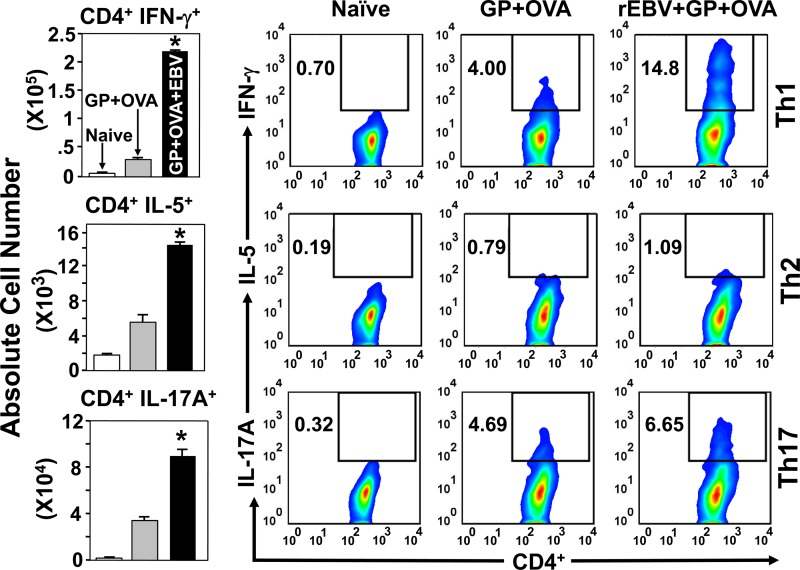
We determined both the absolute numbers and percentages of representative Th1, Th2, and Th17 CD4+ T cells that had infiltrated the lungs of mice vaccinated with rEBV plus CpG and nonvaccinated mice at 9 days postchallenge (Fig. 4). FACS/ICS methods were used to gate for IFN-γ-, IL-5-, and IL-17A-producing subsets of activated CD4+ T cells. The absolute numbers of CD4+ IFN-γ+ CD44+ T cells detected in the infected lungs at 9 days after challenge were approximately 2-fold higher in vaccinated than in nonvaccinated mice. Although fewer CD4+ IL-5+ CD44+ T cells had infiltrated the lungs of both groups of mice at 9 days postchallenge than representative Th1 cells, the absolute numbers of these Th2 cells were approximately 3-fold higher in vaccinated than in nonvaccinated mice. Activated CD4+ IL-17A+ CD44+ T cells also infiltrated the lungs of mice vaccinated with rEBV plus CpG in significantly higher numbers than those of nonvaccinated mice (approximately 3-fold) at 9 days postchallenge. Intracellular staining combined with flow cytometry was employed to examine the percentages of the same T cell phenotypes relative to the total activated CD4+ T cells which had infiltrated the infected lungs after 9 days (Fig. 4). In mice vaccinated with rEBV plus CpG, the percentages of representative Th1 and Th17 cells were markedly higher than the percentage of Th2 cells. In addition, the percentage of IFN-γ-producing Th1 cells was consistently higher than that of IL-17A-producing Th17 cells at 9 days postchallenge. The lung homogenates of nonvaccinated, infected mice revealed significantly lower percentages of the three T cell phenotypes than those of vaccinated mice.
Fig 4.
FACS analysis of IFN-γ-, IL-5-, and IL-17A-expressing Th1, Th2, and Th17 cells, respectively, in lung homogenates derived from mice vaccinated with rEBV plus CpG compared to those derived from nonvaccinated transgenic mice. The latter were immunized with the CpG adjuvant alone. Mice were sacrificed at 9 days postchallenge (DPC). Naïve (untreated) mice were included for determination of the baseline of numbers of corresponding lung-associated immune cells. The absolute numbers of lung-infiltrated T helper cell phenotypes are shown as bar graphs. Asterisks indicate significantly higher absolute numbers of the responsive T cell phenotypes in lungs of vaccinated than in nonvaccinated mice. Error bars indicate standard deviations. Percentages of gated, specific cytokine-producing T helper cells per lung were determined by intracellular cytokine staining and are indicated as insets in each panel. The results reported are representative of three independent experiments.
Mice vaccinated with rEBV plus CpG revealed a significant reduction of fungal burden but marginally increased survival compared to nonvaccinated mice.
Transgenic mice were challenged intranasally with a potentially lethal inoculum of C. posadasii spores (approximately 40 to 50 viable cells). Mice vaccinated by the subcutaneous route with rEBV plus CpG showed a significant reduction of CFU in their lung homogenates (median CFU = 6.5 log10) compared to nonvaccinated mice (median = 7.7 log10) at 9 days postchallenge (P < 0.001; data not shown). A trend was also evident indicating that the numbers of CFU in the spleens of vaccinated mice were lower but not significantly different from the numbers of CFU detected in the spleens of nonvaccinated mice after 9 days. Survival studies were conducted over a period of 3 weeks postchallenge. Control mice immunized with CpG alone and inoculated intranasally showed a steep mortality plot between 13 and 16 days postchallenge (Fig. 5). Mice vaccinated with rEBV plus CpG, on the other hand, revealed an increase in survival between 14 and 20 days after challenge, but this difference was not statistically significant compared to the results seen with the nonvaccinated mice (P = 0.05). These results are representative of three separate fungal burden and survival studies conducted under the same conditions.
Fig 5.
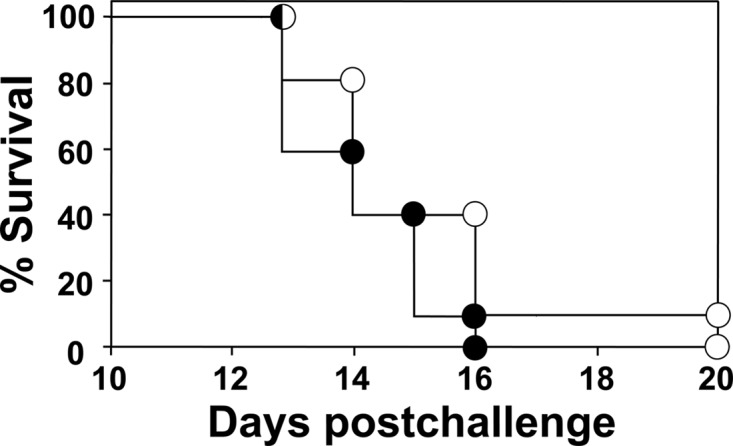
HLA-DR4 transgenic mice vaccinated with rEBV plus CpG (open circles) showed a trend to increased survival between 14 and 20 days after intranasal challenge with 40 spores of C. posadasii compared to nonvaccinated mice immunized with CpG alone (closed circles), but the difference between the survival plots was not statistically significant (P = 0.05). The results are representative of three separate experiments.
Immunization of transgenic mice with the recombinant epitope vaccine plus CpG induced a pattern of immunological response which correlated with protection against coccidioidomycosis in the mouse model (22, 23, 50). However, results of survival studies indicated that durable host protection was not achieved. We speculated that our choice of adjuvant may have at least partly accounted for the limited success of the recombinant protein vaccination protocol. Specifically, we proposed that immunization with the epitope-based vaccine plus CpG ODN may not be optimal for promotion of the Th1 and Th17 signal pathways, which have been reported to be essential for vaccine immunity to Coccidioides infection (22, 50).
Glucan particles were evaluated as an alternate adjuvant and antigen delivery system.
We chose to evaluate the application of glucan particles to our immunization procedure as a replacement of the CpG ODN adjuvant, since vaccination with GPs has recently been reported to combine enhanced cellular immune response with efficient antigen delivery to the host (19, 20). Hydrated GPs are fairly uniform in size (approximately 4 μm diameter) and small enough to be engulfed by host phagocytes (Fig. 6A). Thin sections of particles loaded with rEBV plus OVA plus tRNA and stained with uranyl acetate revealed that most of the protein/nucleic acid complex is contained within the hollow cavity of the GPs (Fig. 6B). Although uranyl acetate preferentially binds to nucleic acids, uranyl salts also bind to both negatively and positively charged amino acids (18). The uranyl acetate-stained GPs revealed deposits of electron-dense material primarily in the hollow center of the particles. Protein separation of the antigen-loaded GPs by gel electrophoresis confirmed that both rEBV (24 kDa) and OVA (43 kDa) had been encapsulated in the hydrated particles and that each immunization with 1 × 108 glucan particles contained approximately 10 μg of rEBV as predicted (Fig. 6C).
Fig 6.
Light and electron microscopic images of hydrated glucan particles which had been loaded with the vaccine protein (rEBV) plus OVA and tRNA are shown in panels A and B, respectively. The thin-sectioned glucan particle (GP) shown in panel B was chemically fixed and stained with uranyl acetate. (C) The protein content of GPs loaded with OVA versus rEBV plus OVA was revealed by SDS-PAGE as shown in lanes L2 and L3, respectively. Lane L1 contained 5 μg of purified rEBV represented by the 24-kDa Coomassie-stained band. The optical density of the band with matching molecular size in lane L3 was approximately twice that of the rEBV band in lane L1. The bars in panels A and B represent 4 μm and 2 μm, respectively.
IFN-γ ELISPOT assays revealed a significantly enhanced recall response of CD4+ T cells from mice immunized with antigen-loaded GPs compared to rEBV plus CpG.
CD4+ T cells isolated from HLA-DR4 mice immunized with GPs which had been loaded with 10 μg of the vaccine protein (rEBV plus GP plus OVA) revealed an approximately 10-fold-higher recall response to a representative synthetic epitope peptide (Pep1-P1) than immune T lymphocytes obtained from mice vaccinated with rEBV plus CpG (cf. Fig. 3 and 7). Both sources of immune T cells showed a higher in vitro response to Pep1-P1 than to the control peptide, and CD4+ cells isolated from mice immunized with CpG or GP plus OVA alone showed minimal in vitro recall responses.
Splenocytes isolated from mice vaccinated with rEBV plus GP plus OVA revealed enhanced secretion of IL-17A and IFN-γ upon in vitro stimulation.
Results of the in vitro assay summarized in Table 3 were obtained under the same conditions as our evaluation of the recall response of immune splenocytes isolated from mice vaccinated with rEBV plus CpG and from nonvaccinated, infected mice reported in Table 2. Concentrations of the same set of cytokines in Table 3 were examined after in vitro stimulation with either OVA or rEBV. Immune splenocytes isolated from vaccinated mice responded to the presence of rEBV but not OVA during incubation, indicating that OVA in the GP preparation did not significantly contribute to the priming of host immune cells following vaccination and Coccidioides infection. A marked increase in secretion of IL-6, IL-17A, IL-2, and IFN-γ was observed in splenocyte supernatants isolated from mice vaccinated with the preparation of glucan particles containing rEBV compared to nonvaccinated mice upon in vitro stimulation with rEBV. This same trend was reported in Table 2. However, immune splenocytes obtained from infected mice vaccinated with rEBV plus GP plus OVA secreted markedly higher concentrations of IL-17A and IFN-γ but lower concentrations of IL-2 upon in vitro stimulation with rEBV than immune splenocytes obtained from mice vaccinated with rEBV plus CpG. In both assays, only a modest increase in IL-10 production in the supernatants of immune splenocytes stimulated with rEBV was recorded. A surprising observation was the elevated production of IL-6 and IFN-γ by splenocytes isolated from infected control mice immunized with GP plus OVA and stimulated in vitro with rEBV (Table 3). We suggest that the β-glucan component of GPs contributed to nonspecific inflammatory cell activation, which could explain the high levels of production of these two cytokines. IFN-γ production in this case could also have been a dominant recall response of activated T cells which recognized epitopes of the rEBV protein presented during Coccidioides infection.
Table 3.
Concentrations (pg/ml) of cytokines secreted in vitro by rEBV- versus OVA-stimulated immune splenocytes isolated from nonvaccinated mice versus mice vaccinated with rEBV plus GP plus OVA and sacrificed at 7 days postchallengea
| Secreted cytokine |
In vitro stimulation of immune splenocytes from nonvaccinated and vaccinated mice |
|||||||
|---|---|---|---|---|---|---|---|---|
| OVA (1 μg/ml) |
rEBV (1 μg/ml) |
OVA (10 μg/ml) |
rEBV (10 μg/ml) |
|||||
| Nonvaccinated | Vaccinated | Nonvaccinated | Vaccinated | Nonvaccinated | Vaccinated | Nonvaccinated | Vaccinated | |
| Proinflammatory | ||||||||
| IL-6 | 110.3 ± 7.1 | 74.5 ± 2.6 | 1,614.1 ± 54.9 | 3,391.6 ± 141.5* | 192.4 ± 9.6 | 97.1 ± 3.3 | 2,004.2 ± 88.3 | 2,925.2 ± 213* |
| IL-17A | 404.9 ± 41.7 | 160.7 ± 2.8 | 258.5 ± 13.2 | 1,317.1 ± 73.6* | 974.6 ± 21.8 | 331.4 ± 29.9 | 153.4 ± 6.6 | 1,460.7 ± 99.3* |
| Regulatory | ||||||||
| IL-10 | 26.8 ± 2.3 | 29.4 ± 1.5 | 102.1 ± 1.3 | 190.9 ± 16.9* | 36.5 ± 2.1 | 30.8 ± 1.9 | 416.2 ± 6.7 | 803.1 ± 10.1* |
| Th1 type | ||||||||
| IL-2 | 270.6 ± 11.0 | 231.7 ± 3.5 | 32.0 ± 2.5 | 450.4 ± 11.3* | 462.6 ± 20.0 | 270.6 ± 1.3 | 32.9 ± 2.4 | 553.4 ± 28.9* |
| IFN-γ | 91.5 ± 6.6 | 19.3 ± 3.1 | 2,138.1 ± 189.6 | 4,880.4 ± 698.5* | 217.2 ± 8.4 | 23.6 ± 4.5 | 1,853.5 ± 340 | 4,163.7 ± 278* |
| Th2 Type | ||||||||
| IL-4 | 28.3 ± 0.7 | 12.7 ± 0.1 | 9.0 ± 0.3 | 45.2 ± 1.9* | 45.0 ± 2.2 | 15.0 ± 0.4 | 4.0 ± 0.1 | 58.3 ± 1.2* |
| IL-5 | 127.3 ± 4.2 | 44.2 ± 1.2 | 18.3 ± 1.6 | 176.0 ± 5.7* | 208.1 ± 13.3 | 74.3 ± 0.9 | 14.9 ± 1.4 | 161.4 ± 6.2* |
OVA or rEBV protein was added to immune cells in growth medium and the mixture was incubated for 48 h. Nonvaccinated mice were immunized with GP plus OVA alone. Asterisks indicate a significant difference between cytokine concentrations (P < 0.05) in culture supernatants of rEBV-stimulated immune splenocytes derived from nonvaccinated versus vaccinated mice.
Higher numbers of activated CD4+ IFN-γ+ and CD4+ IL-17A+ T cells infiltrated the infected lungs of mice vaccinated with rEBV plus GP plus OVA than infiltrated the lungs of nonvaccinated mice.
The absolute cell numbers and percentages of representative Th1, Th2, and Th17 CD4+ cells that had infiltrated the lungs of mice vaccinated with rEBV plus GP plus OVA at 9 days postchallenge (Fig. 8) correlated with the pattern of activation and cytokine production by immune splenocytes upon in vitro stimulation with rEBV reported in Table 3. Absolute numbers and percentages of CD4+ IFN-γ+ CD44+ and CD4+ IL-17A+ CD44+ T cells were significantly higher in lung homogenates of mice vaccinated with rEBV plus GP plus OVA than in those of nonvaccinated mice immunized with GP plus OVA alone. Markedly lower levels of infiltration of Th2-type CD4+ T cells were observed, which correlates with the relatively low levels of IL-4 and IL-5 production reported in Table 3. Comparison of the data in Fig. 4 and 8 supports our argument that the application of glucan particles to our vaccine formulation substantially increased both Th1 and Th17 cell responses to Coccidioides infection. Naïve mice were included in Fig. 8 for baseline comparison of absolute cell numbers and percentages of infiltration of the selected CD4+ T helper phenotypes.
Fig 8.
FACS analysis of IFN-γ-, IL-5-, and IL-17A-expressing Th1, Th2, and Th17 cells, respectively, in lung homogenates derived from mice vaccinated with rEBV plus GP plus OVA compared to nonvaccinated transgenic mice immunized with GP plus OVA alone. Determinations of absolute numbers and percentages of selected immune CD4+ T cell phenotypes which had infiltrated infected lungs of mice at 9 days postchallenge were conducted as described for Fig. 4. Naïve (untreated) mice were included for determination of baseline numbers of corresponding lung-associated immune cells. Asterisks indicate significantly higher absolute numbers of the responsive T cell phenotypes in lungs of vaccinated compared to nonvaccinated mice. Error bars indicate standard deviations. Percentages of gated, specific cytokine-producing T helper cells per lung are indicated as insets in each panel. The results reported are representative of three independent experiments.
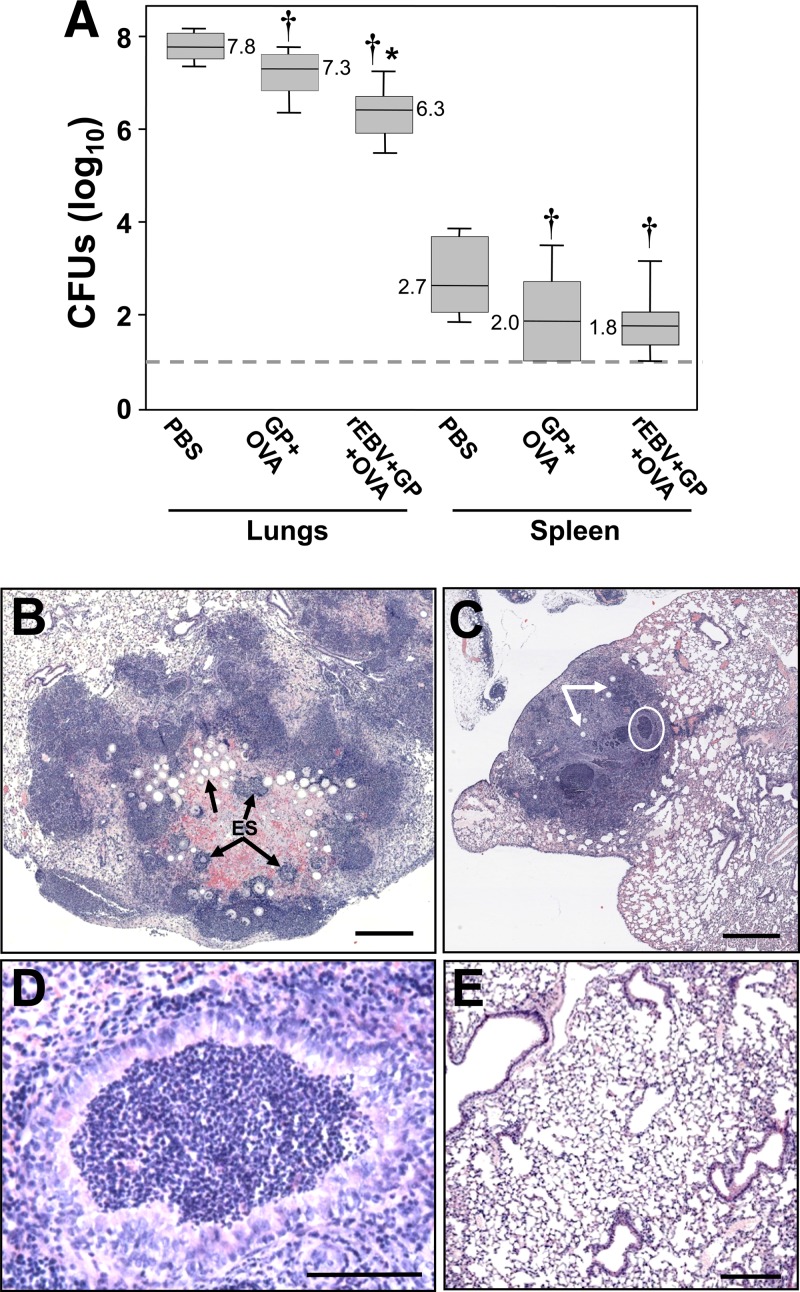
Transgenic mice vaccinated with rEBV plus GP plus OVA showed significantly reduced fungal burden in their lungs and consolidation of infection sites revealed by histopathology.
Mice immunized with the glucan particle preparation containing the rEBV protein revealed a significant reduction of CFU in their lungs at 9 days postchallenge compared to mice immunized with GP plus OVA or PBS (Fig. 9A; median CFU values are indicated). Both the vaccinated mice and the control mice immunized with GP plus OVA showed a lower fungal burden in the spleen than Coccidioides-infected mice which had been injected subcutaneously with PBS alone prior to challenge (P < 0.05). On the other hand, the numbers of CFU in spleen homogenates of the two groups of mice immunized with glucan particle preparations were not significantly different, although a trend was evident that indicated that dissemination of the pathogen to the spleen was reduced in mice vaccinated with rEBV plus GP plus OVA. These results are representative of two separate fungal burden studies conducted under the same conditions.
Fig 9.
Comparison of the fungal burdens (A) and histology results (B to E) of HLA-DR4 transgenic mice vaccinated with rEBV plus GP plus OVA versus nonvaccinated mice. The animals were sacrificed at 9 days postchallenge. Nonvaccinated mice were immunized with GP plus OVA alone or injected with PBS. The boxes in panel A indicate the 25th and 75th percentiles. The horizontal lines within the boxes represent the median CFU. The bars above and below the boxes indicate the 90th and 10th percentiles, respectively. The asterisk in panel A indicates a statistically significant difference between the levels of CFU in the lungs of mice vaccinated with rEBV plus GP plus OVA versus control mice vaccinated with GP plus OVA (P = 0.02). The daggers indicate significant differences in the fungal burden of both the infected mice immunized with GP plus OVA and those vaccinated with rEBV plus pGP plus OVA and the fungal burden of animals injected with PBS followed by intranasal challenge. The dashed line in panel A represents the limit of detection of CFU in organ homogenates. (B and C) The histopathology analyses of Coccidioides-infected lungs of mice immunized with GP plus OVA alone (B) versus transgenic mice vaccinated with rEBV plus GP plus OVA (C) show striking differences in host response. The single arrow in panel B indicates a cluster of young parasitic cells (spherules; preendosporulation stage), while the regions of concentrated inflammatory cells locate sites where mature spherules have undergone endosporulation (ES). The abscess in the lung of a mouse vaccinated with rEBV plus GP plus OVA (C) shows early consolidation of the infection, comparatively few parasitic cells (arrows), and an early stage in the differentiation of an apparent coalescent granuloma (circled). The latter is revealed at higher magnification in panel D. The histology of a corresponding region of the lung of a naïve (untreated) mouse is shown in panel E. Bars in panel B to E represent 500 μm, 1,000 μm, 70 μm, and 250 μm, respectively.
Comparative histopathology of the lungs of mice vaccinated with rEBV plus GP plus OVA and lungs from nonvaccinated mice at 9 days postchallenge (Fig. 9B to D) revealed striking differences in the nature of the host response to infection. Nonvaccinated mice immunized with GP plus OVA alone (Fig. 9B) showed an intense suppurative response to the Coccidioides insult, which is typical of the nonvaccinated murine model (52). Large numbers of spherules were observed in various stages of development. Many of the parasitic cells revealed a large central vacuole (arrow in Fig. 9B), indicative of an early stage of the reproductive cycle. The dense clusters of stained inflammatory cells visible in this section are associated with mature parasitic cells which had released their endospores (endosporulation [ES]). An intense inflammatory cell response to endosporulating spherules has been reported previously (8, 24, 52). Host tissue damage due to inflammatory pathology was evident in the lungs of these nonvaccinated mice, and the immune response resulted in little consolidation of the infection. In contrast, the histopathology of lungs of vaccinated mice revealed a more organized immune response at 9 days postchallenge (Fig. 9C). Comparatively few parasitic cells were typically observed within an abscess (arrows in Fig. 9C), and the host tissue adjacent to the abscess had structural features of normal lung tissue (cf. Fig. 9E). Note that the abscess shown in Fig. 9C contained numerous darkly stained regions composed of concentrated inflammatory cells (e.g., the circled region). At higher magnification, some of these regions are seen to be surrounded by what appears to be a layer of fibroblasts (Fig. 9D). We suggest that the host cell complex shown in Fig. 9D represents an early stage in the formation of a granuloma, which may eventually coalesce with comparable regions of differentiation within the abscess. Coalescent granulomas in histopathology studies of pulmonary tuberculosis and paracoccidioidomycosis have been previously reported (33) and may contribute to protection by forming a region of consolidation and clearance of the pathogen.
Transgenic mice immunized with the vaccine consisting of rEBV plus GP plus OVA showed a significant increase in survival compared to nonvaccinated mice.
The two groups of mice represented in Fig. 10 were challenged intranasally with 50 viable spores. The nonvaccinated mice immunized with GP plus OVA alone were moribund by 10 days postchallenge and began to die by 11 days. The vaccinated mice appeared healthy through this early period after challenge and survived over a significantly longer period than the nonvaccinated mice (P = 0.02). In addition, mice vaccinated with the glucan particle preparation containing the rEBV protein showed a significant increase in survival compared to mice vaccinated with rEBV plus CpG (cf. Fig. 10 and 5, respectively; P = 0.01).
Fig 10.
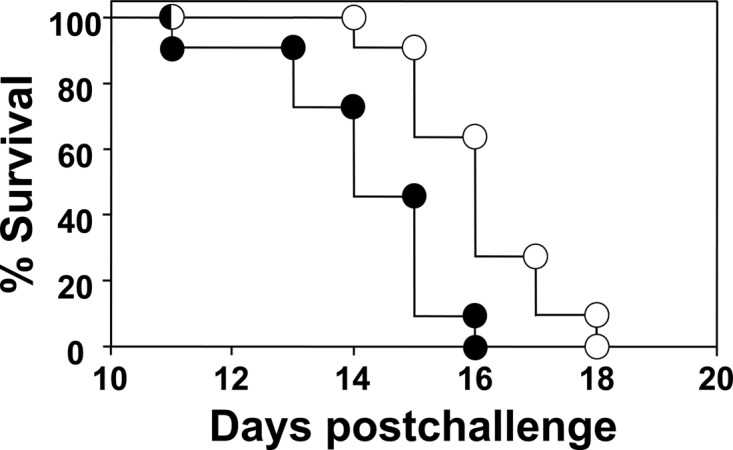
Differences in survival of HLA-DR4 transgenic mice vaccinated with rEBV plus GP plus OVA (open circles) and of nonvaccinated mice immunized with GP plus OVA alone and challenged intranasally with 50 viable spores (closed circles) were statistically significant (P = 0.02). The results are representative of three separate survival experiments.
DISCUSSION
In our previous evaluation of a coccidioidomycosis vaccine which consisted of a mixture of three T cell-reactive proteins (46, 47), we proposed that the superior protection observed in C57BL/6 mice immunized with this multivalent reagent compared to each of the single-protein vaccines was the consequence of presentation of a broader range of epitope peptides to T cell receptors, resulting in a more robust cell-mediated immune response to infection. On the basis of this assumption, we designed experiments to test whether a single polypeptide vaccine construct composed of selected immunodominant T cell epitopes of the mixed recombinant protein vaccine (47) could also protect mice against a potentially lethal intranasal challenge with Coccidioides spores. Selection of epitope peptides for incorporation into this recombinant epitope-based vaccine (rEBV) was initially based upon results of computational analysis of the primary structure of each of the three T cell-reactive proteins. The web-based, freely accessible ProPred algorithm (43) was chosen to identify putative promiscuous epitopes, which were defined as peptides predicted to bind to at least 80% of the MHC II molecules expressed by the 51 HLA-DR alleles represented in the algorithm. For the purpose of validation, we initially used murine ELISPOT assays to confirm that the computationally predicted, synthesized epitope peptides were processed based on their ability to stimulate IFN-γ cytokine production by immune CD4+ T cells in vitro. The immune T cells in the ELISPOT assay initially reported in Table 1 were derived from HLA-DR4 transgenic mice which had been immunized with the combined, full-length recombinant Pep1, Amn1, and Plb proteins as previously reported (47). We have previously argued that, since the HLA-DR allele (DRB1*0401) is the most prevalent DR4 allele in humans (44), this transgenic mouse strain is an appropriate animal model for preclinical evaluation of candidate T cell epitopes destined for incorporation into a human vaccine against coccidioidomycosis (46). Further validation of the ProPred-predicted, promiscuous epitopes was conducted by a sensitive in vitro MHC II binding assay (DELFIA) that permitted selection of peptides with highest affinity for HLA-DR4 (DRB1*0401) molecules (15). Five synthetic epitope peptides selected on the basis of this assay were further evaluated by bioinformatic methods for sequence similarity between the two Coccidioides species, lack of homology with reported human proteins, and absence of nonsynonymous coding SNPs that could result in codon changes. These studies were conducted by appropriate computational analyses of the Broad Institute Coccidioides group genomic database (37, 42) and were essential to ensure that peptides selected for the epitope-based vaccine are conserved between species, unlikely to elicit an autoimmune response by the vaccinated host, and encoded by stable transcripts.
A single-protein construct was designed which included the 5 selected epitopes, linked at their N and C termini to previously reported peptides proposed to promote epitope peptide binding to MHC II and ensure appropriate peptide presentation. The Ii-Key segment of the invariant chain (Ii) was incorporated into the protein construct at the N terminus of each epitope and included the conserved active core (LRMK) plus four downstream mouse-specific residues (LPKS) that have been proposed to link the core to each epitope peptide (21). The Ii-Key peptide component of the immunoregulatory Ii protein has been reported to help direct the vaccine peptides into the antigenic peptide binding site of the MHC II complex (16, 28). In an earlier study, the Ii-Key core was conjugated with the epitope peptide using a flexible, nonnatural amino acid (δ-aminovaleric acid) and the complex was generated by peptide synthesis (28). We chose instead to use a strategy for construction of the epitope-based vaccine which avoided the need for separate peptide synthesis and conjugation and instead permitted versatility in the generation of a multiepitope protein by application of conventional recombinant DNA technology. MHC II molecules bind peptides that are at least 13 residues in length and can be much longer, as described in this study, although typically a 9-residue anchor sequence represents the backbone which associates with the peptide binding groove of the MHC II molecule (27). During processing of the multiepitope vaccine in the endocytic pathway of APCs, there is an obvious need for proper cleavage of the linear array of epitope peptides prior to their presentation to MHC II molecules for delivery to the cell surface. Livingston and coworkers (32) reasoned that separation of epitopes by spacer residues composed of a minimum of 5 amino acids which are not usually found at the anchor positions could facilitate appropriate presentation of tandemly linked epitope peptides by disruption of the binding of junctional epitopes. Those authors provided experimental evidence that a GPGPG spacer performs this function and enables the successful processing of multiepitope vaccine constructs. The five epitope peptides which embodied our recombinant epitope-based vaccine ranged between 20 and 22 residues in length and were separated from each other by a GPGPG spacer, except for Plb-P6 at the C terminus of the vaccine construct. The 21-residue sequence of the vaccine protein upstream of the leading Ii-Key peptide was derived from the plasmid vector used for E. coli transformation and expression of the recombinant polypeptide and included a histidine motif that permitted purification of the rEBV protein by nickel-affinity chromatography.
Initial evaluation of immunoreactivity of the rEBV protein was conducted by combined assessment of patient antibody recognition, cellular immunoassays, and analyses of rEBV-induced cytokine production. All samples of sera from patients with confirmed coccidioidal infection recognized the recombinant protein based on ELISAs of bound IgG-specific antibody, which underscores the relevancy of the recombinant protein for human vaccine development. The outcome of IFN-γ ELISPOT assays of the candidate epitopes indicated that, of the 5 epitope peptides, 4 (Pep1-P1, Pep1-P2, Amn1-P10, and Amn1-P11) were processed by APCs and presented to MHC II complexes, while the C-terminal peptide of rEBV (Plb-P6) was apparently not processed. Partial degradation during bacterial expression of the vaccine protein was ruled out as an explanation, since sequence analysis revealed that the Plb-P6 peptide was intact (data not shown). A possible limiting factor for reactivity of Plb-P6 peptide was that it showed the lowest affinity for human MHC II molecules compared to the other 4 selected epitope peptides examined by DELFIA. However, our previously reported ELISPOT assay confirmed that immune T cells derived from HLA-DR4 transgenic mice vaccinated with the full-length, recombinant Plb protein responded in vitro to the Plb-P6 peptide (47). Another consideration is that the Plb-P6 peptide component of rEBV needed to be flanked at its C terminus with the spacer sequence in order to be processed, as may have the case for the other epitope peptides. Studies are under way to test this possibility. The secretion of cytokines by immune T cells to a large extent defines the functional activity of the lymphocytes (17). The profile of cytokines secreted in vitro by recall stimulation of immune splenocytes obtained from infected mice immunized with rEBV plus CpG suggested that the recombinant vaccine had primed the cells for production of appropriate chemical signals that have previously been shown to induce a protective immune response (22, 23). Th1-type cytokines (IFN-γ and IL-2) were secreted at significantly higher concentrations by rEBV-stimulated splenocytes obtained from mice vaccinated with rEBV plus CpG than by those obtained from nonvaccinated mice. The contributions of activated Th1 cells to host defense against Coccidioides are well established (22, 52). Treatment of susceptible BALB/c mice with recombinant murine IFN-γ significantly protected the animals against systemic challenge, while administration of an anti-IFN-γ monoclonal antibody to resistant DBA/2 mice sharply decreased their ability to control the disease (35). Immunotherapy combining recombinant IFN-γ and IL-12 was effective in ameliorating the course of coccidioidomycosis in BALB/c mice (10). Although limited production of Th2-type cytokines was apparent in the recall assay, activation of the Th2 signal pathway is a consistent feature of vaccine immunity to coccidioidomycosis in mice (22). However, the underlying basis for the occurrence of an amplified Th2 response during infection with Coccidioides has not been clarified (49). IL-6 is a pleiotropic, proinflammatory cytokine produced by an array of immune cells (5). Evidence has been reported that this cytokine participates in a feedback loop to increase the differentiation of Th17 cells (38). In fact, a 10- to 14-fold increase in the concentration of secreted IL-17A correlated with the peak of IL-6 production in supernatants of immune splenocytes from mice vaccinated with rEBV plus CpG compared to nonvaccinated mice. Evidence that activation of the Th17 signal pathway is essential for protection against murine coccidioidomycosis (22, 50, 51) is further discussed below. The relatively low concentrations of IL-10 detected in the supernatants of immune splenocytes compared to IFN-γ are consistent with the suggestion of a dynamic reciprocal relationship between these two cytokines (13). Dampened IL-10 production during early stages of coccidioidal disease may be a factor that contributes to the observed infiltration and expansion of activated CD4+IFN-γ+ T cells in infected lungs of the vaccinated transgenic mice.
FACS analysis of lung homogenates of vaccinated mice indicated that clonal expansion of both Th1 and Th17 cells had occurred by 9 days postchallenge. Significantly higher percentages of activated CD4+ IFN-γ+and CD4+ IL-17A+ T lymphocytes relative to total CD4+ cells that had infiltrated the pulmonary tissue were detected in mice vaccinated with rEBV plus CpG than in nonvaccinated mice. Several lines of evidence indicate that activation of the Th17 signal pathway plays a key role in murine defense against coccidioidomycosis. For example, loss of function of IL-17 receptor A in IL-17ra−/− mice immunized with a protective, live vaccine resulted in a 60% reduction of their survival over 45 days postchallenge compared to 100% survival of mice with intact receptor function and normal Th1 immunity (22). In vitro-activated transgenic CD4+ T cells (Bd 1807) with a polarized Th17 phenotype, when adoptively transferred to mice that lacked endogenous CD4+ cells, conferred protection against coccidioidomycosis (51). Activation of alveolar macrophages and neutrophils by recombinant IL-17 in vitro augmented fungicidal capacity, suggesting that antifungal Th17 cells contribute to protection by recruitment and activation of innate cells (50). It appears that a vaccine against lung infection with Coccidioides should promote early differentiation and activation of Th17 cells if it is to be effective.
On the basis of results of our combined in vitro and ex vivo immunoassays of the vaccine protein, we anticipated a positive outcome of protection studies of HLA-DR4 mice immunized subcutaneously with rEBV plus CpG. In fact, a significant reduction of CFU in the lungs of vaccinated mice was observed at 9 days postchallenge compared to those of the nonvaccinated animals. Although a trend was also evident that mice vaccinated with rEBV plus CpG survived longer than nonvaccinated mice, the difference between the respective survival plots was not statistically significant. The humanized mouse strain used in this study has proven to be highly susceptible to pulmonary infection and dissemination of Coccidioides to the spleen after intranasal challenge with as few as 30 to 40 spores. Nevertheless, our vaccine formulation of rEBV plus CpG and immunization regimen apparently failed to induce a robust and durable protective response to the fungal infection. We speculated that a possible cause of the deficiency may have been the adjuvant employed (CpG ODN). Adjuvants are recognized to play pivotal roles in enhancement of the magnitude of adaptive immunity as well as the specificity and clonotypic diversity of the responding CD4+ T cell repertoire (6). Glucan particles prepared from isolated yeast cell wall material and composed primarily of particulate β-1,3-glucans have recently been shown to function as both an effective adjuvant and an efficient delivery system (20). β-glucans are recognized by Dectin-1 receptors expressed on the surface of host phagocytes, including dendritic cells (DCs), macrophages, and neutrophils, and have been shown to induce the synthesis of cytokines involved in polarization of the Th17 signal pathway (19). GPs are hollow structures which in the appropriate chemical environment can be induced to take up and retain reproducible amounts of antigenic proteins. The particles are small enough to be readily phagocytosed by DCs via their Dectin-1 receptors followed by innate cell secretion of proinflammatory cytokines (20). Results of our comparison of CD4+ IFN-γ ELISPOT recall assays reported here have indicated that delivery of the rEBV to APCs via glucan particles was much more efficiently processed than delivery by presentation of the vaccine protein admixed with CpG. In addition, high in vitro production of IL-17A and IFN-γ by immune splenocytes isolated from infected mice which had been vaccinated with rEBV plus GP plus OVA was observed upon recall stimulation with rEBV but not with OVA. This specificity of the recall response was maintained even though the amount of OVA loaded into the glucan particle preparation was more than twice that of the recombinant vaccine protein. Moreover, this rEBV-specific, in vitro cytokine response of immune splenocytes was dramatically enhanced in mice vaccinated with the GP preparation compared to mice vaccinated with rEBV plus CpG. Correlated with these results was a significant increase in numbers of CD4+ IFN-γ+ and CD4+ IL-17A+ T cells which had infiltrated the lungs of infected mice vaccinated with rEBV plus GP plus OVA compared to mice immunized with rEBV plus CpG. Protection studies also showed that the HLA-DR4 mice immunized with the glucan particle preparation and challenged intranasally with Coccidioides revealed a significantly better outcome than mice vaccinated with rEBV plus CpG based on comparisons of pathogen clearance, histopathology, and survival results. Particularly striking was the difference between the two groups in the results of histopathology analysis of the lungs 9 days after challenge (not shown). Paraffin sections revealed increased consolidation of the lung infections in mice immunized with rEBV plus GP plus OVA compared to animals vaccinated with rEBV plus CpG.
In conclusion, we argue that both the nature of the primed immunological response of the humanized mouse to early stages of Coccidioides infection and the degree of protection achieved by our first-generation, glucan particle-delivered epitope-based vaccine indicate sufficient promise to warrant further investigations. Current studies are focused on modification of several key features of the recombinant protein construct and immunization protocol which we believe will significantly improve the protective efficacy of the vaccine. The observed degree of vaccine immunity was essentially the result of APC presentation of only four T cell epitopes. The epitope-based vaccine construct permits incorporation of alternative and/or additional epitope peptides with relative ease. We have validated our earlier prediction of the existence of a reservoir of Coccidioides parasitic cell wall-extractable, T cell-reactive proteins which contain promiscuous T cell epitopes (47). Selection of additional peptide vaccine candidates using criteria developed in this study is under way. Immune splenocytes derived from mice vaccinated with the glucan particle preparation induced a significantly lower IL-2 response upon in vitro recall stimulation than those derived from mice immunized with rEBV admixed with the CpG ODN adjuvant. IL-2 receptor signaling has been shown to play an important role in Th1 effector cell differentiation (29). We plan to add CpG ODN to the current GP preparation and evaluate its effect on protection. Examination of factors which improve loading and delivery of the vaccine protein in glucan particles and evaluation of different antigen dosages are in progress. Finally, alternate routes of immunization are under investigation. Intranasal (i.n.) immunization has been shown to promote Th17-biased immune responses, which have been previously reported to contribute to the effectiveness of a vaccine against coccidioidomycosis (54). On the other hand, i.n. immunization poses a risk of antigen delivery to the central nervous system and recent studies have indicated that sublingual vaccination is a safer route (39). Further development of an epitope-based vaccine against coccidioidomycosis should both help to elucidate underlying mechanisms of immune recognition and yield a potentially promising protective reagent against this respiratory disease (53).
ACKNOWLEDGMENTS
Support for this work was provided by a Public Health Service grant (AI-071118) from the National Institute of Allergy and Infectious Disease awarded to G.T.C. and an Institutional Research Training Fellowship (AI-007271-23) awarded to B.J.H. Additional support was provided by the Margaret Batts Tobin Foundation, San Antonio, TX.
The colony of HLA-DR4 transgenic mice was derived from a breeding pair kindly provided by Thomas Forsthuber at the University of Texas at San Antonio. We thank Natalia Castro-Lopez and Michael Bellecourt for their technical assistance in the determinations of fungal burden in Coccidioides-infected mice. We also thank Craig M. Rundbaken, for the supply of test sera from patients with confirmed coccidioidomycosis.
Footnotes
Published ahead of print 4 September 2012
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altshuler D, et al. 2000. A SNP map of the human genome generated by reduced representation shotgun sequencing. Nature 407:513–516 [DOI] [PubMed] [Google Scholar]
- 3. Ampel NM. 2009. Coccidioidomycosis: a review of recent advances. Clin. Chest Med. 30:241–251 [DOI] [PubMed] [Google Scholar]
- 4. Ampel NM. 2009. Coccidioidomycosis: what's old and what's new. J. Invasive Fungal Infect. 3:70–75 [Google Scholar]
- 5. Barton BE. 1997. IL-6: insights into novel biological activities. Clin. Immunol. Immunopathol. 85:16–20 [DOI] [PubMed] [Google Scholar]
- 6. Baumgartner CK, Malherbe LP. 2011. Antigen-driven T-cell repertoire selection during adaptive immune responses. Immunol. Cell Biol. 89:54–59 [DOI] [PubMed] [Google Scholar]
- 7. Brown GD, Denning DW, Levitz SM. 2012. Tracking human fungal infections. Science 336:647. [DOI] [PubMed] [Google Scholar]
- 8. Cole GT, Hurtgen BJ, Hung C-Y. September 2012. Progress toward a human vaccine against coccidioidomycosis. Curr. Fungal Infect. Rep. doi:10.1007/s12281-012-0105-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cole GT, Sekiya T, Kasai R, Yokoyama T, Nozawa Y. 1979. Surface ultrastructure and chemical composition of the cell walls of conidial fungi. Exptl. Mycol. 3:132–156 [Google Scholar]
- 10. Cox RA, Magee DM. 2004. Coccidioidomycosis: host response and vaccine development. Clin. Microbiol. Rev. 17:804–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Delgado N, et al. 2003. A recombinant β-1,3-glucanosyltransferase homolog of Coccidioides posadasii protects mice against coccidioidomycosis. Infect. Immun. 71:3010–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dierberg KL, et al. 2012. Donor-derived organ transplant transmission of coccidioidomycosis. Transpl. Infect. Dis. 14:300–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fierer J. 2007. The role of IL-10 in genetic susceptibility to coccidioidomycosis on mice. Ann. N. Y. Acad. Sci. 1111:236–244 [DOI] [PubMed] [Google Scholar]
- 14. Fisher MC, Koenig GL, White TJ, Taylor JW. 2002. Molecular and phenotypic description of Coccidioides posadasii sp. nov., previously recognized as the non-California population of Coccidioides immitis. Mycologia 94:73–84 [PubMed] [Google Scholar]
- 15. Forsthuber TG, et al. 2001. T cell epitopes of human myelin oligodendrocyte glycoprotein identified in HLA-DR4 (DRB1*0401) transgenic mice are encephalitogenic and are presented by human B cells. J. Immunol. 167:7119–7125 [DOI] [PubMed] [Google Scholar]
- 16. Germain RN. 2011. Uncovering the role of invariant chain in controlling MHC class II antigen capture. J. Immunol. 187:1073–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Han Q, et al. 2012. Polyfunctional responses by human T cells result from sequential release of cytokines. Proc. Nat. Acad. Sci. 109:1607–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hayat MA. 2000. Principles and techniques of electron microscopy: biological applications, 4th ed. Cambridge University Press, New York, NY [Google Scholar]
- 19. Huang H, et al. 2012. Relative contributions of Dectin-1 and complement to immune responses to paticulate β-glucans. J. Immunol. 189:312–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang H, Ostroff GR, Lee CK, Specht CA, Levitz SM. 2010. Robust stimulation of humoral and cellular immune responses following vaccination with antigen-loaded β-glucan particles. mBio 1:pii:e00164-10. doi:10.1128/mBio.00164-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Humphreys RE, et al. 2000. Increasing the potency of MHC class II-presented epitopes by linkage to Ii-key peptide. Vaccine 18:2693–2697 [DOI] [PubMed] [Google Scholar]
- 22. Hung C-Y, Gonzalez A, Wüthrich M, Klein BS, Cole GT. 2011. Vaccine immunity to coccidioidomycosis occurs by early activation of three signal pathways of T helper cell response (Th1, Th2, and Th17). Infect. Immun. 79:4511–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hung C-Y, et al. 2012. An agonist of human complement fragment C5a enhances vaccine immunity against Coccidioides infection. Vaccine 30:4681–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hung C-Y, et al. 2005. Metalloproteinase of Coccidioides posadasii contributes to evasion of host defenses. Infect. Immun. 73:6689–6703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hung C-Y, Yu J-J, Seshan KR, Reichard U, Cole GT. 2002. A parasitic phase-specific adhesin of Coccidioides immitis contributes to the virulence of this respiratory fungal pathogen. Infect. Immun. 70:3443–3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ito K, et al. 1996. HLA-DR4-IE chimeric class II transgenic, murine class II-deficient mice are susceptible to experimental allergic encephalomyelitis. J. Exp. Med. 183:2635–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Janeway CA, Travers P, Walport M, Capra JD. 1999. Immunobiology: the immune system in health and disease, 4th ed Garland Publishing, New York, NY [Google Scholar]
- 28. Kallinteris NL, et al. 2005. Enhanced CD4+ T-cell response in DR4-transgenic mice to a hybrid peptide linking the Ii-key segment of the invariant chain to the melanoma gp100(48-58) MHC class II epitope. J. Immunother. 28:352–358 [DOI] [PubMed] [Google Scholar]
- 29. Khoruts A, Mondino A, Pape KA, Reiner SL, Jenkins MK. 1998. A natural immunological adjuvant enhances T cell clonal expansion through a CD28-dependent, interleukin (IL)-2-independent mechanism. J. Exp. Med. 187:225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li H, Ruan J, Durbin R. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18:1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li K, Yu J-J, Hung C-Y, Lehmann PF, Cole GT. 2001. Recombinant urease and urease DNA of Coccidioides immitis elicit an immunoprotective response against coccidioidomycosis in mice. Infect. Immun. 69:2878–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Livingston B, et al. 2002. A rational strategy to design multiepitope immunogens based on multiple Th lymphocyte epitopes. J. Immunol. 168:5499–5506 [DOI] [PubMed] [Google Scholar]
- 33. Loures FV, et al. 2011. MyD88 signaling is required for efficient innate and adaptive immune responses to Paraciccidioides brasiliensis infection. Infect. Immun. 79:2470–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Madsen LS, et al. 1999. A humanized model for multiple sclerosis using HLA-DR2 and a human T-cell receptor. Nat. Genet. 23:343–347 [DOI] [PubMed] [Google Scholar]
- 35. Magee DM, Cox RA. 1995. Roles of gamma interferon and interleukin-4 in genetically determined resistance to Coccidioides immitis. Infect. Immun. 63:3514–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Masannat FY, Ampel NM. 2010. Coccidioidomycosis in patients with HIV-1 infection in the era of potent antiretroviral therapy. Clin. Infect. Dis. 50:1–7 [DOI] [PubMed] [Google Scholar]
- 37. Neafsey DE, et al. 2010. Population genomic sequencing of Coccidioides fungi reveals recent hybridization and transposon control. Genome Res. 20:938–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Onishi RM, Gaffen SL. 2010. Interleukin-17 and its target genes: mechanisms of interleukin-17 function in disease. Immunology 129:311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pedersen G, Cox RA. 2012. The mucosal vaccine quandary: intranasal vs. sublingual immunization against influenza. Hum. Vaccin. Immunother. 8:689–693 [DOI] [PubMed] [Google Scholar]
- 40. Proft T, Hilbert H, Layh-Schmitt G, Herrmann R. 1995. The proline-rich p65 protein of Mycoplasma pneumoniae is a component of the Triton X-100-insoluble fraction and exhibits size polymorphism in the strains M129 and FH. J. Bacteriol. 177:3370–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rosa D, Ribeiro S, Cunha-Neto E. 2010. CD4+ T cell epitope discovery and rational vaccine design. Arch. Immunol. Ther. Exp. (Warsz.) 58:121–130 [DOI] [PubMed] [Google Scholar]
- 42. Sharpton TJ, et al. 2009. Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Res. 19:1722–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Singh H, Raghava GP. 2001. Propred: prediction of HLA-DR binding sites. Bioinformatics 17:1236–1237 [DOI] [PubMed] [Google Scholar]
- 44. Southwood S, et al. 1998. Several common HLA-DR types share largely overlapping peptide binding repertoires. J. Immunol. 160:3363–3373 [PubMed] [Google Scholar]
- 45. Taborda CP, Juliano MA, Puccia R, Franco M, Travassos LR. 1998. Mapping of the T-cell epitope in the major 43-kilodalton glycoprotein of Paracoccidioides brasiliensis which induces a Th-1 response protective against fungal infection in BALB/c mice. Infect. Immun. 66:786–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tarcha EJ, Basrur V, Hung CY, Gardner MJ, Cole GT. 2006. A recombinant aspartyl protease of Coccidioides posadasii induces protection against pulmonary coccidioidomycosis in mice. Infect. Immun. 74:516–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tarcha EJ, Basrur V, Hung CY, Gardner MJ, Cole GT. 2006. Multivalent recombinant protein vaccine against coccidioidomycosis. Infect. Immun. 74:5802–5813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Travassos L, Rodrigues E, Iwai L, Taborda C. 2008. Attempts at a peptide vaccine against paracoccidioidomycosis, adjuvant to chemotherapy. Mycopathologia 165:341–352 [DOI] [PubMed] [Google Scholar]
- 49. Wüthrich M, Deepe G, Klein B. 2012. Adaptive immunity to fungi. Annu. Rev. Immunol. 30:115–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wüthrich M, et al. 2011. Vaccine-induced protection against 3 systemic mycoses endemic to North America requires Th17 cells in mice. J. Clin. Invest. 121:554–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wüthrich M, et al. 2011. ATCR transgenic mouse reactive with multiple systemic dimorphic fungi. J. Immunol. 187:1421–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xue JM, et al. 2009. A genetically engineered live attenuated vaccine against coccidioidomycosis. Infect. Immun. 77:3196–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang L, et al. 2012. TEPITOPEpan: extending TEPITOPE for peptide binding prediction covering over 700 HLA-DR molecules. PLoS One 7:e30483 doi:10.1371/journal.pone.0030483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zygmunt BM, Rharbaoui F, Groebe L, Guzman CA. 2009. Intranasal immunization promotes Th17 immune responses. J. Immunol. 183:6933–6938 [DOI] [PubMed] [Google Scholar]