Abstract
Conditional expression of a gene is a powerful tool to study its function and is typically achieved by placing the gene under the control of an inducible promoter. There is, however, a dearth of such inducible systems in Myxococcus xanthus, a well-studied prokaryotic model for multicellular development, cell differentiation, motility, and light response and a promising source of secondary metabolites. The few available systems have limitations, and exogenously based ones are unavailable. Here, we describe two new, versatile inducible systems for conditional expression of genes in M. xanthus. One employs isopropyl-β-d-thiogalactopyranoside (IPTG) as an inducer and is inspired by those successfully applied in some other bacteria. The other requires vanillate as an inducer and is based on the system developed originally for Caulobacter crescentus and recently adapted for mammalian cells. Both systems are robust, with essentially no expression in the absence of an inducer. Depending on the inducer and the amounts added, expression levels can be modulated such that either system can conditionally express genes, including ones that are essential and are required at high levels such as ftsZ. The two systems operate during vegetative growth as well as during M. xanthus development. Moreover, they can be used to simultaneously induce expression of distinct genes within the same cell. The conditional expression systems we describe substantially expand the genetic tool kit available for studying M. xanthus gene function and cellular biology.
INTRODUCTION
The ability to tightly regulate the expression of a gene of interest is both a valuable and a powerful genetic tool to assess gene function, especially that of genes essential for cell survival and growth. There is, however, a paucity of such controllable expression systems in the Gram-negative soil bacterium Myxococcus xanthus, a model prokaryotic system used to investigate molecular mechanisms involved in multicellular development (23), in coordinated cell movements (24, 32, 49), and in social behavior (44). M. xanthus is also used to study cellular responses to external signals such as light and their regulation at the level of signal transduction and gene expression (5–7) and is a potential source of bioactive secondary metabolites (45). Most gene function analyses in M. xanthus have relied upon tools such as transposon insertions of transcriptional reporters (25), nonpolar in-frame gene deletions (46), and constitutive overexpression of genes (29, 34, 47). Only very recently has an autonomous replicating plasmid been reported in M. xanthus (50), and its applicability remains to be explored.
Systems for conditional expression have been described in M. xanthus, but each has its own drawbacks. Two are light-based inducible systems. One of these is based on the light-inducible PcarQRS promoter, whose very low activity in the dark is enhanced ∼60-fold in the light (26). The other light-based inducible system employs the M. xanthus PB promoter, which is repressed by vitamin B12 in the dark but is activated by light (10). The action of B12 in the dark requires the CarH repressor, and a host strain harboring deletions not only of the endogenous carH gene but also of carA, which encodes a CarH paralog that also represses PB (33, 35), is necessary in order to employ this conditional gene expression system. Repression of PB by B12 in the dark is tight, and the B12/light-based inducible system provided a clear demonstration that rpoN, dksA, and cdnL are essential genes in M. xanthus (9, 10). However, efforts to generate a strain with regulatable expression of ftsZ using the B12/light-based system were futile, since ftsZ expression from PB was not sufficient to attain the high FtsZ levels that appear to be required in vivo (10). A limitation of both light-inducible systems is the requirement for light, which has been reported to impede multicellular fruiting body development (27). Light is also capable of provoking cellular damage and possibly other changes (6). An inducible system was very recently reported that employs a copper-responsive promoter in which expression is undetected in the absence of copper but increases to high levels linearly with the copper concentration (14). The system works well for copper at ≤0.5 mM during vegetative growth and at ≤0.06 mM for fruiting body formation, but higher copper levels affect cell growth and fruiting body development, while even small amounts of copper can interfere somewhat with social and adventurous motility (14). Moreover, besides toxicity, other issues concerning the use of copper to control expression of genes include collateral effects on cell physiology due to induction of other genes, such as those for copper homeostasis and carotenogenesis (13, 31).
In the ideal conditional gene expression system, the inducer itself would have negligible effects on normal cell growth and development. Isopropyl-β-d-thiogalactopyranoside (IPTG) has often served as such an innocuous inducer in various bacteria and has also been tried previously in M. xanthus. An IPTG-based system using the M. xanthus pilA promoter (PpilA) combined with a lac operator (lacO) was integrated at the pilA chromosomal locus, and the gene for the Escherichia coli LacI repressor was supplied at a distinct chromosomal phage Mx8 attachment site (21). However, the high level of basal expression from the promoter limited its use and made it unsuitable to test for essential genes. Also, the need for chromosomal integration of two plasmids in this system was a constraint, since it implied the necessity of using up two of the available M. xanthus antibiotic selection markers: kanamycin (Km) and tetracycline (Tc). In this report, the design of a new IPTG-based inducible system for conditional expression in M. xanthus is described and its applicability demonstrated. In parallel, we also report the design and use of an alternative inducible system in M. xanthus based on the vanillate-inducible system, first developed for Caulobacter crescentus (40) and, very recently, employed in mammalian cells and mice (11). We show that the IPTG- and vanillate-inducible systems can both generate M. xanthus strains that conditionally express several essential genes, including ftsZ, which is required, as mentioned earlier, at high levels in vivo, and can also be used to study genes involved in M. xanthus development. With both systems, gene expression is practically undetected in the absence of an inducer and is medium to high with an inducer present, depending on the amounts of inducer added and the system employed. Each system requires only one plasmid for chromosomal integration, with either Kmr or Tcr as a selectable marker. Moreover, the two can be used simultaneously and independently of each other in a given cell.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli strain DH5α (15) was used for plasmid constructions and was grown at 37°C in Luria broth medium supplemented with the appropriate antibiotics. M. xanthus was grown at 33°C in rich Casitone-Tris (CTT) medium or else in Tris-phosphate-magnesium (TPM) for multicellular development (3). Media were supplemented with inducer (0.5 mM vanillate or 1 mM IPTG unless specified otherwise) or antibiotic (40 μg/ml Km or 10 μg/ml Tc), as required. Motility and sporulation assays were performed as described elsewhere (17).
Construction of strains and plasmids.
Plasmids, M. xanthus strains, and the primers used in this study are listed in Table 1, Table 2, and Table S1 in the supplemental material, respectively. Several of the integration plasmids used in the IPTG- and vanillate-based gene expression systems are shown schematically in Fig. S1 in the supplemental material, and the complete sequences of their multicloning sites (MCS) are shown in Fig. S2 in the supplemental material. Standard protocols and commercially available kits were used in the preparation and manipulation of chromosomal and plasmid DNA. All constructs were verified by DNA sequencing. Plasmids were introduced into M. xanthus cells by electroporation, and integration of the plasmids by homologous recombination was selected on CTT plates containing the appropriate antibiotic and/or by negative selection via a galK gene that confers sensitivity to galactose (Gals).
Table 1.
Relevant plasmidsa
| Plasmid | Relevant genotype or description | Source |
|---|---|---|
| pMR3431 | M. xanthus 1.38-kb-PIPTG-MCS_A, Tcr | This study |
| pMR3435 | M. xanthus 1.38-kb-PIPTG::lacZ, Tcr | This study |
| pMR3487 | M. xanthus 1.38-kb-PIPTG-MCS_A-PR4::lacI, Tcr | This study |
| pMR3489 | M. xanthus 1.38-kb-PIPTG::lacZ-PR4::lacI, Tcr | This study |
| pMR3553 | M. xanthus 1.38-kb-PvanR::vanR-Pvan-MCS_C, Tcr | This study |
| pMR3556 | M. xanthus 1.38-kb-PIPTG::dksA-PR4::lacI, Tcr | This study |
| pMR3558 | M. xanthus 1.38-kb-PvanR::vanR-Pvan::cdnL, Tcr | This study |
| pMR3560 | M. xanthus 1.38-kb-PvanR::vanR-Pvan-MCS_B, Kmr | This study |
| pMR3561 | M. xanthus 1.38-kb-PvanR::vanR-Pvan-MCS_D-eyfp, Kmr | This study |
| pMR3562 | M. xanthus 1.38-kb-PvanR::vanR-Pvan-MCS_E-eyfp, Tcr | This study |
| pMR3564 | M. xanthus 1.38-kb-PvanR::vanR-Pvan::lacZ, Kmr | This study |
| pMR3596 | M. xanthus 1.38-kb-PIPTG::cdnL-PR4::lacI, Tcr | This study |
| pMR3629 | M. xanthus 1.38-kb-PR3-4::vanR-Pvan-MCS_C, Tcr | This study |
| pMR3632 | M. xanthus 1.38-kb-PvanR::vanR-Pvan::lacZ, Tcr | This study |
| pMR3635 | M. xanthus 1.38-kb-PvanR::vanR-Pvan::ftsZ, Tcr | This study |
| pMR3636 | M. xanthus 1.38-kb-PIPTG::ftsZ-PR4::lacI, Tcr | This study |
| pMR3638 | M. xanthus 1.38-kb-PR3-4::vanR-Pvan::lacZ, Tcr | This study |
| pMR3639 | M. xanthus 1.38-kb-PR3-4::vanR-Pvan::lacZ, Kmr | This study |
| pMR3653 | M. xanthus 1.38-kb-PR3-4::vanR-Pvan::ftsZ-eyfp, Kmr | This study |
| pMR3679 | M. xanthus 1.38-kb-PR3-4::vanR-Pvan-MCS_B, Kmr | This study |
| pMR3690 | M. xanthus MXAN_0018-MXAN_0019-PR3-4::vanR-Pvan-MCS_F, Kmr | This study |
| pMR3691 | M. xanthus MXAN_0018-MXAN_0019-PR3-4::vanR-Pvan-MCS_G, Tcr | This study |
| pMR3692 | M. xanthus MXAN_0018-MXAN_0019-PR3-4::vanR-Pvan::lacZ, Kmr | This study |
| pMR3697 | M. xanthus MXAN_0018-MXAN_0019-PR3-4::vanR-Pvan::ftsZ-eyfp, Kmr | This study |
Other plasmids, precursors to those listed here, are described in the text.
Table 2.
Relevant strainsa
| Strain | Integrated plasmid(s) | Relevant genotype or description | Reference or source |
|---|---|---|---|
| DK1050 | M. xanthus wild type | (37) | |
| DK1622 | M. xanthus wild type | (22) | |
| DK6204 | M. xanthus DK1622 ΔmglAB | (16) | |
| MR1981 | pMR3635 | M. xanthus DK1622 1.38-kb-PvanR::vanR-Pvan::ftsZ | This study |
| MR1982 | pMR3635 | M. xanthus DK1622 1.38-kb-PvanR::vanR-Pvan::ftsZ ΔftsZ | This study |
| MR2155 | pMR3435 | M. xanthus DK1622 1.38-kb-PIPTG::lacZ | This study |
| MR2161 | pMR3489 | M. xanthus DK1622 1.38-kb-PIPTG::lacZ-PR4::lacI | This study |
| MR2169 | pMR3556 | M. xanthus DK1622 1.38-kb-PIPTG::dksA-PR4::lacI dksA | This study |
| MR2170 | pMR3556 | M. xanthus DK1622 1.38-kb-PIPTG::dksA-PR4::lacI ΔdksA | This study |
| MR2173 | pMR3558 | M. xanthus DK1622 1.38-kb-PvanR::vanR-Pvan::cdnL | This study |
| MR2174 | pMR3558 | M. xanthus DK1622 1.38-kb-PvanR::vanR-Pvan::cdnL ΔcdnL | This study |
| MR2193 | pMR3596 | M. xanthus DK1622 1.38-kb-PIPTG::cdnL-PR4::lacI cdnL | This study |
| MR2194 | pMR3596 | M. xanthus DK1622 1.38-kb-PIPTG::cdnL-PR4::lacI ΔcdnL | This study |
| MR2195 | pMR3636 | M. xanthus DK1622 1.38-kb-PIPTG::ftsZ-PR4::lacI ftsZ | This study |
| MR2196 | pMR3636 | M. xanthus DK1622 1.38-kb-PIPTG::ftsZ-PR4::lacI ΔftsZ | This study |
| MR2199 | pMR3489 | M. xanthus DK1622 1.38-kb-PIPTG::lacZ-PR4::lacI | This study |
| pMR3697 | MXAN_0018-MXAN_0019-PR3-4::vanR-Pvan::ftsZ-eyfp | ||
| MR2453 | pMR3564 | M. xanthus DK1050 1.38-kb-PvanR::vanR-Pvan::lacZ | This study |
| MR2466 | pMR3632 | M. xanthus DK1050 1.38-kb-PvanR::vanR-Pvan::lacZ | This study |
| MR2468 | pMR3638 | M. xanthus DK1050 1.38-kb-PR3-4::vanR-Pvan::lacZ | This study |
| MR2469 | pMR3639 | M. xanthus DK1050 1.38-kb-PR3-4::vanR-Pvan::lacZ | This study |
| MR2477 | pMR3639 | M. xanthus DK1622 1.38-kb-PR3-4::vanR-Pvan::lacZ | This study |
| MR2479 | pMR3653 | M. xanthus DK1050 1.38-kb-PR3-4::vanR-Pvan::ftsZ-eyfp | This study |
| MR2491 | pMR3692 | M. xanthus DK1050 MXAN_0018-MXAN_0019-PR3-4::vanR-Pvan::lacZ | This study |
Other strains, precursors to those listed here, are described in the text.
Constructs for the IPTG-inducible system.
The sequence containing the IPTG-inducible PIPTG promoter (see Fig. S3 in the supplemental material) was synthesized and cloned into plasmid pUC57 by Genscript USA Inc. From this, the 673-bp fragment containing PIPTG was released by digestion with PstI and EcoRI, purified, and introduced into these sites in pMR2915 (9) to generate pMR3431. The lacZ sequence was PCR amplified with primers lacZ-XbaI and lacZ-BamHI, using plasmid pDAH274 as the template (38), and cloned into XbaI-BamHI-digested plasmid pMR2700 (34) to generate pMR3183. A 3,390-bp lacZ fragment obtained from pMR3183 as described above was inserted into XbaI-SmaI-digested pMR3431 to generate pMR3435, which was then introduced into DK1622 to isolate strain MR2155 by selection on Tc plates. To construct strain MR2161, first, the 96-bp fragment corresponding to PR4, the fourth of four tandem rRNA promoters at the rrnD locus (spanning positions −71 to +25 relative to the transcription start site), was PCR amplified using prRNA5-RI and prRNA3-XhoI as primers (see Table S1 in the supplemental material) and genomic DNA from DK1622 as the template, and the product was purified and digested with EcoRI and XhoI. A second fragment corresponding to the lacI gene was PCR amplified using plasmid pDR111 (4) as the template with lacI_XhoI_DR111 and lacI_NdeI_DR111 as primers, and the product was purified and digested with XhoI and NdeI. The two fragments were cloned into pMR3431 cut with EcoRI and NdeI to obtain pMR3487, into whose XbaI-SmaI sites the lacZ gene fragment isolated as described above was cloned to construct pMR3489. Introducing pMR3489 into DK1622 yielded strain MR2161.
To generate strains MR2169 and MR2170, we digested pMR3085 (10) with XbaI to release the dksA gene fragment and then inserted it into XbaI-digested pMR3487 to obtain pMR3556 (PIPTG::dksA-PR4::lacI). Plasmid pMR3556 was introduced into a dksA/ΔdksA merodiploid (generated by electroporating pMR3254 [10] into DK1622), and the resulting strain was grown for several generations with 1 mM IPTG and no Km and plated on CTT plates supplemented with 1% galactose and 1 mM IPTG to select for the loss of the Gals marker. This evicts vector DNA bearing either wild-type (wt) dksA or the ΔdksA allele by intramolecular recombination events. The resulting Galr Kms cells were diagnosed by PCR to isolate strains harboring the inducible PIPTG-dksA construct and either wild-type dksA (MR2169) or the ΔdksA allele (MR2170). Likewise, the cdnL gene fragment released by cutting pMR3015 (9) with XbaI was cloned into XbaI-digested pMR3487 to generate pMR3596 (PIPTG::cdnL-PR4::lacI). This was introduced into a cdnL/ΔcdnL merodiploid obtained by electroporating plasmid pMR2873 (9) into DK1622. Strains containing PIPTG-cdnL and wild-type cdnL (MR2193) or the ΔcdnL allele (MR2194) were obtained as described above for dksA. Finally, ftsZ from the XbaI digestion of pMR3168 (10) was cloned into pMR3487 to obtain pMR3636 (PIPTG::ftsZ-PR4::ftsZ), which was introduced into a ftsZ/ΔftsZ merodiploid obtained by electroporating pMR3165 (10) into DK1622. Strains with PIPTG-ftsZ and either wild-type ftsZ (MR2195) or the ΔftsZ allele (MR2196) were obtained from MR1980 (DK1622-derived ftsZ/ΔftsZ merodiploid strain with pMR3636 integrated at its chromosome) using the same procedures as described earlier.
Constructs for the vanillate-inducible system.
The PvanR::vanR-Pvan segment obtained from a HindIII-NdeI digestion of pBVMCS-6 (40) was cloned into these sites in pMCS-5 vector (40) to generate pMCSV-5. The 1.38-kb DNA fragment for plasmid integration into the M. xanthus chromosome by homologous recombination was released by HindIII digestion of pMR2700 (34) and introduced into HindIII-digested pMCSV-5 to generate pMR3553 (with Tcr). The 1.38-kb-PvanR::vanR-Pvan fragment from the NotI-NdeI pMR3553 digest cloned into these sites in pMCS-2 (40) yielded pMR3560 (with Kmr). An ∼1-kb fragment corresponding to the 5′ end of lacZ (PCR amplified from pMR3183 with primers 9_LacZ.for and lacZSEQ2 and digested with NdeI and ClaI) and the remaining ∼2.5-kb portion of lacZ (from a ClaI-EcoRI digestion of pMR3183) were cloned into pMR3560 cut with NdeI and EcoRI to generate pMR3564. Introducing pMR3564 into M. xanthus wild-type strain DK1050 generated MR2453. The 1.38-kb-PvanR::vanR-Pvan::lacZ fragment from the NotI-NdeI digestion of pMR3564 cloned into these sites in pMCS-5 yielded pMR3632, which was then integrated into DK1050 to generate strain MR2466. The 236-bp PR3–4 fragment was PCR amplified from DK1622 genomic DNA using as primers 20_rRNA-1.for and 21_rRNA-3.rev and cloned into the NcoI site of pMCSV-5 to generate pMR3598, in which PR3–4 (in addition to PvanR) drives transcription of vanR. Into HindIII-digested pMR3598 we cloned the 1.38-kb fragment obtained earlier to generate pMR3629. The lacZ fragment obtained by treating pMR3564 with NheI, followed by partial NdeI digestion, was cloned into the NdeI-NheI sites of pMR3629 to obtain pMR3638, which when introduced into DK1050 yielded strain MR2468. Cloning into pMCS-2 the ∼3-kb 1.38-kb-PR3–4::vanR-Pvan segment obtained from digesting pMR3629 with NotI and EcoRI generated pMR3679, while cloning the 1.38-kb-PR3–4::vanR-Pvan::lacZ fragment from the NotI-NdeI digestion of pMR3638 produced pMR3639. Introducing pMR3639 into DK1050 and DK1622 generated strains MR2469 and MR2477, respectively.
M. xanthus ftsZ, PCR amplified from pMR3168 (10) using 1_FtsZ.for and 2_FtsZ.rev as primers, was purified, cut with NdeI and KpnI, and cloned into pRVYFPC-2 (40) to generate pMR3507. The NdeI-NheI digestion of pMR3507 yielded the ftsZ-eyfp coding sequence for cloning into NdeI-NheI-cut pMR3639 (which also led to removal of the lacZ segment). Chromosomal integration of the resulting pMR3653 (1.38-kb-PR3–4::vanR-Pvan::ftsZ-eyfp) in DK1050 yielded strain MR2479. The ∼1.3-kb MXAN_0018-MXAN_0019 segment was PCR amplified from DK1622 genomic DNA using 33_Ori.for and 34_Ori.rev as primers, treated with NdeI and XbaI, and cloned into pET15b (Novagen) to generate pMR3647. An internal EcoRI site in pMR3647, 210 bp upstream of the first ATG in MXAN_0018-MXAN_0019, was eliminated by a single base pair change introduced using overlap extension PCR. The resulting PCR product was purified, digested with HindIII, and cloned into HindIII-digested pMR3638 (which causes removal of the 1.38-kb fragment) to obtain pMR3677. From this, the MXAN_0018-MXAN_0019-PR3–4::vanR-Pvan::lacZ insertion, isolated by NotI-NheI digestion, was cloned into these sites in pMCS-2 to generate pMR3692, which when introduced into DK1050 yielded strain MR2491. The 2.8-kb fragment corresponding to MXAN_0018-MXAN_0019-PR3–4::vanR-Pvan from NotI-NdeI-EcoRV treatment of pMR3677 cloned into NotI-NdeI-digested pMCS-2 yielded pMR3690. Cloning ftsZ-eyfp into the NdeI-NheI sites of pMR3690 (derived from pMR3653) generated pMR3697, which when introduced into strain MR2161 resulted in MR2199. The MXAN_0018-MXAN_0019-PR3–4::vanR-Pvan fragment inserted into pMCS-5 cut with NotI and NdeI yielded pMR3691. The 1.38-kb-PvanR::vanR-Pvan fragment (from the pMR3553 NotI-NdeI digest) cloned into pVYFPC-2 and pVYFPC-5 (40) yielded pMR3561 and pMR3562, respectively.
A cdnL fragment PCR amplified with primers cdnL-NdeI and cdnL-BamHI, using DK1050 genomic DNA as the template, was cloned into the NdeI-BamHI sites in pMR3553 to generate pMR3558 (1.38-kb-PvanR::vanR-Pvan::cdnL), which was introduced into the DK1622-derived cdnL/ΔcdnL merodiploid strain to generate MR1947. MR1947 was grown for several generations in the presence of 0.5 mM vanillate and no Km and then plated on 1% galactose–0.5 mM vanillate CTT plates to select for the loss of the vector Gals marker. The Galr Kms colonies obtained were diagnosed by PCR to isolate strains containing Pvan-cdnL and either wild-type cdnL (MR2173) or the ΔcdnL allele (MR2174). Using DK1622 genomic DNA and primers ftsZ-NdeI and ftsZ-EcoRI, ftsZ was PCR amplified, purified, cut with NdeI and EcoRI, and cloned into these sites in pMR3553 to obtain pMR3635 (1.38-kb-PvanR::vanR-Pvan::ftsZ). Introducing pMR3635 into the DK1622-derived ftsZ/ΔftsZ merodiploid strain produced MR1979, from which strains with the vanillate-inducible Pvan-ftsZ and either wild-type ftsZ (MR1981) or the ΔftsZ allele (MR1982) were generated as described above for cdnL.
β-Galactosidase activity and X-Gal agarose overlay assays.
β-Galactosidase activity was qualitatively assessed on CTT plates containing 40 μg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and measured (in nanomoles of o-nitrophenyl β-d-galactoside hydrolyzed per minute per milligram of protein) from liquid cultures or from cells undergoing development using a SpectraMax 340 microtiter plate reader (Molecular Devices) as previously described (9, 10).
For the X-Gal agarose overlay assay, low-melting agarose (Pronadisa) at a final concentration of 5 mg/ml was added to filter-sterilized KPN buffer (0.5 M potassium phosphate buffer [pH 7.0], made by mixing 305 mM K2HPO4, 195 mM KH2PO4, 6% of dimethyl formamide, and 0.1% sodium dodecyl sulfate) and heated to ∼60 to 70°C in a microwave. X-Gal at 0.5 mg/ml and β-mercaptoethanol at 0.05% were added to the warm solution, and 10 ml of the warm solution was evenly poured on the surface of the plate. Once the agar solidified, the plates were incubated at 33°C and scanned after 2 h (for the vanillate-based system) or 4 h (for the IPTG-based system), which is sufficient for blue color development if lacZ expression occurs.
Microscopy.
Samples (100 μl) of cultures taken at different times during growth (monitored using optical density at 550 nm [OD550]) were mixed with the fluorescent dye 4′-6-diamino-2-phenylindole (DAPI; 350-nm excitation maximum, 461-nm emission maximum) to achieve a final concentration of 1 ng/μl. A 1.5-μl drop of this mixture, or of myxospore suspensions (obtained by ultrasonic disruption of fruiting bodies), was immobilized on 1% agarose (Pronadisa) slices prepared in TPM medium and visualized with Nikon Eclipse 80i microscope equipped with a Nikon Plan Apo VC 100×/1.4 differential interference contrast (DIC) objective and a Hamamatsu ORCA-AG charge-coupled-device camera. Images were processed with Metamorph version 4.5 (Universal Imaging Group) and Photoshop CS3 10.0 (Adobe Systems).
RESULTS
Design of an IPTG-inducible system for conditional gene expression in M. xanthus.
Our design of a new IPTG-based inducible gene expression system for M. xanthus that would exhibit zero or very low basal expression in the absence of IPTG was inspired by the Bacillus subtilis hyperspank system (4). The latter has a hybrid promoter-lacO operator consisting of a modified SPO1 bacteriophage promoter (36) flanked by two lacO sequences, with the LacI repressor constitutively expressed from the E. coli lacI gene under the control of a penicillinase promoter (48). In our system, the hybrid promoter-lacO operator corresponds to the construct lacO-PR3-lacO (here termed PIPTG) (Fig. 1A; see also Fig. S3 in the supplemental material), where PR3 is the third of the four tandem rRNA promoters at the rrnD locus in M. xanthus that we have used in earlier studies (34). PIPTG was introduced into a plasmid bearing a Tcr selection marker and a 1.38-kb M. xanthus DNA fragment (the segment between positions 8498672 and 8500048 in the DK1622 genome), with no promoter activity, which allows plasmid integration into the chromosome by homologous recombination and which we have employed in previous work (10). The resulting plasmid was pMR3431 (see Fig. S1 in the supplemental material). Inserting the lacZ gene into the multicloning site (MCS) immediately downstream of PIPTG in pMR3431 generated plasmid pMR3435, in which the PIPTG-lacZ reporter fusion facilitates measurement of PIPTG activity in vivo (plasmid I1 in Fig. 1C). pMR3435 was introduced into M. xanthus wild-type DK1622 by electroporation to generate strain MR2155. A starter liquid culture of MR2155 grown to the stationary phase was diluted into fresh rich (CTT) medium either lacking or containing 1 mM IPTG to an optical density at 550 nm (OD550) of 0.1 and grown for 15 to 20 h to a final OD550 of 0.9 to 1.0. Levels of PIPTG-lacZ expression, from the ß-galactosidase activity, were identical whether or not IPTG was present (Fig. 1C), consistent with M. xanthus lacking an endogenous LacI repressor that could bind to the lacO sites and repress PIPTG.
Fig 1.
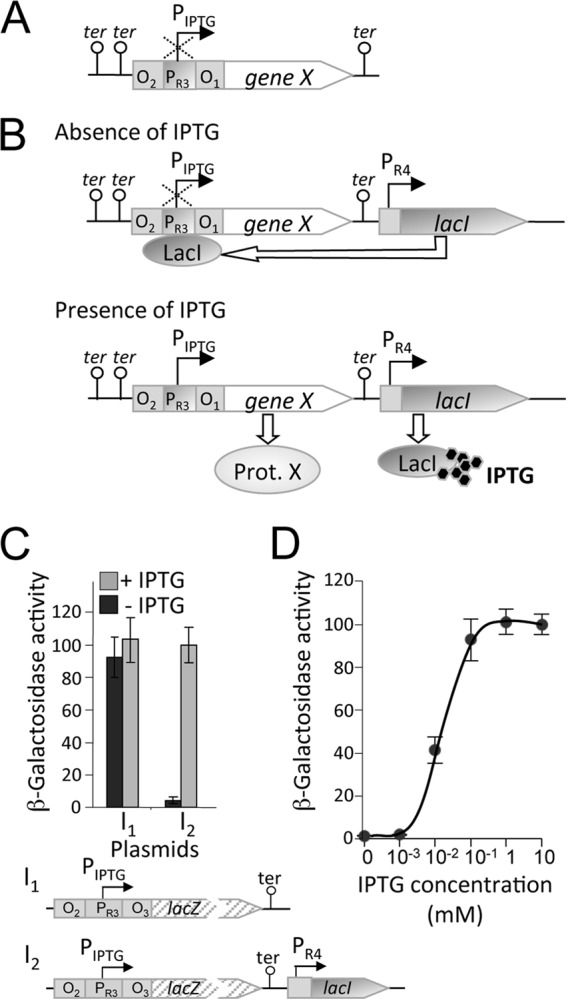
Design of an IPTG-inducible gene expression system for M. xanthus. (A) Schematic showing the design of the PIPTG hybrid promoter-operator of the IPTG-based system. O2 and O1 indicate the flanking lacO operators, PR3 is an rRNA promoter (see the text for details), “ter” represents a transcriptional terminator, and “gene X” is any desired gene placed under the control of PIPTG. (B) The IPTG-based system depicted in panel A was also supplied with the lacI gene under the control of the rRNA PR4 promoter for constitutive expression of the LacI repressor. Binding of LacI to the lacO operators would turn off expression from PIPTG in the absence of IPTG. IPTG, when present, would bind to LacI to prevent it from binding to the operator and allow expression of “gene X” from PIPTG. (C) Expression of lacZ (as “gene X”) from PIPTG, in terms of β-galactosidase-specific activity, with and without 1 mM IPTG, as indicated, in strains without lacI (I1) or with lacI (I2). (D) Reporter lacZ expression (measured in terms of specific β-galactosidase activity) from PIPTG in strain MR2161, which contains the constitutively expressed lacI gene (see the text), as a function of IPTG concentration. In panels C and D, the means of the results of three independent experiments and the standard deviations of the means (error bars) are shown.
Is PIPTG activity then affected if a constitutively expressing copy of the lacI gene is introduced in the strain as described above? To address this question, we generated plasmids pMR3487 and pMR3489, which not only contain PIPTG and PIPTG-lacZ, respectively, as with pMR3431 and pMR3435, but also harbor a copy of the E. coli lacI gene driven from PR4, the fourth of the four tandem rRNA promoters of M. xanthus rrnD, to constitutively produce the LacI repressor (plasmid I2 in Fig. 1C). We introduced pMR3489 into wild-type strain DK1622 by electroporation and examined lacZ activity in the resulting strain, MR2161, as before. In the absence of IPTG, essentially no lacZ activity was detected (Fig. 1C). In contrast, the presence of 1 mM IPTG induced lacZ activity at a level that was comparable to that in MR2155, which lacks LacI (Fig. 1C). Thus, we can conclude that when no inducer (IPTG) is available, the constitutively produced LacI binds to the lacO operators and effectively represses PIPTG, whereas IPTG, when supplied, binds to LacI, prevents operator binding, and allows transcription to proceed from PIPTG.
The dependence of PIPTG expression on the IPTG concentration was examined next. MR2161 grown in the absence of IPTG to an OD550 of 0.8 to 0.9 was diluted (to an OD550 of 0.1) into different culture flasks containing 10 ml of fresh medium with IPTG at increasing concentrations from 0 to 10 mM. After growth to an OD550 of 0.9 to 1.1, samples were collected from each culture to measure lacZ activity. PIPTG-lacZ expression increased with IPTG concentration to a maximum at 1 mM, without any further rise at higher IPTG levels (Fig. 1D). PIPTG expression is therefore enhanced in a dose-dependent manner by IPTG at concentrations of up to 1 mM. Our design of an IPTG-based system for conditional expression of genes in M. xanthus thus appears to fulfill the requirements of tight repression and dose-dependent expression in the absence and presence, respectively, of an inducer.
Conditional expression of essential genes in M. xanthus using the IPTG-inducible system.
We tested the applicability of the IPTG-based system to conditionally express genes that are essential for M. xanthus viability such as dksA, cdnL, and ftsZ (10). Thereby, we were also able to compare it with the B12/light-based system, which was suitable for dksA and cdnL but not for ftsZ, as noted earlier. For all three genes we used a common strategy, depicted in Fig. S4 in the supplemental material. The target gene was cloned into the MCS of pMR3487 to place it under the control of the PIPTG promoter, resulting in plasmids pMR3556, pMR3596, and pMR3636 (for dksA, cdnL, and ftsZ, respectively). Each of these Tcr-generated plasmids was electroporated into a distinct Kmr Gals merodiploid strain, which bears the native as well as a deleted copy of the gene of interest (see Fig. S4 in the supplemental material), and transformants with the plasmid integrated at the 1.38-kb locus were selected on Tc plates. The Kmr Gals merodiploid recipient strain was previously obtained by integrating a plasmid with ∼1-kb each of the upstream and the downstream flanking sequences of the gene of interest but with the gene replaced by a precisely complete in-frame deletion. Haploids were generated by growing the Kmr Gals Tcr strain for several generations in the absence of Km but with 1 mM IPTG present to ensure that the gene is expressed from PIPTG, followed by selection on plates containing galactose supplemented with 1 mM IPTG. Several individual colonies from the resulting Galr Tcr transformants (which would be Kms) were analyzed by PCR to differentiate those that retained at the native chromosomal locus the wild-type allele versus the deleted one (25% to 50% of the colonies retained the deleted allele; see Fig. S4 in the supplemental material). Thus, strains MR2169, MR2193, and MR2195 (denoted with “wt”) contain dksA, cdnL, and ftsZ, respectively, under the control of PIPTG as well as the same gene at the native locus, while strains MR2170, MR2194, and MR2196 (denoted with “Δ”) contain the PIPTG-controlled dksA, cdnL, and ftsZ, respectively, but have the deleted allele at the native locus. The six strains were then tested for growth on plates with or without 1 mM IPTG. All of them grew well on plates under permissive conditions when IPTG was present (Fig. 2A, left panel). However, under restrictive conditions when IPTG was absent, strains with the gene under PIPTG control as the only functional copy showed severely restricted growth, in marked contrast to those in which the wild-type allele was also present at the native locus (Fig. 2A, right panel). These results thus confirm previous results obtained using the B12/light-based conditional expression system showing that dksA and cdnL are essential in M. xanthus (10). Importantly, they show that ftsZ is also essential and, moreover, can be expressed from PIPTG at levels sufficient for viable cellular growth. In other words, our IPTG-based inducible system is suitable to construct strains that conditionally express essential genes, including those, such as ftsZ, whose products are required at high levels in vivo.
Fig 2.
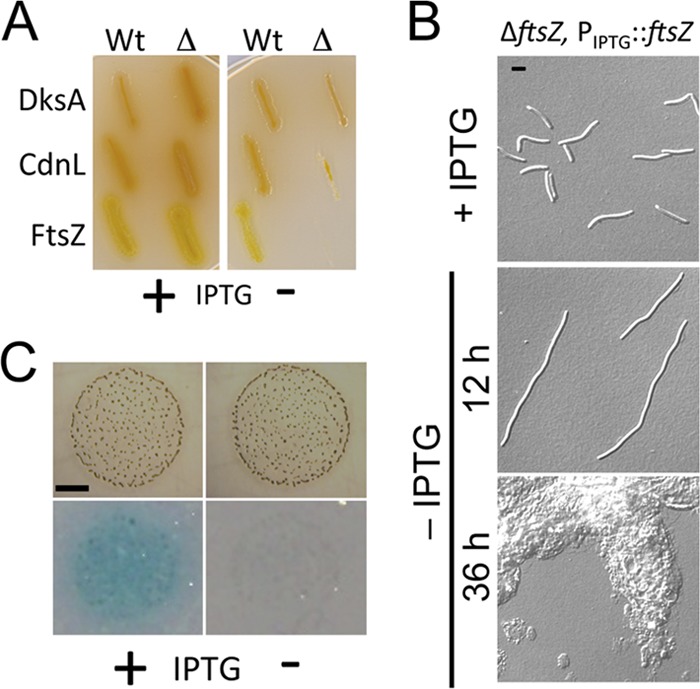
Conditional expression of genes in M. xanthus using the IPTG-inducible system. (A) Conditional expression of the essential M. xanthus genes dksA, cdnL, and ftsZ, using the IPTG-inducible system. Growth on plates containing 1 mM IPTG (left panel) or lacking IPTG (right panel) is shown for M. xanthus wild-type (Wt) strains MR2169, MR2193, and MR2195, which contain the native copy of dksA, cdnL, and ftsZ, respectively, and a second copy of the same gene expressed from the inducible PIPTG promoter; growth is also shown for deletion (Δ) strains MR2170, MR2194, and MR2196, which also harbor the gene under the control of the inducible PIPTG promoter but have the native copy replaced by the corresponding deleted allele (alleles ΔdksA, ΔcdnL, and ΔftsZ, respectively). Note the impaired growth of the “Δ” strains in the absence of IPTG. (B) DIC microscopy images of strain MR2196 (ΔftsZ, PIPTG::ftsZ) grown in the presence of 1 mM IPTG (top panel) and 12 and 36 h after IPTG removal (middle and bottom panels, respectively). Bar, 5 μm. (C) IPTG-induced expression of PIPTG::lacZ during M. xanthus fruiting body development triggered by starvation. The images correspond to fruiting body aggregates (formed after 24 h of incubation) of MR2161 bearing the inducible PIPTG::lacZ (as described for Fig. 1D) on TPM agar plates with 1 mM IPTG (“+”; left panels) or with no IPTG (“−”; right panels) prior to the X-Gal agarose overlay assay (top panels) and when subjected to the assay (bottom panels), as described in the text. Note the blue hue that developed on the IPTG-containing plate with the X-Gal overlay (after 4 h incubation), consistent with PIPTG::lacZ induction under these conditions. Bar, 2 mm.
In cells that conditionally express DksA or CdnL, depleting either of these proteins has been shown to impair cell division and produce filamentous cells (9, 10) and this is also observed if expression of these proteins is restricted using the IPTG-based system (not shown). Depleting FtsZ, an essential cell division protein, would also be expected to lead to a filamentous phenotype. To examine this, MR2196, in which FtsZ is expressed only when IPTG is available, was grown to an OD550 of 0.8 to 0.9, first in the presence of IPTG. Then, cells were pelleted, washed extensively with medium alone to remove IPTG, resuspended in fresh medium, and divided into two volumes. One was grown with no IPTG and the other with 1 mM IPTG present for several hours. Under a microscope, cells from cultures after a 12-h growth appeared normal if IPTG was present (Fig. 2B, top panel) but had the characteristic filamentous phenotype associated with lack of FtsZ and inability to divide (Fig. 2B, middle panel). The considerable amount of cell debris present after 36 h of FtsZ depletion (Fig. 2B, bottom panel) indicated that prolonged growth with no FtsZ caused cell death and lysis. Measurements at OD550 yielded consistent results, with impaired growth occurring in the absence of IPTG and hence FtsZ (see Fig. S5 in the supplemental material).
Conditional gene expression using the IPTG-inducible system during M. xanthus development.
M. xanthus cells starved at a high cell density on a solid surface undergo a multicellular developmental program that involves temporal and spatially coordinated changes in gene expression and cell differentiation to form spore-filled fruiting bodies. This developmental program depends on the ability of cells to move by gliding, for which they use two genetically distinct systems: adventurous motility (A) of single cells and social motility (S) of cells in groups (23, 32, 39). A valuable genetic tool would therefore be one that allows conditional expression of genes during fruiting body development (and motility). Hence, we tested this with our IPTG-inducible system. MR2161, which contains PIPTG-lacZ, was grown to an OD550 of 0.9, and the cells after several washes with TPM medium were concentrated 10-fold. From this, a 10-μl volume was spotted on TPM agar plates with or without 1 mM IPTG and incubated at 33°C. After 24 h of incubation, discrete cell aggregates were observed on plates with or without IPTG (Fig. 2C, top panels), indicating that the presence of IPTG does not interfere with development or, as shown in the assays represented in Fig. S6 in the supplemental material, with motility or sporulation. When the plates were subjected to the X-Gal agarose overlay assay (see Materials and Methods), a significant blue color developed after a 4-h incubation around cell aggregates only on plates containing IPTG (Fig. 2C, bottom panels). Thus, this IPTG-based inducible system can conditionally express genes during M. xanthus development.
A vanillate-based system for regulatable gene expression in M. xanthus.
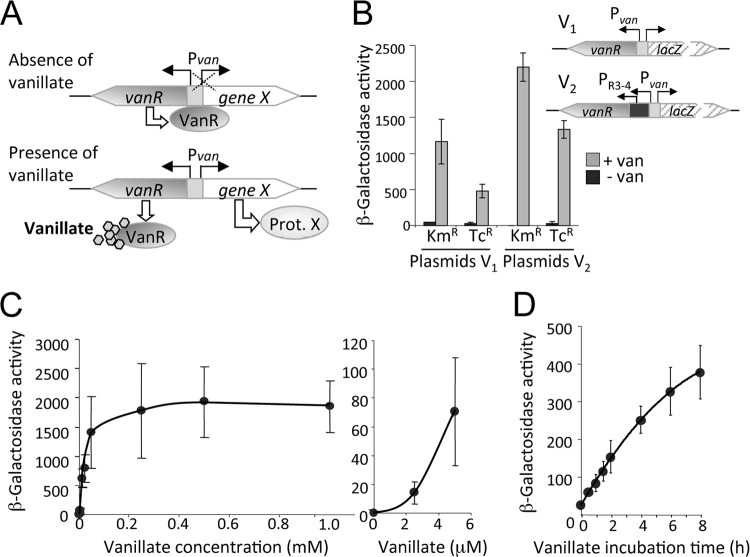
We explored in parallel the applicability in M. xanthus of a vanillate-inducible system that was developed for the bacterium C. crescentus (40) and, very recently, was employed (with suitable modifications) in studies of mammalian cells and mice (11). The system was developed based on the ability of C. crescentus to use vanillate, an intermediate from lignin cleavage, as a carbon source. The first step for vanillate utilization is removal of the O-methyl group by a two-subunit monooxygenase composed of a terminal oxygenase (VanA) and a ferredoxin-like reductase (VanB), both encoded by the vanAB operon regulated by VanR, a GntR-type transcriptional repressor, whose gene is transcribed divergently from the vanAB operon (40). The 126-bp vanR-vanAB intergenic region contains the vanAB promoter, Pvan, and two copies of a perfect inverted repeat (5′-ATTGGATCCAAT-3′), one preceding the −35 promoter region of Pvan and the other overlapping the −10 region and the transcription initiation (+1) site. Binding of VanR to these repeats blocks vanAB expression in the absence of vanillate (40, 43). In C. crescentus, a gene of interest cloned following the 126-bp vanR-vanAB intergenic region, where Pvan is located, is thus repressed in the absence of vanillate but expressed upon adding vanillate to the growth medium (Fig. 3A) (40).
Fig 3.
The vanillate-inducible gene expression system for M. xanthus. (A) Schematic showing how expression of a candidate gene (“gene X”), placed under the control of the vanillate promoter, Pvan, and located in the vanR-vanAB intergenic region, is turned off by the VanR repressor binding to its operator at Pvan in the absence of vanillate. VanR is transcribed constitutively and in the direction opposite to that of Pvan. Vanillate, when present, binds to VanR, preventing binding of its operator and allowing transcription of gene X from Pvan. (B) Expression of Pvan::lacZ with and without 0.5 mM vanillate after 15 to 20 h, shown in terms of the β-galactosidase-specific activity measured in three independent experiments. Plasmids V1 (pMR3564, Kmr; pMR3632, Tcr) contain only the vanR-vanAB intergenic region for vanR expression, while plasmids V2 (pMR3639, Kmr; pMR3638, Tcr) express vanR constitutively from PR3–4 (see the text). (C) Pvan::lacZ expression in the presence of vanillate at higher (left panel) or lower (right panel) concentrations for strain MR2469 (with PR3–4::vanR). (D) Time dependence of Pvan::lacZ expression in the presence of 0.5 mM vanillate for strain MR2453 (with PvanR::vanR). In panels B, C, and D, β-galactosidase-specific activities correspond to the means and standard deviations of the means (error bars) of the results of three independent measurements of samples collected from cultures with an OD550 of 0.9 to 1.1.
We tested if the vanillate-based system is suitable for M. xanthus by constructing two plasmids with a reporter lacZ probe immediately downstream of Pvan in the 126-bp vanR-vanAB intergenic region, with the vanR gene present upstream transcribed in the direction opposite that of lacZ. The plasmids also contained the 1.38-kb DNA fragment for plasmid integration into the M. xanthus chromosome by homologous recombination and had as a selection marker either Kmr (plasmid pMR3564) or Tcr (plasmid pMR3632) (plasmids V1 in Fig. 3B, right panel). We generated two other plasmids, pMR3639 (Kmr) and pMR3638 (Tcr), with an additional 220-bp regulatory region containing the PR3 and PR4 promoters (denoted as PR3–4) of M. xanthus rrnD mentioned earlier to constitutively express vanR in case its natural promoter failed to function in M. xanthus (plasmids V2 in Fig. 3B, right panel). Each of the four plasmids was separately introduced into wild-type M. xanthus DK1050 by electroporation, resulting in four different strains (MR2453, MR2466, MR2468, and MR2469). A starter liquid culture of each strain was grown to the stationary phase and then diluted to an OD550 of 0.1 into fresh CTT medium with or without 0.5 mM vanillate and grown for 15 to 20 h to a final OD550 of 0.9 to 1.0, following which lacZ expression levels were assessed. In all cases, the presence of vanillate increased β-galactosidase activity, which was consistently higher in the Kmr than in the Tcr strains (Fig. 3B). Moreover, Pvan-lacZ activity was greater in the strain with VanR expressed from PR3–4 than that seen with the natural vanR promoter for the same selection marker (Fig. 3B). In the absence of vanillate, Pvan-lacZ expression was very low and was not detected for the Kmr strain with VanR expressed from PR3–4. We have no explanation for the differences between the Kmr and Tcr strains or for the differences seen when VanR was expressed from PR3–4 versus the natural promoter. It is, however, clear from the data that the vanillate-based inducible system works in M. xanthus, where it can therefore be used to conditionally express genes of interest. Moreover, the finding that different levels in gene expression were seen that depended on the construct used expands the available plasmid choices for low-, medium-, or high-level expression.
We next tested how the vanillate-based system in M. xanthus depends on vanillate concentration and the incubation time with the inducer. For this, liquid cultures of MR2469 (the Kmr strain with Pvan-lacZ and PR3–4-vanR) at OD550 = 0.1, and with different concentrations of vanillate, were grown to an OD550 of 0.9 to 1.0 followed by β-galactosidase activity quantification. The β-galactosidase activity was found to increase with vanillate concentration, reaching a maximum at 0.5 mM, after which it remained steady in the presence of up to 1 mM vanillate (Fig. 3C, left). Even low levels of vanillate (2.5 to 5 μM) activated Pvan-lacZ to some extent (Fig. 3C, right). Based on these data, in all further experiments we employed vanillate at 0.5 mM, a concentration that is also typically used in C. crescentus (42). Finally, Pvan-lacZ activity induced by 0.5 mM vanillate increased with time over an 8-h period as monitored in MR2453, the Kmr strain with vanR expressed from its natural promoter (Fig. 3D).
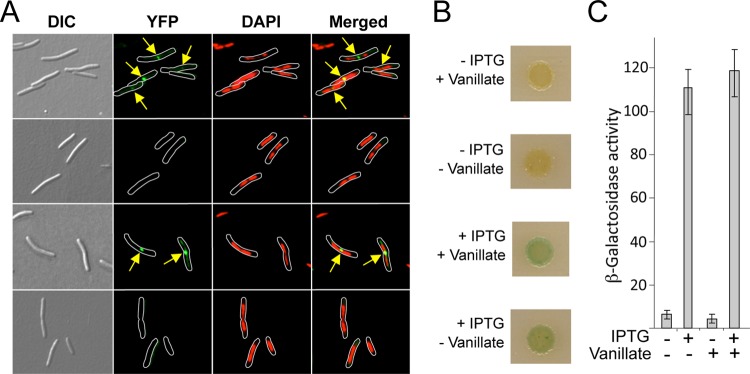
To examine vanillate-inducible gene expression using fluorescence microscopy, we constructed plasmid pMR3653, in which Pvan controls the expression of M. xanthus ftsZ fused to eyfp. FtsZ, the bacterial tubulin homolog, forms a ring in the midcell region, whose constriction culminates in cell division (1, 8, 30). Consequently, FtsZ fused to a fluorescent protein is frequently observed at the midcell or the cell division plane of growing cells (2, 28, 41, 42). Introducing pMR3653 into wild-type M. xanthus DK1050 yielded strain MR2479. Exponentially growing MR2479 was distributed into two aliquots, and 0.5 mM vanillate was added to one of them. After further growth for 1 h, samples from the two cultures were collected and examined using a fluorescence microscope. Fluorescent dots in the cell division plane, corresponding to FtsZ-yellow fluorescent protein (FtsZ-YFP), can be observed in most cells grown with 0.5 mM vanillate present, whereas no YFP fluorescence was detected in cells grown without vanillate (Fig. 4; see also Fig. S7 in the supplemental material). This shows that the vanillate-based regulatable system can express proteins fused to fluorescent tags in M. xanthus at levels sufficient for detection by fluorescence microscopy and confirms the midcell localization of FtsZ in M. xanthus, as seen in most other bacteria.
Fig 4.
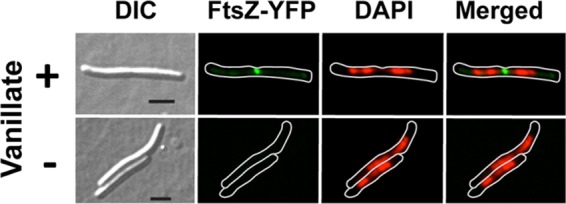
Images obtained using DIC, FtsZ-YFP fluorescence (green), and DAPI fluorescence (red) and a merged image of the last two (green and red) for strain MR2479 grown for 1 h in the presence of 0.5 mM vanillate (top panels) or without vanillate (bottom panels). For the fluorescence images, a white line was drawn bordering the cell boundary. Bars, 5 μm.
Conditional expression of essential M. xanthus genes using the vanillate-inducible system.
The same strategy as described earlier for conditionally expressing essential genes using the IPTG-inducible system (see Fig. S4 in the supplemental material) was used to test if the vanillate-based system is suitable for generating M. xanthus strains that conditionally express such genes. Plasmids were constructed to express cdnL or ftsZ from Pvan by cloning each of these into the MCS of the vanillate-inducible system in which VanR is expressed from its natural promoter (pMR3558 and pMR3635, respectively). Each of these Tcr plasmids thus generated was then electroporated into the previously obtained Kmr Gals merodiploid strain bearing the corresponding native and deleted copy of the gene of interest, and the transformants with the plasmid integrated into the chromosome at the 1.38-kb locus were selected on Tc plates (see Materials and Methods; see also Fig. S4 in the supplemental material). The Kmr Gals Tcr transformants were then grown for several generations with no Km but with 0.5 mM vanillate present to ensure expression of the gene from Pvan and were then selected on galactose plates containing 0.5 mM vanillate. Several of the individual Galr Tcr colonies were analyzed by PCR to distinguish those with the wild-type allele (“wt” strains MR2173 and MR1981, which have the native as well as a Pvan-driven copy of cdnL and ftsZ, respectively), from those with the deleted allele (“Δ” strains MR2174 and MR1982, respectively, which have only the Pvan-driven copy of cdnL and ftsZ). All four strains grew on plates with 0.5 mM vanillate (Fig. 5A, left panel). However, when vanillate was absent, growth was significantly impaired in the “Δ” strains, whose only copy of cdnL or ftsZ is the one conditionally expressed from Pvan, but not in the “wt” strains, in which the endogenous gene is also present (Fig. 5A, right panel). Thus, mutants of M. xanthus conditionally expressing cdnL or ftsZ that are viable under permissive growth conditions but not under restrictive conditions can be generated using the vanillate-inducible system. As shown with the IPTG-inducible system (Fig. 2B; see also Fig. S5 in the supplemental material), depletion of FtsZ by restricting vanillate availability in the medium produced aberrant filamentous cells (Fig. 5B) and impaired growth (see Fig. S5 in the supplemental material).
Fig 5.
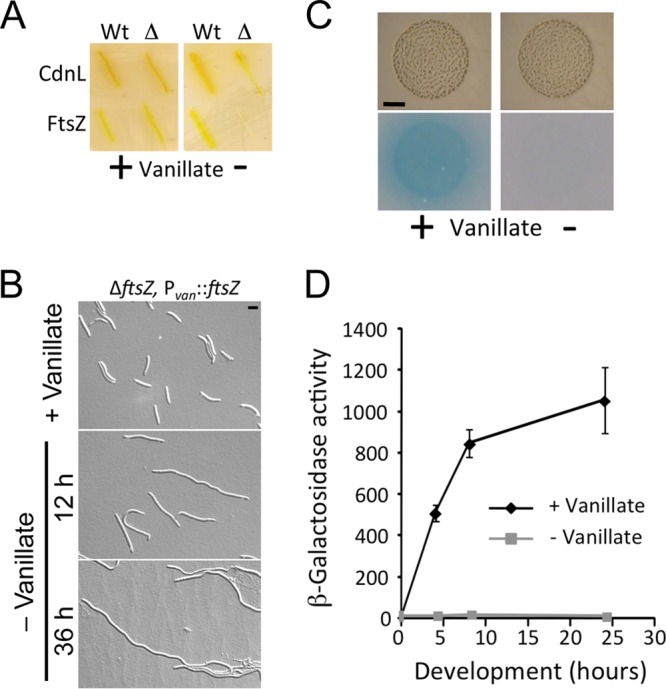
Vanillate-induced gene expression in M. xanthus. (A) Conditional expression of the essential M. xanthus genes cdnL and ftsZ, using the vanillate-inducible system. Growth on plates containing 0.5 mM vanillate (left panel) or lacking vanillate (right panel) is shown. Strains below “Wt” correspond to MR2173 (with cdnL at the native locus and an additional copy under the control of the vanillate-inducible Pvan promoter) and MR1981 (with one native and one Pvan-controlled ftsZ copy). Strains below “Δ” also have the Pvan-controlled gene of interest but also the deleted allele at the native locus (MR2174 and MR1982 for cdnL and ftsZ, respectively). Note the lack of growth of the “Δ” strains in the absence of vanillate. (B) DIC microscopy images for strain MR1982 (ΔftsZ, Pvan::ftsZ) grown in the presence of 0.5 mM vanillate (top panel) and 12 and 36 h after vanillate removal (middle and bottom panels, respectively). Bar, 5 μm. (C) Vanillate-induced expression of Pvan::lacZ during M. xanthus fruiting body development triggered by starvation. The images show fruiting body aggregates (formed after 24 h of incubation) of MR2477 harboring the inducible Pvan::lacZ (as described for Fig. 3B) on TPM agar plates containing 0.5 mM vanillate (“+”; left panels) or lacking vanillate (“−”; right panels) prior to (top panels) and after (bottom panels) the X-Gal agarose overlay assay, as described in the text. Note the blue hue that developed on the vanillate-containing plate with the X-Gal overlay (after 2 h of incubation), consistent with Pvan::lacZ being induced under these conditions. Bar, 2 mm. (D) Vanillate-induced reporter lacZ expression during development for strain MR2477. Cell spots undergoing development on TPM agar plates with or without 0.5 mM vanillate were collected at the times indicated and assayed for specific β-galactosidase activity (the means of the results of three independent experiments and the standard deviations of the means [error bars] are shown).
The vanillate-inducible system for conditional gene expression during M. xanthus development.
The presence of 0.5 mM vanillate in the media, as with IPTG, did not impair motility or sporulation in M. xanthus (see Fig. S6 in the supplemental material). Thus, to test if the inducible Pvan promoter also functions during fruiting body development, we grew strain MR2477 (with the reporter Pvan-lacZ integrated in its chromosome) to an OD550 of 0.9, harvested the cells, washed them with TPM medium, and, after concentrating them 10-fold, spotted 10 μl onto TPM agar plates with or without 0.5 mM vanillate. After 24 h of incubation at 33°C, both plates clearly showed numerous fruiting body aggregates (Fig. 5C, top panels). The X-Gal agarose overlay assay, after incubation for 2 h at 33°C, resulted in blue color development over the entire zone containing the aggregates, consistent with lacZ expression, only in the plate supplemented with 0.5 mM vanillate (Fig. 5C, bottom panels). As shown in Fig. 5D, whereas essentially no β-galactosidase-specific activity was detected in the absence of vanillate over the 24-h period assayed for development, this activity was strongly induced by vanillate, reaching significant levels even as early as 4 h into development. Thus, vanillate-induced conditional gene expression can also operate during M. xanthus development.
Simultaneous use of the IPTG- and vanillate-inducible systems to conditionally express distinct genes in M. xanthus.
We next examined if the IPTG- and vanillate-inducible systems, each of which is suitable for the conditional expression of a gene of interest in M. xanthus, can be used simultaneously to conditionally express distinct genes. If so, this would be of considerable value as a tool to study gene interactions. In both inducible systems described here, the plasmid vectors employed contained the 1.38-kb M. xanthus DNA fragment mentioned earlier to allow plasmid integration into the chromosome by homologous recombination, and the resulting transformants appeared to be normal in growth and cellular morphology (10). To simultaneously use the two inducible systems, a distinct selection marker and a second alternative site for chromosomal integration of the plasmid vector are necessary. In the available genome sequence (12), we chose a 1.31-kb DNA stretch corresponding to the intergenic region (positions 21290 to 22602 in the sequenced DK1622 genome) between two open reading frames annotated as MXAN_0018 (encoding a putative Ser-Thr kinase) and MXAN_0019 (encoding a hypothetical protein) transcribed in opposite directions and tested whether or not it is suitable as a possible region for a chromosomal integration site. This DNA stretch (designated MXAN_0018-MXAN_0019) (Table 1) was PCR amplified from genomic DNA and inserted into a plasmid vector that contains the vanillate-inducible Pvan-lacZ reporter with the Kmr selection marker to generate plasmid pMR3692. This plasmid was electroporated into the M. xanthus wild-type strain DK1050, and its integration in the chromosome in the intergenic region between MXAN_0018 and MXAN_0019 yielded Kmr strain MR2491. We found that plasmid integration in MR2491 has no apparent effect on cell growth and morphology compared to that of the wild type. MR2491 was grown with or without 0.5 mM vanillate to an OD550 of 0.9 to 1.0 (from an initial OD550 of 0.1), and samples were taken for measuring β-galactosidase-specific activity. Reporter Pvan-lacZ expression levels were high when induced by 0.5 mM vanillate but negligible in the absence of vanillate (see Fig. S8A in the supplemental material). Conditional expression from the Pvan promoter is thus maintained in MR2491. Hence, we next checked if this could be simultaneously used with the inducible PIPTG promoter in M. xanthus.
Plasmid pMR3489, described earlier, contains the PIPTG-lacZ reporter, the Tcr selection marker, and the 1.38-kb M. xanthus DNA for chromosomal integration of the plasmid, while pMR3697 harbors Pvan-ftsZ-eyfp, the Kmr selection marker, and the MXAN_0018-MXAN_0019 DNA for chromosomal integration. The two plasmids were sequentially electroporated into the DK1622 wild-type strain, and their integration at the two corresponding chromosomal loci generated Kmr Tcr strain MR2199. The normal motility, cell aggregation, and sporulation of MR2199 (see Fig. S8B to D in the supplemental material) indicated that DNA integration at MXAN_0018-MXAN_0019 (as with the 1.38-kb locus) had no effect on these processes. After growing to an OD550 of 0.9 to 1.0, MR2199 was diluted to an OD550 of 0.1 as two distinct cultures, one with and one without 1 mM IPTG, and grown overnight to an OD550 of 0.7. Each culture was then split into two volumes, one of which was supplemented with 0.5 mM vanillate. This resulted in four different culture conditions: no IPTG or vanillate, 1 mM IPTG but no vanillate, 0.5 mM vanillate but no IPTG, and 1 mM IPTG as well as 0.5 mM vanillate. After a 1-h incubation, samples were drawn from each of the four cultures for measuring β-galactosidase activity or for fluorescence microscopy. FtsZ-YFP fluorescence, localized in the midcell region, was observed in cells grown with 0.5 mM vanillate but not in those grown without vanillate, whether or not 1 mM IPTG was also present (Fig. 6A). On the other hand, when samples from the four cultures were concentrated 10-fold and spotted on plates with or without 1 mM IPTG and/or 0.5 mM vanillate, the cell spots developed a blue color after a 24-h incubation at 33°C only when IPTG was present, irrespective of the presence or absence of vanillate (Fig. 6B). Accordingly, significant reporter PIPTG-lacZ activity was observed in the presence of 1 mM IPTG but not in its absence, whether or not 0.5 mM vanillate was also available (Fig. 6C). Overall, these data indicate that the IPTG- and vanillate-based gene expression systems can be used simultaneously yet independently of each other to conditionally express distinct genes in the same M. xanthus cell.
Fig 6.
The IPTG- and vanillate-based systems can be simultaneously used to conditionally express distinct genes in an independent manner. (A) Images captured using DIC, FtsZ-YFP fluorescence (green), and DAPI fluorescence (red) and merged DAPI FtsZ-YFP images (green and red) for strain MR2199 grown with and without 1 mM IPTG and/or 0.5 mM vanillate as indicated on the right. A white outline of the cell boundary is drawn for easy comparison with the DIC image. Yellow arrows indicate FtsZ-YFP foci. (B) Images from drops of the previous cell cultures described above captured using CTT X-Gal plates with or without 1 mM IPTG and with or without 0.5 mM vanillate (indicated on the left) after a 24-h incubation. Note the presence of the blue colony coloring on plates with IPTG whether or not vanillate is present. (C) Expression of the PIPTG::lacZ reporter from the cultures described in the panel A legend showing its dependence on IPTG but not on vanillate as described for panel B. The β-galactosidase-specific activities correspond to the means and standard deviations of the means (error bars) of the results of three independent measurements of samples collected from cultures with an OD550 of 0.9 to 1.1.
DISCUSSION
M. xanthus is a widely used model system for the study of several important biological processes in bacteria, and tools for its genetic manipulation are available. Nonetheless, the design and development of new genetic tools are worthwhile, especially if these provide an advantage over existing systems or complement them. One particularly powerful tool to study gene function, and a means to obtain conditional mutants of genes essential for viability, is the ability to express or to repress such genes in a tightly regulatable manner. This can be achieved by placing the gene of interest under the control of an inducible promoter that, ideally, should have the following features. First, with such a system the gene must be expressed at levels that are high enough for function in vivo in the presence of the inducer but are negligible when the inducer is absent. Second, it is desirable that the system operates during different growth phases such as during vegetative growth as well as during the development of M. xanthus. Third, the inducer used should act specifically only on the promoter that it induces but be otherwise neutral with respect to the other normal cellular processes. Fourth, the system should be easy to use and manipulate. Finally, it would be particularly useful if such a system could be combined with a second, equally optimal system that employs a different inducer to simultaneously and independently induce expression of distinct target genes. Limitations in one or more of these features in the available regulatable gene expression systems for M. xanthus prompted us to design and develop two new systems that combine all of the desired features described above to conditionally express genes in this bacterium. The first, an IPTG-inducible system, has a design similar to the hyperspank system developed for B. subtilis (4) in having two lac operator sequences flanking a promoter that, for M. xanthus, corresponds to one of tandem rrnD promoters and a lacI gene that constitutively expresses the LacI repressor. The second system we describe is vanillate inducible and was adapted for M. xanthus from that developed for C. crescentus (40). This system is supplied with the vanillate-inducible Pvan promoter that includes the operator and a constitutively expressing gene for the C. crescentus VanR repressor, with transcription from Pvan blocked by VanR binding to the operator but induced by the presence of vanillate. Also, given the lack of autonomous replicating plasmids for M. xanthus, vectors used in both inducible systems include DNA segments that allow chromosomal integration.
IPTG, a nonhydrolyzable lactose mimic, is the most commonly used inducer for conditional gene expression in bacteria and can be used, as in other bacteria, without any adverse effects on the normal growth and development of M. xanthus (21, 26). Vanillate, a natural aromatic plant compound produced from the lignin cleavage, has no apparent deleterious effects on cell physiology in C. crescentus (18–20, 40, 42), and this also appears to be the case in M. xanthus. As a licensed food additive regularly consumed by humans via flavored convenience food, fresh vegetables, and fruits, vanillate is expected to be a safe trigger molecule for transgene expression, and indeed the vanillate-based system was successfully adapted for controlling transgene expression in mammalian cells and mice, as reported recently (11). Thus, both IPTG and vanillate are convenient as innocuous inducers for conditional gene expression in M. xanthus.
We have shown that both the IPTG- and vanillate-based systems respond to their respective inducers in a dose-dependent manner, and in both cases there is essentially no expression in the absence of the inducer. Based on reporter lacZ activity, expression from Pvan was generally greater than from PIPTG, the IPTG-inducible promoter. Pvan-lacZ expression also appeared to be higher in constructs with Kmr for selection than in those with Tcr. In constructs with Kmr, constitutively expressing the VanR repressor at high levels from the strong PR3–4 promoter achieved zero Pvan-lacZ expression with no inducer (vanillate) and, interestingly, a more enhanced expression in the presence of vanillate relative to constructs with VanR expressed from its natural promoter. Although the reasons for this are not known (it could be that PR3–4 recruits more RNA polymerase than the natural vanR promoter to the vicinity of Pvan, resulting in greater lacZ expression), the Pvan-inducible system in which VanR is expressed from PR3–4 clearly offers a larger operative range for the conditional expression of candidate genes. The IPTG- and vanillate-based systems are both suitable to generate M. xanthus mutant strains that conditionally express essential genes. In particular, with either system we could generate M. xanthus mutants that conditionally express ftsZ at the high intracellular levels required to grow in the presence but not in the absence of the inducer (IPTG or vanillate). The vanillate-based system in C. crescentus enabled not only the generation of conditional mutants of essential genes such as dnaA, divL, and divK but also the study of subcellular protein localization (18–20, 42). In M. xanthus, the vanillate-based system was also suitable for subcellular protein localization studies using fluorescence microcopy as shown for FtsZ-YFP, as well as for generating conditionally expressing mutants. Finally, both the IPTG- and vanillate-based systems induce reporter lacZ gene expression during starvation-triggered fruiting body formation, indicating that the two systems can be employed during vegetative growth or development for conditional gene expression.
In sum, we have designed IPTG- and vanillate-based systems with an extensive set of vectors that can be easily employed in M. xanthus. These vectors contain polylinker regions with unique restriction sites into which the gene of interest can be cloned for its inducible expression. The vectors have one or the other of two antibiotic resistance markers available for M. xanthus (Km and Tc) and can integrate into the chromosome at two alternative sites. This facilitates the use of both systems simultaneously for the controlled expression of distinct genes independently in M. xanthus without additional modification of the host strain. Vectors are available that conditionally express the target gene product fused to fluorescent proteins (e.g., a C-terminal YFP fusion) that are suitable for and allow cellular localization studies. The IPTG- and vanillate-based systems that we have described are thus ideally suited to use as genetic tools for studying M. xanthus gene function and biology.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants BFU2009-12445-C02-01 (to M.E.-A.), BFU2011-255422 (to A.A.I.), and RYC-2009-04190 (to A.A.I.) from the Ministerio de Ciencia e Innovación-Spain and cofinanced by the European Union (FEDER). J.A.-R. and A.G.-G. were supported by Ph.D. fellowships from the Ministerio de Ciencia e Innovación-Spain and F.G.-H. by a research contract funded by grant BFU2009-12445-C02-01 (to M.E.-A.).
We thank S. Padmanabhan for critical input on the manuscript, J. A. Madrid for technical support, and Lotte Søgaard-Andersen for providing strain DK6204.
Footnotes
Published ahead of print 24 August 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Adams DW, Errington J. 2009. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7: 642– 653 [DOI] [PubMed] [Google Scholar]
- 2. Ben-Yehuda S, Losick R. 2002. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109: 257– 266 [DOI] [PubMed] [Google Scholar]
- 3. Bretscher AP, Kaiser D. 1978. Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J. Bacteriol. 133: 763– 768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Britton RA, et al. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 184: 4881– 4890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Elías-Arnanz M, Fontes M, Padmanabhan S. 2008. Carotenogenesis in Myxococcus xanthus: a complex regulatory network, p 211–225 In Whitworth DE. (ed), Myxobacteria: multicellularity and differentiation. ASM Press, Washington, DC [Google Scholar]
- 6. Elías-Arnanz M, Padmanabhan S, Murillo FJ. 2011. Light-dependent gene regulation in nonphototrophic bacteria. Curr. Opin. Microbiol. 14: 128– 135 [DOI] [PubMed] [Google Scholar]
- 7. Elías-Arnanz M, Padmanabhan S, Murillo FJ. 2010. The regulatory action of the myxobacterial CarD/CarG complex: a bacterial enhanceosome? FEMS Microbiol. Rev. 34: 764– 778 [DOI] [PubMed] [Google Scholar]
- 8. Erickson HP, Anderson DE, Osawa M. 2010. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 74: 504– 528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. García-Moreno D, et al. 2010. CdnL, a member of the large CarD-like family of bacterial proteins, is vital for Myxococcus xanthus and differs functionally from the global transcriptional regulator CarD. Nucleic Acids Res. 38: 4586– 4598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. García-Moreno D, et al. 2009. A vitamin B12-based system for conditional expression reveals dksA to be an essential gene in Myxococcus xanthus. J. Bacteriol. 191: 3108– 3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gitzinger M, et al. 2012. The food additive vanillic acid controls transgene expression in mammalian cells and mice. Nucleic Acids Res. 40: e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goldman BS, et al. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. U. S. A. 103:15200– 15205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gómez-Santos N, Perez J, Sanchez-Sutil MC, Moraleda-Munoz A, Munoz-Dorado J. 2011. CorE from Myxococcus xanthus is a copper-dependent RNA polymerase sigma factor. PLoS genetics 7: e1002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gómez-Santos N, et al. 2012. Comprehensive set of integrative plasmid vectors for copper-inducible gene expression in Myxococcus xanthus. Appl. Environ. Microbiol. 78:2515– 2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166: 557– 580 [DOI] [PubMed] [Google Scholar]
- 16. Hartzell P, Kaiser D. 1991. Upstream gene of the mgl operon controls the level of MglA protein in Myxococcus xanthus. J. Bacteriol. 173: 7625– 7635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Higgs PI, Merlie JP., Jr 2008. Myxococcus xanthus: cultivation, motility, and development, p 465–478 In Whitworth DE. (ed), Myxobacteria: multicellularity and differentiation. ASM Press, Washington, DC [Google Scholar]
- 18. Iniesta AA, Hillson NJ, Shapiro L. 2010. Cell pole-specific activation of a critical bacterial cell cycle kinase. Proc. Natl. Acad. Sci. U. S. A. 107: 7012– 7017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Iniesta AA, Hillson NJ, Shapiro L. 2010. Polar remodeling and histidine kinase activation, which is essential for Caulobacter cell cycle progression, are dependent on DNA replication initiation. J. Bacteriol. 192: 3893– 3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Iniesta AA, Shapiro L. 2008. A bacterial control circuit integrates polar localization and proteolysis of key regulatory proteins with a phospho-signaling cascade. Proc. Natl. Acad. Sci. U. S. A. 105: 16602– 16607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jelsbak L, Kaiser D. 2005. Regulating pilin expression reveals a threshold for S motility in Myxococcus xanthus. J. Bacteriol. 187: 2105– 2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaiser D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 76: 5952– 5956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaiser D, Robinson M, Kroos L. 2010. Myxobacteria, polarity, and multicellular morphogenesis. Cold Spring Harb. Perspect. Biol. 2: a000380 doi:10.1101/cshperspect.a000380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Konovalova A, Petters T, Sogaard-Andersen L. 2010. Extracellular biology of Myxococcus xanthus. FEMS Microbiol. Rev. 34:89– 106 [DOI] [PubMed] [Google Scholar]
- 25. Kroos L, Kaiser D. 1984. Construction of Tn5 lac, a transposon that fuses lacZ expression to exogenous promoters, and its introduction into Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 81: 5816– 5820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Letouvet-Pawlak B, Monnier C, Barray S, Hodgson DA, Guespin-Michel JF. 1990. Comparison of beta-galactosidase production by two inducible promoters in Myxococcus xanthus. Res. Microbiol. 141: 425– 435 [DOI] [PubMed] [Google Scholar]
- 27. Li S, Lee BU, Shimkets LJ. 1992. csgA expression entrains Myxococcus xanthus development. Genes Dev. 6: 401– 410 [DOI] [PubMed] [Google Scholar]
- 28. Ma X, Ehrhardt DW, Margolin W. 1996. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 93: 12998– 13003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martinez-Canamero M, Munoz-Dorado J, Farez-Vidal E, Inouye M, Inouye S. 1993. Oar, a 115-kilodalton membrane protein required for development of Myxococcus xanthus. J. Bacteriol. 175: 4756– 4763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mingorance J, Rivas G, Velez M, Gomez-Puertas P, Vicente M. 2010. Strong FtsZ is with the force: mechanisms to constrict bacteria. Trends Microbiol. 18: 348– 356 [DOI] [PubMed] [Google Scholar]
- 31. Moraleda-Muñoz A, Pérez J, Fontes M, Murillo FJ, Muñoz-Dorado J. 2005. Copper induction of carotenoid synthesis in the bacterium Myxococcus xanthus. Mol. Microbiol. 56: 1159– 1168 [DOI] [PubMed] [Google Scholar]
- 32. Nan B, Zusman DR. 2011. Uncovering the mystery of gliding motility in the myxobacteria. Annu. Rev. Genet. 45: 21– 39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ortiz-Guerrero JM, Polanco MC, Murillo FJ, Padmanabhan S, Elias-Arnanz M. 2011. Light-dependent gene regulation by a coenzyme B12-based photoreceptor. Proc. Natl. Acad. Sci. U. S. A. 108: 7565– 7570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pérez-Marín MC, López-Rubio JJ, Murillo FJ, Elías-Arnanz M, Padmanabhan S. 2004. The N terminus of Myxococcus xanthus CarA repressor is an autonomously folding domain that mediates physical and functional interactions with both operator DNA and antirepressor protein. J. Biol. Chem. 279: 33093– 33103 [DOI] [PubMed] [Google Scholar]
- 35. Pérez-Marín MC, Padmanabhan S, Polanco MC, Murillo FJ, Elías-Arnanz M. 2008. Vitamin B12 partners the CarH repressor to downregulate a photoinducible promoter in Myxococcus xanthus. Mol. Microbiol. 67: 804– 819 [DOI] [PubMed] [Google Scholar]
- 36. Quisel JD, Burkholder WF, Grossman AD. 2001. In vivo effects of sporulation kinases on mutant Spo0A proteins in Bacillus subtilis. J. Bacteriol. 183: 6573– 6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ruiz-Vázquez R, Murillo FJ. 1984. Abnormal motility and fruiting behavior of Myxococcus xanthus bacteriophage-resistant strains induced by a clear-plaque mutant of bacteriophage Mx8. J. Bacteriol. 160: 818– 821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scanlan DJ, Bloye SA, Mann NH, Hodgson DA, Carr NG. 1990. Construction of lacZ promoter probe vectors for use in Synechococcus: application to the identification of CO2-regulated promoters. Gene 90: 43– 49 [DOI] [PubMed] [Google Scholar]
- 39. Søgaard-Andersen L, et al. 2003. Coupling gene expression and multicellular morphogenesis during fruiting body formation in Myxococcus xanthus. Mol. Microbiol. 48: 1– 8 [DOI] [PubMed] [Google Scholar]
- 40. Thanbichler M, Iniesta AA, Shapiro L. 2007. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 35: e137 doi:10.1093/nar/gkm818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thanbichler M, Shapiro L. 2006. Chromosome organization and segregation in bacteria. J. Struct. Biol. 156:292– 303 [DOI] [PubMed] [Google Scholar]
- 42. Thanbichler M, Shapiro L. 2006. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126: 147– 162 [DOI] [PubMed] [Google Scholar]
- 43. Tropel D, van der Meer JR. 2004. Bacterial transcriptional regulators for degradation pathways of aromatic compounds. Microbiol. Mol. Biol. Rev. 68: 474– 500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Velicer GJ, Vos M. 2009. Sociobiology of the myxobacteria. Annu. Rev. Microbiol. 63: 599– 623 [DOI] [PubMed] [Google Scholar]
- 45. Weissman KJ, Muller R. 2010. Myxobacterial secondary metabolites: bioactivities and modes-of-action. Nat. Prod. Rep. 27: 1276– 1295 [DOI] [PubMed] [Google Scholar]
- 46. Wu SS, Kaiser D. 1996. Markerless deletions of pil genes in Myxococcus xanthus generated by counterselection with the Bacillus subtilis sacB gene. J. Bacteriol. 178: 5817– 5821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu SS, Wu J, Kaiser D. 1997. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol. Microbiol. 23: 109– 121 [DOI] [PubMed] [Google Scholar]
- 48. Yansura DG, Henner DJ. 1984. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 81: 439– 443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Y, Ducret A, Shaevitz J, Mignot T. 2012. From individual cell motility to collective behaviors: insights from a prokaryote, Myxococcus xanthus. FEMS Microbiol. Rev. 36: 149– 164 [DOI] [PubMed] [Google Scholar]
- 50. Zhao JY, et al. 2008. Discovery of the autonomously replicating plasmid pMF1 from Myxococcus fulvus and development of a gene cloning system in Myxococcus xanthus. Appl. Environ. Microbiol. 74: 1980– 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.