Abstract
Uropathogenic Escherichia coli (UPEC) is responsible for the majority of urinary tract infections (UTI). To cause a UTI, UPEC must adhere to the epithelial cells of the urinary tract and overcome the shear flow forces of urine. This function is mediated primarily by fimbrial adhesins, which mediate specific attachment to host cell receptors. Another group of adhesins that contributes to UPEC-mediated UTI is autotransporter (AT) proteins. AT proteins possess a range of virulence properties, such as adherence, aggregation, invasion, and biofilm formation. One recently characterized AT protein of UPEC is UpaH, a large adhesin-involved-in-diffuse-adherence (AIDA-I)-type AT protein that contributes to biofilm formation and bladder colonization. In this study we characterized a series of naturally occurring variants of UpaH. We demonstrate that extensive sequence variation exists within the passenger-encoding domain of UpaH variants from different UPEC strains. This sequence variation is associated with functional heterogeneity with respect to the ability of UpaH to mediate biofilm formation. In contrast, all of the UpaH variants examined retained a conserved ability to mediate binding to extracellular matrix (ECM) proteins. Bioinformatic analysis of the UpaH passenger domain identified a conserved region (UpaHCR) and a hydrophobic region (UpaHHR). Deletion of these domains reduced biofilm formation but not the binding to ECM proteins. Despite variation in the upaH sequence, the transcription of upaH was repressed by a conserved mechanism involving the global regulator H-NS, and mutation of the hns gene relieved this repression. Overall, our findings shed new light on the regulation and functions of the UpaH AT protein.
INTRODUCTION
Uropathogenic Escherichia coli (UPEC) is the major cause of urinary tract infections (UTI) in humans. Infection of the urinary tract by UPEC involves initial adherence to uroepithelial cells to resist the hydrodynamic forces of urine flow and subsequent inflammation triggered by host and bacterial cell signaling pathways. UPEC can also invade bladder epithelial cells, replicate, and establish biofilm-like intracellular bacterial communities (IBCs) that contain large numbers of bacteria (3, 20, 49, 55).
UPEC possesses a range of virulence factors, including adhesins (e.g., type 1 and P fimbrial), toxins (e.g., hemolysin), and siderophore-mediated systems for iron acquisition (e.g., enterobactin, salmochelin, aerobactin), that enable them to colonize the urinary tract and cause disease (37, 62, 80). Adhesion represents a critical initial step for UPEC colonization of the urinary tract and is mediated primarily by fimbriae. However, UPEC also produces a number of autotransporter (AT) proteins that contribute with varying degrees to adherence, aggregation, and biofilm formation (1, 2, 72, 74). Several AT proteins have been characterized from UPEC, and these include the secreted toxin Sat (19, 23, 41), the phase-variable outer membrane protein antigen 43 (Ag43) (35, 72), the trimeric AT protein UpaG (74), and the surface-located UpaB, UpaC, and UpaH proteins (1, 2).
AT proteins represent a major group of Gram-negative bacterial secreted proteins and share several common features: an N-terminal signal sequence, a passenger (α) domain that is either anchored to the cell surface or released into the external milieu, and a translocation (β) domain that resides in the outer membrane (28, 33). AT proteins were originally thought to possess structural properties that facilitate their independent transport across the bacterial membrane system and final routing to the cell surface (29). However, it has been shown that accessory factors, such as the β-barrel assembly machinery (Bam) complex (also known as the YaeT or Omp85 complex), as well as periplasmic chaperones, such as SurA, Skp, and DegP, are required for the secretion of some AT proteins (32, 52, 56–58, 60, 76). Most recently, a new translocation and assembly module (TAM) that promotes the efficient secretion of AT proteins in proteobacteria has been described (63). The passenger domain of AT proteins is the most divergent in sequence and confers the functional characteristics of the protein. It is frequently associated with virulence (29).
The most diverse subgroup of AT proteins is the adhesin-involved-in-diffuse-adherence (AIDA-I) type (29, 78). Of the 11 putative AT-encoding genes that have been identified in the sequenced genome of the prototype UPEC strain CFT073, 7 belong to this subgroup. Ag43 represents the best-characterized UPEC AIDA-I-type AT protein. Ag43 (encoded by the flu gene) is a self-recognizing AT protein that contributes to E. coli biofilm formation (13, 35, 61, 79). Ag43 expression by UPEC is associated with IBC formation (3) and long-term colonization of the mouse urinary tract (72).
Recently we identified and characterized the UpaH AT protein from the UPEC strain CFT073 (2). UpaH is the largest AIDA-I-type AT protein characterized from E. coli and contributes to biofilm formation and bladder colonization in a mouse UTI model. In this study, we investigated upaH gene expression in different UPEC isolates and identified the global regulator H-NS as a transcriptional repressor. Using hns gene deletion mutants, we demonstrated that upaH transcription is derepressed in multiple UPEC backgrounds. While we found the transcription of upaH to be regulated similarly in different UPEC strains, we showed that the coding sequence contains significant variation, particularly in the passenger-encoding domain. This sequence variation results in UpaH protein variants with distinct biofilm-forming properties but a conserved ability to bind extracellular matrix proteins. Bioinformatic analysis of the UpaH passenger domain also identified conserved and hydrophobic regions, and we have shown that these regions contribute to biofilm formation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Cells were routinely grown at 28°C or 37°C on solid or in liquid lysogeny broth (LB) medium (8) supplemented with the appropriate antibiotics: kanamycin (Kan, 50 μg/ml), chloramphenicol (Cam, 30 μg/ml), or ampicillin (Amp, 100 μg/ml). For growth under defined conditions, M9 minimal medium supplemented with 0.2% glucose was used (59).
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference |
|---|---|---|
| E. coli K-12 strains | ||
| OS56 | MG1655flu attB::bla-gfp Gfp+ Ampr | 65 |
| OS56(pBAD) | OS56 pBAD/myc-HisA | 77 |
| OS56(pUpaHCFT073) | pUpaHCFT073 in OS56, Ampr Kanr | 2 |
| OS56(pUpaHM161) | pUpaHM161 in OS56, Ampr Kanr | This study |
| OS56(pUpaHM357) | pUpaHM357 in OS56, Ampr Kanr | This study |
| OS56(pUpaHM369) | pUpaHM369 in OS56, Ampr Kanr | This study |
| OS56(pUpaHIA2) | pUpaHIA2 in OS56, Ampr Kanr | This study |
| OS56(pUpaHHR) | pUpaHCFT073 with no hydrophobic region in OS56, Ampr Kanr | This study |
| OS56(pUpaHCR) | pUpaHCFT073 with no conserved region in OS56, Ampr Kanr | This study |
| E. coli UPEC strains | ||
| CFT073 | Wild-type UPEC isolate | 47 |
| CFT073upaH | CFT073upaH::kan Kanr | 2 |
| CFT073hns | CFT073c1701::kan Kanr | 1 |
| CFT073lacIZ | CFT073lacIZ::cam Camr | 1 |
| CFT073lacIZ upaH::lacZ-zeo | CFT073lacIZ::cam upaH::lacZ-zeo Camr Zeor | This study |
| CFT073lacIZ upaH::lacZ-zeo hns::kan | CFT073lacIZ::cam upaH::lacZ-zeo c1701::kan Camr Zeor Kanr | This study |
| CFT073lacIZ upaH::lacZ-zeo hns::kan pBR322 | CFT073lacIZ::cam upaH::lacZ-zeo c1701::kan pBR322 Camr Zeor Kanr Ampr | This study |
| CFT073lacIZ upaH::lacZ-zeo hns::kan pH-NS | CFT073lacIZ::cam upaH::lacZ-zeo c1701::kan pH-NS, Camr Zeor Kanr Ampr | This study |
| CFT073lacIZ upaH::lacZ-zeo c0421::kan | CFT073lacIZ::cam upaH::lacZ-zeo c0421::kan Camr Zeor Kanr | This study |
| CFT073lacIZ upaH::lacZ-zeo c1699::kan | CFT073lacIZ::cam upaH::lacZ-zeo c1699::kan Camr Zeor Kanr | This study |
| CFT073lacIZ upaH::lacZ-zeo c2091::kan | CFT073lacIZ::cam upaH::lacZ-zeo c2091::kan Camr Zeor Kanr | This study |
| CFT073lacIZ upaH::lacZ-zeo c2411::kan | CFT073lacIZ::cam upaH::lacZ-zeo c2411::kan Camr Zeor Kanr | This study |
| CFT073lacIZ upaH::lacZ-zeo c3218::kan | CFT073lacIZ::cam upaH::lacZ-zeo c3218::kan Camr Zeor Kanr | This study |
| CFT073lacIZ upaH::lacZ-zeo c3244::kan | CFT073lacIZ::cam upaH::lacZ-zeo c3244::kan Camr Zeor Kanr | This study |
| CFT073lacIZ upaH::lacZ-zeo c3744::kan | CFT073lacIZ::cam upaH::lacZ-zeo c3744::kan Camr Zeor Kanr | This study |
| CFT073lacIZ upaH::lacZ-zeo c4864::kan | CFT073lacIZ::cam upaH::lacZ-zeo c4864::kan Camr Zeor Kanr | This study |
| CFT073lacIZ upaH::lacZ-zeo c5054::kan | CFT073lacIZ::cam upaH::lacZ-zeo c5054::kan Camr Zeor Kanr | This study |
| M161 | Wild-type UPEC isolate | 2 |
| M161upaH | M161upaH::kan Kanr | 2 |
| M161hns | M161hns::kan Kanr | This study |
| M357 | Wild-type UPEC isolate | 2 |
| M357upaH | M357upaH::kan Kanr | 2 |
| M357hns | M357hns::kan Kanr | This study |
| M369 | Wild-type UPEC isolate | This study |
| M369upaH | M369upaH::kan Kanr | This study |
| M369hns | M369hns::kan Kanr | This study |
| IA2 | Wild-type UPEC isolate | 11 |
| IA2upaH | IA2upaH::kan Kanr | This study |
| IA2hns | IA2hns::kan Kanr | This study |
| Plasmids | ||
| pBR322 | Ampr Tetr | 9 |
| pH-NS | pBR322 c1701 (H-NS) from CFT073, Ampr | 1 |
| pKOBEG | pSC101 ts (replicates at 30°C), araC, arabinose-inducible λ-redγβα operon, Camr | 10 |
DNA manipulations and genetic techniques.
DNA techniques were performed as described by Sambrook and Russell (59). The isolation of plasmid DNA was carried out using the QIAprep spin miniprep kit (Qiagen). Restriction endonucleases were used according to the manufacturer specifications (New England BioLabs). Chromosomal DNA purification was performed using the DNeasy blood and tissue kit (Qiagen). Oligonucleotides were purchased from Sigma (Australia or France). All PCRs were performed with the Expand high-fidelity polymerase system (Roche) or Phusion (Finnzymes) as described by the manufacturers. The in vitro transposon mutagenesis employed was performed as previously described using the primers Kan-2 FP-1 (5′-ACCTACAACAAAGCTCTCATCAACC) and Kan-2 RP-1 (5′-GCAATGTAACATCAGAGATTTTGAG) (2). DNA sequencing was performed using the ABI BigDye v3.1 kit (ABI) by the Australian Equine Genetics Research Centre (University of Queensland, Brisbane, Australia).
Construction of plasmids.
The plasmids pUpaHM161, pUpaHM357, pUpaHM369, and pUpaHIA2 contain the upaH gene from strains M161, M357, M369, and IA2, respectively, and were created by PCR amplification using the primers 520 (5′-CGCGCTCGAGATAATAAGGAATTATTATGCAAAGGAAAACTCTATTGTCG) and 521 (5′-CGCGCAAGCTTTTAGAATGTTTTCTTGAAGT), digestion with XhoI-HindIII, and insertion into the XhoI-HindIII-digested vector pBAD/myc-HisA. A final modification of the plasmid was made by inserting a kanamycin resistance-encoding gene into the HindIII site. Resistance to kanamycin was required to facilitate the transformation of these plasmids into the flu-negative gfp-positive K-12 strain OS56. In the pUpaH vectors, the expression of upaH is under the control of the arabinose-inducible araBAD promoter (24).
Construction of deletions and mutants.
Mutants containing deletions in the upaH gene were constructed by λ-red-mediated homologous recombination as previously described (2, 14). We created lacZ reporter transcriptional fusions or interrupted the regulator genes in strain CFT073 using homologous recombination mediated by λ-red recombinase expressed from the pKOBEG plasmid (15) and either a one-step PCR procedure with 40-bp homology arms for recombination or a three-step PCR procedure with 500-bp homology arms for recombination (15) (http://www.pasteur.fr/recherche/unites/Ggb/matmet.html). lacZ reporter transcriptional fusions were constructed using the primers upaH-lacZzeo-Long-5 (5′-GGGTATCCGACCTTCTTTATATTCGCCAGAAGGATTTATTATGACCATGATTACGGATTCACTG-3′) and upaH-lacZeo-Long-3 (5′-CCTGTATCAGCGTTGCGGTTTATGTATTACATGCTAAAAATCAGTCCTGCTCCTCGGCCACGAA-3′). lacZ mutants were confirmed via PCR using the primers upstream region (upaH.ext-5 5′-CAGGGGCGGGTAGGGGAAACAT-3′ and lacZATG + 100-3 5′-GGGGGATGTGCTGCAAGGCGATTAAG-3′) and downstream region (zeo.verif-5 5′-ACGAACTTCCGGGACGCCTCCG-3′ and upaH.ext-3 5′-CCATGACCATCCCCCATCAT-3′). The primers were designed and used as described previously to mutate the upaH gene (2) and the set of regulatory genes examined (1). All mutants were confirmed via PCR and sequencing using the primers Gene.ext 5′ and Gene.ext 3′, either as a primer set or individually in combination with internal primers designed to bind within the inserted DNA fragments.
Immunoblotting and glycoprotein staining.
A polyclonal anti-UpaH serum raised against a portion of the passenger domain of UpaH from strain CFT073 was used for UpaH detection (2). For immunoblotting, whole-cell lysates were subjected to SDS-PAGE using NuPAGE Novex 3 to 8% Tris-acetate precast gels with NuPAGE Tris-acetate running buffer and subsequently transferred to polyvinylidene difluoride (PVDF) microporous membrane filters using the iBlot dry-blotting system as described by the manufacturer (Invitrogen). Serum raised against the passenger subunit of UpaH was used as the primary serum, and the secondary antibody was alkaline phosphatase-conjugated anti-rabbit IgG. Sigma fast 5-bromo-4-chloro-3-indolylphosphate (BCIP)/nitroblue tetrazolium (NBT) was used as the substrate in the detection process. Glycoprotein staining was performed using the Pierce glycoprotein staining kit per the manufacturer's instructions.
Biofilm assays.
Flow cells were inoculated with optical density at 600 nm (OD600)-standardized cultures pregrown overnight in glucose M9 minimal medium. For E. coli OS56 biofilms, kanamycin was added as required for the maintenance of plasmids. Biofilm development was monitored by confocal scanning laser microscopy 16 h or 24 h after inoculation. For analysis of the flow cell biofilms, we collected 6 z-stacks for each strain and analyzed them by using the COMSTAT software program (31). All experiments were performed in duplicate. COMSTAT data were analyzed using the nonparametric Kruskal-Wallis test within the Minitab (v14) software package. P values of <0.05 were considered significant.
Extracellular matrix protein binding assays.
Bacterial binding to extracellular matrix (ECM) proteins was performed in a microtiter plate enzyme-linked immunosorbent assay (ELISA) essentially as described previously (74). Microtiter plates (MaxiSorp; Nunc) were coated overnight at 4°C with MaxGel human extracellular matrix (10 μg/ml) or with 2 μg of the following ECM proteins: collagen IV, collagen V, fibronectin, fibrinogen, laminin, and bovine serum albumin (BSA) (Sigma-Aldrich). The wells were washed twice with Tris-buffered saline (TBS) (137 mM NaCl, 10 mM Tris [pH 7.4]) and then blocked with TBS-2% milk for 1 h. After being washed with TBS, 200 μl of washed and standardized (OD600 = 0.1) OS56 (containing pUpaH plasmids) or OS56(pBAD) was added, and the plates were incubated at 37°C for 2 h. After being washed to remove nonadherent bacteria, adherent cells were fixed with 4% paraformaldehyde, washed, and incubated for 1 h with anti-E. coli serum (Meridian Life Sciences Inc., catalog no. B65001R) diluted 1:500 in 0.05% TBS-Tween and 0.2% milk, washed, and incubated for 1 h with a secondary anti-rabbit-conjugated horseradish peroxidase antibody (diluted 1:1,000) (Sigma-Aldrich, catalog no. A6154). After a final wash, the adhered bacteria were detected by adding 150 μl of 0.3 mg/ml ABTS [2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid)] (Sigma-Aldrich) in 0.1 M citric acid (pH 4.3) and activated with 1 μl/ml 30% hydrogen peroxide, and the absorbance at 405 nm was read. The mean absorbance readings were compared with negative control readings for OS56(pBAD) using two-sample t tests within the Minitab v14 software package. P values of <0.05 were considered significant.
Investigation of hydrophobic and conserved regions of UpaH.
The hydrophobicity of the N-terminal 300 amino acids (aa) of UpaH was analyzed using the Kyte-Doolittle scale in ProtScale (21, 38). To examine the function of the hydrophobic and conserved regions in the α-domain of UpaH, the plasmids pUpaHHR (lacking nucleotides 193 to 267) and pUpaHCR (lacking nucleotides 846 to 1416) were constructed. For the construction of pUpaHHR, the primers UpaHHRF (5′-GCGCGCTCGAGATAATAAGGAATTCTTATGCAAAGGAAAACTCTA-3′) and UpaHHRR (5′-GCGCGTGATCATCATCGTCGTTGACTACAGAGGAGCATTCAG-3′) were used to PCR amplify the N-terminal region of the upaH gene from pUpaHCFT073 without the predicted hydrophobic region. This PCR product was then digested with XhoI/BclI and ligated with an XhoI/BclI-digested pUpaHCFT073 plasmid. For the construction of pUpaHCR, the overlapping oligonucleotides UpaHCRF (5′-TCGAGATAATAAGGAATTCTTATGCAAAGGAAAACTCTATTGTCGGCCTGTATTGCATTAGCTCTGAGTGGTCAGGGTTGGGCGGCAGATATTACAGAACACAA-3′) and UpaHCRR (5′-TTGTGTTCTGTAATATCTGCCGCCCAACCCTGACCACTCAGAGCTAATGCAATACAGGCCGACAATAGAGTTTTCCTTTGCATAAGAATTCCTTATTATC-3′) were resuspended in 100 mM potassium acetate and 30 mM HEPES to a concentration of 100 μM, combined, heated to 99°C, and allowed to anneal by cooling at room temperature. The overlapping oligonucleotides were designed to create ends corresponding to digestion with XhoI/AleI upon annealing, and the double-stranded product was ligated with the XhoI/AleI-digested pUpaHCFT073 plasmid. The resultant pUpaHHR and pUpaHCR plasmids were screened via EcoRI digestion, confirmed via sequencing, and transformed into E. coli strain OS56 for phenotypic assays.
Microscopy and image analysis.
An anti-UpaH-specific serum was used for immunofluorescence microscopy essentially as described previously with the exception that an Alexa Fluor 350 goat anti-rabbit IgG A11046 (Life Technologies) secondary antibody was used (2, 74). Immunofluorescence microscopy and image acquisition were performed on a scanning confocal laser microscope (Axioplan2; Zeiss) equipped with detectors and filters for monitoring of the Alexa Fluor 350. Microscopic observations of biofilms and image acquisition were performed on a scanning confocal laser microscope (LSM510 META; Zeiss) equipped with detectors and filters for the monitoring of green fluorescent protein (GFP). Vertical cross-sections through the biofilms were visualized using the Zeiss LSM image examiner. The images were further processed for display by using Photoshop software (Adobe, Mountain View, CA).
Cell surface hydrophobicity assay.
The microbial adhesion to hydrocarbons (MATH) assay was performed essentially as described previously (44). Briefly, overnight cultures were washed twice, resuspended in saline, and fixed to an OD600 of 0.3. In quadruplicate, 5-ml bacterial suspensions were overlaid with 0.4 ml n-hexadecane and mixed on a platform shaker at 37°C (400 rpm, 15 min). After 15 min of equilibration at room temperature, the lower aqueous phase was removed, and the OD600 was measured. The hydrophobicity index (HPBI) was calculated as (ODinitial − ODfinal)/ODinitial. The results were analyzed by the Kruskal-Wallis test.
β-Galactosidase assays.
β-Galactosidase assays were performed essentially as described previously (46). Briefly, the strains carrying lacZ fusions were grown on LB plates for 16 h and then inoculated into LB medium. After 16 to 18 h of growth, the culture was diluted in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 50 mM β-mercaptoethanol, 10 mM KCl, 1 mM MgSO4 [pH 7]), 0.004% SDS and chloroform were added, and the samples were vortexed to permeabilize the cells. The samples were incubated at 28°C, and the reaction was initiated by the addition of o-nitrophenyl-β-d-galactopyranoside (ONPG). The reactions were stopped with the addition of sodium bicarbonate, and the enzymatic activity was assayed in quadruplicate for each strain by measuring the absorbance at 420 nm. Where required, β-galactosidase activity was also observed on LB agar plates containing 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal).
Electrophoretic mobility shift assay.
Gel shift assays were performed essentially as described previously (5). A DNA mixture comprising the PCR-amplified upaH promoter region and TaqI-SspI-digested pBR322 at an equimolar ratio was incubated at room temperature for 15 min with increasing amounts of native purified H-NS protein (a gift from S. Rimsky) in 30 μl of a reaction mixture containing 40 mM HEPES (pH 8), 60 mM potassium glutamate, 8 mM magnesium aspartate, 5 mM dithiothreitol, 10% glycerol, 0.1% octylphenoxypolyethoxyethanol, 0.1 mg/ml BSA (H-NS-binding buffer). DNA fragments and DNA-protein complexes were resolved by gel electrophoresis (0.5× Tris-borate-EDTA [TBE], 3% molecular screening [MS] agarose gel run at 50 V at 4°C) and visualized after staining with ethidium bromide.
DNA curvature prediction and DNA PAGE analysis.
The upaH promoter region was analyzed in silico using BendIt, a program that enables the prediction of a curvature-propensity plot calculated with DNase I-based parameters (75). The curvature is calculated as a vector sum of dinucleotide geometries (roll, tilt, and twist angles) and expressed as degrees per helical turn (10.5°/helical turn = 1°/bp). Experimentally tested curved motifs produce curvature values of 5 to 25°/helical turn, whereas straight motifs give values below 5°/helical turn. The upaH 250-bp promoter region was amplified using the primers upaH.pro.ext-5 + 250 (5′-TTGATTTATACCGTTTTTTCATTA-3′), upaH.pro.ext-5 + 250B (5′-TTGGTTTATACCGTTTTTTCATTA-3′), upaH.pro.ext-5 + 250C (5′-TGGTTTATACCGTTTTTTTCATTA-3′), and upaH.pro.ext-3-1ATG (5′-AATAAATCCTTCTGGCGAATATAA-3′) and its intrinsic curvature was assessed by comparing its electrophoretic mobility with that of a 100-bp DNA ladder (New England BioLabs) on a 0.5× TBE, 7.5% PAGE gel at 4°C.
Evolutionary and sequence analysis.
The prevalence of the upaH gene was analyzed in 61 complete and 247 draft E. coli genome sequences available in the NCBI database. The N-terminal 300-amino-acid sequence was extracted from 139 strains and included 51 different sequences from complete and intact genes. The sequences were compared and aligned using Muscle, and their phylogenetic relationship was inferred using the neighbor-joining method in MEGA5 (71). The genome context was analyzed and drawn using Easyfig (67).
Nucleotide sequence accession numbers.
The complete nucleotide sequences of the upaH gene from strains M161, M357, M369, and IA2 have been deposited in the NCBI database (BankIt submission ID 1482470, accession no. JN709850 through JN709854).
RESULTS
UpaH is expressed in multiple UPEC strains.
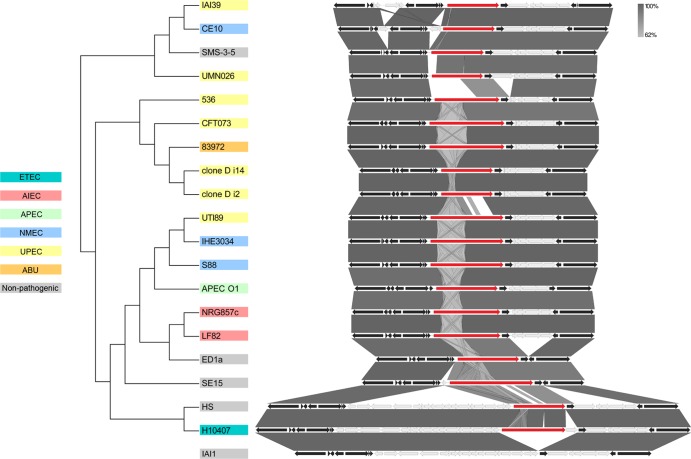
We previously demonstrated the expression of UpaH in three UPEC strains (CFT073, M161, and M357) via Western blot analysis employing UpaH-specific antibodies. In all three strains, the sizes of the UpaH protein as assessed by SDS-PAGE analysis were similar (2). Moreover, we showed that the upaH gene cloned and sequenced from strain CFT073 encodes a protein that is 2,845 amino acids long (2). In contrast, bioinformatic analyses of the complete E. coli genome sequences available in the NCBI database showed that despite being in the same genetic context, there was considerable variation in the sizes of upaH and the sequence of the repetitive region (Fig. 1). For example, the largest predicted UpaH open reading frame (ORF) is 3,161 amino acids long (present in SE15), while the smallest predicted UpaH ORF is 1,940 amino acids long (present in UMN026) (Fig. 1). These differences could be partially due to errors in the sequence of the large and highly repetitive region within the passenger-encoding domain of UpaH, and we investigated this further by examining additional UPEC strains for UpaH expression. SDS-PAGE and Western blot analyses with UpaH antibodies indicated a significant variation in the size of UpaH in the previously described UPEC strain IA2 (11), as well as in M369 (a UPEC strain from our laboratory collection), compared to the size of UpaH in the UPEC strains CFT073, M161, and M357 (Fig. 2A). Notably, the apparent molecular mass of the UpaH protein from strain IA2 was approximately 200 kDa smaller than the UpaH protein from strain CFT073. The identity of UpaH was confirmed via the construction of isogenic upaH deletion mutants (referred to as M369upaH and IA2upaH); no bands were observed in Western blot analyses of these upaH-null strains with the UpaH antibody (Fig. 2A). To examine if the variation in the size of UpaH was due to glycosylation, we tested for the glycosylation of UpaH in all five UPEC strains using a commercially available glycoprotein-staining kit. In this assay, we saw no evidence of the glycosylation of UpaH (data not shown). Thus, while the UpaH protein is expressed by multiple UPEC strains, there are significant differences in the sizes of the protein from different strains.
Fig 1.
Evolutionary relationships and genome context of UpaH. The evolutionary history of UpaH (red) was inferred using the neighbor-joining method, and its genome context was compared in 19 E. coli strains. The bootstrap consensus tree was drawn to scale, and the evolutionary distances were computed using the p-distance method in MEGA5 (70). The genome context and gene variability were evaluated using complete and annotated E. coli genome sequences in Easyfig (67). Core (black) and variable (gray) genes are compared to those in the upaH-negative E. coli strain IAI1.
Fig 2.
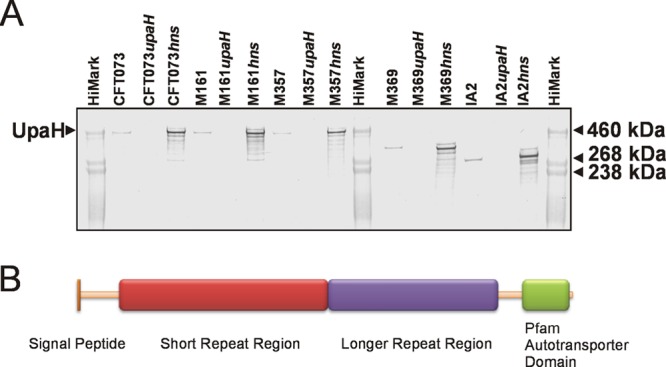
(A) Western blot analysis of UpaH production in whole-cell lysates prepared from E. coli strains CFT073, M161, M357, M369, and IA2 and their respective upaH or hns mutants. (B) Schematic illustration of the domain organization of UpaH from E. coli. Indicated are the signal peptide, the small repeat region, the longer repeat region, and the transmembrane β-domain. The repeat regions contain the majority of the sequence differences identified in the UpaH variants.
Cloning and sequencing of upaH gene variants.
To examine the functional consequences of UpaH size variation, the upaH gene was PCR amplified from the UPEC strains M161, M357, M369, and IA2 and cloned as a transcriptional fusion downstream of the tightly regulated araBAD promoter in the pBAD/myc-HisA expression vector to generate the plasmids pUpaHM161, pUpaHM357, pUpaHM369, and pUpaHIA2, respectively. Due to the very large (>2-kb) and highly repetitive region that exists in the passenger-encoding domain of the upaH gene, an in vitro transposon mutagenesis approach was employed to generate a library of overlapping transposon mutants, which enabled sequencing across this region of each gene. The overall lengths of the upaH genes were different and ranged from 7,581 to 8,538 bp. Translation of these sequences resulted in predicted UpaH proteins ranging in size from 2,527 to 2,846 amino acids (aa); predicted molecular masses ranged from 258 to 288 kDa (Table 2).
Table 2.
Analysis of UpaH variants found in different UPEC strains
| Origin of upaH (UPEC strain) | Size (bp) | Size (aa) | Predicted molecular mass (kDa) | No. of acidic amino acids | % acidic amino acids | No. of repeats predicted by RADAR | No. of repeatsa |
|---|---|---|---|---|---|---|---|
| CFT073 | 8,538 | 2,846 | 288 | 376 | 13.2 | 50 | 50 |
| M161 | 8,538 | 2,846 | 288 | 378 | 13.3 | 50 | 52 |
| M357 | 8,463 | 2,821 | 287 | 375 | 13.3 | 47 | 47 |
| M369 | 7,818 | 2,606 | 265 | 331 | 12.7 | 40 | 41 |
| IA2 | 7,581 | 2,527 | 258 | 323 | 12.8 | 36 | 37 |
Number of repeats present using the degenerative repeat motif (XGXXXXGXXAXXXXXGXXXXXX) based on the repeats identified in UpaHCFT073.
In silico sequence analysis of UpaH variants reveals high sequence divergence.
A bioinformatic approach was employed to compare the amino acid sequences of the five UpaH variants. Analyses using InterProScan (82) and the Simple Modular Architecture Design Tool (SMART) (40) identified the presence of a characteristic signal peptide, parallel β-helix repeats (PbH1) and the SCOP d1dbga from the pectin lysase-like superfamily (known to have a predominant secondary structure of right-handed parallel β-helix topology) within the passenger domain of each UpaH variant. The presence of an AT β-domain was also confirmed in the C-terminal translocation domain of each variant (Fig. 2B).
The amino acid sequences of the five UpaH variants were aligned using ClustalW and displayed an amino acid sequence identity of 53% across the full length of the protein. The highest percentage sequence identity was observed between the 300 amino acids in the N- and C-terminal regions of each protein (93% and 92% identity, respectively) (see Fig. S1 in the supplemental material). Conversely, more extensive amino acid sequence variation was observed in the remainder of the protein (43%). Different numbers of the 24- to 25-aa short degenerative repeat motif previously described in UpaHCFT073 were identified in the remainder of the protein, ranging from 39 repeats in UpaHIA2 to 52 repeats in UpaHM161 (Table 2). A similar repeat configuration was also identified after an independent analysis of each UpaH variant using the Rapid Automated Detection and Alignment of Repeats (RADAR) (26) program (Table 2). The region immediately following these short repeats exhibited the greatest sequence variation and contained a series of longer, more disordered, repeats as predicted by RADAR (Fig. 2B; see also Fig. S1).
UpaH variants mediate different levels of biofilm formation.
In order to assess the possible differences in function between the UpaH variants, the upaH gene was PCR amplified from strains CFT073, M161, M357, M369, and IA2, cloned into the pBAD-myc-HisA vector, and expressed in the E. coli K-12 flu gfp-tagged strain OS56 (65). This strain is unable to mediate the classical cell aggregation and biofilm phenotypes associated with Ag43 expression (46). Western blot analyses using UpaH-specific antiserum detected the UpaH protein in whole-cell lysates prepared from the E. coli strain OS56 containing pUpaHCFT073, pUpaHM161, pUpaHM357, pUpaHM369, and pUpaHIA2 following growth in LB containing 0.2% arabinose. In all cases, the size of the UpaH protein expressed from each plasmid construct matched that observed in the analysis of the corresponding wild-type strain (data not shown). Consistent with a previous report for UpaHCFT073 (2), the size of the UpaH protein variants was significantly larger than the corresponding predicted molecular mass and ranged from 290 to 460 kDa. A similar level of UpaH expression was detected from each of the plasmid constructs in the E. coli strain OS56 (data not shown).
The ability to mediate biofilm formation after expression in the E. coli OS56 strain was assessed for each of the UpaH variants under dynamic conditions using the continuous-flow chamber model. This system permits monitoring of the bacterial distribution within an evolving biofilm at the single-cell level due to the combination of scanning confocal laser microscopy and GFP-tagged OS56 cells. In this assay, all of the UpaH variants promoted significantly increased biofilm growth at 24 h compared to that of the E. coli OS56(pBAD/myc-HisA) negative control after induction with arabinose (Fig. 3A and B). Comparative analysis of the biofilm formed by each of the UpaH variants revealed significant differences in level of biovolume (P < 0.001), substratum coverage (P = 0.002), surface area-to-biovolume ratio (P < 0.001), vertical spreading (P < 0.001), mean thickness (P = 0.006), and biofilm roughness (P = 0.002) (Fig. 3A and B). Similar results were also obtained following 16 h of biofilm growth (data not shown). Taken together, these results demonstrate that the UpaH variants mediate different levels of biofilm formation when expressed from the same vector system (in the same host background), and we attribute this to differences in their passenger-domain sequence.
Fig 3.
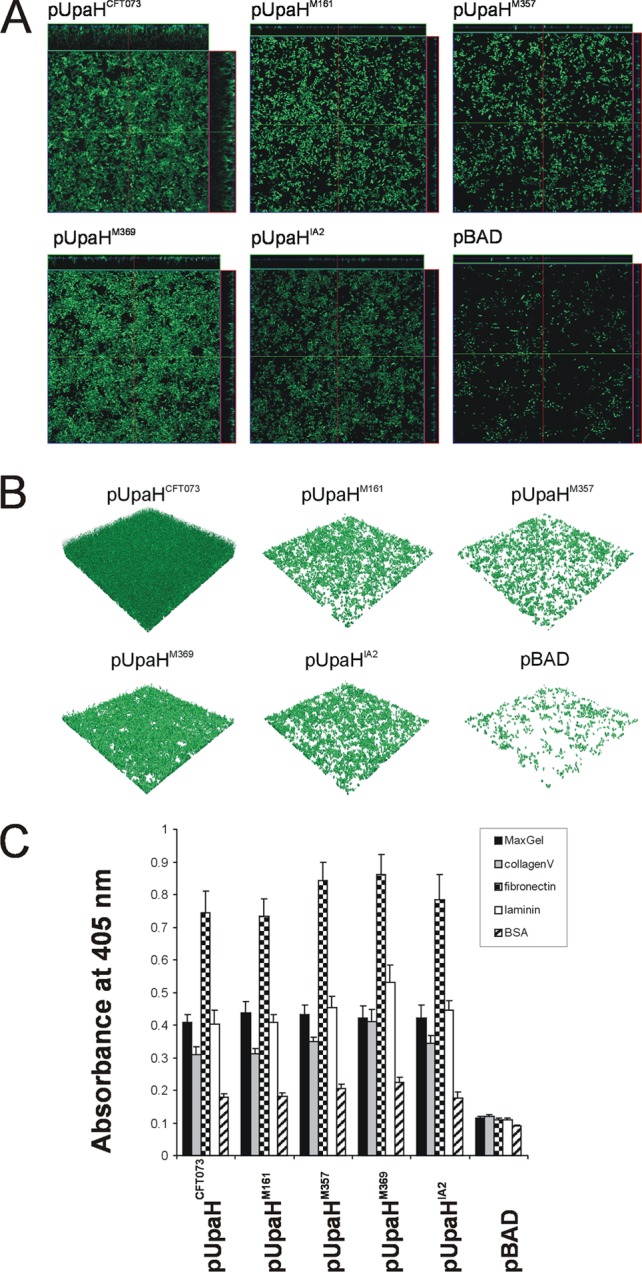
UpaH variants mediate different levels of biofilm formation in E. coli. (A) Overhead-view and (B) 3D-reconstructed fluorescence micrographs of biofilms formed under continuous flow in the dynamic-flow-chamber assay by E. coli OS56 strains expressing different UpaH variants (pUpaHCFT073, pUpaHM161, pUpaHM357, pUpaHM369, pUpaHIA2, or pBAD). Biofilm development was monitored via scanning confocal laser microscopy of the GFP-tagged E. coli strain OS56 cells induced with arabinose. The images in panel A are representative horizontal sections collected within each biofilm and vertical sections (the right panel and above each larger panel, representing the yz plane and the xz plane, respectively). Under 24 h of continuous flow, UpaH variants promoted biofilm growth with significant differences in level of biovolume (P < 0.001), substratum coverage (P = 0.002), vertical spreading (P = 0.001), surface area-to-biovolume ratio (P < 0.001), mean thickness (P = 0.005), and biofilm roughness (P = 0.001). Measurements were analyzed using COMSTAT and the Kruskal-Wallis test. These results demonstrate that UpaH variants resulted in different levels of biofilm formation when expressed from the same plasmid and in the same host background. (C) UpaH variants that mediate E. coli binding to MaxGel, collagen V, fibronectin, and laminin in ELISA-based binding to the same levels. The bar charts represent the average absorbance measurements at 405 nm of 4 independent experiments plus the standard errors of the mean (SEM). The mean absorbance readings were compared with negative control readings for OS56(pBAD).
UpaH mediates adherence to extracellular matrix proteins.
We employed an ELISA-based method to study the adhesive properties of the UpaH variants to ECM proteins following expression in the E. coli OS56 strain. Initially, we tested the ability of the UpaH variants to mediate adherence to MaxGel human ECM (Sigma-Aldrich), a commercially available mixture of ECM components, including collagens, laminin, fibronectin, tenascin, elastin, proteoglycans, and glycosaminoglycans. All of the UpaH variants promoted significant binding to MaxGel (Fig. 3C). To more specifically examine this adherence, we then tested the binding of the UpaH variants to a specific set of ECM proteins. The UpaH variants all promoted binding to collagen V, fibronectin, and laminin but not to collagen IV, fibrinogen, or BSA (Fig. 3C and data not shown). Thus, UpaH promotes specific binding to some ECM proteins, and this adherence property is conserved among different variants.
Identification and mutation of hydrophobic and conserved regions of UpaH.
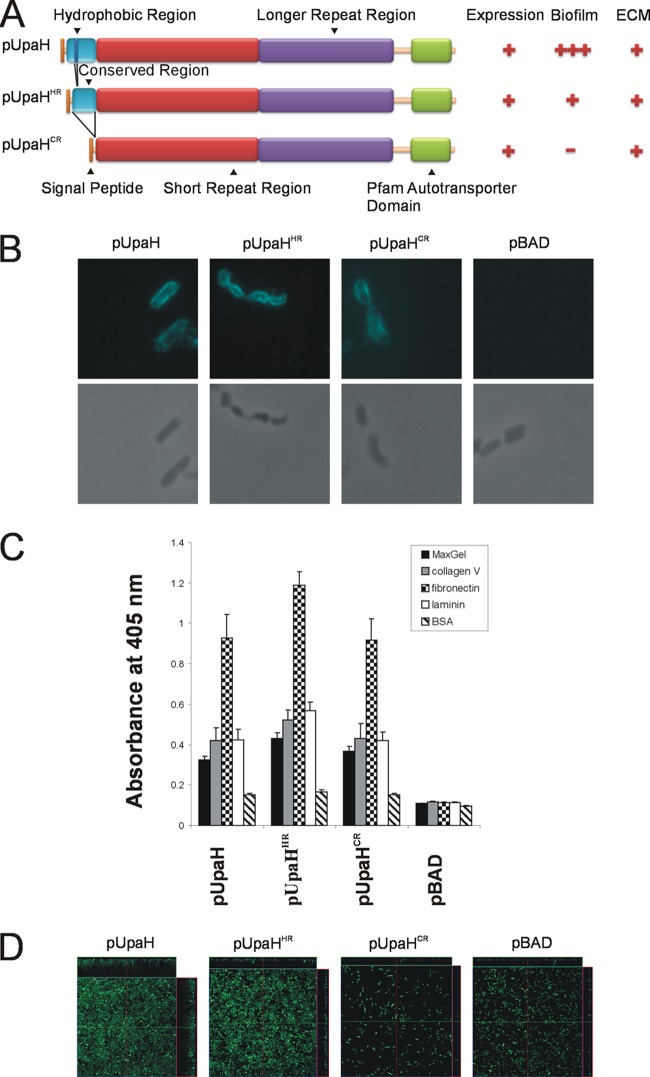
Binding to ECM molecules has also been demonstrated for several other AT proteins, including YadA, a trimeric AT adhesin from Yersinia enterocolitica (4, 25, 27, 68, 69, 71). YadA possesses the ability to mediate adherence to ECM proteins such as fibronectin, laminin, and collagen. YadA has a hydrophobic domain of 22 amino acids (corresponding to amino acids 55 to 76 of the mature protein) that when deleted results in the loss of collagen binding (66, 69). We investigated the N-terminal 300-amino-acid region of UpaH and identified a hydrophobic segment of 25 amino acids (corresponding to residues 44 to 68 of the mature UpaHCFT073 protein) that is highly conserved in all of the variants examined in this study (Fig. 4A; see also Fig. S1 in the supplemental material). We hypothesized that this region may confer the binding of UpaH to collagen V and other ECM proteins and investigated this further by modifying the upaH gene in pUpaHCFT073 through deletion of the hydrophobic region (pUpaHHR, lacking amino acids 44 to 68 of the mature protein) or the majority of the conserved region (pUpaHCR, lacking amino acids 6 to 192 of the mature protein) (Fig. 4A). The CFT073 UpaHHR and UpaHCR proteins were produced in equivalent amounts as determined by Western blot analyses of whole-cell lysates (data not shown) and effectively translocated to the cell surface of E. coli strain OS56, as demonstrated by immunofluorescence microscopy performed with UpaH-specific antisera on E. coli strains OS56(pUpaHHR), OS56(pUpaHCR), and OS56(pUpaH; control) following induction with arabinose during growth in LB (Fig. 4B). No reaction was seen with E. coli strain OS56(pBAD) cells (Fig. 4B).
Fig 4.
(A) Schematic illustration of the domain organization of UpaH, UpaHHR, and UpaHCR and a summary of phenotypic results. Indicated are the signal peptide, the hydrophobic region, the conserved region, the small repeat region, the longer repeat region, and the transmembrane β-domain. (B) Immunofluorescence (top) or phase-contrast (bottom) microscopy employing UpaH-specific antiserum against the cells of the specified E. coli strains. Strains were grown in the presence of 0.2% arabinose. The overnight cultures were fixed and incubated with anti-UpaH serum followed by incubation with goat anti-rabbit IgG coupled to Alexa Fluor 350. Positive reactions indicating surface localization of UpaH, UpaHHR, and UpaHCR were detected. (C) UpaH, UpaHHR, and UpaHCR mediate E. coli binding to MaxGel, collagen V, fibronectin, and laminin in ELISA-based binding to the same levels. The bar charts represent average absorbance measurements at 405 nm of 3 independent experiments plus the SEM. The mean absorbance readings were compared with negative control readings from strain OS56(pBAD). (D) UpaH mutants deleted for the hydrophobic or conserved region that mediates different levels of biofilm formation in E. coli. Shown are the overhead views of biofilms formed under continuous flow by E. coli OS56 strains expressing pUpaHCFT073, pUpaHHR, pUpaHCR, or pBAD. Biofilm development was monitored via scanning confocal laser microscopy of the GFP-tagged E. coli strain OS56 cells. The images are representative horizontal sections collected within each biofilm and vertical sections (the right panel and above each larger panel, representing the yz plane and the xz plane, respectively). Under 24 h of continuous flow, the UpaH mutants affected biofilm growth with significant differences in level of biovolume (P = 0.001), substratum coverage (P = 0.003), vertical spreading (P = 0.001), mean thickness (P = 0.002), and biofilm roughness (P = 0.001). Measurements were analyzed using COMSTAT and the Kruskal-Wallis test. These results demonstrate that expression of pUpaHCFT073, pUpaHHR, and pUpaHCR resulted in different levels of biofilm formation when expressed from the same plasmid and in the same host background.
Mutation of the hydrophobic and conserved regions of UpaH affects biofilm formation.
To examine the phenotype of E. coli cells expressing UpaHHR or UpaHCR, we tested the ability of E. coli strains OS56(pUpaH), OS56(pUpaHHR), and OS56(pUpaHCR) to bind to ECM proteins and form a biofilm. In the ECM assay, the three strains bound at similar levels to MaxGel, collagen V, fibronectin, and laminin, demonstrating that the ability to bind to ECM proteins is not dependent on these regions of UpaH (Fig. 4C). However, in the flow cell assay, comparative analyses of the biofilm formed by each strain revealed significant differences in level of biovolume (P = 0.001), substratum coverage (P = 0.003), vertical spreading (P = 0.001), mean thickness (P = 0.002), and biofilm roughness (P = 0.001) (Fig. 4D). In the flow cell, pUpaH formed a thick biofilm, as previously observed (Fig. 3A and B and 4D) (2). The expression of UpaHHR resulted in reduced vertical spreading but maintained clear substratum coverage in comparison to the overexpression of pUpaH, while expression of UpaHCR did not promote any biofilm growth (Fig. 4D). Thus, the ability to mediate biofilm formation is dramatically affected by both the hydrophobic and the conserved regions of UpaH.
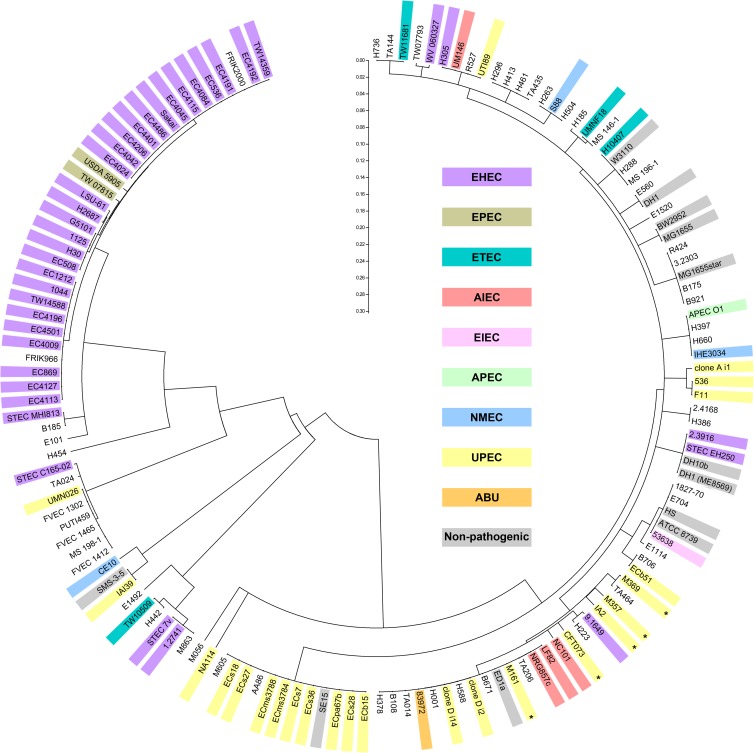
A high degree of sequence variation is present in UpaH sequences.
The conserved N-terminal 300 amino acids identified in this study were used to investigate the variation in UpaH sequences present in the E. coli genomes available in the NCBI database. This region was chosen to avoid the repetitive region due to the inherent difficulties of sequencing long stretches of repetitive DNA, the problems associated with aligning these sequences, and the high error rate for repetitive sequences in draft genomes. Interestingly, when the N-terminal 300 amino acids of 143 UpaH homologues (5 obtained via sequencing in this study and 138 from the NCBI database) were analyzed using MEGA5 (70), the enterohemorrhagic E. coli (EHEC) and extraintestinal pathogenic E. coli (ExPEC) (UPEC, asymptomatic bacteriuria [ABU], neonatal meningitis E. coli [NMEC]) sequences were grouped roughly into separate clades (Fig. 5). There are significant differences between these two clades; for example, the N-terminal 300 amino acids of the strains 1.2741 (EHEC clade) and CFT073 (ExPEC clade) share 38% sequence identity. The difference between these two clades extends throughout the majority of the passenger domain (see Fig. 1, UMN026 versus 536). As an example, the entire passenger domain (1 to 2,527 aa) of UpaH from strains CFT073 and 1.2741 contains 38.3% sequence identity, compared to 84.3% sequence identity in the predicted β-barrel domain. The clades identified in the alignment of the N-terminal 300 amino acids also accurately reflect the branches present in the tree generated from 19 UpaH sequences obtained from complete genomes that contain an intact open reading frame (Fig. 1 and 5).
Fig 5.
Evolutionary relationships of UpaH. The evolutionary relationship of UpaH was inferred using the N-terminal 300 amino acids of 143 upaH-positive E. coli strains. The neighbor-joining method was used, and the bootstrap consensus tree was taken to represent the evolutionary history of the taxa analyzed in MEGA5 (70). The branches corresponding to partitions reproduced in less than 50% of the bootstrap replicates are collapsed. The tree is drawn to scale, and the distances were computed using the p-distance method. The E. coli strains are color coded according to their pathotypes, and the UPEC strains used in this study are indicated by an asterisk.
Transcription of the upaH variant alleles is regulated by the H-NS silencing protein.
The observation that UpaH variants possess domains with conserved or divergent functional roles prompted us to examine the regulation of these different upaH alleles at the transcriptional level. To facilitate this, we inserted a lacZ reporter as a transcriptional fusion to the promoter of the upaH gene on the chromosome of CFT073lacI-Z to generate the strain CFT073lacI-Z upaH::lacZ-zeo. We subsequently created specific gene deletions in CFT073lacI-Z upaH::lacZ-zeo by λ-red-mediated homologous recombination in a series of known/putative regulatory genes: c0421 (virF-like), c1699 (rpoS), c1701 (hns), c2091 (virF-like), c2411 (hns-like), c3218 (stpA), c3244 (luxS; AI-2), c3744 (virF-like), c4864 (cpxR; periplasmic stress), and c5054 (soxR; oxidative stress). Each of these mutants was then assayed for β-galactosidase activity to assess the effect of the deletion of each regulatory gene on transcription from the upaH promoter. The CFT073lacI-Z upaH::lacZ-zeo hns mutant exhibited a 32-fold increase in β-galactosidase activity compared to that of CFT073lacI-Z upaH::lacZ-zeo, while no significant differences were observed for the other specific gene deletion mutants (Fig. 6A and data not shown). No color heterogeneities on X-Gal plates were observed among the CFT073lacI-Z upaH::lacZ-zeo colonies or subsequent mutants, indicating that the expression of UpaH is apparently not subjected to phase variation. The complementation of the CFT073lacI-Z upaH::lacZ-zeo hns mutant with an hns-containing plasmid (pH-NS) resulted in a significant reduction in β-galactosidase activity (Fig. 6A). Thus, transcription of the upaH gene is derepressed in a CFT073 hns mutant.
Fig 6.
H-NS represses upaH expression. (A) β-Galactosidase assay of upaH::lacZ-zeo reporter fusions in strain CFT073, isogenic hns deletion mutant, complemented mutant, and complement vector control. (B) Electrophoretic band shift of the amplified 250-bp upaH promoter and the bla promoter from TaqI-SspI-digested pBR322 DNA in the presence of increasing concentrations of H-NS (0.05, 0.1, 0.2, 0.5, 1, and 2 μM). The pBR322 fragments not containing the bla-promoter (313/315 bp and 475 bp) were not shifted by H-NS. (C) Curvature-propensity plot of the sequence (250 bp) upstream of the upaH gene from CFT073(PupaHCFT073) and M369(PupaHM369) showing the predicted regions of curved DNA. (D) Curved DNA PAGE gel of the amplified 250-bp promoters PupaHCFT073 and PupaHM369.
To investigate whether H-NS influences gene expression by direct binding to the promoter region of the upaH gene, we performed electrophoretic mobility shift assays. The 250-bp PCR product was mixed with TaqI-SspI-digested pBR322 DNA, which contains the bla promoter previously shown to be bound by H-NS (5, 43, 83), incubated with increasing concentrations of purified H-NS protein and subsequently visualized by gel electrophoresis. The 250-bp upaH promoter region and the fragment containing the bla promoter were retarded in their mobility by the addition of 0.5 μM H-NS (Fig. 6B). The pBR322 fragments not containing the bla promoter were not influenced by H-NS at these concentrations, indicating that H-NS binds with specificity. These results suggest that H-NS binds to the regulatory region of the upaH gene by recognizing a DNA region within 250 bp upstream of the ATG translation start codon. Comparative analyses of the upaH promoter region from UPEC strains CFT073, M161, M357, M369, and IA2 revealed several nucleotide differences (data not shown). An in silico-generated curvature-propensity plot calculated with DNase I-based parameters for the upaH variants suggested that the promoter regions of upaH may adopt a curved conformation, consistent with its ability to bind to H-NS. The curvature-propensity plot for upaH from strains CFT073, M161, M357, and IA2 revealed an almost identical curved conformation, while the upaH promoter from M369 contained an additional peak in the curvature-propensity plot caused by a single G-to-A change at ATG-168 (Fig. 6C and data not shown). To demonstrate this curvature experimentally, we amplified the 250-bp PCR products containing the predicted upaH promoter regions and examined them by polyacrylamide gel electrophoresis at 4°C. This method has been used previously to demonstrate DNA curvature (73, 81). Consistent with our in silico analysis, we found that both 250-bp upaH promoter regions displayed retarded gel electrophoretic mobility compared to noncurved DNA standards, with the mobility of the upaH promoter region from M369 impeded more than the corresponding region from CFT073 (Fig. 6D).
The DNA curvature identified in the promoter region of the upaH variants prompted us to examine the effect of H-NS on UpaH production from CFT073, as well as UPEC strains M161, M357, M369, and IA2. Isogenic hns deletion mutants were constructed for all five UPEC strains and UpaH production was examined via Western blot analysis. The loss of H-NS from strains CFT073, M161, M357, M369, and IA2 resulted in the increased expression of UpaH (with evidence of protein degradation) (Fig. 2A). The overexpression of UpaH also resulted in multiple smaller bands presumed to be breakdown products, consistent with previous reports (Fig. 2A) (2). Thus, H-NS negatively regulates the expression of upaH in UPEC, and this property is conserved despite variation in the upaH coding region.
DISCUSSION
Bacterial adhesins mediate attachment to host tissues and provide the first critical step in colonization. UPEC possesses a range of different adhesins, many of which belong to the AT class of proteins. We recently described the identification and characterization of the UpaH AT protein from UPEC CFT073 (2). Here we show that the sequences of UpaH vary between different UPEC strains and that this sequence variation is associated with differences in the abilities of UpaH proteins to mediate biofilm formation. In contrast, all of the UpaH variants examined in this study retained a conserved ability to mediate binding to ECM proteins.
UpaH is the largest AT protein described from UPEC. UpaH from CFT073 (UpaHCFT073) is a 2,845-amino-acid protein that comprises a typical AT domain structure: an N-terminal signal sequence, a passenger domain, and a C-terminal translocation domain. The passenger domain of UpaHCFT073 contains 50 imperfect repeats, each of which comprises 24 to 25 amino acid residues (2). In this study, we cloned and sequenced the upaH gene from four additional UPEC strains. This required the generation of libraries of overlapping transposon mutants to sequence across the highly repetitive passenger-encoding domain of each upaH variant. The sizes of the upaH genes from the UPEC strains varied; this was reflected by differences in the numbers of short repeat sequences in each variant. Short repeat sequences are common to the AIDA-I AT group and have been analyzed within the passenger domain of the TibA, AIDA-I, and Ag43 AT proteins from E. coli (6, 12, 30, 34, 78). AIDA-I contains 21 repeats built up of 19 to 20 amino acid residues followed, after a short interruption, by an additional 10 repeats. The repeat structure of Ag43 is less well defined and more variable in length (12 to 20 amino acids). Like these proteins, the repeating sequence pattern in the passenger domain of the UpaH variants suggests that this region is predominantly β-helical in structure. The variable repeat sequence structure in each of the upaH variants also indicates that they are prone to internal expansion or collapse while maintaining gene integrity. The region immediately following these repeats consisted of longer, more disordered, repeats that contained less variation in size but more variation in sequence. The evolution of the upaH gene may be driven by amplification/deletion of repeats, resulting in various numbers of repeat sequences, as well as acquisition/loss of whole blocks of sequences, resulting in new domains.
We observed a significant difference in the abilities of the UpaH variants to mediate biofilm formation in a dynamic flow cell system. These differences did not correspond to the number of short repeat sequences in each variant; UpaHCFT073 (50 short repeats) and UpaHM369 (40 short repeats) promoted the strongest biofilm, while UpaHM161 (50 short repeats), UpaHIA2 (36 short repeats), and UpaHM357 (47 short repeats) all produced a biofilm that was more diffuse in structure. This result is in contrast to recent studies on the Hap and Cha AT proteins from Haemophilus, where large deletions or expansion of the repetitive region from each protein negatively affects bacterial aggregation (45, 64). A difference in the ability to mediate biofilm formation has also been reported for the two Ag43 variants of CFT073. In the flow cell system, Ag43a mediates strong biofilm formation compared to Ag43b (72). It is possible that specific residues within the repeat sequences of UpaH and Ag43 contribute to differences in the abilities of both proteins to mediate biofilm formation. However, direct demonstration of this by site-directed mutagenesis represents a major challenge for proteins, like UpaH, that contain very large repeats.
A conserved property of all of the UpaH variants examined in this study was the ability to bind to ECM proteins. Significant UpaH-mediated binding was observed to collagen V, fibronectin, and laminin. Binding to ECM molecules has also been demonstrated for other AT proteins, including Ag43, AIDA-I, UpaG, and YadA (27, 39, 54, 69, 71, 74). The YadA trimeric AT adhesin from Yersinia enterocolitica has been extensively characterized (4, 25, 27, 68, 69, 71). YadA possesses multiple virulence-associated properties, including the ability to mediate adherence to ECM proteins, such as fibronectin, laminin, and collagen. YadA has a hydrophobic domain of 22 amino acids (corresponding to amino acids 55 to 76 of the mature protein) that when deleted results in the loss of collagen binding (66, 69). Interestingly, UpaH also contains a hydrophobic region of 25 amino acids (corresponding to residues 44 to 68 of the UpaHCFT073 protein) in its N terminus that is highly conserved in all of the variants examined in this study. This region is also highly conserved (88 to 100% amino acid sequence identity) in all 90 of the UpaH sequences from the ExPEC clade compared to in the EHEC clade (39 to 64% amino acid sequence identity), and we hypothesized that it may confer binding of UpaH to collagen V and other ECM proteins. However, we showed that the deletion of this region (UpaHHR), or even a larger conserved segment spanning this region (UpaHCR), did not reduce ECM binding. Although cells overexpressing UpaH exhibited a strong hydrophobicity index compared to cells that did not express UpaH, the deletion of the HR or CR regions also did not significantly reduce cell hydrophobicity (data not shown). Thus, it is possible that the conserved ECM binding mediated by UpaH, UpaHHR, and UpaHCR may be associated with an overall change in cell surface hydrophobicity. We note that we cannot rule out the possibility that the binding of UpaH to ECM proteins is mediated by the short repeat sequences in the passenger domain. Indeed, a similar structural configuration built by multiple short repeat sequences has been shown to mediate the binding of the Candida albicans adhesin Als5p to fibronectin (53).
In contrast to ECM proteins, the UpaHHR and UpaHCR proteins exhibited a reduced capacity to promote biofilm formation. This may be a direct effect resulting from deletion of these regions. Alternatively, the deletion of the HR or CR may alter the folding of the short and/or long repeat regions of UpaH, thus altering the tertiary structure of the protein. We also note that the HR and CR overlap, such that deletion of the CR also removes the HR. Indeed, the expression of UpaHCR did not promote biofilm formation in the flow cell assay.
A role for H-NS in the regulation of the upaH gene was demonstrated in several independent experiments. At the transcriptional level, a significant increase in upaH promoter activity was observed in a CFT073lacI-Z upaH lacZ-zeo hns mutant. Consistent with this result, we also observed an increase in the production of the UpaH protein by CFT073 and five other UPEC strains in an hns mutant background. H-NS is a histone-like DNA-binding protein that shows affinity for A-T-rich and bent nucleation sites on DNA (17, 18). Previous studies have shown that mutation of the hns gene in the UPEC strain 536 results in the derepression of multiple virulence factors, including alpha-hemolysin, iron uptake systems, and fimbriae (48). Our data demonstrate that H-NS acts as a repressor of upaH transcription, most likely through direct binding to a region comprising the 250 bp upstream of the upaH open reading frame. The modulation of H-NS expression may represent a mechanism by which UPEC controls the coordinated expression of virulence genes during colonization of the urinary tract. Indeed, analysis of the microarray data from Muller et al. (48) showed that transcription of the truncated E. coli K-12 upaH gene was approximately 17 times higher in an hns mutant. Furthermore, in the UPEC strain 536 (which contains an intact upaH gene), upaH transcription was approximately five times higher in an hns mutant (48). Interestingly, H-NS has been shown to exert the opposite effect on the transcription of aidA (which encodes AIDA-I), where it acts as an activator (7). We have also previously shown that upaH is transcribed by CFT073 voided in the urine of mice in our UTI model (2). It is possible that the transcription of upaH is coordinated with other H-NS-repressed adhesin genes. Indeed, H-NS also represses the expression of the recently described upaC gene from the UPEC strain CFT073, which encodes an AT protein associated with adherence to ECM proteins (1). H-NS is also involved with the regulation of type 1-fimbria expression (16). In addition, several cryptic E. coli K-12 chaperone-usher fimbrial operons that have orthologs in UPEC genomes were also recently shown to be repressed by H-NS (36). This suggests that in UPEC, H-NS may coordinate the expression of multiple virulence factors.
H-NS has a role in nucleoid organization and acts as a global regulator of gene expression (17). In the Salmonella enterica serovar Typhimurium, H-NS represses the transcription of multiple virulence genes, in particular those located on A-T-rich pathogenicity islands (22, 42, 51). Related enteric bacteria also utilize the levels of histone-like nucleoid structuring proteins to constrain virulence gene expression (17, 50). Further investigation is required to determine if specific conditions encountered by UPEC in the urinary tract lower intracellular levels of H-NS, or perhaps alter its binding affinity, to increase the expression of UpaH. Such conditions would also derepress other H-NS-regulated genes and thus lead to the coordinated expression of multiple virulence factors.
The bioinformatic analysis of upaH from multiple genome-sequenced strains revealed that the gene is always found in the same genetic context. Despite this, the sequences of the upaH genes were shown to vary between different strains. To explore this sequence variability of UpaH independently of the repeat regions, we identified a 300-amino-acid nonrepetitive region in the N-terminal regions of 143 strains and compared these nonrepetitive regions. Using this sequence, we showed that UpaH variants can be grouped into defined clades. Several of the UpaH variants that clade separately were analyzed and shown to vary with respect to their capacity to mediate biofilm formation. Further analysis of UpaH functions could be targeted to the study of variants from distinct clades (e.g., UMN026, IAI39). It is also noteworthy that the 300-amino-acid sequences of the conserved region of UpaH proteins differed significantly between ExPEC and EHEC pathotypes. This suggests the possibility that UpaH variation may be associated with pathotype-specific functions and host tissue tropism. This remains to be determined, as does the integrity of the upaH gene in EHEC, which is disrupted by an insertion element or a frameshift mutation in many genome-sequenced strains.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sylvie Rimsky for providing the purified native H-NS protein.
This work was supported by grants from the Australian National Health and Medical Research Council (631654), the Australian Research Council (DP1097032), the Institut Pasteur, the Network of Excellence EuroPathoGenomics, and the European Community (LSHB-CT-2005-512061). M.A.S. is supported by an ARC future fellowship (FT100100662).
Footnotes
Published ahead of print 17 August 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Allsopp LP, et al. 2012. Molecular characterization of UpaB and UpaC, two new autotransporter proteins of uropathogenic Escherichia coli CFT073. Infect. Immun. 80:321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allsopp LP, et al. 2010. UpaH is a newly identified autotransporter protein that contributes to biofilm formation and bladder colonization by uropathogenic Escherichia coli CFT073. Infect. Immun. 78:1659–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson GG, et al. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301:105–107 [DOI] [PubMed] [Google Scholar]
- 4.Balligand G, Laroche Y, Cornelis G. 1985. Genetic analysis of virulence plasmid from a serogroup 9 Yersinia enterocolitica strain: role of outer membrane protein P1 in resistance to human serum and autoagglutination. Infect. Immun. 48:782–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beloin C, Dorman C. 2003. An extended role for the nucleoid structuring protein H-NS in the virulence gene regulatory cascade of Shigella flexneri. Mol. Microbiol. 47:825–838 [DOI] [PubMed] [Google Scholar]
- 6.Benz I, Schmidt MA. 1992. AIDA-I, the adhesin involved in diffuse adherence of the diarrhoeagenic Escherichia coli strain 2787 (O126:H27), is synthesized via a precursor molecule. Mol. Microbiol. 6:1539–1546 [DOI] [PubMed] [Google Scholar]
- 7.Benz I, van Alen T, Bolte J, Wormann ME, Schmidt MA. 2010. Modulation of transcription and characterization of the promoter organization of the autotransporter adhesin heptosyltransferase and the autotransporter adhesin AIDA-I. Microbiology 156:1155–1166 [DOI] [PubMed] [Google Scholar]
- 8.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolivar F, et al. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95–113 [PubMed] [Google Scholar]
- 10.Chaveroche MK, Ghigo JM, d'Enfert C. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clegg S. 1982. Cloning of genes determining the production of mannose-resistant fimbriae in a uropathogenic strain of Escherichia coli belonging to serogroup O6. Infect. Immun. 38:739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cote JP, Mourez M. 2011. Structure-function analysis of the TibA self-associating autotransporter reveals a modular organization. Infect. Immun. 79:1826–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danese PN, Pratt LA, Dove SL, Kolter R. 2000. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37:424–432 [DOI] [PubMed] [Google Scholar]
- 14.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derbise A, Lesic B, Dacheux D, Ghigo JM, Carniel E. 2003. A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunol. Med. Microbiol. 38:113–116 [DOI] [PubMed] [Google Scholar]
- 16.Donato GM, Lelivelt MJ, Kawula TH. 1997. Promoter-specific repression of fimB expression by the Escherichia coli nucleoid-associated protein H-NS. J. Bacteriol. 179:6618–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorman CJ. 2007. H-NS, the genome sentinel. Nat. Rev. Microbiol. 5:157–161 [DOI] [PubMed] [Google Scholar]
- 18.Dorman CJ. 2004. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2:391–400 [DOI] [PubMed] [Google Scholar]
- 19.Dutta PR, Cappello R, Navarro-Garcia F, Nataro JP. 2002. Functional comparison of serine protease autotransporters of Enterobacteriaceae. Infect. Immun. 70:7105–7113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eto DS, Sundsbak JL, Mulvey MA. 2006. Actin-gated intracellular growth and resurgence of uropathogenic Escherichia coli. Cell. Microbiol. 8:704–717 [DOI] [PubMed] [Google Scholar]
- 21.Gasteiger E, et al. 2003. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groisman EA, Ochman H. 1997. How Salmonella became a pathogen. Trends Microbiol. 5:343–349 [DOI] [PubMed] [Google Scholar]
- 23.Guyer DM, Henderson IR, Nataro JP, Mobley HL. 2000. Identification of sat, an autotransporter toxin produced by uropathogenic Escherichia coli. Mol. Microbiol. 38:53–66 [DOI] [PubMed] [Google Scholar]
- 24.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heesemann J, Eggers C, Schroder J. 1987. Serological diagnosis of yersiniosis by immunoblot technique using virulence-associated antigen of enteropathogenic Yersiniae. Contrib. Microbiol. Immunol. 9:285–289 [PubMed] [Google Scholar]
- 26.Heger A, Holm L. 2000. Rapid automatic detection and alignment of repeats in protein sequences. Proteins 41:224–237 [DOI] [PubMed] [Google Scholar]
- 27.Heise T, Dersch P. 2006. Identification of a domain in Yersinia virulence factor YadA that is crucial for extracellular matrix-specific cell adhesion and uptake. Proc. Natl. Acad. Sci. U. S. A. 103:3375–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henderson IR, Cappello R, Nataro JP. 2000. Autotransporter proteins, evolution and redefining protein secretion. Trends Microbiol. 8:529–532 [DOI] [PubMed] [Google Scholar]
- 29.Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala'Aldeen D. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henderson IR, Owen P. 1999. The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and oxyR. J. Bacteriol. 181:2132–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heydorn A, et al. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407 [DOI] [PubMed] [Google Scholar]
- 32.Ieva R, Bernstein HD. 2009. Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc. Natl. Acad. Sci. U. S. A. 106:19120–19125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jose J, Jahnig F, Meyer TF. 1995. Common structural features of IgA1 protease-like outer membrane protein autotransporters. Mol. Microbiol. 18:378–380 [DOI] [PubMed] [Google Scholar]
- 34.Kajava AV, Steven AC. 2006. The turn of the screw: variations of the abundant beta-solenoid motif in passenger domains of type V secretory proteins. J. Struct. Biol. 155:306–315 [DOI] [PubMed] [Google Scholar]
- 35.Klemm P, Hjerrild L, Gjermansen M, Schembri MA. 2004. Structure-function analysis of the self-recognizing antigen 43 autotransporter protein from Escherichia coli. Mol. Microbiol. 51:283–296 [DOI] [PubMed] [Google Scholar]
- 36.Korea CG, Badouraly R, Prevost MC, Ghigo JM, Beloin C. 2010. Escherichia coli K-12 possesses multiple cryptic but functional chaperone-usher fimbriae with distinct surface specificities. Environ. Microbiol. 12:1957–1977 [DOI] [PubMed] [Google Scholar]
- 37.Korea CG, Ghigo JM, Beloin C. 2011. The sweet connection: solving the riddle of multiple sugar-binding fimbrial adhesins in Escherichia coli: multiple E. coli fimbriae form a versatile arsenal of sugar-binding lectins potentially involved in surface-colonisation and tissue tropism. Bioessays 33:300–311 [DOI] [PubMed] [Google Scholar]
- 38.Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105–132 [DOI] [PubMed] [Google Scholar]
- 39.Laarmann S, Schmidt MA. 2003. The Escherichia coli AIDA autotransporter adhesin recognizes an integral membrane glycoprotein as receptor. Microbiology 149:1871–1882 [DOI] [PubMed] [Google Scholar]
- 40.Letunic I, Doerks T, Bork P. 2009. SMART 6: recent updates and new developments. Nucleic Acids Res. 37:D229–D232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lievin-Le Moal V, et al. 2011. Secreted autotransporter toxin (Sat) triggers autophagy in epithelial cells that relies on cell detachment. Cell. Microbiol. 13:992–1013 [DOI] [PubMed] [Google Scholar]
- 42.Lucchini S, et al. 2006. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2:e81 doi:10.1371/journal.ppat.0020081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lucht JM, Dersch P, Kempf B, Bremer E. 1994. Interactions of the nucleoid-associated DNA-binding protein H-NS with the regulatory region of the osmotically controlled proU operon of Escherichia coli. J. Biol. Chem. 269:6578–6586 [PubMed] [Google Scholar]
- 44.Martin MA, Pfaller MA, Massanari RM, Wenzel RP. 1989. Use of cellular hydrophobicity, slime production, and species identification markers for the clinical significance of coagulase-negative staphylococcal isolates. Am. J. Infect. Control 17:130–135 [DOI] [PubMed] [Google Scholar]
- 45.Meng G, Spahich N, Kenjale R, Waksman G, St Geme JW., III 2011. Crystal structure of the Haemophilus influenzae Hap adhesin reveals an intercellular oligomerization mechanism for bacterial aggregation. EMBO J. 30:3864–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria, vol 1 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 47.Mobley HL, et al. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58:1281–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muller CM, et al. 2006. Role of histone-like proteins H-NS and StpA in expression of virulence determinants of uropathogenic Escherichia coli. J. Bacteriol. 188:5428–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mysorekar IU, Hultgren SJ. 2006. Mechanisms of uropathogenic Escherichia coli persistence and eradication from the urinary tract. Proc. Natl. Acad. Sci. U. S. A. 103:14170–14175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Navarre WW, McClelland M, Libby SJ, Fang FC. 2007. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 21:1456–1471 [DOI] [PubMed] [Google Scholar]
- 51.Navarre WW, et al. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236–238 [DOI] [PubMed] [Google Scholar]
- 52.Purdy GE, Fisher CR, Payne SM. 2007. IcsA surface presentation in Shigella flexneri requires the periplasmic chaperones DegP, Skp, and SurA. J. Bacteriol. 189:5566–5573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rauceo JM, et al. 2006. Threonine-rich repeats increase fibronectin binding in the Candida albicans adhesin Als5p. Eukaryot. Cell 5:1664–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reidl S, Lehmann A, Schiller R, Salam Khan A, Dobrindt U. 2009. Impact of O-glycosylation on the molecular and cellular adhesion properties of the Escherichia coli autotransporter protein Ag43. Int. J. Med. Microbiol. 299:389–401 [DOI] [PubMed] [Google Scholar]
- 55.Rosen DA, Hooton TM, Stamm WE, Humphrey PA, Hultgren SJ. 2007. Detection of intracellular bacterial communities in human urinary tract infection. PLoS Med. 4:e329 doi:10.1371/journal.pmed.0040329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rossiter AE, et al. 2011. The essential {beta}-barrel assembly machinery complex components BamD and BamA are required for autotransporter biogenesis. J. Bacteriol. 193:4250–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruiz-Perez F, et al. 2009. Roles of periplasmic chaperone proteins in the biogenesis of serine protease autotransporters of Enterobacteriaceae. J. Bacteriol. 191:6571–6583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruiz-Perez F, Henderson IR, Nataro JP. 2010. Interaction of FkpA, a peptidyl-prolyl cis/trans isomerase with EspP autotransporter protein. Gut Microbes 1:339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 60.Sauri A, et al. 2009. The Bam (Omp85) complex is involved in secretion of the autotransporter haemoglobin protease. Microbiology 155:3982–3991 [DOI] [PubMed] [Google Scholar]
- 61.Schembri MA, Hjerrild L, Gjermansen M, Klemm P. 2003. Differential expression of the Escherichia coli autoaggregation factor antigen 43. J. Bacteriol. 185:2236–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schubert S, Rakin A, Fischer D, Sorsa J, Heesemann J. 1999. Characterization of the integration site of Yersinia high-pathogenicity island in Escherichia coli. FEMS Microbiol. Lett. 179:409–414 [DOI] [PubMed] [Google Scholar]
- 63.Selkrig J, et al. 2012. Discovery of an archetypal protein transport system in bacterial outer membranes. Nat. Struct. Mol. Biol. 19:506–510, S1 [DOI] [PubMed] [Google Scholar]
- 64.Sheets AJ, St Geme JW., III 2011. Adhesive activity of the Haemophilus cryptic genospecies cha autotransporter is modulated by variation in tandem Peptide repeats. J. Bacteriol. 193:329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sherlock O, Schembri MA, Reisner A, Klemm P. 2004. Novel roles for the AIDA adhesin from diarrheagenic Escherichia coli: cell aggregation and biofilm formation. J. Bacteriol. 186:8058–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Skurnik M, Wolf-Watz H. 1989. Analysis of the yopA gene encoding the Yop1 virulence determinants of Yersinia spp. Mol. Microbiol. 3:517–529 [DOI] [PubMed] [Google Scholar]
- 67.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tahir YE, Kuusela P, Skurnik M. 2000. Functional mapping of the Yersinia enterocolitica adhesin YadA. Identification of eight NSVAIG-S motifs in the amino-terminal half of the protein involved in collagen binding. Mol. Microbiol. 37:192–206 [DOI] [PubMed] [Google Scholar]
- 69.Tamm A, et al. 1993. Hydrophobic domains affect the collagen-binding specificity and surface polymerization as well as the virulence potential of the YadA protein of Yersinia enterocolitica. Mol. Microbiol. 10:995–1011 [DOI] [PubMed] [Google Scholar]
- 70.Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tertti R, Skurnik M, Vartio T, Kuusela P. 1992. Adhesion protein YadA of Yersinia species mediates binding of bacteria to fibronectin. Infect. Immun. 60:3021–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ulett GC, et al. 2007. Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect. Immun. 75:3233–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ussery DW, Higgins CF, Bolshoy A. 1999. Environmental influences on DNA curvature. J. Biomol. Struct. Dyn. 16:811–823 [DOI] [PubMed] [Google Scholar]
- 74.Valle J, et al. 2008. UpaG, a new member of the trimeric autotransporter family of adhesins in uropathogenic Escherichia coli. J. Bacteriol. 190:4147–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vlahovicek K, Kajan L, Pongor S. 2003. DNA analysis servers: plot.it, bend.it, model.it and IS. Nucleic Acids Res. 31:3686–3687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wagner JK, Heindl JE, Gray AN, Jain S, Goldberg MB. 2009. Contribution of the periplasmic chaperone Skp to efficient presentation of the autotransporter IcsA on the surface of Shigella flexneri. J. Bacteriol. 191:815–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wells TJ, et al. 2008. EhaA is a novel autotransporter protein of enterohemorrhagic Escherichia coli O157:H7 that contributes to adhesion and biofilm formation. Environ. Microbiol. 10:589–604 [DOI] [PubMed] [Google Scholar]
- 78.Wells TJ, Totsika M, Schembri MA. 2010. Autotransporters of Escherichia coli: a sequence based characterisation. Microbiology 156:2459–2469 [DOI] [PubMed] [Google Scholar]
- 79.Wells TJ, Tree JJ, Ulett GC, Schembri MA. 2007. Autotransporter proteins: novel targets at the bacterial cell surface. FEMS Microbiol. Lett. 274:163–172 [DOI] [PubMed] [Google Scholar]
- 80.Wiles TJ, Kulesus RR, Mulvey MA. 2008. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp. Mol. Pathol. 85:11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yamada H, Muramatsu S, Mizuno T. 1990. An Escherichia coli protein that preferentially binds to sharply curved DNA. J. Biochem. 108:420–425 [DOI] [PubMed] [Google Scholar]
- 82.Zdobnov EM, Apweiler R. 2001. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848 [DOI] [PubMed] [Google Scholar]
- 83.Zuber F, Kotlarz D, Rimsky S, Buc H. 1994. Modulated expression of promoters containing upstream curved DNA sequences by the Escherichia coli nucleoid protein H-NS. Mol. Microbiol. 12:231–240 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.