Abstract
The small multidrug resistance (SMR) transporter protein EmrE in Escherichia coli is known to confer resistance to toxic antiseptics classified as quaternary cation compounds (QCCs). Naturally derived QCCs synthesized during metabolic activities often act as osmoprotectants, such as betaine and choline, and participate in osmotic homoestasis. The goal of this study was to determine if EmrE proteins transport biological QCC-based osmoprotectants. Plasmid-encoded copies of E. coli emrE and the inactive variant emrE-E14C (emrE with the E→C change at position 14) were expressed in various E. coli strains grown in either rich or minimal media at various pHs (5 to 9) and under hypersaline (0.5 to 1.0 M NaCl and KCl) conditions to identify changes in growth phenotypes induced by osmoprotectant transport. The results demonstrated that emrE expression reduced pH tolerance of E. coli strains at or above neutral pH and when grown in hypersaline media at or above NaCl or KCl concentrations of 0.75 M. Hypersaline growth conditions were used to screen QCC osmoprotectants betaine, choline, l-carnitine, l-lysine, l-proline, and l-arginine. The study identified that betaine and choline are natural QCC substrates of EmrE.
INTRODUCTION
Quaternary ammonium compounds (QACs) and more generally quaternary cation compounds (QCCs) describe a highly diverse range of chemicals with the structure XR4+ where, X+ is a permanently charged cation (typically cationic N or P) and each R is any type of acyl chain or aromatic hydrocarbon. Anthropogenic QCCs serve a variety of purposes as medical antimicrobials (benzalkonium and cetylpyridnium), industrial surfactants (tetraphenyl phosphonium), and lipophilic dyes or stains (ethidium and acriflavin). Anthropogenically derived QCCs are often highly toxic to organisms due to their membrane-disrupting activity. Biologically synthesized QCCs are also produced by both prokaryotes and eukaryotes and naturally accumulate under osmotic stress and particular physiological conditions in the cell. Naturally occurring QCCs, such as betaine, choline, and carnitine, and amino acids, such as glycine, lysine, serine, and proline, play an important role as osmoprotectants during cellular osmotic stress (8, 9, 18, 30, 32) and participate in bacterial intracellular pH regulation (as reviewed in reference 7).
Members of the small multidrug resistance (SMR) protein family (26) confer resistance to a variety of lipophilic toxins, primarily QCCs and DNA interchelating dyes, within Archaea and Bacteria via proton motive force-energized efflux (16, 20, 34). SMR protein members are distinct from other secondary active multidrug resistance transporters based on their short length (∼100 to 140 amino acids), resulting in proteins composed of only 4 transmembrane (TM) segments. These transporters are present on a variety of mobile genetic elements, primarily in the 3′ conserved regions of class 1 to 3 integrons and multidrug resistance plasmids, in addition to the host chromosome (as reviewed in references 4 and 29). In general, members of the SMR family form a 4-TM α-helix monomer (as reviewed in reference 4) within the plasma membrane that functions as a homo-oligomer, where the minimum oligomeric subunit is a homodimer (23). SMR transport activity and ligand binding are known to involve a highly conserved (98%) (5) active site Glu residue within the first TM strand, and mutations to this residue eliminate host resistance to QCCs (16, 24, 37). In Escherichia coli, EmrE (ethidium multidrug resistance protein E) (28) is considered to be the archetype of the SMR protein family. EmrE is the most-characterized SMR protein member and has been shown to transport/interact with the broadest range of QCC substrates in comparison to all other subclass members (as reviewed in reference 4).
The involvement or influence of EmrE protein during bacterial cellular activities unrelated to multidrug resistance is poorly understood and uncharacterized. There are many biological QCC metabolites and intermediates that can potentially serve as the “natural” QCC substrates for SMR proteins. SMR protein participation in these activities has been speculated (5, 25) but has not been experimentally confirmed.
The goal of this study was to determine if the SMR protein EmrE participates in biological regulation of QCC-based osmoprotectants. If SMR proteins transport naturally occurring QCCs, such as betaine or choline, overaccumulation of these proteins at increasing pH and/or osmotic stress should reduce host tolerance to these conditions by exporting the QCC it needs to survive the stress. To test this hypothesis, we examined the influence of E. coli EmrE overaccumulation in various E. coli strains grown at increasing pH and under osmotic conditions. The growth of E. coli K-12 transformed with plasmids encoding active or inactive copies of emrE in media at various pH values (5–9) and at increasing salt (NaCl or KCl) concentrations (0.25 M to 1.0 M) was examined to identify growth conditions altered by SMR proteins. Based on the outcome of these assays, a variety of QCC-based osmoprotectants, including betaine, carnitine, choline, arginine, and lysine, were selected for growth phenotype screening assays. The addition of osmoprotectants to the culture media of the wild-type E. coli K-12 strain and those lacking QCC osmoprotectant biosynthesis genes, betA and betB, expressing active and inactive forms of emrE genes, was used to identify osmoprotectant substrates of SMR transporters. The results of this approach identified that betaine and choline are substrates of EmrE and revealed that only EmrE proteins participate in osmotic regulation. The implications of this study may suggest that the inheritance and spread of this multidrug transporter family within bacteria are influenced by other environmental factors in addition to anthropogenic drug exposure.
MATERIALS AND METHODS
Materials and strains used in this study.
All E. coli strains used in this study (BW25113, JW0531, JW5738, JW0303, and JW0304) were provided by the single-gene-knockout Keio Collection constructed through a collaboration between the Institute of Advanced Bioscience at Keio University and the Nara Institute of Science and Technology in Japan (1) (see Table S1 in the supplemental material). The nucleotide primers used for PCR experiments were synthesized by Sigma (St. Louis, MO) and Integrated DNA Technology (Coralville, IA). Growth medium compounds and chemicals were supplied by Sigma, EMD, and BD Biosciences.
SMR gene cloning and mutagenesis.
The E. coli SMR gene emrE was cloned into the ampicillin-resistant expression vector pMS119EH (pEmrE) as described in reference 33. Plasmid DNA was isolated using a Fermentas Gene Jet spin column kit (Fermentas Canada, Inc., Ontario, Canada). Replacement of the active site glutamate codon E14C was generated using the QuikChange II site-directed mutagenesis kit (Stratagene, CA) with the forward and reverse nucleotide primers for each SMR gene provided in Table S1 in the supplemental material. All plasmid constructs were confirmed by DNA sequencing from the PtacI promoter of pMS119EH (PtacI primer 5′ CTG TTG ACA ATT AAT CAT CGG CTC GTA TAA TG 3′) (see Table S2 in the supplemental material). Sequenced SMR plasmids were transformed into the E. coli K-12 Keio collection strains listed in Table S2 (1).
Reporter protein accumulation from “leaky” PtacI promoter expression.
In all experiments, cloned SMR gene expression was found to occur at sufficient levels from “leaky” expression from the PtacI promoter of pMS119EH based on the accumulation quantities of a hexahistidyl-tagged emrE reporter protein to generate a growth phenotype. Chemical induction of SMR gene overexpression by isopropyl β-d-1-thiogalactopyranoside (IPTG) at final concentrations ranging from 0.001 to 0.1 mM was immediately toxic to the cultures and led to immediate reductions and/or arrest of cell growth based on optical density (OD) measurements.
To determine the relative levels of gene expression and protein accumulation from leaky PtacI promoter expression, a C-terminal myc epitope-hexahistidinyl (His6) fusion tag was added to cloned emrE in the pMS119EH vector (pEmrE-my-His6) in a two step PCR and with the same restriction sites maintained. After DNA sequencing, the translated C-terminal sequence of the myc-His6 fusion tag (starting at residue H110 of EmrE) corresponded to NH2-HLEFEAYVEQK LISEEDLNSAVDHHHHHH-CO2H. Western dot blot analysis, as described by Chan et al. (10), was used to determine EmrE-myc-His6 protein accumulation in E. coli strains grown in the different culture media, pHs, and hypersaline conditions used in this work. His6-tagged EmrE-myc-His6 protein accumulation levels within all E. coli strains tested were colorimetrically detected using the INDIA His probe-horseradish peroxidase (HRP)-conjugated antibody (Pierce-Endogen, Thermo Fisher) and quantified using the Kodac gel logic imaging system and Kodac 1D version 3.6.6 software. EmrE-myc-His6 protein quantities (μg) were determined from the mean spot intensity using a standard curve of 1/4 to 1/256 dilutions of His6-tagged control protein (0.825 mg/ml), and values from the growth experiments are provided in Table S3 in the supplemental material. Overall, the relative quantities of tagged protein were similar (1.1- to 1.7-fold difference in calculated protein in μg) to the amounts of protein reported by de Boer et al. for the same PtacI promoter using E. coli galactokinase assays (12).
pH tolerance of SMR-transformed E. coli strains.
E. coli K-12 strains BW25113 (wild-type) and JW0531 (ΔemrE) transformed with each SMR vector (and empty vector control) were used for pH tolerance experiments (see Table S1 in the supplemental material). Plasmid-transformed strains were inoculated from frozen dimethyl sulfoxide stocks and grown overnight (16 h) at 37°C in Luria-Bertani (LB) medium (1% [wt/vol] yeast extract, 0.5% [wt/vol] tryptone, 0.5% [wt/vol] NaCl, 0.01% [wt/vol] glucose) containing 100 μg/ml ampicillin buffered to pH 7 using 50 mM phosphate buffer. Overnight (16-h) cultures were diluted to 1.5 absorbance units at the optical density at 600 nm (OD600) and used as an inoculant for pH tolerance experiments. Two types of media were used in pH growth experiments: LB medium and M9 minimal salts medium (1.3% [wt/vol] NaH2PO4 · 7H2O, 0.3% [wt/vol] K2HPO4, 0.05% [wt/vol] NaCl, 0.1% NH4Cl, 1.6 ×10−5% [wt/vol] MgSO4, 9.0 ×10−7% [wt/vol] CaCl2, 0.00015% [wt/vol] thiamine) supplemented with 0.01% (wt/vol) glucose as the primary carbon source. The media for E. coli strains transformed with a vector also contained 100 μg/ml ampicillin to ensure plasmid maintenance. All media were buffered to pH values of 4.3, 5.0, 6.0, 7.0, 8.0, and 9.0 using 50 mM phosphate buffer. All pH values were confirmed immediately before culture inoculation using a Beckman Φ 720 pH meter with an Accumet 1.5-in. microelectrode (Thermo Fisher Scientific) with an error of ±0.15 pH unit and/or confirmed using Whatman type CF pH 0-to-14 indicator strips.
pH tolerance experiments were performed by dilution (10−3) of overnight (16-h) cell cultures into 3.5 ml of M9 or LB medium at pH values of 4.3 to 10. Cell growth experiments were performed in plastic-stoppered autoclaved glass cuvettes that provided sufficient headspace (1/2 the volume of the cuvette) to ensure aerobic growth. All cultures were incubated at 37°C in a shaking incubator (210 to 250 rpm). Cultures were monitored at OD600 every hour up to 16 h and again at 24 h. OD values were measured directly in glass cuvettes using an Ocean Optics DH-2000-BAL UV-Vis-NIR light source spectrophotometer and baseline corrected with each medium type prior to measurement. The most acidic (pH 4.3) and basic (pH 10.0) pH values failed to culture viable cells and were excluded from the results of this study. (The cell viability experiments are described in the next section.) A minimum of three independently inoculated pH growth trials for each set of transformed E. coli strains were performed and used to calculate average OD600 values. For this study, two-tailed t tests with P values of ≤0.05 were considered to be significantly different.
The pH of the culture medium used in pH growth experiments was monitored during growth curve assays to determine if growth phenotypes were attributed to pH fluctuations. pH growth curves performed at neutral to alkaline values (pH 7 to 9) demonstrated relatively constant (±0.35 pH unit) pH values in rich LB medium and in M9 minimal medium (±0.45 pH unit) until the late log phase (OD600 > 0.8 units) and/or stationary phase was reached, where pH values gradually decreased over time. The reduction in pH at stationary phase was attributed to the increase in metabolites accumulating in the medium.
Osmotic hypersaline tolerance of SMR-transformed E. coli strains.
The E. coli strains (BW25113, and JW0531, and JW5738) transformed with SMR plasmids used in pH tolerance growth experiments were examined for their ability to grow under hypersaline conditions (see Tables S1 and S2 in the supplemental material). The starting cultures were diluted/concentrated from LB overnight cultures to a final OD600 of 1.5 U and then diluted 10−3 into sterile glass cuvettes containing 3.5 ml of pH 7.0 50 mM sodium phosphate-buffered LB or M9 medium supplemented with 20 mM glucose in the presence or absence of NaCl or KCl salts. Hyperosmotic growth experiments involved the addition of NaCl or KCl salts to LB or M9 medium at final concentrations of 0.25, 0.5, 0.75, and 1 M. Cell cultures were incubated in a shaking incubator (210 to 250 rpm) at 37°C, where growth curve OD600 measurements were monitored every hour up to 16 h and at 24 h, as described for pH tolerance experiments. Average OD600 values for each transformed strain were calculated from a minimum of three growth trials, and statistical analysis of growth differences was determined as described for pH tolerance experiments.
Under certain conditions in which cell growth was arrested based on OD600 measurements, cell viability experiments were performed during growth curve experiments to determine if cells were capable of growth (viable). Cells corresponding to ∼0.1 OD600 unit were selected from growth curve assay experiments and plated onto LB agar to determine the number of CFU. Cells were deemed viable if CFU values were similar to values determined for the control growth curve experiment based on the number of CFU/OD600 unit. In general, plates with 10 colonies or less were considered nonviable.
SMR overexpression screening experiments to identify QCC osmoprotectant substrates.
Based on the results of pH and hyperosmotic growth experiments, hypersaline tolerance experiment conditions were selected for further screening of various QCCs. SMR plasmid-transformed E. coli strain BW25113 (wild type) cells were grown in either M9 or LB medium buffered to pH 7.0 (50 mM phosphate buffer) in the presence of 0.75 and 1.0 M NaCl or KCl. Osmoprotectants, betaine, choline, l-carnitine, l-proline, l-arginine, and l-lysine were filter sterilized (0.2-μm-pore filter) in 0.5 M stock solutions and added to culture media at final concentrations of 5 mM or 10 mM. Culture growth was monitored by OD600 as described for pH tolerance and hypersaline experiments, and averaged OD600 values were calculated from a minimum of three independent trials. Statistical significance of growth differences was determined as described for pH tolerance experiments.
Since the outcome of the osmoprotectant screens identified that betaine and choline were potential substrates, SMR plasmids were also transformed into E. coli K-12 strains JW0303 (ΔbetA) and JW0304 (ΔbetB) for further osmotic tolerance experiments (see Table S1 in the supplemental material). Hyperosmotic growth curve experiments with SMR plasmid-transformed E. coli strains JW0303 and JW0304 in the presence and absence of 10 mM betaine or choline in pH 7.0 buffered M9 and LB media were performed. OD600 measurements were determined as described above for the osmotic tolerance experiments, and an average of at minimum three independently inoculated growth trials were used in this analysis. The outcomes of these experiments provided in the Results section are reported as mean OD600 fold difference of each plasmid-transformed E. coli strain. This value represents the fold change in OD600 growth of a plasmid-transformed strain (based on mean OD600 values) between two different growth conditions. In almost all cases, the mean fold difference is the mean OD600 value of a plasmid-transformed strain grown in a specific hypersaline medium (LB or M9) in the presence of 10 mM osmoprotectant divided by its mean OD600 value in hypersaline medium only at a single time point.
RESULTS
Only emrE gene expression decreases host strain pH tolerance at neutral and alkaline ranges.
The first objective of this study was to determine if the E. coli SMR multidrug transporter emrE participates in osmoregulation. Before beginning a screen to identify biological QCC osmoprotectants of SMR transporters, it was essential to determine if SMR-transformed E. coli strains, such as the wild-type strain BW25113 or the SMR gene deletion strain JW0531 (ΔemrE), significantly altered its growth phenotype under various osmotic medium alterations. Osmoregulatory metabolites, such as betaine, choline, and carnitine, are known to contribute to pH maintenance and osmotic tolerance of E. coli cells and are generally referred to as osmoprotectants (17, 32). Two types of growth media were examined in this study: the LB rich medium and the M9 selective minimal medium. Both media were crucial for identification and selection of osmotic conditions to screen QCCs in the second part of this study, since M9 medium is completely defined and its osmolarity is 100-fold lower than that of LB medium (M9 medium, 5 mosM [3]; LB medium, 500 mosM [2]), it may identify phenotypes hidden in osmotically enriched LB medium. The amounts of EmrE and EmrE-E14C (coded for by emrE with the E→C change at position 14) protein accumulation within E. coli BW25113 derived from leaky plasmid expression were determined by using a C-terminal hexahistidinyl-myc epitope-tagged EmrE construct in pMS119EH, as described in Materials and Methods. Western dot blotting analysis determined that the leaky PtacI promoter expression of this reporter resulted in consistent amounts of protein accumulation from cultures grown under various osmotic conditions, where values ranged from 0.011 to 0.037 μg per OD600 unit (see Table S3 in the supplemental material).
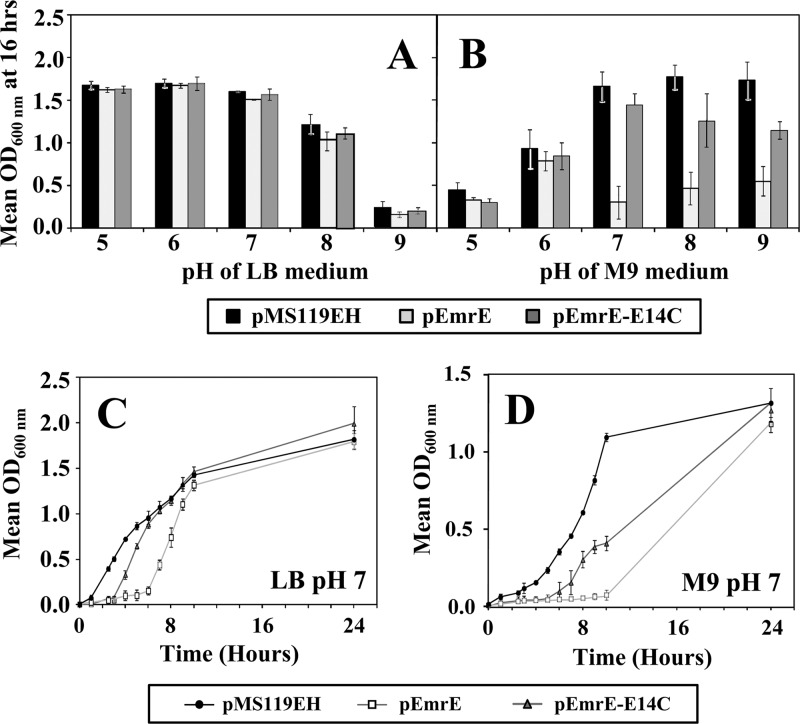
The pH susceptibility of E. coli strain BW25113 and SMR gene deletion strains transformed with plasmids encoding functional copies of emrE or the inactive variant emrE-E14C was used to screen growth phenotype changes. Empty vector transformants were included in these experiments to act as a positive growth control for gain- or loss-of-growth phenotypes that are caused by SMR gene expression. Growth curve experiments were performed for all transformed strains, and growth was measured at 37°C (optimal temperature) over 24 h in LB or M9 medium buffered to pH values of 5 to 9. The outcome of these experiments identified that only wild-type emrE expression resulted in a significant growth reduction in E. coli cells cultured in M9 medium at neutral to basic pH ranges after 12 to 16 h of growth (Fig. 1). This growth phenotype reduction at a pH of ≥7 caused by pEmrE expression in E. coli disappeared after 24 h of growth, and cultures became indistinguishable from the empty control vector or inactive pEmrE-E14C-transformed strains (Fig. 1C and D). This indicates that EmrE activity at neutral to high pH only delays E. coli growth, since all plasmid-transformed strains showed no significant differences in cell viability (data not shown).
Fig 1.
pH susceptibility of E. coli BW25113 strains transformed with SMR plasmids. OD600 values after 16 h of growth in either LB (A) or M9 (B) media at pH values ranging from 5 to 9 are shown. The results for E. coli BW25113 transformed with pMS119EH (black), pEmrE (gray), and active site mutant pEmrE-E14C (dark gray) after 16 h of growth in LB (A) or M9 (B) medium are shown in a bar chart format. Asterisks indicate statistically significant differences (P < 0.01) compared to either the untransformed strain without plasmid or the strain transformed with the empty control vector (pMS119EH). Panels C and D show pH susceptibility growth (OD600) curve experiments with E. coli BW25113 transformed with pMS119EH (circles), pEmrE (squares), and pEmrE-E14C (triangles) grown at 37°C in either LB (C) or M9 (D) medium at pH 7.0 over 24 h.
No significant differences were observed in the pH growth curves determined for plasmid-transformed E. coli strains lacking ΔemrE (JW0531) (see Fig. S1 in the supplemental material), indicating that the deletion of emrE does not alter the growth phenotype from that of the wild-type strain.
EmrE protein accumulation decreases host osmotic tolerance to hypersaline conditions.
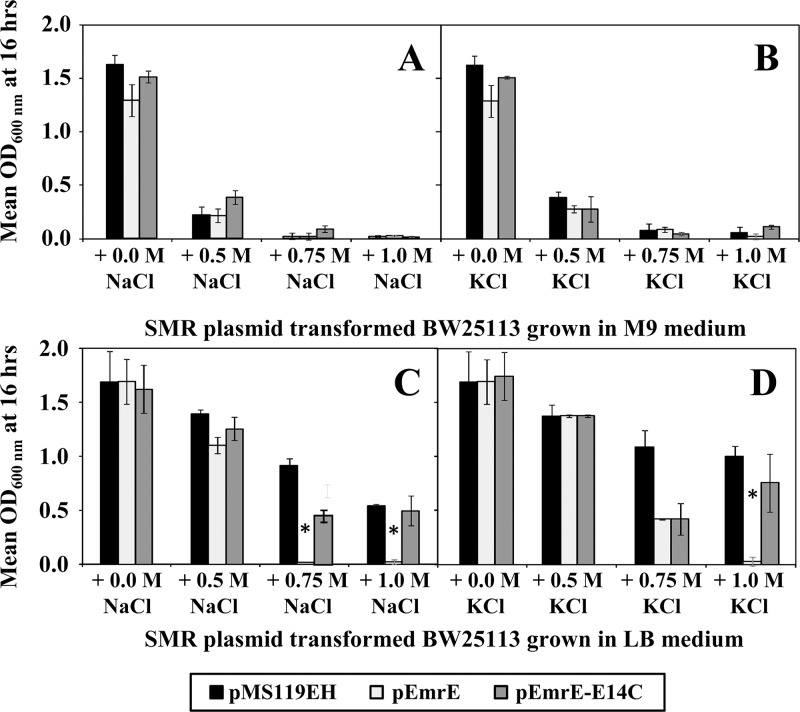
Based on the outcome of the pH growth assays, additional growth curve experiments were performed in LB and M9 media (neutral pH 7.0) at increasingly hypersaline salt concentrations to determine if high osmotic conditions also induce a growth phenotype in the presence of EmrE. Two salts, NaCl and KCl, were selected to determine if the loss-of-growth phenotype in the presence of overaccumulated EmrE is influenced by a particular salt. If EmrE transport activity reduces intracellular QCC-based osmoprotectants, a reduction or loss of growth should be observed to a greater extent in low-osmolarity M9 medium compared to the high-osmolarity LB medium. At hypersaline concentrations of 0.25 M, 0.5 M, 0.75 M, and 1.0 M for NaCl and KCl in LB medium, reduced growth was only observed in pEmrE-transformed strains at salt concentrations above 0.75 M (Fig. 2; see Fig. S2 in the supplemental material). The emrE-E14C inactive variant showed no significant growth differences from the empty vector control (Fig. 2; see Fig. S2). As would be expected for cultures grown without osmoprotectants, the growth of all plasmid-transformed E. coli cultures in either NaCl or KCl hypersaline M9 medium was severely reduced, and the viability of these cultures was eliminated after 6 h of growth at salt concentrations at or above 0.75 M (Fig. 2; see Fig. S2). Similar to pH susceptibility experiments, hypersaline growth experiments using E. coli emrE gene deletion strain JW0531 in LB and M9 media showed no differences from the wild-type strain BW25113 (see Fig. S3 in the supplemental material). This indicates that elimination of emrE genes has no effect on host osmotic tolerance under the growth conditions examined.
Fig 2.
Hypersaline susceptibility of E. coli BW25113 transformed with SMR plasmids. All panels show mean OD600 values from plasmid-transformed E. coli BW25113 strains after 16 h of growth in either M9 (A and B) or LB (C and D) medium at hypersaline concentrations of NaCl (A and C) or KCl (B and D) ranging from 0.0 to 1.0 M salt. In each panel, the mean OD600 values for E. coli BW25113 transformed with pMS119EH (black), pEmrE (gray), and pEmrE-E14C (dark gray) are provided in a bar chart format. Asterisks indicate statistically significant differences (P < 0.01) compared to either the untransformed strain lacking a plasmid (data not shown) or the strain transformed with the empty control vector (pMS119EH).
Screening of various osmoprotectants under hypersaline growth conditions identified that emrE participates in betaine and choline export.
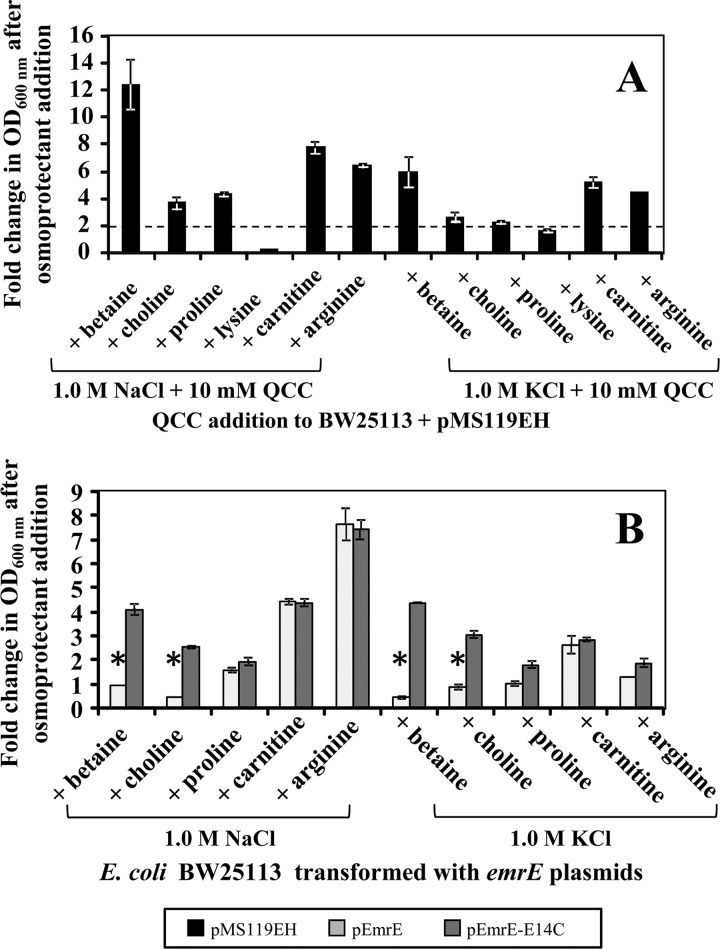
Based on the outcome of the hyperosmotic EmrE susceptibility growth curves, hypersaline M9 medium provides an ideal environment to screen various QCC-based osmoprotectants. The loss-of-growth phenotype in all transformed strains grown in M9 medium at salt concentrations at or above 0.75 M provides an ideal screening background to determine if an osmoprotectant compound is a substrate of EmrE. Previous studies have demonstrated that betaine and choline act as osmoprotectants at concentrations in excess of 5 mM when added to E. coli medium and enhance E. coli growth under hyperosmotic conditions (18). Other compounds, such as l-carnitine (32), and amino acid compounds, such as l-proline (9) and l-lysine (30), can also confer osmoprotection to E. coli based on previous studies. l-Arginine was included as a natural QCC control compound that is not directly involved in osmoprotection but has demonstrated an ability to increase osmolarity as an osmolyte (30). Hence, 6 different QCC-based osmoprotectants, betaine, choline, l-carnitine, l-proline, l-lysine, and l-arginine, were screened (at 10 mM concentration) for their ability to enhance E. coli growth in hypersaline M9 medium (1.0 M NaCl or KCl). Before screening, all 6 osmoprotectant substrates examined in this study were expected to rescue cell growth of pMS119EH-transformed BW25113 by a 2-fold minimum based on the outcome from previous experiments. As shown in Fig. 3A, all hypersaline M9-grown pMS119EH-transformed E. coli cultures grown in the presence of each osmoprotectant demonstrated a ≥2-fold increase in growth (OD600), except in the presence of l-lysine. Increasing the concentration of l-lysine in hypersaline M9 medium above 10 mM (to 100 mM) failed to significantly enhance hypersaline growth tolerance, and l-lysine was excluded from further screening. Betaine conferred the highest fold increase in pMS119EH-transformed E. coli growth (NaCl, 13-fold; KCl, 7-fold) under both hypersaline M9 conditions, indicating that this compound provides the most osmoprotection of all substrates screened.
Fig 3.
Growth phenotype screens of plasmid-transformed E. coli BW25113 in hypersaline M9 medium in the presence of various osmoprotectants. In all panels, the fold change in OD600 (growth) after osmoprotectant addition was determined after 16 h of growth at 37°C. The fold changes in OD600 after osmoprotectant addition in panels A and B are shown for E. coli transformed with plasmids pMS119EH (black), pEmrE (gray), and pEmrE-E14C (dark gray). All panels represent the fold changes in the growth of the culture in M9 medium at 1.0 M salt (NaCl or KCl) with 10 mM osmoprotectant divided from its mean OD600 value in M9 medium at 1.0 M salt only, as indicated on the x axis. Panel A demonstrates the osmoprotection conferred by the addition of 10 mM osmoprotectant (refer to x axis) based on the growth of pMS119EH-transformed E. coli control strains in hypersaline (either 1.0 M NaCl or KCl) M9 medium. The area below the dashed line on this chart indicates any osmoprotectants that failed to exceed the 2.5-fold-cutoff value statistically determined to identify valid osmoprotectants for hypersaline EmrE osmoprotection screens shown in panel B. Panel B shows the results of hypersaline EmrE osmoprotection screens as the fold change in growth (OD600) of E. coli BW25113(pEmrE) (gray) and BW25113(pEmrE-E14C) (dark gray) caused by the addition of 10 mM osmoprotectant (refer to x axis) to M9 medium containing either 1.0 M NaCl or KCl.
Since the remaining 5 osmoprotectants conferred osmotic tolerance to E. coli transformed with the control vector, any osmoprotectant that is a substrate of overaccumulated EmrE should be transported out of E. coli, and the strain should demonstrate a loss-of-growth phenotype in the presence of the osmoprotectant. Hence, the fact that a pEmrE-transformed E. coli strain fails to grow in the presence of the osmoprotectant shows that the osmoprotectant is a substrate of EmrE. In contrast, the strain transformed with the inactive variant of EmrE-E14C (pEmrE-E14C) or empty vector (pMS119EH) should confirm the specificity for the substrate by demonstrating cell growth under the same conditions. As shown in Fig. 3B, hypersaline growth phenotype screening assays of E. coli BW25113 transformed with pEmrE and pEmrE-E14C in the presence of 5 osmoprotectants indicated that only betaine and choline appear to be substrates of EmrE, based on fold change growth differences. Only betaine and choline addition to hypersaline M9 cultures of pEmrE-transformed E. coli cultures resulted in significant fold growth losses (≤1) in comparison to the growth of the inactive variant under the same conditions. The hypersaline screen may also suggest that proline is a potential substrate of EmrE. However, the fold growth change values of pEmrE- and pEmrE-E14C-transformed E. coli were at the threshold of significance, since a loss of growth was only observed for pEmrE-transformed E. coli under hypersaline KCl conditions (Fig. 3B). Repeating the KCl hypersaline EmrE osmoprotectant growth screens at higher concentrations of proline (20 and 30 mM) resulted in similar inconclusive growth phenotypes of pEmrE-transformed E. coli. The remaining two osmoprotectants, carnitine and arginine, showed no osmoprotection phenotype differences since both of the fold growth changes between pEmrE- and pEmrE-E14C-transformed E. coli strains were statistically similar under both hypersaline condition (Fig. 3B).
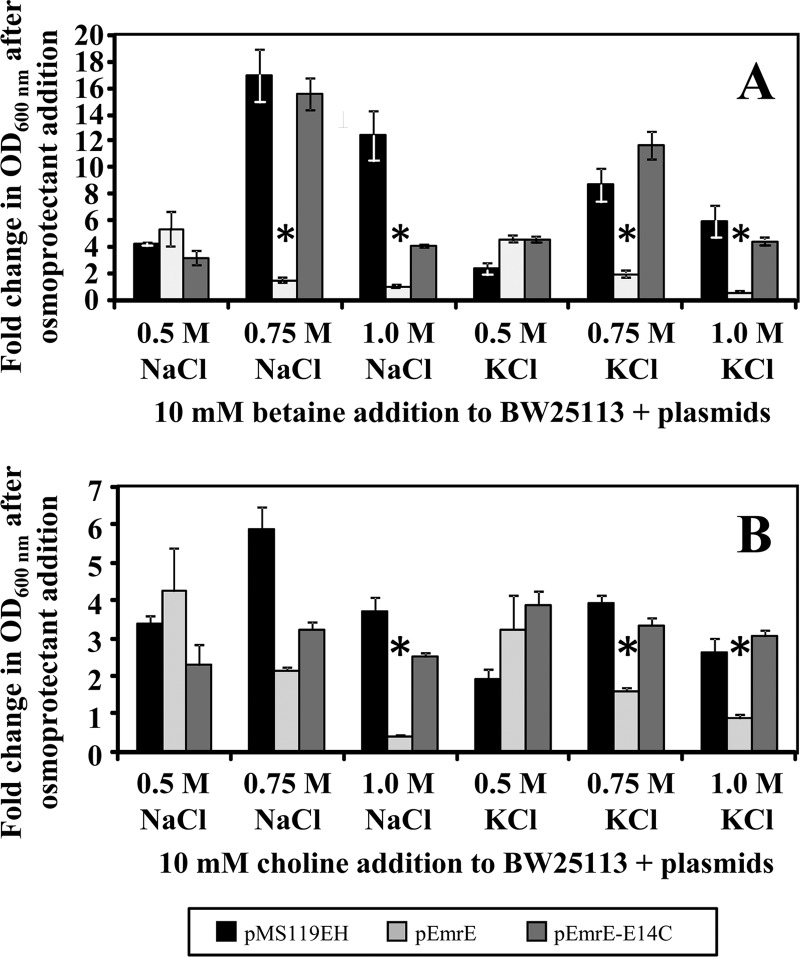
To ensure that betaine and choline were accurately identified from this hypersaline EmrE osmoprotection screen, growth curve experiments were repeated for the wild-type E. coli strain (BW25113) transformed with pEmrE or pEmrE-E14C at lower saline concentration ranges (0.5 to 1.0 M salt) in the presence of the both osmoprotectants to ensure that the growth phenotype was caused by the hyperosmotic condition and was not due to specific osmoprotectant-induced transport (Fig. 4). Similar to the results of the hypersaline susceptibility screening experiments shown in Fig. 3, losses of the growth phenotype in the presence of betaine were only observed at salt concentrations at or above 0.75 M by pEmrE-transformed E. coli cultures (Fig. 4A). Similar results were obtained for the reduced hypersaline choline osmoprotection growth assay of pEmrE-transformed E. coli strains (Fig. 4B). This indicates that hypersaline concentrations of 0.75 M are important to observe growth phenotype differences between the active and inactive EmrE variants. It is important to note that in some cases, the hypersaline growth of pEmrE-E14C-transformed E. coli in the presence of betaine or choline only partially restored the growth phenotypes in comparison to the control pMS119EH-transformed E. coli values (1.0 M NaCl in Fig. 5A and 0.075 M NaCl in Fig. 5B). Since the fold growth changes presented in Fig. 5 (and growth curves in Fig. S3 in the supplemental material) represent mean OD600 differences between cultures grown in hypersaline medium with osmoprotectants and those grown without, reduced growth of EmrE-E14C compared to pMS119EH transformants may reflect partial activity of EmrE-E14C. Partial QCC substrate interactions between E. coli EmrE-E14 variants and QCC substrates have been reported for QCC substrates such as acriflavine and tetraphenylphosphonium in vitro transport experiments (6, 24, 37) and in vivo E. coli QCC growth tolerance/resistance assays (35).
Fig 4.
Mean fold change in growth (OD600) of emrE plasmid-transformed E. coli BW25113 in the presence of betaine and choline. The fold change in OD600 (growth) after osmoprotectant addition is shown on the y axis of both charts and represents the fold change in growth of plasmid-transformed E. coli BW25113 in hypersaline medium after the addition of 10 mM osmoprotectant (refer to x axis) at 16 h of growth at 37°C. In both panels, the fold change in growth after osmoprotectant addition is provided for E. coli BW25113 transformed with pMS119EH (black), pEmrE (gray), or pEmrE-E14C (dark gray) in hypersaline M9 medium at 0.5, 0.75, and 1.0 M NaCl or KCl in the presence and absence of 10 mM betaine (A) or choline (B). Asterisks indicate statistically significant differences (P < 0.01) compared to either the untransformed strain lacking a plasmid (data not shown) or the strain transformed with the empty control vector (pMS119EH).
Fig 5.
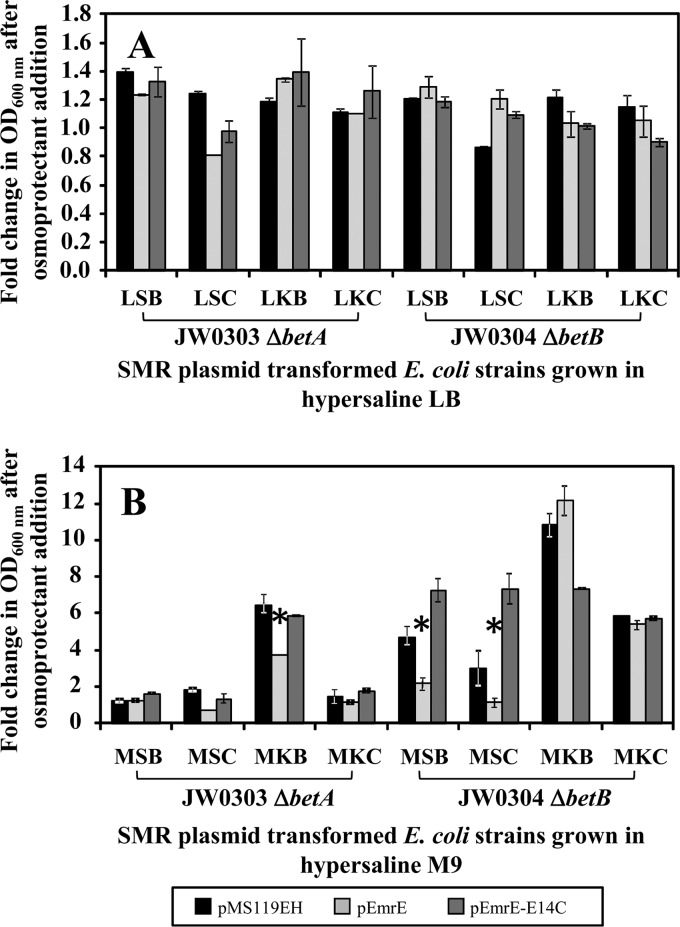
Hypersaline tolerance of emrE plasmid-transformed E. coli JW0303 and JW0304 strains grown in the presence of betaine and choline. On the y axis, both panels show the fold change in OD600 (growth) after osmoprotectant addition to the medium after 16 h of growth in either LB (A) medium or M9 (B) medium at a 1.0 M concentration of NaCl or KCl and 10 mM betaine or choline osmoprotectants. E. coli strains JW0303 (ΔbetA) and JW0304 (ΔbetB) transformed with pMS119EH (black), pEmrE (gray), and active site mutant pEmrE-E14C (dark gray) are shown on the x axis. The three-letter abbreviations provided on the x axes of both panels A and B indicate the culture medium compositions and are defined below. Panel A shows results for plasmid-transformed JW0303 and JW0304 after 16 h of growth in LB medium containing NaCl with betaine (LSB) or choline (LSC) and LB medium containing KCl with betaine (LKB) or choline (LKC). Panel B shows the fold change in growth after osmoprotectant addition to plasmid-transformed JW0303 and JW0304 after 16 h of growth in M9 medium containing NaCl with betaine (MSB) or choline (MSC) and M9 medium containing KCl with betaine (MKB) or choline (MKC). Asterisks indicate statistically significant differences (P < 0.01) compared to either the untransformed strain without plasmid or the strain transformed with the empty control vector (pMS119EH).
Evaluation of EmrE growth phenotypes in choline and betaine biosynthetic null backgrounds.
To confirm that betaine and choline are EmrE substrates, an additional hypersaline growth assay was performed in E. coli K-12 strains JW0303 and JW0304, lacking betaine and choline biosynthetic genes that encode the enzymes choline dehydrogenase (betA) and betaine-aldehyde dehydrogenase (betB), respectively. Previous studies involving the osmotic tolerance of the E. coli ΔbetA and/or ΔbetB mutants identified that the elimination of betaine-aldehyde dehydrogenase activity and/or its gene enhanced cell growth in the presence and absence of osmoprotectants under hyperosmotic conditions in comparison to wild-type E. coli (14, 17, 18). Based on this study, the osmoprotective growth phenotype conferred by betB is due to increased accumulation of intracellular glycine betaine aldehyde (17, 18). By extension, the elimination of choline dehydrogenase (ΔbetA) is expected to increase the intracellular accumulation of osmoprotective choline. To determine if increasing the intracellular concentration of either osmoprotective compound during the same hypersaline growth screens can rescue the emrE growth phenotype, pEmrE and pEmrE-E14C were transformed into either ΔbetA (JW0303) or ΔbetB (JW0304) gene deletion strains. The expected outcome for this experiment was that exogenous addition of betaine or choline to either E. coli ΔbetAB deletion strain in the presence of pEmrE would rescue the EmrE-induced osmotic hypersensitivity of the strain by the additional intracellular accumulation of osmoprotectants. In contrast to wild-type E. coli strains transformed with pEmrE (Fig. 4), Fig. 5A shows a consistent 1- to 1.4-fold increase in growth of all plasmid-transformed E. coli JW0303 and JW0304 strains in hypersaline LB medium in the presence or absence of betaine or choline. Since LB medium has 100-fold greater osmolarity than M9 medium, this result was not surprising for betA and betB deletion strains. Unexpectedly, only E. coli JW0303 or JW0304 strains transformed with pEmrE grown in 0.75 or 1.0 M KCl containing M9 medium supplemented with 10 mM betaine or choline reversed the loss-of-growth phenotype observed for pEmrE-transformed wild-type E. coli experiments (Fig. 5B). M9 medium hypersaline growth experiments involving NaCl containing betaine or choline showed lower fold growth changes of in either E. coli JW0303 or JW0304 transformed with pEmrE in comparison to KCl (Fig. 5B). The difference between NaCl and KCl suggests that EmrE activity may be reduced or inhibited in the presence of NaCl. Although the results from hypersaline M9 medium osmoprotection growth assays suggest that KCl was preferential to NaCl, growth-rescuing phenotypes were observed for E. coli ΔbetA and ΔbetB gene deletion strains overexpressing emrE under hyperosmotic growth conditions (LB medium). This outcome indicates that betaine and choline are substrates of EmrE and when grown in an osmotically limited growth medium (M9) for transport, either omsoprotectant is influenced by particular salts.
DISCUSSION
The results of this study strongly indicate that the expression of the E. coli multidrug transporter emrE reduces the pH and osmotic tolerance of its host by the transport of QCC-based osmoprotectants betaine and choline. Only functional emrE gene expression reduced the growth of E. coli in M9 medium at neutral (pH 7) and alkaline (pH 8 to 9) pH (Fig. 1), and this effect does not eliminate cell viability. Hyperosmotic growth assays of E. coli pEmrE and pEmrE-E14C strains in hypersaline LB and M9 media identified that both media resulted in a significant loss of growth for cultures overexpressing functional emrE only (Fig. 2).
The loss-of-growth phenotype of E. coli overaccumulating EmrE under defined hyperosmotic growth conditions supported our hypothesis that QCC-based osmoprotectants are possible substrates of SMR transporters. E. coli hypersaline growth assays in the presence of various compounds identified that only betaine and choline failed to rescue the loss-of-growth phenotype caused by EmrE overaccumulation (Fig. 3 and 4). Other QCC-based osmoprotectants we examined using the hypersaline osmoprotectant screening method, such as carnitine, proline, and arginine, failed to surpass the thresholds necessary to indicate statistically significant transport (Fig. 3). One exception was hypersaline screening experiments involving the osmoprotectant l-proline. The addition of proline to M9 medium at high KCl concentrations demonstrated a loss-of-growth phenotype in pEmrE-transformed E. coli cultures only (Fig. 3B), suggesting that under particular conditions, proline may be an additional substrate of EmrE. Studies of an unrelated QCC multidrug transporter from Lactobacillus plantarum QacT have also demonstrated an affinity for the transport of osmoprotectants glycine betaine and carnitine but low affinities for proline (15). One explanation for the specific proline affinity in our experiments may be due to the activities of alternative proline efflux mechanisms, such as mechanosensitive channels and its own dedicated transporter, ProP (as reviewed in reference 27). Furthermore, the uptake of betaine has been shown to increase the growth rate of E. coli more than proline uptake and results in intracellular (cytoplasmic) K+ depletion (9). Hence, the intermediate-growth phenotypes of pEmrE-transformed E. coli strain cultures observed in the presence of proline at high KCl concentrations may be due to the activity of other osmotically induced transport systems or due to a low EmrE affinity for proline. It should be noted that the remaining QCC osmoprotectants, l-carnitine and l-arginine, may still be potential substrates of EmrE, but the conditions for their transport could be specifically linked to an as-yet-undetermined conditional physiology or environmental stress.
It is important to note that hypersaline overexpression experiments performed with the same E. coli strain transformed with plasmids expressing SugE, an SMR protein family member with a limited transport profile in comparison to EmrE, did not alter growth phenotypes in comparison to empty-vector controls (unpublished results). The differences between emrE and sugE provide additional evidence supporting functional differences and evolutionary divergence within the SMR family and between multidrug transporters (5).
An important observation from the pH susceptibility growth assays of plasmid-transformed BW25113(pEmrE) strains was the significant lag in cell growth at neutral to alkaline pH ranges in M9 minimal medium (Fig. 1). The observable lag phase in growth was not due to fluctuations in the measurable pH of the M9 medium (which remained relatively constant) as determined at various time points over the course of growth, indicating that the neutral to alkaline pH influences EmrE activity. This suggests that acidic environmental pH ranges may alter the transport function and activity of EmrE in E. coli. Previous studies performed on the Staphylococcus aureus SMR homologue QacC/Smr demonstrated that SMR proteins were reliant on proton motive force to drive QCC export, making SMR proteins secondary active transporters (16, 20). pH-based alterations of EmrE substrate efflux have also been demonstrated in studies involving purified EmrE protein reconstituted into artificial membranes, which revealed changes in QCC transport at acidic pH values (11, 22, 36, 38). Further experimental exploration of pH-induced EmrE substrate transport using this in vivo pH susceptibility screening method may help resolve these issues.
Another important finding from this study was the apparent difference in levels of E. coli host osmoprotection by betaine and choline in the presence of EmrE at hypersaline NaCl and KCl in the presence of betaine or choline (Fig. 3, 4, and 5). The results from this study indicate that hypersaline concentrations of NaCl (>0.75 M) specifically inhibit the osmoprotective growth phenotype of osmotolerant E. coli strains (ΔbetA and ΔbetB) overexpressing emrE in M9 medium in comparison to experiments involving KCl in the presence of betaine or choline (Fig. 5). The inhibition of osmotolerance caused by NaCl may be due to poisoning of the proton relay in pEmrE-transformed E. coli. In this event, Na+ ions may inhibit the functional activity of EmrE by competing with H+ ions that are known to be coupled to EmrE drug efflux activity. This may indicate that the K+ plays a role in EmrE-meditated QCC/osmoprotectant transport activities, and both salts should be examined in future EmrE transport studies.
The influence of pH and salinity on multidrug transporter activity is not unprecedented. Another multidrug transporter involved in QCC efflux, E. coli MdfA, alters host hypersaline tolerance to Na+ and K+ ions and enhances host alkali tolerance (19). Unlike MdfA overaccumulation, EmrE appears to enhance host alkali susceptibility and decrease hypersaline tolerance, suggesting that EmrE may participate in osmotic regulation by eliminating the buildup of these compounds during or after significant osmotic stress. SMR family protein involvement in host osmotic susceptibility may also provide insight into why many multidrug transporters have overlapping QCC substrates. In E. coli, a variety of larger multidrug transporters (12 to 14 transmembrane strands) transport similar QCC substrates recognized and transported by SMR proteins and include the YdhE (MdtK) multidrug and toxic compound extrusion (MATE) transporter (21), the AcrA and AcrB complex from the root nodulation and cell division (RND) family (31), and the EmrA and EmrB transporter complex of the major facilitator superfamily (MFS) (13). The functional redundancy of QCC resistance conferred by diverse transporter families may be select for a particular transporter to be active under a particular physiological condition and for EmrE may involve participation in an osmotic downshock. The presence of multiple multidrug transporters with similar substrate recognitions profiles often masks phenotypes derived solely from emrE (29, 31) and may explain why single-gene deletions of emrE in E. coli did not significantly alter hyperosmotic tolerance (see Fig. S1 and S2 in the supplemental material).
The hyperosmotic susceptibility screening method used for this study indicates that betaine and choline are biologically relevant QCC substrates of EmrE. Other substrates screened herein, such as carnitine, proline, and arginine, were not identified as osmoprotectant substrates of EmrE under the conditions examined in this study. In addition to their osmoprotective roles, the impact of betaine and choline transport by EmrE may suggest that this particular SMR subclass (SMP) member may regulate osmotic regulation by rapidly removing betaine and choline osmoprotectants when E. coli cells no longer reside under hyperosmotic growth conditions. In conclusion, this screening method has identified that betaine and choline are specific QCC osmoprotectant substrates of EmrE based on an assay of various candidates. This method will provide a useful nontoxic biologically relevant strategy to screen for other biological substrates of additional EmrE protein, other SMR homologues, or other unrelated multidrug transporters suspected of cation efflux.
Supplementary Material
ACKNOWLEDGMENTS
We thank Shannon Sarro, Komal Sidhu, Karen Duncalf, and Jason Burt for experimental assistance and Veerle De Wever for helpful manuscript discussions.
Funding for this work was provided by operating and accelerator grants from the National Science and Engineering Research Council (NSERC) to R.J.T.
Footnotes
Published ahead of print 31 August 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio Collection. Mol. Syst. Biol. 2: 2006– 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baldwin WW, Kubitschek HE. 1984. Evidence for osmoregulation of cell growth and buoyant density in Escherichia coli. J. Bacteriol. 159: 393– 394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baldwin WW, Myer R, Kung T, Anderson E, Koch AL. 1995. Growth and buoyant density of Escherichia coli at very low osmolarities. J. Bacteriol. 177: 235– 237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bay DC, Rommens KL, Turner RJ. 2008. Small multidrug resistance proteins: a multidrug transporter family that continues to grow. Biochim. Biophys. Acta 1778: 1814– 1838 [DOI] [PubMed] [Google Scholar]
- 5. Bay DC, Turner RJ. 2009. Diversity and evolution of the small multidrug resistance protein family. BMC Evol. Biol. 9: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bay DC, Turner RJ. 2012. Spectroscopic analysis of small multidrug resistance protein EmrE in the presence of various quaternary cation compounds. Biochim. Biophys. Acta 1818: 1318– 1331 [DOI] [PubMed] [Google Scholar]
- 7. Booth IR. 1985. Regulation of cytoplasmic pH in bacteria. Microbiol. Rev. 49: 359– 378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bourot S, et al. 2000. Glycine betaine-assisted protein folding in a lysA mutant of Escherichia coli. J. Biol. Chem. 275: 1050– 1056 [DOI] [PubMed] [Google Scholar]
- 9. Cayley S, Lewis BA, Record MT., Jr 1992. Origins of the osmoprotective properties of betaine and proline in Escherichia coli K-12. J. Bacteriol. 174: 1586– 1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chan CS, Zlomislic MR, Tieleman DP, Turner RJ. 2007. The TatA subunit of Escherichia coli twin-arginine translocase has an N-in topology. Biochemistry 46: 7396– 7404 [DOI] [PubMed] [Google Scholar]
- 11. Curnow P, Lorch M, Charalambous K, Booth PJ. 2004. The reconstitution and activity of the small multidrug transporter EmrE is modulated by non-bilayer lipid composition. J. Mol. Biol. 343: 213– 222 [DOI] [PubMed] [Google Scholar]
- 12. de Boer HA, Comstock LJ, Vasser M. 1983. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc. Natl. Acad. Sci. U. S. A. 80: 21– 25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Elkins CA, Mullis LB. 2007. Substrate competition studies using whole-cell accumulation assays with the major tripartite multidrug efflux pumps of Escherichia coli. Antimicrob. Agents Chemother. 51: 923– 929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Falkenberg P, Strom AR. 1990. Purification and characterization of osmoregulatory betaine aldehyde dehydrogenase of Escherichia coli. Biochim. Biophys. Acta 1034: 253– 259 [DOI] [PubMed] [Google Scholar]
- 15. Glaasker E, Heuberger EH, Konings WN, Poolman B. 1998. Mechanism of osmotic activation of the quaternary ammonium compound transporter (QacT) of Lactobacillus plantarum. J. Bacteriol. 180: 5540– 5546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grinius LL, Goldberg EB. 1994. Bacterial multidrug resistance is due to a single membrane protein which functions as a drug pump. J. Biol. Chem. 269: 29998– 30004 [PubMed] [Google Scholar]
- 17. Lamark T, Styrvold OB, Strom AR. 1992. Efflux of choline and glycine betaine from osmoregulating cells of Escherichia coli. FEMS Microbiol. Lett. 75: 149– 154 [DOI] [PubMed] [Google Scholar]
- 18. Landfald B, Strom AR. 1986. Choline-glycine betaine pathway confers a high level of osmotic tolerance in Escherichia coli. J. Bacteriol. 165: 849– 855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lewinson O, Padan E, Bibi E. 2004. Alkalitolerance: a biological function for a multidrug transporter in pH homeostasis. Proc. Natl. Acad. Sci. U. S. A. 101: 14073– 14078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Littlejohn TG, et al. 1992. Substrate specificity and energetics of antiseptic and disinfectant resistance in Staphylococcus aureus. FEMS Microbiol. Lett. 74: 259– 265 [DOI] [PubMed] [Google Scholar]
- 21. Long F, Rouquette-Loughlin C, Shafer WM, Yu EW. 2008. Functional cloning and characterization of the multidrug efflux pumps NorM from Neisseria gonorrhoeae and YdhE from Escherichia coli. Antimicrob. Agents Chemother. 52: 3052– 3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miller D, et al. 2009. In vitro unfolding and refolding of the small multidrug transporter EmrE. J. Mol. Biol. 393: 815– 832 [DOI] [PubMed] [Google Scholar]
- 23. Morrison EA, et al. 2012. Antiparallel EmrE exports drugs by exchanging between asymmetric structures. Nature 481: 45– 50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Muth TR, Schuldiner S. 2000. A membrane-embedded glutamate is required for ligand binding to the multidrug transporter EmrE. EMBO J. 19: 234– 240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Paulsen IT, Brown MH, Skurray RA. 1996. Proton-dependent multidrug efflux systems. Microbiol. Rev. 60: 575– 608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paulsen IT, et al. 1996. The SMR family: a novel family of multidrug efflux proteins involved with the efflux of lipophilic drugs. Mol. Microbiol. 19: 1167– 1175 [DOI] [PubMed] [Google Scholar]
- 27. Poolman B, Glaasker E. 1998. Regulation of compatible solute accumulation in bacteria. Mol. Microbiol. 29: 397– 407 [DOI] [PubMed] [Google Scholar]
- 28. Purewal AS. 1991. Nucleotide sequence of the ethidium efflux gene from Escherichia coli. FEMS Microbiol. Lett. 66: 229– 231 [DOI] [PubMed] [Google Scholar]
- 29. Schuldiner S. 2009. EmrE, a model for studying evolution and mechanism of ion-coupled transporters. Biochim. Biophys. Acta 1794: 748– 762 [DOI] [PubMed] [Google Scholar]
- 30. Shahjee HM, Banerjee K, Ahmad F. 2002. Comparative analysis of naturally occurring L-amino acid osmolytes and their D-isomers on protection of Escherichia coli against environmental stresses. J. Biosci. 27: 515– 520 [DOI] [PubMed] [Google Scholar]
- 31. Tal N, Schuldiner S. 2009. A coordinated network of transporters with overlapping specificities provides a robust survival strategy. Proc. Natl. Acad. Sci. U. S. A. 106: 9051– 9056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Verheul A, Wouters JA, Rombouts FM, Abee T. 1998. A possible role of ProP, ProU and CaiT in osmoprotection of Escherichia coli by carnitine. J. Appl. Microbiol. 85: 1036– 1046 [DOI] [PubMed] [Google Scholar]
- 33. Winstone TL, Duncalf KA, Turner RJ. 2002. Optimization of expression and the purification by organic extraction of the integral membrane protein EmrE. Protein Expr. Purif. 26: 111– 121 [DOI] [PubMed] [Google Scholar]
- 34. Yerushalmi H, Lebendiker M, Schuldiner S. 1995. EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J. Biol. Chem. 270: 6856– 6863 [DOI] [PubMed] [Google Scholar]
- 35. Yerushalmi H, Lebendiker M, Schuldiner S. 1996. Negative dominance studies demonstrate the oligomeric structure of EmrE, a multidrug antiporter from Escherichia coli. J. Biol. Chem. 271: 31044– 31048 [DOI] [PubMed] [Google Scholar]
- 36. Yerushalmi H, Schuldiner S. 2000. A common binding site for substrates and protons in EmrE, an ion-coupled multidrug transporter. FEBS Lett. 476: 93– 97 [DOI] [PubMed] [Google Scholar]
- 37. Yerushalmi H, Schuldiner S. 2000. An essential glutamyl residue in EmrE, a multidrug antiporter from Escherichia coli. J. Biol. Chem. 275: 5264– 5269 [DOI] [PubMed] [Google Scholar]
- 38. Yerushalmi H, Schuldiner S. 2000. A model for coupling of H(+) and substrate fluxes based on “time-sharing” of a common binding site. Biochemistry 39: 14711– 14719 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.