Abstract
Poly(3-hydroxybutyrate) (PHB) granules are covered by a surface layer consisting of mainly phasins and other PHB granule-associated proteins (PGAPs). Phasins are small amphiphilic proteins that determine the number and size of accumulated PHB granules. Five phasin proteins (PhaP1 to PhaP5) are known for Ralstonia eutropha. In this study, we identified three additional potential phasin genes (H16_B1988, H16_B2296, and H16_B2326) by inspection of the R. eutropha genome for sequences with “phasin 2 motifs.” To determine whether the corresponding proteins represent true PGAPs, fusions with eYFP (enhanced yellow fluorescent protein) were constructed. Similar fusions of eYFP with PhaP1 to PhaP5 as well as fusions with PHB synthase (PhaC1), an inactive PhaC1 variant (PhaC1-C319A), and PhaC2 were also made. All fusions were investigated in wild-type and PHB-negative backgrounds. Colocalization with PHB granules was found for all PhaC variants and for PhaP1 to PhaP5. Additionally, eYFP fusions with H16_B1988 and H16_B2326 colocalized with PHB. Fusions of H16_B2296 with eYFP, however, did not colocalize with PHB granules but did colocalize with the nucleoid region. Notably, all fusions (except H16_B2296) were soluble in a ΔphaC1 strain. These data confirm that H16_B1988 and H16_B2326 but not H16_B2296 encode true PGAPs, for which we propose the designation PhaP6 (H16_B1988) and PhaP7 (H16_B2326). When localization of phasins was investigated at different stages of PHB accumulation, fusions of PhaP6 and PhaP7 were soluble in the first 3 h under PHB-permissive conditions, although PHB granules appeared after 10 min. At later time points, the fusions colocalized with PHB. Remarkably, PHB granules of strains expressing eYFP fusions with PhaP5, PhaP6, or PhaP7 localized predominantly near the cell poles or in the area of future septum formation. This phenomenon was not observed for the other PGAPs (PhaP1 to PhaP4, PhaC1, PhaC1-C319A, and PhaC2) and indicated that some phasins can have additional functions. A chromosomal deletion of phaP6 or phaP7 had no visible effect on formation of PHB granules.
INTRODUCTION
Survival of microorganisms during periods of starvation depends on their ability to accumulate reserve materials in times of surplus of nutrients. Many prokaryotes are able to accumulate reserve materials such as cyanophycin (C+N reserve), polyhydroxyalkanoates (PHAs) (C reserve), and polyphosphate (phosphate reserve) in the form of inert osmotic inclusions. Poly(3-hydroxybutyrate) (PHB) is the most abundant PHA type among bacteria, and PHB contents of 90% of the cellular dry weight have been reported. Biosynthesis and biodegradation of PHB and related PHAs have been studied in several labs during the last 25 years (6, 8, 11, 12, 16, 17, 24, 27, 33, 44, 45). It turned out that PHB granules in vivo consist of an amorphous polymer core and a complexly organized proteinaceous surface layer. Meanwhile, at least five types of PHB granule-associated proteins (PGAPs) have been described for Ralstonia eutropha, most of which are present in multiple isoforms. These are (i) PHB synthases (PhaCs), (ii) PHB depolymerases [PhaZ(a/d)s], (iii) PHB oligomer hydrolases (PhaZb/PhaZc or PhaYs), (iv) phasins (PhaPs), and (v) regulatory proteins (PhaRs). However, the biochemical details of the processes that take place during formation, homeostasis, and reutilization of PHB granules still are not well understood. One reason for this lack of knowledge might be that not all proteins that are involved in PHB metabolism have been identified. Several phasin proteins have been described for R. eutropha in previous studies (35, 51). In recent work, we identified a new phasin (PhaP5) and the first member of a new (sixth) type of PGAP in R. eutropha (PhaM) that is responsible for anchoring PHB granules with the nucleoid (30, 31). PhaM interacts with the PHB synthase PhaC1, PhaP5, and the DNA of the nucleoid and is involved in distribution of PHB granules to daughter cells during cell division. Interaction of PhaM with PhaC1 and with PHB was recently independently confirmed in J. Stubbe's lab (7) by copurification of PhaM with (streptavidin-tagged) PhaC1 and PHB. The other new PGAP, PhaP5, represented a new phasin and strongly interacted with most other phasins and with PhaM. In this study, we chose another approach to identify potentially new, not-yet-identified PHB players in R. eutropha: inspection of the amino acid sequences of phasins revealed the presence of a common sequence motif, a so-called phasin 2 motif, in all five phasins of R. eutropha. Consequently, we searched the R. eutropha genome for additional coding sequences corresponding to phasin 2 motifs and investigated the three positive hits for relevance in PHB metabolism. Two of them turned out to be true PGAPs (phasins).
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacteria and plasmids of this study are shown in Table 1. Escherichia coli strain JM109 was used in cloning experiments and was grown on Luria broth (LB) supplemented with the appropriate antibiotics. R. eutropha strains were routinely grown on nutrient broth (NB) medium at 30°C. A 0.2% (wt/vol) concentration of sodium gluconate was added as indicated in the next paragraph to promote PHB accumulation. Alternatively, PHB granule formation was followed in mineral salts medium with 0.5% (wt/vol) fructose.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli JM109 | Cloning strain | |
| E. coli S17-1 | Conjugation strain | 41 |
| R. eutropha H16 | Wild-type strain | DSMZ 428 |
| R. eutropha PHB-4 | PHB-negative mutant of R. eutropha H16 | 39 |
| R. eutropha H16 ΔphaC1 | Chromosomal deletion of phaC1 | This study |
| R. eutropha H16 ΔphaP1 | Chromosomal deletion of phaP1 | 35 |
| R. eutropha H16 ΔphaP6 | Chromosomal deletion of phaP6 | This study |
| R. eutropha H16 ΔphaP7 | Chromosomal deletion of phaP7 | This study |
| Plasmids | ||
| pBBR1MCS2-PphaC-eyfp-c1 | Universal vector for construction of fusions C terminal to eyfp under the control of the phaCAB promoter | 30 |
| pBBR1MCS2-PphaC-eyfp-n1 | Universal vector for construction of fusions N terminal to eyfp under the control of the phaCAB promoter | 31 |
| pBBR1MCS-2-PphaC-eyfp-phaP1 | C-terminal fusion of PhaP1 to eYFP | This study |
| pBBR1MCS-2-PphaC-eyfp-phaP2 | C-terminal fusion of PhaP2 to eYFP | 30 |
| pBBR1MCS-2-PphaC-eyfp-phaP3 | C-terminal fusion of PhaP3 to eYFP | This study |
| pBBR1MCS-2-PphaC-eyfp-phaP4 | C-terminal fusion of PhaP4 to eYFP | 30 |
| pBBR1MCS-2-PphaC-eyfp-phaP5 | C-terminal fusion of PhaP5 to eYFP | 30 |
| pBBR1MCS-2-PphaC-eyfp-phaP6 | C-terminal fusion of H16_B1988 to eYFP | This study |
| pBBR1MCS-2-PphaC-eyfp-phaP7 | C-terminal fusion of H16_B2326 to eYFP | This study |
| pBBR1MCS-2-PphaC-eyfp-B2296 | C-terminal fusion of H16_B2296 to eYFP | This study |
| pBBR1MCS-2-PphaC-phaP1-eyfp | N-terminal fusion of PhaP1 to eYFP | This study |
| pBBR1MCS-2-PphaC-phaP2-eyfp | N-terminal fusion of PhaP2 to eYFP | This study |
| pBBR1MCS-2-PphaC-phaP3-eyfp | N-terminal fusion of PhaP3 to eYFP | This study |
| pBBR1MCS-2-PphaC-phaP4-eyfp | N-terminal fusion of PhaP4 to eYFP | This study |
| pBBR1MCS-2-PphaC-phaP5-eyfp | N-terminal fusion of PhaP5 to eYFP | 31 |
| pBBR1MCS-2-PphaC-phaP6-eyfp | N-terminal fusion of H16_B1988 to eYFP | This study |
| pBBR1MCS-2-PphaC-phaP7-eyfp | N-terminal fusion of H16_B2326 to eYFP | This study |
| pBBR1MCS-2-PphaC-B2296-eyfp | N-terminal fusion of H16_B2296 to eYFP | This study |
| pBBR1MCS-2-PphaC-phaC1-eyfp | N-terminal fusion of PhaC1 to eYFP | This study |
| pBBR1MCS-2-PphaC-phaC1(C319A)-eyfp | N-terminal fusion of PhaC1(C319A) to eYFP | This study |
| pBBR1MCS-2-PphaC-phaC2-eyfp | N-terminal fusion of PhaC2 to eYFP | This study |
| pLO3 | Suicide vector; Tcr | 23 |
| pLO3 ΔphaC1 | Deletion vector for phaC1; fragments upstream and downstream of the gene were connected via overlap PCR and cloned between SacI and XbaI sites of pLO3 | This study |
| pLO3 ΔphaP6 | Deletion vector for phaP6 (H16_B1988) | This study |
| pLO3 ΔphaP7 | Deletion vector for phaP7 (H16_B2326) | This study |
Generation of PHB-free cells and induction of PHB granule formation.
All steps were performed at 30°C. A single colony of the strain of interest was used for inoculation of a 10-ml NB seed culture (10 ml in a 100-ml Erlenmeyer flask filled with 0.8% [wt/vol] nutrient broth) and was incubated for 24 h. One ml of this seed culture was transferred to 10 ml fresh NB medium and incubated for 24 h on a rotary shaker. For recombinant strains harboring pBBR1MCS-2 derivatives, 150 μg/ml kanamycin was present in the seed cultures. The cells intermediately accumulated PHB granules during this time. After 24 h, the bacteria were in the stationary growth phase, as indicated by shortening of the cells and loss of any Nile red-stainable PHB granules. Occasionally, a few very long cells (<1%) with PHB granules remained. Short cells were free of PHB granules. For monitoring formation of PHB granules, 0.1 volume of a stationary-phase, PHB-free R. eutropha culture was used for inoculation of 0.9 volume of fresh NB medium to which 0.2% sodium gluconate had been added to increase the concentration of a suitable carbon source and to stimulate PHB accumulation. This procedure resulted in the generation of a quasisynchronized culture in which all (living) cells immediately started to multiply and to accumulate PHB. Transmission electron microscopy (TEM) revealed cells with dividing nucleoids within just 5 to 10 min after transfer of the cells to fresh medium. Formation of PHB granules became visible by fluorescence microscopy (Nile red staining) within 5 min after transfer of the cells to fresh medium. Samples were taken at intervals and were immediately examined by microscopy.
Construction of fusion proteins with eYFP.
Fusions of the gene for enhanced yellow fluorescent protein (eyfp) with genes of R. eutropha H16 (Tables 1 and 2) were generally constructed both as N-terminal (pBBR1MCS-2-PphaC-eyfp-c1) and C-terminal (pBBR1MCS-2-PphaC-eyfp-n1) fusions using universal vectors based on the broad-host-range plasmid pBBR1MCS-2. These vectors harbored the constitutively (in R. eutropha) expressed promoter of the phaCAB operon (PphaC) and the coding region of eyfp (for details, see reference 31). The gene of interest was cloned in frame with the eyfp gene between the XhoI and BamHI restriction sites of pBBR1MCS-2-PphaC-eyfp-c1 and between the NdeI and BamHI restriction sites of pBBR1MCS-2-PphaC-eyfp-n1. The phaP3-eyfp fusion was obtained via XhoI/BamHI cloning because of an internal NdeI sequence and included the ribosome binding site of phaP3. All constructs were conjugatively transferred from recombinant E. coli S17-1 to R. eutropha H16, and selection was carried out on mineral salts medium supplemented with 0.5% (wt/vol) fructose and 350 μg ml−1 kanamycin.
Table 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′–3′)a |
|---|---|
| C-terminal fusions to eyfp | |
| phaP1_C_f_XhoI | CCGCTCGAGGCATGATCCTCACCCCGGAACAAGTTGC |
| phaP1_C_r_BamHI | CGGGATCCTCAGGCAGCCGTCGTCTTCTTTGC |
| phaP2_C_f _XhoI | CCGCTCGAGGCATGACTCAGTGGACAGCAG |
| phaP2_C_r_BamHI | CGGGATCCCTACTTTGCAGCTGCCGG |
| phaP3_C_f _XhoI | CCGCTCGAGGCATGTCCCCTTTTATGCCCG |
| phaP3_C_r_BamHI | CGGGATCCTTATTGCTTGGAAGCGCGG |
| phaP4_C_f_XhoI | CCGCTCGAGGCATGACTCAGTGGTCCCCC |
| phaP4_C_r_BamHI | CGGGATCCTTAATTTGCAGCTGCCTTTGAGG |
| phaP5_C_f_XhoI | CCGCTCGAGGCATGGCCACGCCTCCC |
| phaP5_C_r_BamHI | CGGGATCCCTAGCCCTTGGATTTCGGCTTG |
| B1988_C_f_XhoI | CCGCTCGAGGCATGCAACATAAATACCCCTATGCTTGGAGG |
| B1988_C_r_BamHI | CGGGATCCTCATGATGCACGCAGGCGCTTCG |
| B2326_C_f_XhoI | CCGCTCGAGGCATGATGACCGCAATGCAAACCTTCTGG |
| B2326_C_r_BamHI | CGGGATCCTCATGCCGGCCGCGGGGTTTCG |
| B2296_C_f_XhoI | CCGCTCGAGGCATGCGTATGAAACGCCAGCCAGCATCG |
| B2296_C_r_BamHI | CGGGATCCTCATGCGCGACGGGTCCGCTCG |
| N-terminal fusions to eyfp | |
| phaC1_N_f_NdeI | GGGAATTCCATATGGCGACCGGCAAAGG |
| phaC1_N_r_BamHI | CGGGATCCCCTGCCTTGGCTTTGACGTATCG |
| phaC2_N_f_NdeI | GGGAATTCCATATGAGGATCGACGACAGGACG |
| phaC2_N_r_BamHI | CGGGATCCCCGGGTTCCAGCACGTAGGTACC |
| phaP1_N_f_NdeI | GGGAATTCCATATGATCCTCACCCCGGAACAAGTTGC |
| phaP1_N_r_BamHI | CGGGATCCCCGGCAGCCGTCGTCTTCTTTGCC |
| phaP2_N_f_NdeI | GGGAATTCCATATGACTCAGTGGACAGCAGAGCAATGC |
| phaP2_N_r_BamHI | CGGGATCCCCCTTTGCAGCTGCCGGAGACGC |
| phaP3_N_f_XhoI | CCGCTCGAGCCACAGGAGATTTTGCATGTCCCCTTTTATGC |
| phaP3_N_r_BamHI | CGGGATCCCCTTGCTTGGAAGCGCGGGGC |
| phaP4_N_f_NdeI | GGGAATTCCATATGACTCAGTGGTCCCCCGAGC |
| phaP4_N_r_BamHI | CGGGATCCCCATTTGCAGCTGCCTTTGAGGCC |
| phaP5_N_f_NdeI | GGGAATTCCATATGGCCACGCCTCCCAATCC |
| phaP5_N_r_BamHI | CGGGATCCCCGCCCTTGGATTTCGGCTTGAGC |
| B1988_N_f_NdeI | GGGAATTCCATATGCAACATAAATACCCCTATGCTTGGAGG |
| B1988_N_r_BamHI | CGGGATCCCCTGATGCACGCAGGCGCTTCG |
| B2326_N_f_NdeI | GGGAATTCCATATGATGACCGCAATGCAAACCTTCTGG |
| B2326_N_r_BamHI | CGGGATCCCCTGCCGGCCGCGGGGTTTCG |
| B2296_N_f_NdeI | GGGAATTCCATATGCGTATGAAACGCCAGCCAGCATCG |
| B2296_N_r_BamHI | CGGGATCCCCTGCGCGACGGGTCCGCTCG |
| phaC1(C319A) eyfp fusion | |
| phaC1_C319A_fwd | CGTGCTCGGCTTCGCCGTGGGCGGCAC |
| phaC1_C319A_rev | GTGCCGCCCACGGCGAAGCCGAGCAC |
| Chromosomal deletions | |
| phaC1upstreamF | CGATCAGAGCTCTGCGCGCGCTCGGCTTCAGCCTTGC |
| phaC1upstreamR | GCCGTTAATTAAGCCGGATTTGATTGTCTCTCTGCCGTCACTATTCG |
| phaC1downstreamF | CGGCTTAATTAACGGCCGCTTGCATGAGTGCCGGCGTGCGTCATGC |
| phaC1downstreamR | GCTCTAGACCCTTCCTTATTTGCGCTCGACTGCCAGCGCCACG |
| phaP6upstreamF | TGAAGGCGGAGCTCTCGCTCGCCATGG |
| phaP6upstreamR | GCCGTTAATTAAGCCGTGCACCATTTCTTTTGGTTTTCTGCGCTGG |
| phaP6downstreamF | CGGCTTAATTAACGGCAGGCTGCCGGGCCCGGGCGCATGC |
| phaP6downstreamR | GCTCTAGACCACATCGGGCCTGCGCGTGC |
| phaP7upstreamF | CGATCAGAGCTCACGGCGGCACCCTCTTCCTCG |
| phaP7upstreamR | GCCGTTAATTAAGCCGCACGGGAATCTCCGGTGGTGGGC |
| phaP7downstreamF | CGGCTTAATTAACGGCCCCGGCCGCCCCCGCCTTGCGC |
| phaP7downstreamR | GCTCTAGAGGCAGTACGCCGGGAGGCATCAGC |
Underlining indicates restriction sites; bold type indicates start and stop codons.
Construction of chromosomal knockouts.
A precise chromosomal deletion of the phaC1 gene was constructed in R. eutropha H16 using the sacB-sucrose selection method (15% sucrose used for selection) and pLO3 as a deletion vector as described elsewhere (23, 31). The same method was used to generate chromosomal deletions of phaP6 or phaP7 of R. eutropha H16. The genotype of the resulting deletion mutant was verified by PCR of the respective genomic region and determination of its DNA sequence. Only clones with the correct DNA sequence were used.
Site-directed mutagenesis of phaC1 (C319A).
PCR was performed with PrimeStar polymerase. Insertion of mutations into the coding sequence of phaC1 was done using overlap extension PCR with primers given in Table 2. E. coli cells were chemically transformed with plasmid DNA by standard procedures. The success of introduced mutations was confirmed by DNA sequencing.
Other methods.
Quantitative analysis of PHA content was done by gas chromatography analysis after acid methanolysis of lyophilized cells, as described in reference 3. DNA manipulation and construction of plasmids were done by standard molecular biological methods and according to supplier's instructions. Protein levels were routinely determined by the Bradford method (2). Polyacrylamide gel electrophoresis was performed under denaturing (sodium dodecyl sulfate) and reducing (mercaptoethanol) conditions. Gels were stained with Coomassie brilliant blue G250 or with silver. Western blot analysis was performed using commercial polyclonal antibodies raised against green fluorescent protein. Fluorescence microscopy of Nile red-stained cells was done as described previously (14), except that ethanol was replaced by dimethyl sulfoxide (DMSO) as the cosolvent in some experiments (1 μg Nile red/ml DMSO).
RESULTS
Localization of the PHB synthase PhaC1 in the presence and absence of PHB.
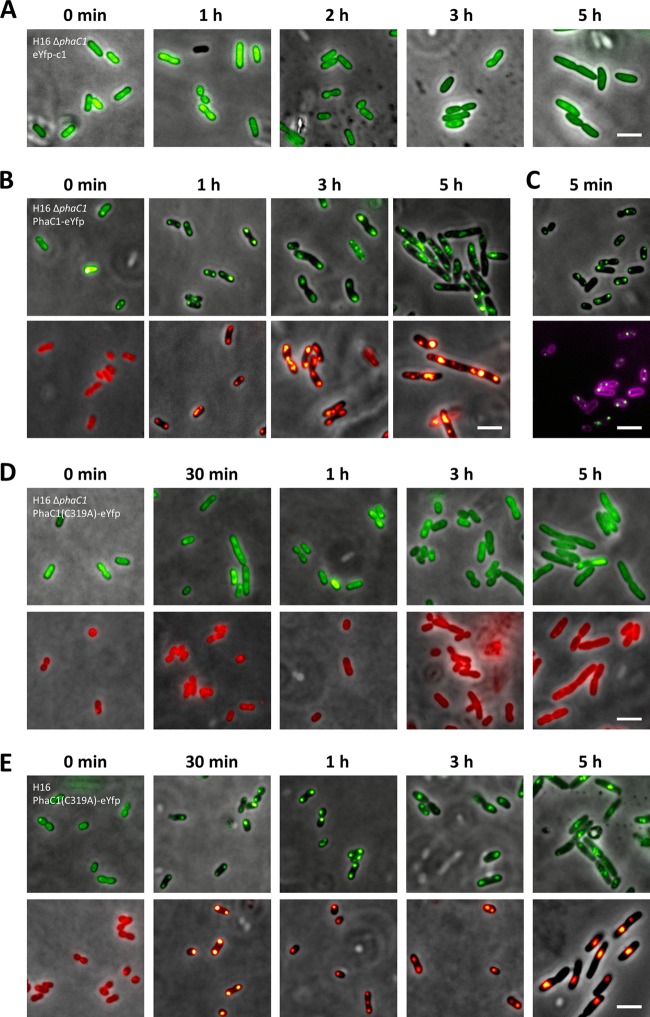
The PHB synthase PhaC1 is the key enzyme of PHB synthesis in R. eutropha (28, 36, 37, 40, 42, 44, 47, 54). It is generally accepted that PhaC1 is attached to the surface of PHB granules in vitro and in vivo: (i) PHB synthase activity copurifies with isolated PHB granules in R. eutropha and in all other PHA-accumulating organisms that have been looked at, (ii) transmission electron microscopy (TEM) data of R. eutropha H16 using PhaC1-specific antibodies and immunogold-labeled anti-antibodies clearly showed a surface localization of PHB synthase (10), and (iii) very recently, Cho and coworkers copurified strep-tagged-PhaC1 together with an ∼350-kDa PHB molecule (7). Correct localization of fusions with the green fluorescent protein and its color variants is sometimes complicated by the formation of fluorescent inclusion bodies (49) that could be mixed up with PHA granules. Therefore, construction of fusions of enhanced yellow fluorescent protein (eYFP) with PHB synthase and confirmation of localization in the presence and absence of PHB granules would be a good control to test if fusions with eYFP result in correct subcellular localization in R. eutropha. A C-terminal fusion of PhaC1 with eYFP was constructed and expressed in R. eutropha PHB-4 (a chemically induced PHB-free mutant because of a stop codon in the reading frame of phaC1) (25) and in a precise ΔphaC1 deletion mutant of R. eutropha H16. The phaC1-eyfp fusion—like all fusions in this study—was cloned in a modified pBBR1MCS-2 vector under the control of the constitutive (in R. eutropha) phaC1 promoter (see Materials and Methods). When R. eutropha H16 ΔphaC1 was cultivated under conditions permissive for PHB accumulation, PHB granules were not observed, showing that PhaC1 is essential for PHB granule formation. Cells of R. eutropha strains (wild-type or PHB-negative background; images only for the ΔphaC1 strain are shown) that harbored pBBR1MCS2-PphaC-eyfp-c1 for constitutive expression of eYFP always showed more or less homogeneously distributed yellow-green fluorescence and confirmed that eYFP is a soluble protein in R. eutropha independent of the presence or absence of PHB (Fig. 1A). Occasionally, fluorescence in the central region of the cells, where the nucleoid should be located, appeared slightly less intense, suggesting that eYFP concentration in the nucleoid region can be lower than that in other parts of the cell. Next, we investigated PHB granule formation and eYFP fluorescence of R. eutropha ΔphaC1 (or strain PHB-4; images not shown) that harbored a plasmid conferring constitutive PhaC1-eYFP expression. Formation of PhaC1-eYFP protein was confirmed by Western blot analysis (data not shown). PhaC1-eYFP was homogeneously distributed in the absence of PHB. When PHB-free cells of this strain were transferred to conditions permissive for PHB accumulation, the cells accumulated PHB granules again. PhaC1-eYFP fluorescence condensed to foci that colocalized with PHB granules (Nile red stained). These results confirmed that the fusion of the PHB synthase PhaC1 to eYFP is catalytically active (Fig. 1B). The number and size of the granules were undistinguishable from those in the wild type. Interestingly, PHB granule formation and binding of PhaC1-eYFP fluorescence to PHB granules turned out to be a very rapid process: the first visible granules with attached PhaC1-eYFP appeared within only 5 min after transfer of PHB-free cells to conditions permissive for PHB accumulation. Most cells had one or two PHB granules after 5 min (Fig. 1C).
Fig 1.
Localization of eYFP and PhaC1-eYFP in R. eutropha H16 and in R. eutropha H16 ΔphaC1. (A) eYFP-c1 was expressed in R. eutropha H16 ΔphaC1 in the absence and presence of conditions permissive for PHB accumulation. Homogeneous localization of eYFP in the cytoplasm was also found in the wild-type H16 and PHB-4 strains of R. eutropha (data not shown). (B) PHB-free (stationary-phase) cells of R. eutropha H16 ΔphaC1 expressing PhaC1-eYFP were transferred to conditions permissive for PHB accumulation (NB plus 0.2% [wt/vol] gluconate). Samples were taken at the indicated time points and examined by microscopy for eYFP fluorescence (top) and for PHB granules after staining with Nile red (bottom). (C) Cells were also stained with membrane-specific FM4-64 dye. (D and E) Time course of PHB granule formation and localization of PhaC1(C319A)-eYFP in R. eutropha ΔphaC1 (D) and R. eutropha H16 (E). Bars, 3 μm.
In order to distinguish between insoluble but still fluorescent inclusion bodies of eYFP fusions and true PHB-granule bound fusions, we constructed a catalytically inactive PhaC1-eYFP variant by exchanging the active site Cys319 of PhaC with alanine. We investigated the subcellular localization of expressed PhaC1-C319A–eYFP in R. eutropha H16 and in R. eutropha ΔphaC1. The fusion protein clearly colocalized with PHB granules in the wild type, confirming that the mutation had no influence on binding to PHB granules. When PhaC1-C319A–eYFP was expressed in the ΔphaC1 mutant, fluorescence was homogeneously distributed under all culture condition, including those under which the wild type would have accumulated PHB (Fig. 1D and E). These controls confirm that colocalization of PhaC1-eYFP foci with PHB granules is not an artifact caused by inclusion body formation.
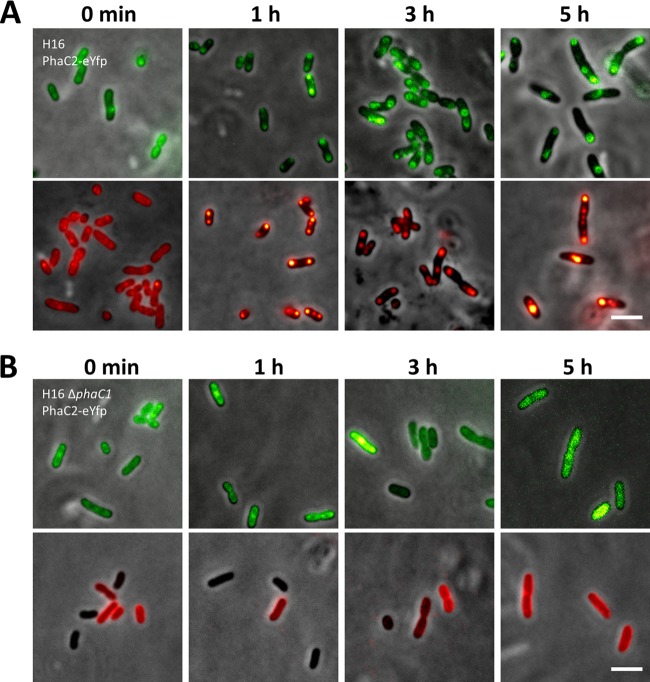
Analysis of the R. eutropha genome had revealed the presence of a second PHB synthase gene, phaC2 (32), that is assumed to be catalytically inactive. To determine whether the inactivity of PhaC2 is caused by insufficient binding to PHB, we constructed a PhaC2-eYFP fusion and investigated its subcellular localization and PHB granule formation in the wild type and in the ΔphaC1 mutant. The PhaC2-eYFP fusion was able to specifically bind to PHB granules in the wild type (Fig. 2A). Expression in a PHB-negative background confirmed the absence of PHB synthase activity, as the PhaC2-eYFP fluorescence was homogeneously distributed in the cytoplasm independent of the physiological situation of the cells, and PHB granule formation was not observed (Fig. 2B). A previous analysis suggested that PhaC2 could compensate for the mutation in PhaC1 in the chemically induced PHB-negative mutant R. eutropha PHB-4 (29). However, when we transferred phaC2-eyfp (with the constitutive phaC1 promoter) to R. eutropha PHB-4, we did not find evidence for PHB granule formation under culture conditions permissive for PHB accumulation (data not shown).
Fig 2.
Localization of PhaC2-eYFP in R. eutropha H16 (A) and R. eutropha H16 ΔphaC1 (B). Experiments were performed as described in the legend to Fig. 1. Bars, 3 μm.
Localization of phasins PhaP1 to PhaP5 in the absence and presence of PHB.
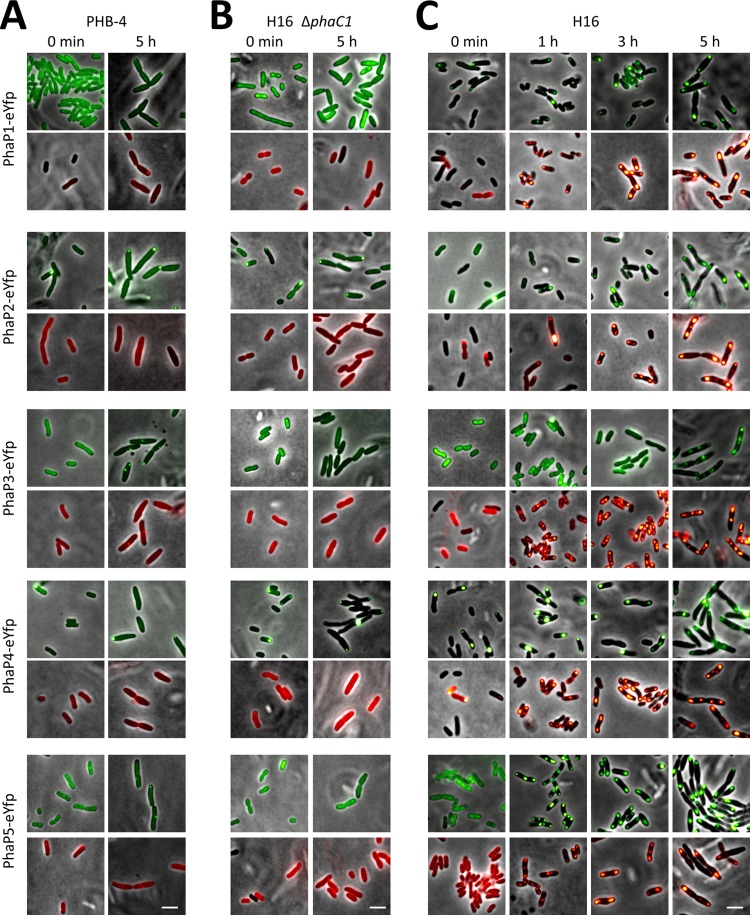
The results shown above confirmed that proteins fused to eYFP can be localized in R. eutropha. Next, we were interested in the subcellular localization of previously described or postulated phasins in R. eutropha. Phasins are small PHA granule-associated proteins (PGAPs) and have been found in all PHA-accumulating bacteria that have been investigated for the presence of phasins. Phasins are also present in PHB-accumulating Archaea (5). In R. eutropha, PhaP1 is the most abundant phasin and can contribute to approximately 5% of the protein fraction during PHB accumulation (51). Colocalization of PhaP1 with PHB has been previously shown (7, 26, 51). Three other phasins (PhaP2, PhaP3, and PhaP4) have been postulated from the sequenced R. eutropha genome (32) on the basis of sequence similarity to PhaP1 (22, 35), and recently, we identified a fifth phasin (PhaP5) via its interaction with PhaP2 in a two-hybrid approach (30). For most phasins, subcellular localization in vivo under defined culture conditions in the absence and presence of PHB has not yet been investigated systematically. We therefore constructed C-terminal and N-terminal fusions of all five phasin genes (phaP1 to phaP5) with eyfp and cloned them under the control of the constitutively expressed phaC1 promoter. All 10 constructs were transferred to R. eutropha PHB-4, to the R. eutropha ΔphaC1 deletion mutant, and to the wild-type strain R. eutropha H16. Western blot analysis revealed that fusion proteins of the expected size were produced in R. eutropha strains (data not shown). In general, no significant differences between N- and C-terminal fusions were found, except that fusions of eYFP to the C termini of phasins generally resulted in stronger fluorescence. Therefore, only images of C-terminal fusions are shown here. First, we determined subcellular localization in R. eutropha PHB-4 and in the defined ΔphaC1 strain under conditions in which the wild type would accumulate PHB. For this, stationary-phase NB-grown cells of R. eutropha PHB-4 and of R. eutropha ΔphaC1—NB-grown wild-type cells have no PHB in late stationary phase—were transferred to fresh medium supplemented with 0.2% gluconate and were incubated at 30°C. Samples were taken within the first 5 h of growth and investigated for eYFP fluorescence. PhaC1-expressing cells start to form PHB granules within 5 min under these conditions (Fig. 1C). All R. eutropha PHB-4 and ΔphaC1 cells expressed the respective fusion proteins, and the eYFP-phasin fluorescence was generally homogeneously distributed in the cells at time zero when no PHB was present in any strain (Fig. 3A and B). Expression of PhaP2-eYFP and PhaP4-eYFP fusions was notably strong, and cells occasionally contained fluorescent foci near the cell poles, probably because of the high self-oligomerization potential of PhaP2 and PhaP4, as shown in our previous work (30). Formation of fluorescent foci was also increased, but many cells were still free of foci. PHB granule formation was not detected in any of these cells. Rarely, cells had faint foci in the Nile red channel, probably caused by cross talk of fluorescence between the GFP and Nile red channels. Formation of fluorescent foci was less pronounced in cells expressing the respective N-terminal fusions (i.e., eYFP-PhaP; images not shown), probably because the expression of N-terminal fusions was lower than the expression of C-terminal fusions; PHB was not detectable in these strains. Most fusions formed fluorescent foci in the PHB-4 strain near the cell poles or in the area of future septum formation. Remarkably, these cell pole-localized foci were observed after only a few hours of growth under conditions permissive for PHB accumulation. However, focus formation was only rarely observed in the ΔphaC1 strain. Apparently, formation of fluorescent foci in strain PHB-4 is an artifact of this strain. We assume that inclusion body formation of highly expressed eYFP fusion protein can be a consequence of targeting insoluble proteins to places of proteolytic activity. ClpCP protease is located near the cell poles and at midcell in B. subtilis (19). In summary, fusions of PhaP1, PhaP2, PhaP3, PhaP4, and PhaP5 with eYFP were all constitutively expressed in both PHB-negative strains (PHB-4 and ΔphaC1) and were mostly localized in soluble form in the cytoplasm in the ΔphaC1 strain.
Fig 3.
Localization of phasins PhaP1, PhaP2, PhaP3, PhaP4, and PhaP5 fused to eYFP in R. eutropha PHB-4 (A), H16 ΔphaC1 (B), and H16 (C). Experiments were performed as described in the legend to Fig. 1. Bar, 3 μm.
Different results were obtained when subcellular localization of the constructed phasin-eYFP fusions was followed in a PHB-positive background during the course of PHB accumulation (Fig. 3C). The fusion proteins of all five phasins were constitutively expressed. At time zero, most cells (>95%) were free of PHB, and fusions (PhaP1-eYFP to PhaP5-eYFP and eYFP-PhaP1 to eYFP-PhaP5) were homogeneously distributed in PHB-free wild-type H16 cells. Occasionally, occurrence of inclusion bodies at time zero was observed (PhaP1-eYFP, PhaP2-eYFP, PhaP4-eYFP, eYFP-PhaP1, and eYFP-PhaP3) (Fig. 3C). When the cells were transferred to conditions permissive for PHB accumulation (NB plus 0.2% gluconate), all strains accumulated PHB, which was visible after staining of the cells with Nile red. PHB granule formation started within 10 min (images not shown). Fusions of PhaP1, PhaP2, PhaP4, and PhaP5 with eYFP colocalized with PHB granules at any time. Note that Nile red-stained PHB granules and colocalized eYFP-phasin fluorescence appeared in more than 95% of all cells and were larger in diameter than putative fluorescent inclusions in the PHB-negative background. These results confirmed that PhaP1, PhaP2, PhaP4, and PhaP5 are true phasins. Notably, PhaP3-eYFP remained soluble 1 h and 3 h after transfer to conditions permissive for PHB accumulation. After 5 h of incubation, however (and at later time points [images not shown]), PhaP3-eYFP (and eYFP-PhaP3) colocalized with PHB granules. Apparently, binding of PhaP3-eYFP to PHB granules is weak and can be prevented during the early stages of PHB accumulation.
Screening of the R. eutropha genome for potential other phasin genes.
Comparison of the amino acid sequences of the phasin proteins PhaP1 to PhaP5 showed that PhaP1 to PhaP4 have strong sequence similarities, with 38 to 70% identity. However, the similarity of PhaP5 to the other phasins was only poor (13 to 17% amino acid identity). Apparently phasins can differ widely in amino acid sequence. This fact raised the question of whether other phasins in R. eutropha could be present that cannot be identified by sequence similarity (e.g., BLAST search). Closer inspection of the five known phasin sequences revealed that all of them have a common so-called phasin 2 motif that is characterized not by a specific amino acid sequence but by a region enriched in hydrophobic amino acids (Fig. 4). Phasin 2 motifs are typical of phasins in PHB-accumulating bacteria, whereas phasin 1 motifs are present in phasins of species that accumulate medium-chain-length PHAs (PHAMCL). We assume that the presence of such a phasin 2 motif could be indicative of subcellular localization of the expressed protein at the surface of PHB granules. When we screened the translated genome sequence of R. eutropha H16 for the presence of phasin 2 motifs, we identified three genes in addition to the previously known five phasin genes, namely, H16_B1988, H16_B2296, and H16_B2326. Two of them (H16_B1988 and H16_B2326) code for hypothetical proteins consisting of 207 amino acids (22.7 kDa) and 142 amino acids (16.4 kDa), respectively (Table 3). These values are almost in the same range as those for PhaP1 to PhaP5 (16 to 20 kDa). The amino acid sequence of the H16_B2296 gene product comprised 342 amino acids, and the deduced molecular mass therefore is significantly larger that those of the other two deduced proteins (38.1 kDa). The H16_B2296 gene product has similarities to LysR-type transcriptional regulators.
Fig 4.
Alignment (ClustalX 2.1) of phasin 2 motifs (KEGG database) of PhaP1-4, PhaP5 (H16_B1934), PhaP6 (H16_B1988), PhaP7 (H16_B2326), and H16_B2296.
Table 3.
Overview of known or postulated PGAPs in R. eutropha
| Protein | Mass (kDa) | Putative functiona | Evidenceb | Transcriptionc,e | Remarksd,e | In vivo interaction(s)e | Reference(s) |
|---|---|---|---|---|---|---|---|
| PhaC1 | 64.3 | PHB synthase | Exp | C (++) | Key enzyme | PhaM | 10, 28, 40, 42, this study |
| PhaC2 | 65.4 | PHB synthase | Similarity | − | Function unknown; catalytically inactive | 32, this study | |
| PhaP1 | 20.0 | Phasin | Exp | A (+++) | Major phasin | Other phasins | 4, 26, 29, 30, 51, this study |
| PhaP2 | 20.2 | Phasin | Similarity, exp | A (+) | Minor phasin | PhaP4, PhaP5, other phasins | 30, 35, this study |
| PhaP3 | 19.5 | Phasin | Similarity, exp | (+) | Minor phasin, reg? | Other phasins, PhaZa1 | 30, 35, this study |
| PhaP4 | 20.2 | Phasin | Similarity, exp | A (++) | Minor phasin | PhaP2, other phasins | 4, 30, 35, this study |
| PhaP5 | 15.7 | Phasin | Similarity, exp | A (+) | Minor phasin | PhaP2-4 PhaM, (PhaR, PhaZa1) | 30, 31, 35, this study |
| PhaP6 | 22.7 | Phasin | Phas. mot., exp | A (+) | Minor phasin | This study | |
| PhaP7 | 16.4 | Phasin | Phas. mot., exp | A (+) | Minor phasin | This study | |
| PhaR | 21.0 | Regulator | Exp | C (++) | Regulator | PhaP5 | 30 |
| PhaZa1 | 47.3 | iPHB depolym | Exp | A (++) | Depolymerase | PhaP3 | 13, 38, 50 |
| PhaZa2 | 44.8 | iPHB depolym | Similarity | A (++) | Putative depolymerase | 4, 52 | |
| PhaZa3 | 45.2 | iPHB depolym | Similarity | A (+) | Putative depolymerase | 4, 52 | |
| PhaZa4 | 27.3 | iPHB depolym | Similarity | Putative depolymerase | 4, 32 | ||
| PhaZa5 | 45.2 | iPHB depolym | Similarity | A (+) | Putative depolymerase | 4, 32 | |
| PhaZd1 (PhaZ6) | 39.2 | iPHB depolym | Exp | High dep activity | 1, 4, 32 | ||
| PhaZd2 (PhaZ7) | 38.4 | iPHB depolym | Similarity | A (+) | Putative depolymerase | 4, 32 | |
| PhaZb (PhaY1) | 74.3 | Oligomer hydr | Exp | A (+) | 20 | ||
| PhaZc (PhaY2) | 31.6 | Oligomer hydr | Exp | 21 | |||
| PhaM | 26.6 | Phasin+ PHB localization | Exp | C (++) | Binds to DNA | PhaC1, PhaP5 | 31 |
iPHB depolym, intracellular PHB depolymerase; hydr, hydrolase.
Exp, experimentally verified function; phas. mot., phasin 2 motif.
C, constitutive expression; A, increased expression under PHB accumulation conditions; −, not detectable; +, low; ++, medium; +++, high.
reg, regulation; dep, depolymerase.
An empty cell indicates that the parameter is not known.
Localization of H16_B1988, H16_B2296, and H16_B2326.
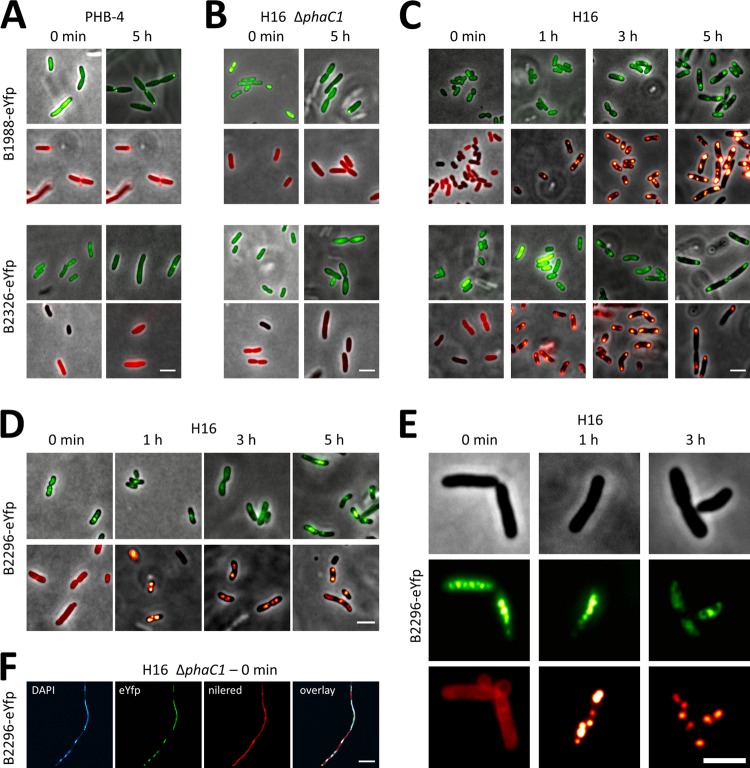
To investigate if the three identified proteins with phasin 2 motif were attached to PHB granules in vivo, N- and C-terminal fusions of the coding sequences of H16_B1988, H16_B2296, and H16_B2326 with eYFP were constructed using the modified pBBR1MCS-2 vector and cloned under the control of the constitutive phaC1 promoter. The respective constructs were transferred to R. eutropha PHB-4, R. eutropha ΔphaC1, and the wild-type R. eutropha strain H16 via conjugation. Transconjugants harboring the plasmid coding for the eYFP-H16_B2296 construct were not obtained in several independent conjugation experiments. We assume that expression of eYFP-H16_B2296 is lethal in R. eutropha, as was found for the PhaM-eYFP fusion (31). The transconjugant with the C-terminal fusion (H16_B2296-eYFP) was obtained without problems. Formation of the respective fusion proteins was verified by Western blot analysis in all cases (data not shown).
Determination of subcellular localization of eYFP fusions (N and C terminal) with H16_B1988 and with H16_B2326 resulted in the same results for all four constructs: the fusion proteins were generally homogeneously distributed in R. eutropha ΔphaC1 independent of the time point of sampling. Similar results were obtained in R. eutropha PHB-4 with the exception that H16_B1988-eYFP formed cell pole-localized inclusion bodies similar to those found for some of the phasin-eYFP fusions discussed above (Fig. 2 and Fig. 5A and B). Homogeneous fluorescence was also obtained in R. eutropha H16 at time zero and after 1 to 2 h of conditions permissive for PHB accumulation (Fig. 5C). Since R. eutropha produces several PHB granules in the first hour on NB medium supplemented with 0.2% gluconate, both H16_B1988 and H16_B2326 gene products apparently are not PHB granule bound at the very early stages of PHB accumulation. Remarkably, however, a colocalization of the H16_B1988-eYFP fusion and the H16_B2326-eYFP fusion with PHB granules was found at later time points (5 h) of growth permissive for PHB production. This indicated that H16_B1988 and H16_B2326 have the capacity to specifically bind to PHB granules but that binding to PHB was prevented at the early phases of PHB accumulation. We conclude that the H16_B1988 and H16_B2326 gene products are phasins, and we suggest that they be designated PhaP6 (H16_B1988) and PhaP7 (H16_B2326).
Fig 5.
Localization of phasins H16_B1988 (PhaP6), H16_B2326 (PhaP7), and H16_B2296 each fused to eYFP in R. eutropha. Experiments were performed as described in the legend to Fig. 1. Localization of H16_B1988-eYFP and H16_B2326-eYFP in R. eutropha PHB-4 (A), H16 ΔphaC1 (B), and H16 (C) and localization of H16_B2296-eYFP in R. eutropha H16 (D and E) and H16 ΔphaC1 (F) are shown. Note that colocalization of H16_B2296-eYFP with PHB granules (Nile red stain) was not observed, as shown by single-channel images from individual cells stained with Nile red (E). In contrast, laser scanning microscopy (F; channels as indicated) confirmed that H16_B2296-eYFP colocalizes with the DAPI-stained nucleoid. R. eutropha H16 and mutant strains expressing H16_B2296-eYFP were impaired in cell growth and in cell division, as indicated by slower growth and unusual elongation of some cells. Bars, 3 μm (A to E) and 10 μm (F).
Precise deletions in the phaP6 and phaP7 coding sequences were constructed in R. eutropha H16 and investigated for a phenotype in PHB metabolism. However, no differences in wild-type cells were detected with respect to cellular growth or number and localization of PHB granules. A weak phenotype was observed in a chromosomal ΔphaP1 R. eutropha H16 background in which PhaP6-eYFP or PhaP7-eYFP fusions were expressed: the cells with PhaP6-eYFP or PhaP7-eYFP fusions formed only one or two unusual small granules exclusively at or near the cell poles (occasionally a third PHB granule was formed at midcell) under PHB-permissive conditions This phenomenon was less pronounced in the ΔphaP1 strain without an expressed phasin fusion. However, the strong effect of PhaP1 overexpression on the number and size of PHB granules (51) was not detected for PhaP6- or PhaP7-overexpressing cells. Expression of eYFP alone in the ΔphaP1 strain (control) was always soluble as in the wild type, regardless of the absence or presence of PHB.
Analysis of subcellular localization revealed that H16_B2296-eYFP is located in the cytoplasm and in the center of the cells independent of the presence or absence of PHB in R. eutropha H16 and of the growth phase, similar to results obtained with PhaM (Fig. 5D and E). Colocalization with PHB granules was, however, not observed. Staining of the cells with DAPI (4,4′,6′-diamidino-2-phenylindole) revealed colocalization of H16_B2296-eYFP with the nucleoid region in a high fraction of the cells (Fig. 5F). We conclude that H16_B2296 apparently is not a PHB granule-bound protein but probably is a DNA-bound regulator protein, as predicted by its annotation; the function of the phasin 2 motif in H16_B2296 is unknown.
DISCUSSION
Several models of PHB granule biosynthesis are currently under discussion for the model organism of PHB research, R. eutropha H16. The so-called micelle model assumes that soluble (cytoplasmic) PHB synthase (PhaC1 dimers) starts to produce the hydrophobic PHB molecule if the concentration of the synthase substrate (3-hydroxybutyryl coenzyme A [3-hydroxybutyryl-CoA]) is sufficiently high. Because of the low solubility of PHB, nascent polymer chains aggregate and form micelle-like structures, with PhaC molecules sitting on the polymer surface (43, 44), to which phasins and other PGAPs are attached as the granule grows. The budding model assumes that the PHB synthase is located in or is attached to the cytoplasmic membrane and that the growing PHB chain is liberated into the membrane, leading to granule formation in the cytoplasm membrane. Once PHB granules have reached a specific size, the granules bleb out, and phasins and other PGAPs become attached to the granules. Several studies were performed in the lab of A. Sinskey and J. Stubbe and in our lab in the past addressing the question of where exactly in the cell nascent PHB granules are produced: TEM studies showed that PHB granules of cells that were cultivated under conditions permissive for PHB accumulation but not permissive for growth (high C, low N) were often located more or less in the center of the cells in close proximity to dark-stained “mediation elements” (46, 48). Contradictory results were found by fluorescence microscopy investigations, which suggested that PHB granules were synthesized in the cell periphery and/or near the cell pole area (14, 15, 18). Recently, we identified a new PGAP, PhaM, that specifically binds to PHB and to DNA (31). We showed that PhaM is necessary for equal distribution of PHB granules to daughter cells during cell division, similar to PhaF in PHAMCL-accumulating Pseudomonas putida (9). In this study, we observed formation of the first visible granules with attached PhaC1-eYFP within only 5 min after transfer of PHB-free cells to conditions permissive for growth and PHB accumulation (Fig. 1C). In this experiment, the cells were additionally stained with FM4-64, a dye that specifically stains the cell membrane. Most cells had one or two PHB granules associated with PhaC1-eYFP foci after 5 min (Fig. 1C). Notably, cells that had formed two PHB granules always had one PHB granule in each cell half. PHB granules and PhaC1-eYFP fluorescence were often located in neighborhood of the membrane and in the vicinity of the cell poles or area of future septum formation. However, we did not observe a colocalization of PhaC1-eYFP with the cytoplasm membrane, as suspected previously (16). We also never observed cells with two PHB granules in one cell half and none in the other half. Apparently, the initiation sites for PHB granule formation are not randomly distributed in the cell. This implies that the binding of PHB or PhaM to the nucleoid is not random, but a molecule that ensures specific attachment of the PHB granule to certain parts of the nucleoid is not known. In our previous study, PhaM turned out to bind unspecifically to DNA (31). Another explanation for the fact that two PHB granules were not located together in one cell half could be that the local concentration of the PHB synthase substrate (3-hydroxybutyryl-CoA) is low in the vicinity of already-formed PHB granules. Together with the early observations of Sinskey and Stubbe's group (46, 48), our data suggest that PHB granules are connected to the bacterial nucleoid and that the mediation elements represent the nucleoids of the bacteria. Because of the use of quasisynchronized cells in our experiments and transfer of PHB-free cells to conditions permissive for both growth and PHB accumulation (high C and high N), the first granules became attached to the dividing chromosome, resulting in the impression that PHB granules are located at the periphery and/or cell pole. In contrast, in the experiments described by the Sinskey-Stubbe lab, cells of a seed culture were transferred to a PHB production medium that contained high C but almost no nitrogen (0.01%) to promote PHB accumulation (46, 48). Since the first sample was not taken before 2.5 h of incubation, the nitrogen source should have been consumed, resulting in an arrest of nucleoid and cell division. If newly synthesized PHB granules become attached to the nucleoid via PhaM and the nucleoid does not divide any more, the driving force that separates PHB granules to the daughter cells is abolished. This will result in the impression of a more random localization of PHB granules, as observed by Tian and coworkers (46, 48). Recently, the Stubbe lab published a paper reporting the use of electron cryotomography to show that PHB granules are more or less statistically distributed in R. eutropha cells (1a). Again, the culture condition used in that study (2% fructose, 0.01 NH4Cl, inoculation ratio of 1:2.5, first sample after 2.5 h) are unlikely to allow cell proliferation at the time point of sampling. These data clearly exclude the budding model but do not exclude a scaffolding model with the nucleoid as the scaffold, as already discussed by Cho et al. (7). From these considerations, the apparent contradictions between our results on PHB granule localization and the data published by the Sinskey-Stubbe group can be explained by the use of different culture conditions and different time points of sampling.
The aim of this study was to find out whether some PGAPs of PHB granules in the model organism of PHB research, R. eutropha H16, are still unidentified. Since proteome analysis of isolated PHB granules gives many false-positive results because of artificial binding of proteins to PHB upon cell disruption (our unpublished data), proteome analysis of isolated PHB granules did not appear to be a good method to find new PGAPs. Our recently employed method of two-hybrid analysis resulted in the identification of two novel PGAPs (PhaP5 and PhaM) (30, 31). However, a few dozen presumably false-positive hits (data not shown) were also found, highlighting the limitation of this method for the identification of true new PGAPs. We were therefore interested in a faster and easier method for the identification of new PGAPs. We noticed that all currently known phasin proteins (PhaP1 to PhaP5) of R. eutropha harbored the so-called phasin 2 motif. This motif was also present in three other hypothetical proteins of the R. eutropha genome, namely, the H16_B1988, H16_B2296, and H16_B2326 gene products (Fig. 4). While two of the three proteins had molecular masses (16 and 23 kDa) similar to those of known phasins (16 to 20 kDa), the other candidate (H16_B2296) was significantly larger (38 kDa) and showed similarities to LysR transcriptional regulator proteins. We speculated that the H16_B2296 protein could be involved in binding of PHB granules to DNA, similar to PhaR (34, 53) or PhaM (31). However, fusion analysis with eYFP excluded the possibility that the H16_B2296 gene product binds to PHB granules. Therefore, it appears unlikely that the H16_B2296 gene product is involved in PHB metabolism. The reason why it has a phasin 2 motif remains obscure. The phasin 2 motif of H16_B2296 is the shortest among all phasins. It should be mentioned that not all PGAPs but only phasins sensu stricto have a phasin 2 motif. All other PGAPs (such as PhaCs, PhaZs, and PhaM) do not have this sequence motif. Therefore, it is unlikely that the phasin 2 motif represents a PHB-binding domain.
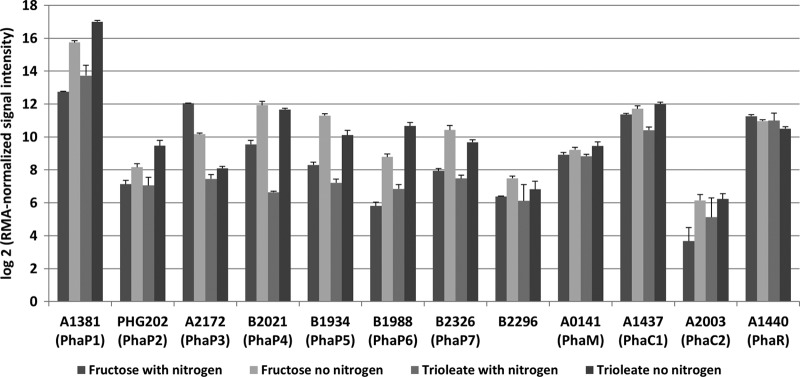
The function of most phasins in R. eutropha is unknown. Only for PhaP1 is a clearly detectable effect of a deletion of the respective gene known. Deletion of phaP1 results in formation of only one or two big PHB granules instead of 8 to 12 medium-sized granules in the wild type. Moreover, deletion of phaP1 enhanced degradation of PHB both in vivo (22) and in vitro (unpublished data). Deletion of other phasins (encoded by phaP2 to phaP7) or overexpression of phaP6 or phaP7 has no pronounced effect on size or number of accumulated PHB granules as determined by light microscopy (22, 30; also this study). Interestingly, overexpression of PhaP5, PhaP6, or PhaP7 resulted in cell pole-localized PHB granules and indicated that these phasins can have additional properties compared to PhaP1 to PhaP4. A comparison of global gene expression of R. eutropha grown under conditions permissive for PHB accumulation was performed previously (4). Analysis of the published transcriptome data revealed that transcription of phaP6 and phaP7 was upregulated under PHB-permissive conditions (high C, low N) (Fig. 6) in comparison to typical growth conditions (high C, high N). This expression pattern is typical for other phasin genes, such as phaP1, phaP2, phaP4, and phaP5, and is in contrast to phaC1 and phaM, which were constitutively expressed. However, overall expression of phaP1 was significantly higher than that of any of the other PGAP genes (Table 3), confirming that PhaP1 is the major phasin. Since the expression levels of phasins other than PhaP1 are significantly lower than that of PhaP1, they have only limited effects on the number and size of PHB granules in vivo. Similar results have been obtained during analysis of mutants lacking either phaP2, phaP3, or phaP4 (22). The function of the other phasins in PHB homeostasis remains obscure.
Fig 6.
Expression levels of selected R. eutropha genes under growth conditions and PHB-permissive conditions. All values were taken from published transcriptome data (4) and the GEO series accession number GPL10276 (http://www.ncbi.nlm.nih.gov/geo/).
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft.
We thank Simone Reinhardt and Dominik Rais for technical assistance in many cloning experiments.
Footnotes
Published ahead of print 24 August 2012
REFERENCES
- 1. Abe T, Kobayashi T, Saito T. 2005. Properties of a novel intracellular poly(3-hydroxybutyrate) depolymerase with high specific activity (PhaZd) in Wautersia eutropha H16. J. Bacteriol. 187: 6982–6990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a. Beeby M, Cho M, Stubbe J, Jensen GJ. 2012. Growth and localization of polyhydroxybutyrate granules in Ralstonia eutropha. J. Bacteriol. 194: 1092–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- 3. Brandl H, Gross RA, Lenz RW, Fuller RC. 1988. Pseudomonas oleovorans as a source of poly(beta-hydroxyalkanoates) for potential applications as biodegradable polyesters. Appl. Environ. Microbiol. 54: 1977–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brigham CJ, et al. 2010. Elucidation of beta-oxidation pathways in Ralstonia eutropha H16 by examination of global gene expression. J. Bacteriol. 192: 5454–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cai S, et al. 2012. Identification of the haloarchaeal phasin (PhaP) that functions in polyhydroxyalkanoate accumulation and granule formation in Haloferax mediterranei. Appl. Environ. Microbiol. 78: 1946–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen G-Q. 2009. A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry. Chem. Soc. Rev. 38: 2434–2446 [DOI] [PubMed] [Google Scholar]
- 7. Cho M, Brigham CJ, Sinskey AJ, Stubbe J. 2012. Purification of a polyhydroxybutyrate synthase from its native organism, Ralstonia eutropha: implications in the initiation and elongation of polymer formation in vivo. Biochemistry 51: 2276–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Escapa IF, García JL, Bühler B, Blank LM, Prieto MA. 2012. The polyhydroxyalkanoate metabolism controls carbon and energy spillage in Pseudomonas putida. Environ. Microbiol. 14: 1049–1063 [DOI] [PubMed] [Google Scholar]
- 9. Galán B, et al. 2011. Nucleoid-associated PhaF phasin drives intracellular location and segregation of polyhydroxyalkanoate granules in Pseudomonas putida KT2442. Mol. Microbiol. 79: 402–418 [DOI] [PubMed] [Google Scholar]
- 10. Gerngross TU, Reilly P, Stubbe J, Sinskey AJ, Peoples OP. 1993. Immunocytochemical analysis of poly-beta-hydroxybutyrate (PHB) synthase in Alcaligenes eutrophus H16: localization of the synthase enzyme at the surface of PHB granules. J. Bacteriol. 175: 5289–5293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grage K, et al. 2009. Bacterial polyhydroxyalkanoate granules: biogenesis, structure, and potential use as nano-/micro-beads in biotechnological and biomedical applications. Biomacromolecules 10: 660–669 [DOI] [PubMed] [Google Scholar]
- 12. Han J, et al. 2010. Wide distribution among halophilic archaea of a novel polyhydroxyalkanoate synthase subtype with homology to bacterial type III synthases. Appl. Environ. Microbiol. 76: 7811–7819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Handrick R, Reinhardt S, Jendrossek D. 2000. Mobilization of poly(3-hydroxybutyrate) in Ralstonia eutropha. J. Bacteriol. 182: 5916–5918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hermawan S, Jendrossek D. 2007. Microscopical investigation of poly(3-hydroxybutyrate) granule formation in Azotobacter vinelandii. FEMS Microbiol. Lett. 266: 60–64 [DOI] [PubMed] [Google Scholar]
- 15. Jendrossek D. 2005. Fluorescence microscopical investigation of poly(3-hydroxybutyrate) granule formation in bacteria. Biomacromolecules 6: 598–603 [DOI] [PubMed] [Google Scholar]
- 16. Jendrossek D. 2009. Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes). J. Bacteriol. 191: 3195–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jendrossek D, Handrick R. 2002. Microbial degradation of polyhydroxyalkanoates. Annu. Rev. Microbiol. 56: 403–432 [DOI] [PubMed] [Google Scholar]
- 18. Jendrossek D, Selchow O, Hoppert M. 2007. Poly(3-hydroxybutyrate) granules at the early stages of formation are localized close to the cytoplasmic membrane in Caryophanon latum. Appl. Environ. Microbiol. 73: 586–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kirstein J, Strahl H, Molière N, Hamoen LW, Turgay K. 2008. Localization of general and regulatory proteolysis in Bacillus subtilis cells. Mol. Microbiol. 70: 682–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kobayashi T, Shiraki M, Abe T, Sugiyama A, Saito T. 2003. Purification and properties of an intracellular 3-hydroxybutyrate-oligomer hydrolase (PhaZ2) in Ralstonia eutropha H16 and its identification as a novel intracellular poly(3-hydroxybutyrate) depolymerase. J. Bacteriol. 185: 3485–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kobayashi T, Uchino K, Abe T, Yamazaki Y, Saito T. 2005. Novel intracellular 3-hydroxybutyrate-oligomer hydrolase in Wautersia eutropha H16. J. Bacteriol. 187: 5129–5135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuchta K, Chi L, Fuchs H, Pötter M, Steinbüchel A. 2007. Studies on the influence of phasins on accumulation and degradation of PHB and nanostructure of PHB granules in Ralstonia eutropha H16. Biomacromolecules 8: 657–662 [DOI] [PubMed] [Google Scholar]
- 23. Lenz O, Friedrich B. 1998. A novel multicomponent regulatory system mediates H2 sensing in Alcaligenes eutrophus. Proc. Natl. Acad. Sci. U. S. A. 95: 12474–12479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Madison LL, Huisman GW. 1999. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol. Mol. Biol. Rev. 63: 21–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakamura S, Fukui T, Mifune J. 2008. Targeted engineering of Cupriavidus necator chromosome for bio-synthesis of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) from vegetable oil. Can. J. Chem. 86: 621–627 [Google Scholar]
- 26. Neumann L, et al. 2008. Binding of the major phasin, PhaP1, from Ralstonia eutropha H16 to poly(3-hydroxybutyrate) granules. J. Bacteriol. 190: 2911–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pederson EN, McChalicher CWJ, Srienc F. 2006. Bacterial synthesis of PHA block copolymers. Biomacromolecules 7: 1904–1911 [DOI] [PubMed] [Google Scholar]
- 28. Peoples OP, Sinskey AJ. 1989. Poly-beta-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. Identification and characterization of the PHB polymerase gene (phbC). J. Biol. Chem. 264: 15298–15303 [PubMed] [Google Scholar]
- 29. Peplinski K, et al. 2010. Genome-wide transcriptome analyses of the “Knallgas” bacterium Ralstonia eutropha H16 with regard to polyhydroxyalkanoate metabolism. Microbiology 156: 2136–2152 [DOI] [PubMed] [Google Scholar]
- 30. Pfeiffer D, Jendrossek D. 2011. Interaction between poly(3-hydroxybutyrate) granule-associated proteins as revealed by two-hybrid analysis and identification of a new phasin in Ralstonia eutropha H16. Microbiology 157: 2795–2807 [DOI] [PubMed] [Google Scholar]
- 31. Pfeiffer D, Wahl A, Jendrossek D. 2011. Identification of a multifunctional protein, PhaM, that determines number, surface to volume ratio, subcellular localization and distribution to daughter cells of poly(3-hydroxybutyrate), PHB, granules in Ralstonia eutropha H16. Mol. Microbiol. 82: 936–951 [DOI] [PubMed] [Google Scholar]
- 32. Pohlmann A, et al. 2006. Genome sequence of the bioplastic-producing “Knallgas” bacterium Ralstonia eutropha H16. Nat. Biotechnol. 24: 1257–1262 [DOI] [PubMed] [Google Scholar]
- 33. Pötter M, Steinbüchel A. 2006. Biogenesis and Structure of polyhydroxyalkanoate granules. Microbiol. Monogr. 1: 110–136 [Google Scholar]
- 34. Pötter M, Madkour MH, Mayer F, Steinbüchel A. 2002. Regulation of phasin expression and polyhydroxyalkanoate (PHA) granule formation in Ralstonia eutropha H16. Microbiology 148: 2413–2426 [DOI] [PubMed] [Google Scholar]
- 35. Pötter M, et al. 2004. The complex structure of polyhydroxybutyrate (PHB) granules: four orthologous and paralogous phasins occur in Ralstonia eutropha. Microbiology 150: 2301–2311 [DOI] [PubMed] [Google Scholar]
- 36. Rehm BHA. 2003. Polyester synthases: natural catalysts for plastics. Biochem. J. 376: 15–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rehm BHA, Antonio RV, Spiekermann P, Amara AA, Steinbüchel A. 2002. Molecular characterization of the poly(3-hydroxybutyrate) (PHB) synthase from Ralstonia eutropha: in vitro evolution, site-specific mutagenesis and development of a PHB synthase protein model. Biochim. Biophys. Acta 1594: 178–190 [DOI] [PubMed] [Google Scholar]
- 38. Saegusa H, Shiraki M, Kanai C, Saito T. 2001. Cloning of an intracellular poly[d(−)-3-hydroxybutyrate] depolymerase gene from Ralstonia eutropha H16 and characterization of the gene product. J. Bacteriol. 183: 94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schlegel HG, Lafferty R, Krauss I. 1970. The isolation of mutants not accumulating poly-beta-hydroxybutyric acid. Arch. Mikrobiol. 71: 283–294 [DOI] [PubMed] [Google Scholar]
- 40. Schubert P, Steinbüchel A, Schlegel HG. 1988. Cloning of the Alcaligenes eutrophus genes for synthesis of poly-beta-hydroxybutyric acid (PHB) and synthesis of PHB in Escherichia coli. J. Bacteriol. 170: 5837–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Simon R, Priefer U, Pühler A. 1983. A broad host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat. Biotechnol. 1: 784–791 [Google Scholar]
- 42. Slater SC, Voige WH, Dennis DE. 1988. Cloning and expression in Escherichia coli of the Alcaligenes eutrophus H16 poly-beta-hydroxybutyrate biosynthetic pathway. J. Bacteriol. 170: 4431–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stubbe J, Tian J. 2003. Polyhydroxyalkanoate (PHA) hemeostasis: the role of PHA synthase. Nat. Prod. Rep. 20: 445–457 [DOI] [PubMed] [Google Scholar]
- 44. Stubbe J, et al. 2005. Nontemplate-dependent polymerization processes: polyhydroxyalkanoate synthases as a paradigm. Annu. Rev. Biochem. 74: 433–480 [DOI] [PubMed] [Google Scholar]
- 45. Sudesh K, Abe H, Doi Y. 2000. Synthesis, structure and properties of polyhydroxyalkanoates: biological polymers. Prog. Polym. Sci. 25: 1503–1555 [Google Scholar]
- 46. Tian J, et al. 2005. Analysis of transient polyhydroxybutyrate production in Wautersia eutropha H16 by quantitative Western analysis and transmission electron microscopy. J. Bacteriol. 187: 3825–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tian J, Sinskey AJ, Stubbe J. 2005. Detection of intermediates from the polymerization reaction catalyzed by a D302A mutant of class III polyhydroxyalkanoate (PHA) synthase. Biochemistry 44: 1495–1503 [DOI] [PubMed] [Google Scholar]
- 48. Tian J, Sinskey AJ, Stubbe J. 2005. Kinetic studies of polyhydroxybutyrate granule formation in Wautersia eutropha H16 by transmission electron microscopy. J. Bacteriol. 187: 3814–3824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tsumoto K, Umetsu M, Kumagai I, Ejima D, Arakawa T. 2003. Solubilization of active green fluorescent protein from insoluble particles by guanidine and arginine. Biochem. Biophys. Res. Commun. 312: 1383–1386 [DOI] [PubMed] [Google Scholar]
- 50. Uchino K, Saito T, Jendrossek D. 2008. Poly(3-hydroxybutyrate) (PHB) depolymerase PhaZa1 is involved in mobilization of accumulated PHB in Ralstonia eutropha H16. Appl. Environ. Microbiol. 74: 1058–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wieczorek R, Pries A, Steinbüchel A, Mayer F. 1995. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J. Bacteriol. 177: 2425–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. York GM, et al. 2003. Ralstonia eutropha H16 encodes two and possibly three intracellular poly[d-(−)-3-hydroxybutyrate] depolymerase genes. J. Bacteriol. 185: 3788–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. York GM, Stubbe J, Sinskey AJ. 2002. The Ralstonia eutropha PhaR protein couples synthesis of the PhaP phasin to the presence of polyhydroxybutyrate in cells and promotes polyhydroxybutyrate production. J. Bacteriol. 184: 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yuan W, et al. 2001. Class I and III polyhydroxyalkanoate synthases from Ralstonia eutropha and Allochromatium vinosum: characterization and substrate specificity studies. Arch. Biochem. Biophys. 394: 87–98 [DOI] [PubMed] [Google Scholar]