Abstract
Deinococcus radiodurans, a radiation-resistant bacterium, harbors a multipartite genome. Chromosome I contains three putative centromeres (segS1, segS2, and segS3), and ParA (ParA1) and ParB (ParB1) homologues. The ParB1 interaction with segS was sequence specific, and ParA1 was shown to be a DNA binding ATPase. The ATPase activity of ParA1 was stimulated when segS elements were coincubated with ParB1, but the greatest increase was observed with segS3. ParA1 incubated with the segS-ParB1 complex showed increased light scattering in the absence of ATP. In the presence of ATP, this increase was continued with segS1-ParA1B1 and segS2-ParA1B1 complexes, while it decreased rapidly after an initial increase for 30 min in the case of segS3. D. radiodurans cells expressing green fluorescent protein (GFP)-ParB1 produced foci on nucleoids, and the ΔparB1 mutant showed growth retardation and ∼13%-higher anucleation than the wild type. Unstable mini-F plasmids carrying segS1 and segS2 showed inheritance in Escherichia coli without ParA1B1, while segS3-mediated plasmid stability required the in trans expression of ParA1B1. Unlike untransformed E. coli cells, cells harboring pDAGS3, a plasmid carrying segS3 and also expressing ParB1-GFP, produced discrete GFP foci on nucleoids. These findings suggested that both segS elements and the ParA1B1 proteins of D. radiodurans are functionally active and have a role in genome segregation.
INTRODUCTION
The faithful transmission of genetic information from one cell to other cells by accurate genome partitioning is coordinated with cell division in both bacteria (16) and higher organisms (36). In eukaryotes, the actin filaments perform a number of functions associated with chromosome partitioning, the internalization of membrane vesicles, and the formation of the cytokinetic ring (39). In bacteria, genome partitioning occurs mainly by either the pushing or pulling of the duplicated genome toward the cell poles. This involves three core components, (i) an origin-proximal centromere or a similar cis element, (ii) centromere binding, and (iii) P-loop Walker ATPases, which, through polymerization/depolymerization dynamics, provide force leading to the movement of the centromere (16, 19). In the case of the R1 plasmid, it was demonstrated previously that ParR first binds to parC, a centromeric region, on the plasmid DNA, followed by the polymerization of ParM. The force generated due to ParM polymerization is utilized for pushing plasmid molecules toward opposite poles (8, 15, 37). In addition, an alternate genome partitioning system comprised of the parAB operon and its cognate centromeric sequence parS has been reported for certain plasmids and the majority of bacterial chromosomes (3, 20, 49). The nucleation of ParB or its homologues on cognate centromeric sequences (7, 33) and a dynamic change in the polymerization/depolymerization kinetics of cognate ParA-type proteins (19) eventually regulate the segregation of genetic elements. Furthermore, it has been shown that ParAs encoded on some of these plasmids and the majority of the bacterial chromosomes bind nonspecifically with DNA and move dynamically over the nucleoid (1, 9). These ParAs undergo polymerization in the presence of ATP, which, upon an interaction with ParB bound to the centromere, stimulates the intrinsic activity of ParAs, and this results in depolymerization (19, 20, 40). It has been shown that the levels of its intrinsic ATPase activity regulate the polymerization/depolymerization dynamics of ParA. A major understanding of the mechanisms underlying bacterial genome segregation has been obtained from bacteria harboring a single circular chromosome per cell and a low-copy-number plasmid. Recently, the genome sequences of several bacteria from diverse phylogenetic groups have been reported. Some of these bacteria, such as Agrobacterium tumefaciens, Sinorhizobium meliloti (25), Deinococcus radiodurans (52), and the human pathogen Vibrio cholerae (21), harbor multipartite genomes, and both primary and secondary chromosomes in many of these bacteria contain multiple centromeres (34). However, the molecular basis of multipartite genome segregation and the nature of chromosome partitioning systems in these organisms are not clearly known. In the case of Vibrio cholerae, it was shown previously that both chromosomes have distinct replication machineries (12) and that the localization and segregation systems are different (13, 54) as well as chromosome specific (24).
Deinococcus radiodurans is characterized by its extraordinary resistance to several abiotic stresses, including radiation and desiccation (4, 47). This bacterium contains four genetic elements, designated chromosome I (2.65 Mb), chromosome II (412 kb), the megaplasmid (177 kb), and the small plasmid (46 kb) (52). Chromosome I encodes the majority of the proteins essential for the normal growth of this bacterium and functions as the primary chromosome, while other genome replication units encode proteins that contribute largely to secondary phenotypes (35) of this bacterium. Except for the small plasmid, other genome replication units have their own encoded putative “Par” proteins, organized as putative parAB operons. Except for a recent study that characterized the roles of an SMC (structural maintenance of chromosome) protein and the SbcCD complex in chromosome maintenance (6), further studies of the mechanisms of genome maintenance and segregation have not been reported for D. radiodurans and would be worth investigation.
Here, we identified three putative chromosomal-type centromeric sequences on chromosome I of D. radiodurans, designated segS (cis elements involved in genome segregation), and demonstrated the sequence-specific interaction of a putative ParB-type protein (here referred to as ParB1) with segS elements under both in vivo and in vitro conditions. The ParA-type protein of chromosome I (here referred to as ParA1) was characterized as a DNA binding ATPase. Levels of ATPase activity stimulation and ParA1 polymerization/depolymerization dynamics measured by both sedimentation analysis and light scattering were different with the segS1, segS2, and segS3 elements. ParB1-green fluorescent protein (GFP) formed foci on the nucleoids of D. radiodurans and of Escherichia coli harboring recombinant plasmid pDAGS3 but not on the nucleoid of wild-type Escherichia coli. A ΔparB1 mutant of this bacterium showed growth retardation and a significantly high level of anucleation compared to the wild type. E. coli cells harboring an unstable mini-F plasmid cloned with segS elements showed very high levels of stable inheritance of these plasmids into daughter cells. These results suggested that ParB1 binding with segS was sequence specific and that ParA1 could produce the higher-order complex in the presence of segS-ParB1. Furthermore, the polymerization/depolymerization of ParA1 seems to be regulated by levels of its intrinsic ATPase activity in vitro. In vivo results further supported the role of the chromosome I “Par” system, comprised of ParB1, ParA1, and segS elements, in the maintenance of the D. radiodurans genome.
MATERIALS AND METHODS
Bacterial strains and materials.
D. radiodurans (ATCC 13939) was a generous gift from M. Schaefer (46). Cells of the wild type and its derivatives were maintained in TGY (0.5% Bacto tryptone, 0.3% Bacto yeast extract, 0.1% glucose) broth or on agar plates, as required, at 32°C. The E. coli expression vectors pET28a+ and p11559 (28) and their derivatives were maintained in E. coli strain DH5α. Kanamycin (25 μg/ml) and spectinomycin (40 μg ml−1) for E. coli and spectinomycin (75 μg ml−1) for D. radiodurans were used. Standard recombinant DNA techniques were used, as described previously (45). All molecular-biology-grade chemicals were purchased from Sigma Chemical Company; Roche Biochemicals, Germany; New England BioLabs; TaKaRa Bio Inc., Japan; and Bangalore Genei, India.
Bioinformatic analysis.
The genome sequence of D. radiodurans R1 was searched for both a B. subtilis-type centromere sequence (TGTTNCACGTGAAACA) and a P1 plasmid-type centromere (14) by using an NCBI-BLAST search. The consensus boxes similar to the typical P1 element were identified by using Bioedit, version 7, software. The functional motif search was carried out by using standard online bioinformatics tools, as reported previously (10). In brief, the amino acid sequences of DR_0012 and DR_0013, annotated as putative ParB and ParA proteins, respectively, on chromosome I of this bacterium were subjected to a PSI-BLAST search with the Swiss-Prot database, with “genome partitioning proteins” as key words. The sequences of close homology were aligned by T-COFFEE, and the conserved motifs were marked in CLUSTAL-X. The secondary structure was inferred from PSIPRED, JNET, and Prof with the Quick2D server of the Max Planck Institute for Developmental Biology.
Expression and purification of recombinant ParB1 and ParA1.
Genomic DNA of D. radiodurans R1 was prepared as reported previously (2), and open reading frames (ORFs) DR_0012 (ParB1) and DR_0013 (ParA1) were PCR amplified from genomic DNA by using primers dr0012F and dr0012R for the dr0012 (parB1) gene and primers dr0013F and dr0013R for the dr0013 (parA1) gene (see Table S1 in the supplemental material). PCR products were ligated at the NdeI and XhoI sites in pET28a+ to yield pET0012 and pET0013, respectively. Recombinant plasmids were transformed into E. coli BL21(DE3)/pLysS cells, and the recombinant ParA1/ParB1 proteins were purified by nickel affinity chromatography, as described previously (27). In brief, the cells were lysed in Cell-Lytic Express (Sigma Chemical Company), and the pellet-containing majority of the recombinant protein in inclusion bodies was dissolved in buffer B (100 mM NaH2PO4, 10 mM Tris-HCl [pH 8.0], and 8 M urea) and purified by using Ni-nitrilotriacetic acid (NTA) Sepharose according to the manufacturer's protocol (Qiagen, Germany). Partially purified protein was refolded by the serial dilution of urea with a concurrent increase in the dithiothreitol (DTT) concentration and repurified under native conditions using nickel affinity column buffer supplemented with 10% glycerol and 2% ethanol. Finally, the fractions showing more than 99% purity were pooled, dialyzed in buffer (20 mM Tris-HCl, 50 mM NaCl, 1 mM DTT, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 50% glycerol), and stored in small aliquots at −20°C until further use. The mass spectrometric analyses of both these recombinant proteins were carried out commercially (The Centre for Genomic Applications, New Delhi, India), and their identities were confirmed.
DNA-protein interaction studies.
For the DNA binding assay of the ParB1 protein, the 300-bp putative segS1, segS2, and segS3 elements located on chromosome I were PCR amplified from genomic DNA of D. radiodurans by using sequence-specific primers (see Table S1 in the supplemental material). The PCR products were gel purified and kinased with [γ-32P]ATP. Approximately 0.1 pmol labeled substrate was incubated with different concentrations of purified recombinant ParB1 and ParA1, in different combinations, in 20 μl of reaction buffer containing 50 mM Tris-HCl (pH 8.0), 5 mM MnCl2, 75 mM NaCl, and 0.1 mM DTT at 37°C for 10 min. For the competition assay, ParB1 was incubated with seg sequences before the 250-bp competitor DNA was added and was further incubated with or without ParA1 and ATP in different combinations, as per experimental requirements. Mixtures were separated on 6% native PAGE gels, the gels were dried, and autoradiograms were developed with a PhosphorImager (Molecular Dynamics Inc.). For studies of the effect of ParA1 polymerization in the presence of segS and ParB1, the segS elements were incubated with different concentrations of ParB1 and ParA1 separately and in combination, with and without ATP. Because of the formation of larger macromolecular complexes of these proteins with cis elements, the interactions of ParA1, ParB1, and cis elements were analyzed on a 1.2% agarose gel. DNA bands were visualized with ethidium bromide, and the band intensity was quantified by using Gene Genius ImageQuant software (SynGene Inc.), as required. The integrated intensities for bound and unbound fractions per unit area were measured separately, and the DNA fraction bound to the protein was plotted as a function of the protein concentration by using Graphpad Prism 5. The Kd (dissociation constant) for the curve fitting of individual plots was determined with software working on the principle of the least-squares method by applying the formula Y = Bmax × [X]/Kd + [X], where [X] is the protein concentration, Y is the bound fraction, and Bmax is the maximum binding capacity, as described previously (10).
Measurements of ATPase activity.
The ATPase activity of recombinant ParA1 was checked by using a modified protocol described previously (27). In brief, increasing concentrations of purified ParA1 (200 ng to 1,000 ng) were incubated with 2 mM ATP at 37°C for 1 h. To determine the effect of DNA elements on ParA1 activity, ∼1,000 ng of ParA1 was incubated with a preincubated mixture of 200 ng ParB1 and 100 ng DNA at 37°C for 1 h. The reaction was terminated by the addition of a one-quarter volume of malachite green reagent to the mixture, followed by a further incubation for 20 min. The release of Pi was measured at 630 nm and quantified by using standard procedures, essentially as described previously (53). The ATPase activity was calculated as nmol Pi formed per min μg protein−1.
Cloning of segS elements on the mini-F plasmid and plasmid stability studies.
All three segS elements were PCR amplified from genomic DNA of D. radiodurans by using sequence-specific primers (see Table S1 in the supplemental material). The ends of the segS1, segS2, and segS3 PCR products were made blunt with T4 DNA polymerase and cloned separately at the end-filled SexAI site in mini-F plasmid pDAG203 (30). The recombinant plasmids were designated pDAGS1, pDAGS2, and pDAGS3, respectively. Both recombinant plasmids and the pDAG203 vector were separately mobilized into E. coli MG1655 cells. In parallel, the putative parA1-parB1 operon was PCR amplified by using primers CIOF and CIOR and cloned at the BamHI and HindIII sites into pDSW209 (51). Recombinant proteins were expressed in E. coli cells harboring plasmid pDAGS3/pDAG203. The stable inheritance of plasmids pDAGS1, pDAGS2, and pDAGS3 was compared with that of pDAG203 as a control by scoring chloramphenicol (Cm) resistance in daughter cells, as described previously (18). In brief, these cells were grown in the absence of Cm, an antibiotic marker on plasmid pDAG203, and the cells were then selected in the presence of Cm (15 μg ml−1).
Nucleotide sequencing and determination of the plasmid-to-chromosome ratio.
Recombinant plasmids harboring segS1, segS2, and segS3 were sequenced for their respective segS elements by using dideoxy-chain termination chemistry on an ABI 100 sequencer (model 377). The sequences obtained were searched for homologous sequences in databases. E. coli cells harboring these plasmids and the pDAG203 vector were grown in LB medium supplemented with chloramphenicol (15 μg ml−1). Genomic DNA was prepared according to protocols described previously (45). The plasmid-specific cat gene and the chromosome-specific xerC gene were PCR amplified with 20 ng and 150 ng of template from the same pool of genomic DNA, respectively. Products were analyzed on an agarose gel, and the band intensity was estimated densitometrically. The ratio of the yield of cat to the yield of xerC was estimated and analyzed.
Construction of a parB1 deletion mutant in D. radiodurans.
For the generation of a parB1 deletion mutant in D. radiodurans, a suicidal plasmid, pNOK0012, was constructed by using a strategy described previously (26). In brief, the fragments 1 kb upstream and 1 kb downstream of ORF DR_0012 were PCR amplified with primers hari64 and hari65 for upstream fragments and primers hari66 and hari67 for downstream fragments (see Table S1 in the supplemental material) and cloned at the KpnI-EcoRI and BamHI-SacI sites, respectively. The recombinant plasmid thus obtained, pNOK0012, was linearized with XmnI and transformed into D. radiodurans cells. Transformants were maintained through several rounds of subculturing, and the homozygous replacement of parB1 with nptII was ascertained by PCR amplification using internal primers of parB1. Mutant cells completely missing parB1 were taken for subsequent studies. The growths of mutant and wild-type cells were monitored as the optical density at 600 nm.
Preparation of the translational fusion of ParB1 with GFP.
The parB1 gene was PCR amplified by using primers Hari12F and Hari12R (see Table S1 in the supplemental material) and cloned at the N terminus of GFP at compatible sites of pDSW209 (51) to yield pHJ0012. This allowed the generation of a translational fusion of ParB1 with GFP. Subsequently, the gfp-parB1 chimera was PCR amplified from pHJ0012 by using primers GfpF and ParB1R (see Table S1 in the supplemental material). In parallel, p11559 was modified by subcloning a 42-bp linker (5′-CATATGAGATCTAGTACTGAGCTCGTCGACCTTAAGCTCGAG-3′) at its NdeI and XhoI sites, generating NdeI, ScaI, SacI, SalI, AflII, and XhoI cloning sites downstream of the existing promoter, and the resulting plasmid was named pVHS559. The PCR-amplified gfp-parB1 chimera was subcloned at the SacI and XhoI sites of pVHS559 to yield pD12GFP. D. radiodurans cells harboring the vector and recombinant pD12GFP plasmids were grown in TYG broth containing spectinomycin (75 μg ml−1), and the inducible expression of recombinant proteins was ascertained as described previously (28). In brief, the E. coli cells were induced with 500 μM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h, while D. radiodurans cells were induced with 10 mM IPTG for 16 h. The IPTG-inducible expression of the GFP-ParB1 chimera on plasmids pHJ0012 and pD12GFP was confirmed for both E. coli and D. radiodurans, respectively, by immunoblotting using antibodies against GFP (Clontech, TaKaRa, Japan), according to protocols described previously (43).
Microscopy studies.
Fluorescence microscopy of D. radiodurans, its derivatives, and E. coli cells was carried out as described previously (55), using a Zeiss AxioImager (model LSMS10 Meta; Carl Zeiss) equipped with a Zeiss AxioCam HRm camera. For the localization of GFP-ParB1 foci, both E. coli and D. radiodurans R1 cells harboring pD12GFP were induced with IPTG, and the cells were stained with DAPI (4′,6-diamidino-2-phenylindole). Both GFP and DAPI fluorescences were imaged. Micrographs were superimposed for the localization of ParB1-GFP spots on the DAPI-stained nucleoid.
For deletion mutant studies, both ΔparB1 mutant and wild-type cells were grown overnight, and cells were imaged under bright-field and DAPI fluorescences by using a fluorescence microscope (AxioCam model MRC5; Carl Zeiss). A large number of cells from both wild-type and ΔparB1 mutant populations were examined for comparisons of anucleation frequencies at different time intervals of growth. Experiments were repeated to ensure the reproducibility and significance of these data.
Oligomerization studies using sedimentation assays and light scattering.
A sedimentation assay was carried out by using modified protocols described previously (5). In brief, 750 ng ParA1 was incubated with 100 ng segS elements, 200 ng ParB1, and 1 mM ATP in different combinations and permutations in a solution containing 50 mM HEPES (pH 7.6), 50 mM KCl, 5 mM MgCl2, 30 mM sodium acetate, and 100 μg/ml bovine serum albumin (BSA) for 15 min at 25°C. Reaction mixtures were centrifuged at 22,000 × g for 30 min at 4°C, and the proteins were analyzed on 10% SDS-PAGE gels. Protein bands were visualized with silver nitrate, and the ParA1 band intensity was quantified by densitometric scanning using GeneGenius ImageQuant software (SynGene Inc.).
Dynamic light scattering was measured as described previously (5), by using a Malvern 4800 Autosizer employing a 7132 digital correlator, as described previously (50). In brief, ∼1 μM ParA1 was incubated with ∼25 nM ParB1 and ∼5 nM segS centromeric sequences in 400 μl of reaction buffer (50 mM HEPES [pH 7.6], 50 mM KCl, 5 mM MgCl2, 100 μg ml−1 BSA, 30 mM sodium acetate) in the presence and absence of 1 mM ATP, as described in the figure legends. The scattered light intensity was measured at 90° from a 150-μm pinhole at 25°C for 10 s with 10 shots at 10-min intervals using a 25-mW He-Ne laser light source at 633 nm. The data obtained as kilocounts per second were analyzed by using inbuilt software for the machine.
RESULTS AND DISCUSSION
Chromosome I was identified to contain centromeres and encode genome partitioning proteins.
The genome sequence of D. radiodurans was searched for the plasmid-type cis element parS (14) and chromosomal-type centromere sequences, as reported previously for Bacillus subtilis (33). Homologous sequences were examined for features typical of bacterial centromeres. For example, the canonical parS element contains boxes of inverted repeats, which are sequentially organized as A1-B1-IHF (integration host factor) binding site-A2-A3-B2 in the P1 plasmid (14). Analyses showed that chromosome I and chromosome II of D. radiodurans (52) contain boxes of inverted repeats similar to those of typical plasmid-type parS elements (Dp1 and Dp2, respectively) (Fig. 1A). However, the arrangement of these repeats on the chromosomes and the spacing between these boxes were different from the typical structure of parS elements. In addition, both Dp1 and Dp2 lacked conserved IHF binding sites. The precise spacing between the boxes of inverted repeats and the presence of the IHF binding site are the features necessarily required for the productive interaction of parS with ParB. Therefore, the possibility of Dp1 functioning as a centromere for chromosome I segregation was nearly ruled out. On the other hand, chromosome I was found with three putative chromosomal-type centromeres (here referred to as segS) (Fig. 1B) similar to those reported previously for B. subtilis (33) and many other bacteria (34). Since these elements showed some difference in nucleotide sequence, they are designated segS1, segS2, and segS3 (Fig. 1). Previously, it was found that nearly 69% of bacterial chromosomes contain B. subtilis-type chromosomal centromeres (34). Homology searches and comparative analyses of centromeric sequences in the genomes of a large number of bacteria showed the presence of multiple centromeres on both primary and secondary chromosomes (34).
Fig 1.
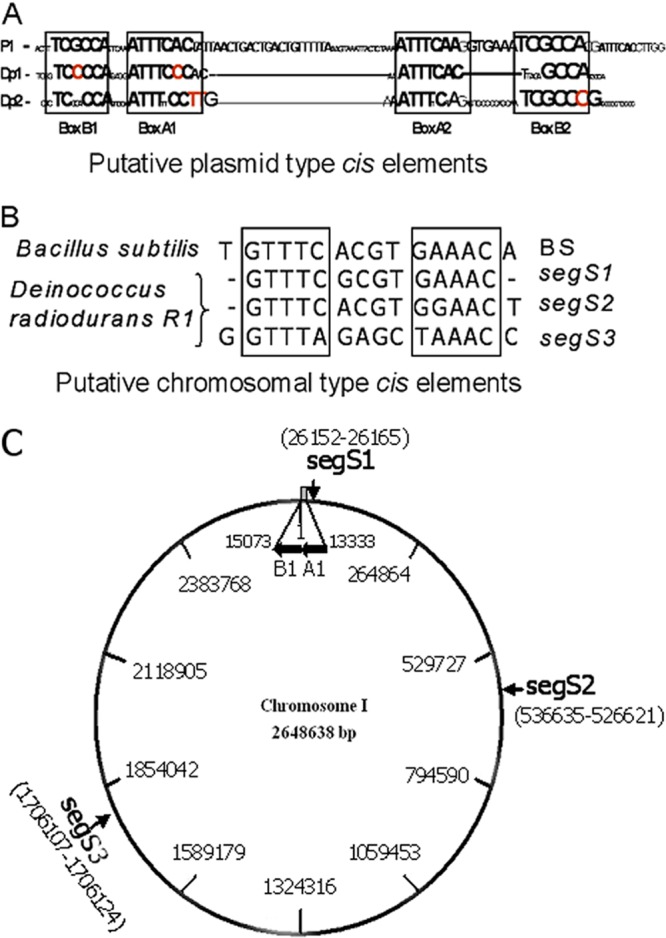
Identification of centromeric regions in the genome of Deinococcus radiodurans. The genome sequence of this bacterium was searched for the presence of typical parS elements, as reported previously for the P1 plasmid and chromosomal-type centromeres in Bacillus subtilis. (A) The inverted repeat boxes characteristic of parS elements were detected in chromosome I (Dp1) and chromosome II (Dp2). (B) Three putative centromeric regions similar to Bacillus subtilis-type chromosomal centromeres were detected in chromosome I and named segS1, segS2, and segS3. (C) These elements are distributed at different locations on circular chromosome I. The ParA1B1 (A1B1) operon is located upstream of the segS1 element, between bp 13333 and 15073.
ORFs DR_0012 and DR_0013 were annotated as a dicistronic operon on chromosome I of D. radiodurans and encode putative ParB (ParB1) and ParA (ParA1) proteins (35), respectively. The amino acid sequences of ParB1 showed similarity with the centromere binding protein Spo0J reported previously for both B. subtilis and Thermus thermophilus (7, 31). ParB1 contains conserved regions (region I, region II, and region III) and ParB-specific boxes (box I and box II) along with HTH motifs (38) (see Fig. S1 in the supplemental material). Similarly, the amino acid sequences of ParA1 showed homology with chromosomal and plasmid-encoded ParAs (19, 22, 32) (see Fig. S2 in the supplemental material) and were configured with the P-loop-type Walker ATPase domain. Both these proteins also contain two conserved arginine residues forming the characteristic DNA binding arginine pairs (32). These analyses suggested that chromosome I of D. radiodurans encodes putative ParA (ParA1)- and ParB (ParB1)-type proteins and contains three putative centromeric sequences (segS). The functional significance of segS elements in genome segregation and their interaction with the cognate putative centromere binding protein ParB1 was further investigated.
ParB1 is characterized as a centromere-specific DNA binding protein.
Both the ParB1 and ParA1 proteins were expressed in E. coli, and recombinant proteins were purified to homogeneity and confirmed by peptide mass fingerprints (PMFs) obtained by mass spectrometry (see Fig. S3 in the supplemental material). The purified recombinant ParB1 protein was checked for the characteristics known for other ParB-type proteins. ParB1 showed strong binding with all three segS elements(segS1, segS2, and segS3) and showed similar Kd values of 398.4 ± 96.32 nM, 486.5 ± 79.41 nM, and 316.1 ± 34.83 nM, respectively (data not shown). The ParB1 interaction with all three segS elements was specific, which remained unaffected even in the presence of a 100-fold-higher molar ratio of nonspecific DNA (Fig. 2). The nearly identical interactions of ParB1 with all three centromeric sequences indicated a strong possibility of functional redundancy among these cis elements, which was investigated independently. Previously, it was shown that ParB binding on chromosomal centromeres flanking the long regions of inverted repeats is advantageous for the interaction of ParA during genome segregation (17, 32). Our results showed that ParB1 could form a specific interaction with segS elements, which suggests the possibility that these elements function like chromosomal-type centromeres for chromosome I segregation in D. radiodurans.
Fig 2.
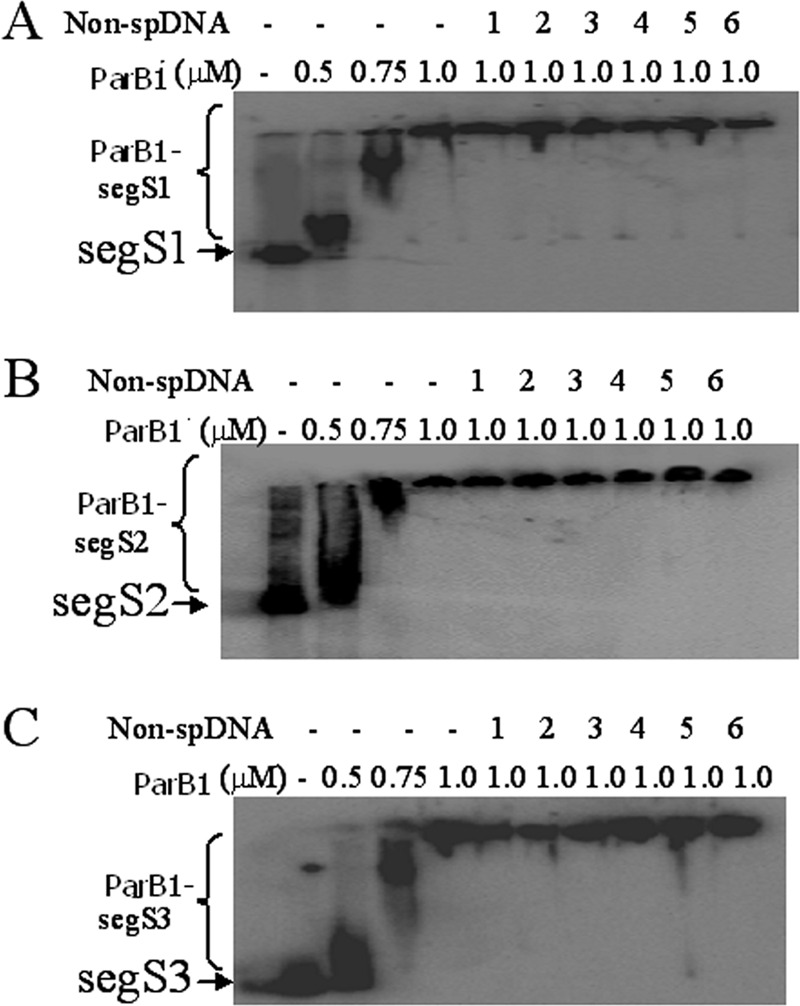
Interaction of ParB1 with segS-type centromeres of chromosome I. All three chromosomal-type centromeres, segS1 (A), segS2 (B), and segS3 (C), were PCR amplified, and approximately 300-bp PCR products of each element were labeled with [γ-32P]ATP and incubated with different concentrations of recombinant ParB1. For competition with 400 bp nonspecific DNA, the segS elements were incubated with ParB1 and chased with 1:1 (lane 1), 1:5 (lane 2), 1:10 (lane 3), 1:20 (lane 4), 1:50 (lane 5), and 1:100 (lane 6) molar ratios of 250 bp of nonspecific dsDNA (Non-spDNA). Products were separated on 5% native PAGE gels and dried, and autoradiograms were developed. Experiments were repeated two times, and reproducible data from a representative experiment are shown.
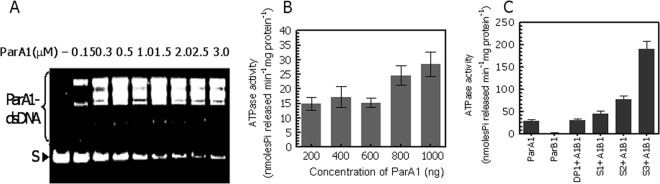
ParA1 is a DNA binding ATPase.
A functional domain search showed that ParA1 contained both Walker ATPase domains and a characteristic arginine pair implicated in the DNA-protein interaction. This indicates the possibility that this protein functions as a DNA binding ATPase. Purified recombinant ParA1 was subsequently checked for its ATPase activity and interactions with DNA. It showed double-stranded-DNA (dsDNA) binding activity in vitro (Fig. 3A), with a Kd value of 211.6 ± 55.79 nM (data not shown). A gel mobility shift assay of ParA1 with a 400-bp dsDNA yielded multiple mobility-retarded DNA bands, suggesting the random and concentration-dependent binding of ParA1 on DNA. ParA1 showed ATPase activity in vitro, which increased with increasing concentrations of protein (Fig. 3B) and was stimulated when both ParB1 and segS elements were coincubated (here referred to as the segS-ParB1 complex) (Fig. 3C). Note that the stimulation of ATPase activity was severalfold higher with segS3 than with segS1 and segS2, which is intriguing. Nevertheless, these results suggested that ParA1 was a dsDNA binding ATPase in vitro, and its activity was stimulated, although to different levels, in the presence of the segS-ParB1 complex. Since ParB1 alone and/or with segS elements did not show ATPase activity, the characteristic increase in the ATPase activity of ParA1 in the presence of both of these elements was interesting. The stimulation of ParA's ATPase activity upon an interaction with ParB or homologues bound on centromeric sequences was reported previously to be one of the factors contributing to the dynamic change in the polymerization/depolymerization kinetics of such proteins (44). Therefore, the functional significance of segS-ParB1-stimulated ATPase activity for the polymerization/depolymerization dynamics of ParA1 was hypothesized and checked.
Fig 3.
Activity characterization of recombinant ParA1. (A) The DNA binding activity of purified ParA1 was checked with 400 bp PCR-amplified nonspecific DNA and increasing concentrations of protein, and products were analyzed on a 1.2% agarose gel. S, supernatant. (B) The ATPase activity of ParA1 was measured with cold ATP, and the release of inorganic phosphate was estimated as a function of the ParA1 concentration. (C) Similarly, the ATPase activities of ParA1, ParB1, and ParA1 plus ParB1 (A1B1) incubated with aberrant parS (Dp1+A1B1), segS1 (S1+A1B1), segS2 (S2+A1B1), and segS3 (S3+A1B1) of chromosome I were measured as described in Materials and Methods, and the release of inorganic phosphorous was estimated.
ParA1 produced a heavier complex with segS elements and ParB1.
Both sedimentation analysis and light scattering approaches were used to check the oligomerization of ParA1. Purified recombinant protein was incubated with segS in the presence and absence of ATP, and the distribution of ParA1 between the pellet and supernatant was monitored. Purified ParA1 reproducibly showed a level of sedimentation which did not change in the presence of ATP (Fig. 4), which may be explained if we assume that ParA1 might be forming shorter polymers independent of dsDNA and ParB1. When ParA1 was incubated with ParB1-segS1 or ParB1-segS2 complexes in the absence of ATP, the amount of ParA1 increased significantly in the pellet (Fig. 4) in the cases of segS2 and segS3 but did not change in the case of segS1. This increase in the concentration of ParA1 in the pellet was relatively higher with segS3 than with segS2, which could be explained by the relatively higher affinity of ParB1 for segS3 than for segS2 (data not shown). In the presence of ATP, however, the levels of ParA1 in the pellet increased in the case of segS1 and continued to be high in the case of segS2, while they decreased significantly in the case of segS3. This showed that although ParB1 forms complexes with all segS elements, the interactions of ParA1 with segS-ParB1 complexes were different in the presence of ATP. Presumably, it dissociates faster from a hypothetical segS3-ParA1B1 complex in the presence of ATP than from similar types of complexes with segS1 and segS2. This result also agrees with an independent observation where the higher mobility of the segS3-ParA1B1 nucleoprotein complex was observed in the presence of ATP (data not shown). The ATP effect on the higher mobility of the segS3 nucleoprotein complex, the higher sedimentation rate of ParA1 in mixtures containing ParA1 incubated with the segS3-ParB1 complex, and the greater stimulation of ATPase activity in the presence of segS3 together suggested a correlation between the stimulation of ATPase activity and the depolymerization of the ParA1 polymer. The ATPase activity stimulation-mediated depolymerization of ParA, which provides the force for the movement of the replicated genome toward the cell poles, was suggested previously (16, 44). Furthermore, sedimentation analysis has also been used as a tool for demonstrating the oligomerization of ParA-type proteins (5), including ParA2 from V. cholerae (24). It has been shown that the conditions that favor the polymerization of SopA/ParA2 also produced the larger pellets of these proteins upon centrifugation.
Fig 4.
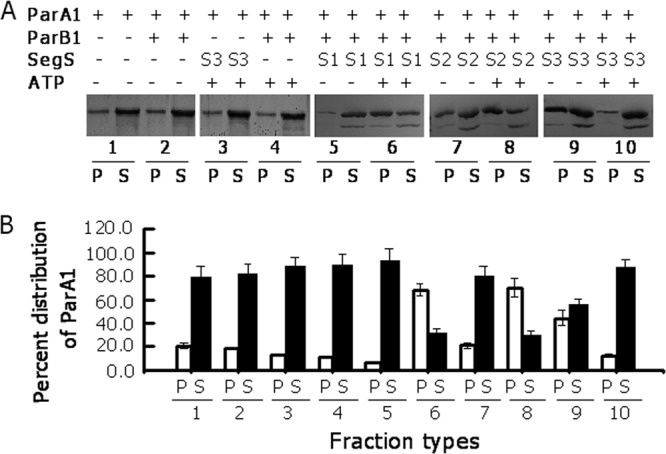
Sedimentation analysis of ParA1 in the presence of its cognate putative chromosome I partitioning components. Recombinant ParA1 was incubated with segS1 (S1), segS2 (S2), segS3 (S3), ParB1, and ATP in different combinations. (A) Mixtures were centrifuged at 22,000 × g for 30 min, and both supernatant (S) and pellet (P) fractions were analyzed by SDS-PAGE. (B) The gel was stained with silver nitrate, and the intensity of the protein band corresponding to ParA1 was estimated by densitometric scanning of the respective gel. Each experiment was repeated three times, and results from one reproducible experiment are shown in panel A and with standard deviations in panel B.
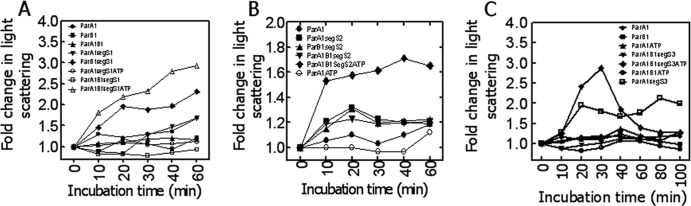
The effect of ATP, segS elements, and ParB1 on the oligomerization of ParA1 was also checked by light scattering analyses. The results showed that ParB1 and ParA1, both individually and together, did not change light scattering significantly. However, the incubation of ParA1 with segS-ParB1 complexes of all three segS elements showed a steady increase in light scattering albeit in the absence of ATP (Fig. 5). However, in the presence of ATP, the increase in light scattering continued for 60 min in the cases of segS1 and segS2. With segS3, however, the initial increase in light scattering for 30 min was followed by a sharp decrease, indicating the possibility of a rapid change in the macromolecular interaction between ParA1 and the segS3-ParB1 complex. The light scattering approach was used previously to determine the dynamic variations in the sizes of macromolecular complexes. An increase in light scattering with time has been attributed to an increase in the sizes of ParA protein polymers (5), while a decrease in light scattering was suggested to be due to the disassembly of the ParA filament (44, 48). Note that ParA, belonging to the type I category, e.g., Soj or pSM19035 delta, polymerizes in the presence of ATP and DNA; the interaction of this complex with Spo0J stimulates the intrinsic ATPase activity of ParA, leading to its depolymerization (40–42). Thus, a rapid decrease in light scattering, a higher gel mobility, and a relatively smaller amount of ParA1 in the pellet in the presence of segS3 and ATP may suggest that the higher ATPase activity of ParA1 upon the interaction with the ParB1-segS3 complex could lead to its rapid depolymerization. The in vitro results obtained so far suggested that ParB1 is a sequence-specific centromere binding protein and that ParA1 is a DNA binding ATPase. The sequence-specific interaction of ParB1 with all three segS elements with nearly similar Kd values indicated a strong possibility of multiple centromere-like sequences functioning in chromosome I partitioning in D. radiodurans.
Fig 5.
Light scattering studies of ParA1 incubated with ParB1 and segS elements in the presence and absence of ATP. Recombinant purified ParA1 (ParA1) and ParB1 (ParB1) were taken alone and together in different combinations with segS1 (A), segS2 (B), and segS3 (C), and ATP and light scattering spectra were recorded as a function of time. For experiments where both ParA1 and ParB1 were incubated with segS elements, ParB1 was preincubated with segS before ParA1 was added. For the ATP effect, the ATP was added last in any given combination. Experiments were repeated at least three times independently. Data presented here are the means of values obtained with 10 shots of 10 s each at every single time point from a representative experiment.
The segS elements and ParA1B1 proteins support the inheritance of the mini-F plasmid in E. coli.
The in vitro characterization of both cis elements and partitioning proteins produced some intriguing observations, such as the observation that segS1 and segS2 elements bound to ParB1 produced a stable complex with ParA1 even in the presence of ATP, while these interactions behaved differently with segS3. This may allow us to speculate that the minor differences in the sequences of these cis elements could make them function differently in vivo. In order to test this hypothesis and the in vivo functions of these elements as centromeres in genome segregation, we employed plasmid stability assays using unstable mini-F plasmid pDAG203 (23). All three segS elements (segS1, segS2, and segS3) were cloned into pDAG203, yielding pDAGS1, pDAGS2, pDAGS3, respectively, and transformed into E. coli. Cells harboring these derivatives were grown to different generations, and the numbers of cells conferring chloramphenicol resistance (a phenotype encoded on pDAG203) were scored. Interestingly, the cells carrying pDAGS1 and pDAGS2 showed a nearly 100% stable inheritance of the respective plasmids independently of ParA1B1 (Fig. 6A), while the stability of plasmid pDAGS3, in terms of cells expressing chloramphenicol resistance, although significantly high even in the absence of ParA1B1 compared to the control, was improved further to nearly 100% when both ParA1 and ParB1 were coexpressed (Fig. 6B). These results indicated that (i) all three segS elements could function like bacterial centromeres, and (ii) segS1 and segS2 could find the partitioning protein complements in an E. coli host, while segS3 recognition by the E. coli complement(s) was partial and required cognate ParA1B1 for its complete inheritance in a dividing population. Previously, a plasmid stability assay using unstable mini-F plasmid pDAG203 was employed for the characterization of genome partitioning systems in other bacteria (11, 18). These results suggested that segS3 functions differently from segS1 and segS2. While the latter elements could support the stable segregation of the mini-F plasmid independently of ParA1B1, the function of segS3 required ParA1B1 for its efficient function in E. coli. The molecular basis for segS1 and segS2 stabilizing the unstable mini-F plasmid independently of its cognate partitioning proteins in E. coli is not clear. However, the possibility of any change in copy number in terms of the amount of plasmid DNA per unit of chromosomal DNA (Fig. 6C and D) and mutations in segS elements (see Fig. S4 in the supplemental material) has been ruled out. Furthermore, the possible mechanisms of how all three segS elements coordinate genome segregation in D. radiodurans will be studied independently.
Fig 6.
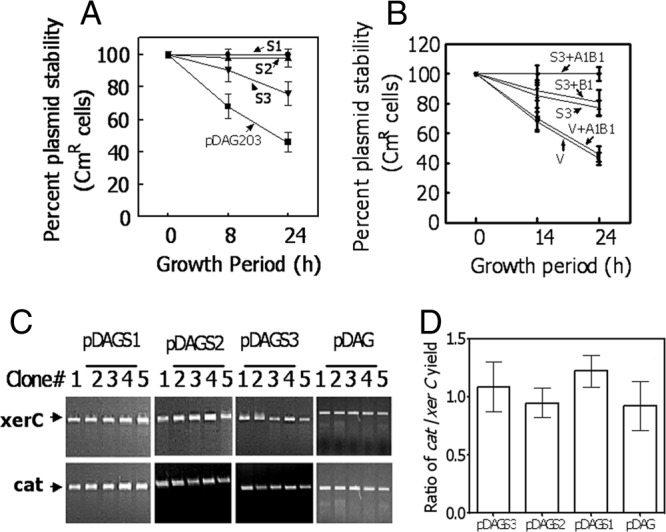
Centromere activity assay of segS elements using a plasmid stability test in Escherichia coli. (A and B) The segS1 (S1), segS2 (S2), and segS3 (S3) elements were cloned into unstable mini-F plasmid pDAG203 (V), and cells carrying recombinant plasmids pDAGS1, pDAGS2, and pDAGS3 and pDAGS3-harboring cells coexpressing either ParB1 (S3+B1) or ParB1 and ParA1 together (S3+A1B1) (B) were checked for plasmid stability as a function of chloramphenicol resistance (A). Data were compared with data for cells carrying vector pDAG203 (V) alone and also coexpressing both ParA1 and ParB1 in the vector background (V+A1B1) (B). (C and D) Genomic DNAs prepared from 5 independent clones (clones 1 to 5) of E. coli each harboring pDAGS1, pDAGS2, pDAGS3, and pDAG203 (pDAG) and relative proportions of plasmid (cat) and genomic (xerC) DNAs were checked for each clone (C), and the ratios of cat to xerC were calculated (D).
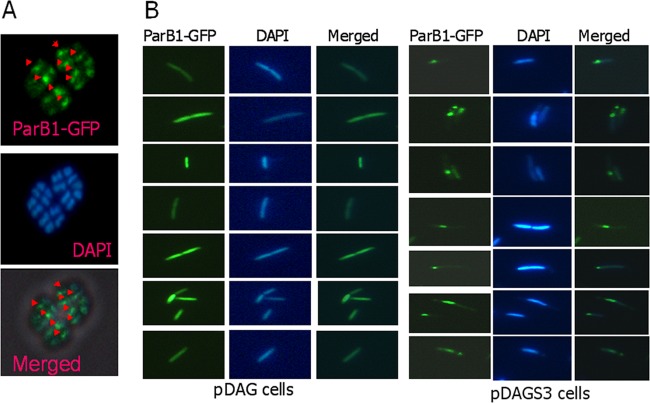
ParB1 interacts with the nucleoid in D. radiodurans, and its deletion results in growth defects and anucleation.
D. radiodurans cells harboring pD12GFP were induced with IPTG, and the synthesis of the GFP-ParB1 fusion protein was confirmed by immunoblotting using GFP antibodies. A microscopic examination of D. radiodurans cells expressing the GFP-ParB1 fusion showed the presence of at least 1 fluorescent focus per cell, which was not observed for wild-type cells harboring the p11559-GFP vector control (data not shown). The overlapping of the micrographs imaged under GFP and DAPI fluorescences showed that GFP-ParB1 formed foci directly on the nucleoids of Deinococcus cells (Fig. 7A), suggesting that ParB1 could bind on the genome of this bacterium. On the other hand, wild-type E. coli cells expressing the GFP-ParB1 fusion did not show GFP-ParB1 focus formation, further indicating that the ParB1 interaction with the genome is specific to sequences present in the genome of D. radiodurans but not E. coli. However, E. coli cells harboring recombinant plasmid pDAGS3, harboring segS3 and also expressing ParB1-GFP, showed discrete focus formation on DNA, which was not observed for cells harboring pDAG203 as a vector control (Fig. 7B). These results suggest that ParB1 is a typical ParB-type protein that nucleates on the segS3 sequence present on a plasmid in E. coli and possibly on the genome of D. radiodurans.
Fig 7.
In vivo interaction of ParB1 with nucleoids containing a segS element(s) in bacteria. D. radiodurans R1 (A) and E. coli (B) cells harboring the pDAG203 vector (pDAG) and pDAGS3 (pDAGS3) were expressed with ParB1-GFP on a plasmid and micrographed for GFP fluorescence (ParB1-GFP), and cells were stained with DAPI. These images were merged to localize the position of the GFP spot on the nucleoid or otherwise, as the case may be.
In order to understand the role of ParB1 in the maintenance of the D. radiodurans genome, the ParB1 gene was deleted from the bacterial genome, and the ΔparB1 mutant was compared with the wild type to determine its growth characteristics and anucleation phenotype under normal growth conditions. Mutant cells showed a significant decrease in turbidity measured at 600 nm compared to the turbidity measured for wild-type cells (Fig. 8A). Previously, the effect of the parAB deletion on similar types of growth defects was observed for other bacteria, e.g., Pseudomonas putida (18). Fluorescent micrographs of mutant cells stained with DAPI showed notable effects of the parB1 deletion on genome maintenance under normal growth conditions. The anucleation frequency in mutant cells was in the range of 8 to 13%, compared to the less than 1% observed for wild-type cells (Table 1). Morphologically, several tetrads were seen as triads, possibly because one of the four cells that lacked a nucleoid did not survive. These triads also showed three nucleoids by DAPI staining, instead of the four nucleoids normally observed for unaffected populations (Fig. 8B). Also, a large number of tetrad populations showed a relatively low density of DNA in one of the four compartments of the tetrads. Similar levels of anucleation were reported previously for other bacteria when the parB gene and/or the parA gene was deleted (29). These results suggest that ParB1 interacts with the nucleoid of this bacterium and has a functional role in genome maintenance. The heterogeneity in cell populations showing anucleation could be attributed to the toroidal packaging of all genome replication units and also to the functional redundancy if contributed by other ParB-type proteins encoded on other replication units in this bacterium.
Fig 8.
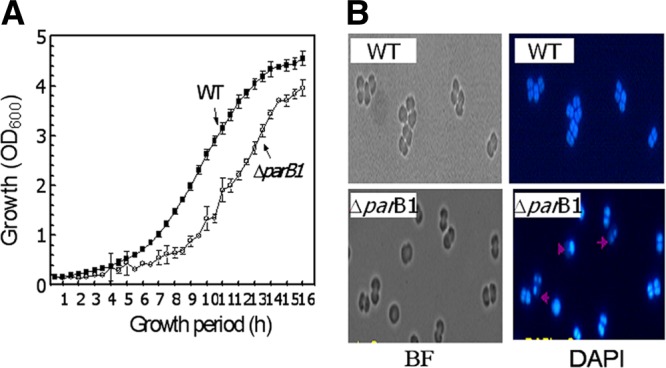
Effect of ParB1 deletion on genome maintenance in Deinococcus radiodurans. (A) The parB1 deletion mutant (ΔparB1) of D. radiodurans was generated, and its growth pattern at an optical density at 600 nm (OD600) was compared with that of D. radiodurans R1 (wild type [WT]). (B) The effect of the parB1 deletion on anucleation was examined microscopically. Cells grown at different time intervals were micrographed under bright-field (BF) fluorescence, and cells were stained with DAPI. The percentage of anucleation was calculated from the number of cells missing a genome out of the total number of cells counted. Micrographs shown are those of wild-type and ΔparB1 mutant cells grown for 10 h under normal growth conditions.
Table 1.
Effect of a parB1 deletion on the anucleation of the genome of D. radiodurans
| Growth period (h) | Wild type |
ΔparB1 mutant |
||
|---|---|---|---|---|
| No. of cells counted | Mean anucleation frequency (%) ± SE | No. of cells counted | Mean anucleation frequency (%) ± SE | |
| 2 | 189 | 0.60 ± 0.12 | 280 | 8.57 ± 1.34 |
| 4 | 203 | 0 | 297 | 9.42 ± 0.98 |
| 6 | 202 | 0.049 ± 0.021 | 258 | 10.07 ± 1.45 |
| 8 | 249 | 0 | 255 | 10.19 ± 1.58 |
| 10 | 295 | 0 | 206 | 13.39 ± 0.87 |
D. radiodurans is known for its extreme phenotypes, including its extraordinary resistance to gamma radiation (47). The genome sequence of this bacterium revealed several interesting facets (52). Among these, the presence of a multipartite genome system, ploidy, and multicopy genes was speculated to be key for its extreme phenotypes. The maintenance of the multipartite genome and ploidy in dividing cells of bacteria has been an interesting question and equally intriguing. Recent studies of V. cholerae, which also has a multipartite genome, showed that both chromosomes I and II of this bacterium encode independent systems of genome segregation. This study has brought forth some interesting findings to suggest that chromosome I of D. radiodurans contains an independently functional partitioning system that includes three chromosomal-type centromeric sequences named segS1 to segS3 and the partitioning proteins ParA1 and ParB1. This study further demonstrated the roles of these elements in the segregation of an unstable mini-F plasmid and genome maintenance in D. radiodurans. Molecular interaction studies between the ParA1/ParB1 and segS elements suggested that ParA1 could form polymers in the presence of dsDNA, as evidenced by sedimentation analyses (Fig. 4) and light scattering studies (Fig. 5). The higher ATPase activity of ParA1 in the presence of segS3-ParB1 than in the presence of centromeric complexes and the higher gel mobility of a hypothetical segS3-ParB1P nucleoprotein complex, but not a segS1-ParB1P or segS2-ParB1P complex, especially in the presence of ATP (data not shown), suggested that ParA1 is a DNA binding protein and that ParA1 polymerization/depolymerization may be regulated by the levels of its intrinsic ATPase activity. Both the DNA binding of ParA1 and its polymerization/depolymerization as a function of its ATPase activity are typical characteristics of ATPases associated with genome segregation (31, 40, 48). Further studies will be required to understand the real-time functioning of segS and ParA1B1 and the oscillation of ParA1 proteins on the genome of D. radiodurans. Results obtained for the ParA1B1-dependent role of segS3 elements in mini-F plasmid stability (Fig. 6) and the nearly 8 to 13% loss of nucleoids in the ΔparB1 mutant of D. radiodurans, in comparison to the 1% loss in normal cells (Table 1), strongly supported the roles of both segS and the “Par” proteins of this bacterium in its genome segregation. The results presented here report for the first time that (i) ParA1 is a DNA binding ATPase; (ii) ParB1 is a sequence-specific centromere binding protein; (iii) the ATPase activity of ParA1 was stimulated in the presence of both ParB1 and dsDNA, which seems to be regulating the dynamics of ParA1 polymerization; and (iv) segS elements function like bacterial centromeres for unstable plasmid segregation, and ParB1 has a role in genome maintenance in D. radiodurans. An attempt to delete segS3 from the genome of D. radiodurans was unsuccessful. Cells approaching the homozygous replacement of segS3 with a selection marker lose viability. This indicates a strong possibility that segS3 acts as a preferred centromere during the ParAB1-mediated partitioning of chromosome I in D. radiodurans. These findings collectively suggested that both segS elements and Par proteins harbored on chromosome I of D. radiodurans are functionally active and play a role in genome segregation and the maintenance of the chromosome in this bacterium.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. K. Apte for his constructive criticism and encouragement while pursuing this work. We thank Sudhir Singh and Swathi Kota for critical reading of the manuscript and their technical comments and P. A. Hassan, Chemistry Division, BARC, for his support in light scattering experiments.
V.K.C. thanks the Department of Atomic Energy, Government of India, for a research fellowship.
Footnotes
Published ahead of print 27 July 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Batt SM, Bingle LEH, Dafforn TR, Thomas CM. 2009. Bacterial genome partitioning: N-terminal domain of IncC protein encoded by broad-host-range plasmid RK2 modulates oligomerisation and DNA binding. J. Mol. Biol. 385:1361–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Battista JR, Park MJ, McLemore AE. 2001. Inactivation of two homologues of proteins presumed to be involved in the desiccation tolerance of plants sensitizes Deinococcus radiodurans R1 to desiccation. Cryobiology 43:133–139 [DOI] [PubMed] [Google Scholar]
- 3. Bignell CR, Thomas CM. 2001. The bacterial ParA-ParB partitioning proteins. J. Biotechnol. 91:1–34 [DOI] [PubMed] [Google Scholar]
- 4. Blasius M, Sommer S, Hubscher U. 2008. Deinococcus radiodurans: what belongs to the survival kit? Crit. Rev. Biochem. Mol. Biol. 43:221–238 [DOI] [PubMed] [Google Scholar]
- 5. Bouet JY, Ah-Seng Y, Benmeradi N, Lane D. 2007. Polymerization of SopA partition ATPase: regulation by DNA binding and SopB. Mol. Microbiol. 63:468–481 [DOI] [PubMed] [Google Scholar]
- 6. Bouthier de la Tour C, et al. 2009. The Deinococcus radiodurans SMC protein is dispensable for cell viability yet plays a role in DNA folding. Extremophiles 13:827–837 [DOI] [PubMed] [Google Scholar]
- 7. Breier AM, Grossman AD. 2007. Whole genome analysis of the chromosome partitioning and sporulation protein SpoOJ (ParB) reveals spreading and origin distal sites on the Bacillus subtilis chromosome. Mol. Microbiol. 64:703–718 [DOI] [PubMed] [Google Scholar]
- 8. Campbell CS, Mullins RD. 2007. In vivo visualization of type II plasmid segregation: bacterial actin filaments pushing plasmids. J. Cell Biol. 179:1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Castaing JP, Bout JY, Lane D. 2008. F plasmid partition depends on interaction of SopA with non-specific DNA. Mol. Microbiol. 70:1000–1011 [DOI] [PubMed] [Google Scholar]
- 10. Das AD, Misra HS. 2011. Characterization of DRA0282 from Deinococcus radiodurans for its role in bacterial resistance to DNA damage. Microbiology 157:2196–2205 [DOI] [PubMed] [Google Scholar]
- 11. Dubarry N, Pasta F, Lane D. 2006. ParABS systems of the four replicons of Burkholderia cenocepacia: new chromosome centromeres confer partition specificity. J. Bacteriol. 188:1489–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Egan ES, Waldor MK. 2003. Distinct replication requirements for the two Vibrio cholerae chromosomes. Cell 114:521–530 [DOI] [PubMed] [Google Scholar]
- 13. Fogel MA, Waldor MK. 2005. Distinct segregation dynamics of the two Vibrio cholerae chromosomes. Mol. Microbiol. 55:125–136 [DOI] [PubMed] [Google Scholar]
- 14. Funnell EB, Gagnier L. 1993. The plasmid partition complex at parS. II. Analysis of ParB protein binding activity and specificity. J. Biol. Chem. 268:3616–3624 [PubMed] [Google Scholar]
- 15. Garner EC, Campbell CS, Mullins RD. 2004. Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science 306:1021–1025 [DOI] [PubMed] [Google Scholar]
- 16. Gerdes K, Howard M, Szardenings F. 2010. Pushing and pulling in prokaryotic DNA segregation. Cell 141:927–942 [DOI] [PubMed] [Google Scholar]
- 17. Ghosh SK, Hajra S, Paek A, Jayaram M. 2006. Mechanisms for chromosome and plasmid segregation. Annu. Rev. Biochem. 75:211–241 [DOI] [PubMed] [Google Scholar]
- 18. Godfrin-Estevenon AM, Pasta F, Lane D. 2002. The parAB gene products of Pseudomonas putida exhibit partition activity in both P. putida and Escherichia coli. Mol. Microbiol. 43:39–49 [DOI] [PubMed] [Google Scholar]
- 19. Hayes F, Barilla D. 2006. The bacterial segrosome: a dynamic nucleoprotein machine for DNA trafficking and segregation. Nat. Rev. Microbiol. 4:133–143 [DOI] [PubMed] [Google Scholar]
- 20. Hazan R, Ben-Yehuda S. 2006. Resolving chromosome segregation in bacteria. J. Mol. Microbiol. Biotechnol. 11:126–139 [DOI] [PubMed] [Google Scholar]
- 21. Heidelberg JF, et al. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hester CM, Lutkenhaus J. 2007. Soj (ParA) DNA binding is mediated by conserved arginines and is essential for plasmid segregation. Proc. Natl. Acad. Sci. U. S. A. 104:20326–20331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hirano M, et al. 1998. Autoregulation of the partition genes of the mini-F plasmid and the intracellular localization of their products in Escherichia coli. Mol. Gen. Genet. 257:392–403 [DOI] [PubMed] [Google Scholar]
- 24. Hui MP, et al. 2010. ParA2, a Vibrio cholerae chromosome partitioning protein, forms left handed helical filaments on DNA. Proc. Natl. Acad. Sci. U. S. A. 107:4590–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kahng LS, Shapiro L. 2003. Polar localization of replicon origins in the multipartite genomes of Agrobacterium tumefaciens and Sinorhizobium meliloti. J. Bacteriol. 185:3384–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Khairnar NP, Kamble VA, Misra HS. 2008. RecBC enzyme overproduction affects UV and gamma radiation survival of Deinococcus radiodurans. DNA Repair 7:40–47 [DOI] [PubMed] [Google Scholar]
- 27. Kota S, Kumar CV, Misra HS. 2010. Characterization of an ATP-regulated DNA-processing enzyme and thermotolerant phosphoesterase in the radioresistant bacterium Deinococcus radiodurans. Biochem. J. 431:149–157 [DOI] [PubMed] [Google Scholar]
- 28. Lecointe F, Coste G, Sommer S, Bailone A. 2004. Vectors for regulated gene expression in the radioresistant bacterium Deinococcus radiodurans. Gene 336:25–35 [DOI] [PubMed] [Google Scholar]
- 29. Lee PS, Grossman AD. 2006. The chromosome partitioning protein Soj (ParA) and Spo0J (ParB) contribute to accurate chromosome partitioning, separation of replicated sister origins and regulation of replication initiation in Bacillus subtilis. Mol. Microbiol. 60:853–869 [DOI] [PubMed] [Google Scholar]
- 30. Lemonnier M, Bouet JY, Libante V, Lane D. 2000. Disruption of the F plasmid partition complex in vivo by partition protein SopA. Mol. Microbiol. 38:493–505 [DOI] [PubMed] [Google Scholar]
- 31. Leonard TA, Butler PJ, Lowe J. 2004. Structural analysis of the chromosome segregation protein Spo0J from Thermus thermophilus. Mol. Microbiol. 53:419–432 [DOI] [PubMed] [Google Scholar]
- 32. Leonard TA, Butler PJ, Lowe J. 2005. Bacterial chromosome segregation: structure and DNA binding of the Soj dimer—a conserved biological switch. EMBO J. 24:270–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin DC, Grossman AD. 1998. Identification and characterization of a bacterial chromosome-partitioning site. Cell 92:675–685 [DOI] [PubMed] [Google Scholar]
- 34. Livny J, Yamaichi Y, Waldor MK. 2007. Distribution of centromere-like parS sites in bacteria: insight from comparative genomics. J. Bacteriol. 189:8693–8703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Makarova KS, et al. 2001. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 65:44–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McIntosh JR, et al. 2008. Fibrils connect microtubule tips with kinetochores: a mechanisms to couple tubulin dynamics to chromosome motion. Cell 135:322–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moller-Jensen J, et al. 2003. Bacterial mitosis: parM of plasmid R1 moves plasmid DNA by an actin-like insertional polymerization mechanism. Mol. Cell 12:1477–1487 [DOI] [PubMed] [Google Scholar]
- 38. Murray H, Ferreira H, Errington J. 2006. The bacterial chromosome segregation protein Spo0J spreads along DNA from parS nucleation sites. Mol. Microbiol. 61:1352–1361 [DOI] [PubMed] [Google Scholar]
- 39. Pollard TD, Cooper JA. 2009. Actin, a central player in cell shape and movement. Science 326:1208–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pratto F, et al. 2008. Streptococcus pyogenes PSM19035 requires dynamic assembly of ATP-bound ParA and ParB on parS DNA during plasmid segregation. Nucleic Acids Res. 36:3676–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pratto F, Suzuki Y, Takeyasu K, Alonso JC. 2009. Single-molecule analysis of protein-DNA complexes formed during partition of newly replicated plasmid molecules in Streptococcus pyogenes. J. Biol. Chem. 284:30298–30306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ptacin JL, et al. 2010. A spindle-like apparatus guides bacterial chromosome segregation. Nat. Cell Biol. 12:791–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rajpurohit YS, Misra HS. 2010. Characterization of a DNA damage-inducible membrane protein kinase from Deinococcus radiodurans and its role in bacterial radioresistance and DNA strand break repair. Mol. Microbiol. 77:1470–1482 [DOI] [PubMed] [Google Scholar]
- 44. Ringgaard S, van Zon J, Howard MK, Gerdes K. 2009. Movement and equipositioning of the plasmids by ParA filament disassembly. Proc. Natl. Acad. Sci. U. S. A. 106:19369–19374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 46. Schafer M, et al. 2000. Systematic study of parameters influencing the action of Rose Bengal with visible light on bacterial cells: comparison between the biological effect and singlet-oxygen production. Photochem. Photobiol. 71:514–523 [DOI] [PubMed] [Google Scholar]
- 47. Slade D, Radman M. 2011. Oxidative stress resistance in Deinococcus radiodurans. Microbiol. Mol. Biol. Rev. 75:133–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Soberon NE, Lioy VS, Pratto F, Volante A, Alonso JC. 2011. Molecular anatomy of the Streptococcus pyogenes pSM19035 partition and segresome complexes. Nucleic Acids Res. 39:2624–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Surtees JA, Funnell BE. 2003. Plasmid and chromosome traffic control: how ParA and ParB drive partition. Curr. Top. Dev. Biol. 56:145–180 [DOI] [PubMed] [Google Scholar]
- 50. Varade D, et al. 2005. Effect of salt on the micelles of cetyl pyridinium chloride. Colloids Surf. A Physicochem. Eng. Asp. 259:95–101 [Google Scholar]
- 51. Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181:508–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. White O, et al. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yakunin AF, et al. 2004. The HD domain of the Escherichia coli tRNA nucleotidyltransferase has 2′,3′-cyclic phosphodiesterase, 2′-nucleotidase, and phosphatase activities. J. Biol. Chem. 279:36819–36827 [DOI] [PubMed] [Google Scholar]
- 54. Yamaichi Y, Fogel MA, McLeod SM, Hui MP, Waldor MK. 2007. Distinct centromere-like parS sites on the two chromosomes of Vibrio spp. J. Bacteriol. 189:5314–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zimmerman JM, Battista JR. 2005. A ring-like nucleoid is not necessary for radioresistance in the Deinococcaceae. BMC Microbiol. 5:17 doi:10.1186/1471-2180-5-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.